Abstract
The prevalence and heterogeneity of Chlamydia trachomatis infections in a cohort of female sex workers in Dakar (Senegal) were determined by using endocervical-swab-based PCR DNA amplification assays. The overall prevalence of cervical chlamydial infection was 28.5% (206 of 722), and most of these infections were asymptomatic. An increased number of sexual partners was significantly associated with infection (adjusted odds ratio [AOR] = 1.37; 95% confidence interval [CI] = 1.06 to 1.77), while the presence of a yeast infection was negatively associated with chlamydial infection (AOR = 0.28; 95% CI = 0.10 to 0.83). Six different C. trachomatis genotypes were identified based on phylogenetic analysis of the omp1 gene sequences. Interestingly, genotype E predominated (47.6%) and was not associated with visible signs of cervical inflammation compared to non-E genotypes (P < 0.05). Overall, the high rate of asymptomatic C. trachomatis infection by genotype E may suggest genotype-specific properties that confer a transmission advantage in this high risk population.
Chlamydia trachomatis is the causative agent of a variety of diseases and syndromes, including trachoma, urogenital infections, chlamydial conjunctivitis, infant pneumonia, and lymphogranuloma venereum (26). Worldwide, C. trachomatis is one of the most common sexually transmitted bacterial pathogens. In women, urogenital C. trachomatis infections can cause a broad spectrum of clinical manifestations, including urethritis, cervicitis, and pelvic inflammatory disease, which if untreated may lead to serious complications, including ectopic pregnancy and tubal infertility. Many women do not seek medical care, since more than 70% of genital infections are asymptomatic (17). From a public health perspective, this leads to the persistence of an undetected C. trachomatis reservoir with significant perpetuation of transmission.
Strains of C. trachomatis are antigenically variant and can be classified into serovars (14, 45). The predominant antigen that defines individual serovars is the major outer membrane protein (MOMP) (6), the main surface antigen of C. trachomatis. The single-copy gene omp1 encodes for MOMP and consists of five regions of conserved sequence that alternate with four variable regions (35, 47). The omp1 variable domains can be used for genotyping isolates due to their extensive genetic variation. There is a high degree of concordance between the serological classification and omp1-based genotypes, with 18 currently identified genotypes (44, 45, 47). These chlamydial serovars and genotypes are classified into three different groups, on the basis of their genetic relatedness and serological cross-reactivity: class B (B/Ba, D/Da, E, L1, and L2), class C (A, C, H, I/Ia, J, K, and L3), and the intermediate group (F and G) (14, 45, 47). Studies have documented the correlation between infecting C. trachomatis strains and disease manifestations, although the biological basis for such associations is not well understood: genotypes A to C are most often associated with trachoma, genotypes L1 to L3 are always associated with lymphogranuloma venereum, while genotypes D to K are primarily associated with urogenital and neonatal infections (26). Researchers have also raised the possibility that there is a correlation between infecting strains and clinical manifestations in genital chlamydial infections (2, 9, 40, 41, 46), where multiple genotypes may contribute to the broad spectrum of symptoms observed in such infections.
Epidemiological studies have shown that sexually transmitted pathogens, including nonulcerative agents such as C. trachomatis, may serve as biological cofactors for human immunodeficiency virus (HIV) seroconversion (15, 22, 30). However, basic data, such as the incidence and prevalence of many sexually transmitted diseases (STDs), which are necessary for obtaining estimates of their impact on sexual transmission of HIV, are relatively scarce in developing countries. In many areas, diagnosis of C. trachomatis genital infection is only performed in selected populations and is often based on the presence of clinical symptoms. Considering the high rate of asymptomatic chlamydial infection, particularly in women, a substantial “silent” or undetected epidemic of C. trachomatis infections could put this population at significant risk for HIV infection. In addition, there is currently limited data about C. trachomatis genotype distribution in asymptomatic infections, and it is unknown if genotypes differ in the risk they represent for subsequent HIV infection.
For over a decade, our laboratory has studied HIV-1, HIV-2, and other STDs in a cohort of registered female commercial sex workers (FCSW) in Dakar, Senegal (20, 39). We want to better understand the biological impact of inflammatory STDs and their potential role as risk factors for HIV infection in this high-risk population. This study was therefore undertaken to determine the prevalence and potential risk factors for C. trachomatis genital infection in this cohort of FCSW by using a highly sensitive PCR-based diagnostic assay. In addition, this study was intended to describe the distribution of C. trachomatis genotypes in this population and to assess any association between C. trachomatis genotypes and genital tract pathology.
MATERIALS AND METHODS
Study population and data collection.
An open cohort of self-registered FCSW in Dakar, Senegal, was established in 1985 to study the epidemiology and natural history of HIV-1 and HIV-2. Briefly, after informed consent and enrollment, basic demographic data, and sexual, reproductive, and medical histories were obtained as previously described (20). FCSW were provided with regular medical examinations, free medication, safe sex education, and free condoms. Every 2 to 3 months, a physical and gynecological exam, laboratory diagnosis for common STDs and HIV, and collection of behavioral data were performed. In addition, from June 1996 to January 1997, for the purpose of this study, endocervical swabs were collected from every enrolled FCSW coming to the clinic for their routine physical examination.
HIV serodiagnosis and STD diagnostic assays.
Serum samples were evaluated by immunoblotting to HIV-1 (IIIb-Molt) and HIV-2 (MS-U937) viral proteins as previously described (20). Dually reactive samples were confirmed by type-specific PCR amplification of proviral DNA from both HIV-1 and HIV-2 (31). Neisseria gonorrhoeae was detected by the presence of intracellular diplococci on a Gram-stained smear of cervicovaginal swabs and/or by culture on chocolate medium supplemented with isovitalex, colistin, and nystatin, with or without vancomycin, and incubation in CO2-enriched atmosphere. The identification of yeast, Trichomona vaginalis, and Gardenerella vaginalis was made by direct microscopic observation of wet mounts and/or Gram staining of genital secretion smears. Syphilis diagnosis was performed by serologic methods, including the rapid plasma reagin (RPR) test and the T. pallidum hemoagglutination assay (TPHA). A positive RPR in the presence of a reactive TPHA was identified as an active case of syphilis.
Collection of endocervical swabs and DNA extraction.
During routine pelvic examination, the physical aspect of the cervix was recorded (healthy, friable, or inflamed), and two cotton swabs of the cervical os were obtained. The swabs were placed into a sterile 15-ml collection tube containing 2 ml of 1× phosphate-buffered saline and transported to the laboratory for further processing. Upon arrival, the swabs were vortexed vigorously, rotated 360° against the inside of the collection tube to remove as much fluid as possible, and then discarded. The swab fluid was centrifuged at 2,500 rpm for 10 min, the supernatant was removed, and the cell pellet resuspended in 150 μl of 1% sodium dodecyl sulfate solution. The suspension was boiled for 15 min, and the resulting lysate was extracted twice with an equal volume of phenol-chloroform and once with pure chloroform. DNA was precipitated overnight in 100% ethanol, washed once with 70% ethanol, dried and resuspended in 50 μl of Tris-EDTA buffer. Every sample was subjected to PCR amplification of a 500-bp fragment of the human β-globin gene as a quality control measure. This process evaluated the efficacy of the DNA extraction and confirmed the presence of amplifiable DNA. Samples that tested negative twice for β-globin amplification were not further studied.
C. trachomatis diagnosis by PCR.
To detect the presence of C. trachomatis in the cervical DNA samples, a 201-bp fragment of the bacterial endogenous plasmid was amplified (diagnostic PCR). It is estimated that 10 copies of endogenous plasmid are present per elementary body (27). The plasmid fragment was amplified by using primers CTP1 and CTP2 and according to the protocol of Lan et al. (25). The PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide and confirmed by hybridization on a nylon membrane with a radiolabeled internal oligonucleotide probe CTP3 (25). DNA extracted from a C. trachomatis serovar L2 culture was used as a positive control, while water served as a negative control. Both positive and negative controls were included in all amplification reactions and subsequent hybridizations.
omp1 amplification, purification, and sequencing.
Cervical DNA samples positive by diagnostic PCR were selected for omp1 genotyping. A 988-bp fragment of the omp1 gene was amplified by nested PCR by using primer pairs SERO1A and SERO2A and VD42 and PCTM3 and amplification conditions as described previously (24). The amplified product was purified by gel electrophoresis and purification columns (Quiaquick gel extraction kit; Quiagen, Inc., Chatsworth, Calif.). Sequences of the omp1 VD1, -2, -3, and -4 were obtained by direct dye terminator cycle sequencing by using Taq polymerase (Perkin-Elmer, Applied Biosystem Division, Foster City, Calif.) and an automatic sequencer ABI 373 (Perkin-Elmer). All samples were sequenced in both directions by using primers 3 and 5 (4) for VD1 and VD2 and primers MVF3 (10) and VD42 (24) for VD3 and VD4. When necessary, inserts were cloned into pCR2.1 vector (TOPO TA Cloning Kit and TOP10 One Shot Cells; Invitrogen, San Diego, Calif.). Plasmid preparation for double-stranded DNA sequencing was performed by alkaline lysis and purification on DEAE columns (Quiagen Plasmid Minikit; Quiagen, Inc.).
Genotyping and phylogenetic analysis of C. trachomatis strains.
A phylogenetic tree was constructed by using the concatenated sequences of VD1, -2, -3, and -4 from our field strains and from 17 prototypic C. trachomatis genotypes that included other previously described clinical strains (1, 8, 11, 16, 18, 28, 29, 32, 34–36, 47, 48). Multiple alignment and phylogenetic analysis were performed with the CLUSTAL package (CLUSTAL X) (38). All variable domain sequences were submitted to GenBank (accession numbers AF178221 to AF178304).
Statistical analysis.
For the analysis, chlamydial infection was defined as cervical DNA samples that were positive by diagnostic PCR and confirmed to be positive by Southern blot hybridization. A univariate analysis of association with chlamydial infection was performed on all potential risk factors by using the chi-square test. All significant variables and potential confounders were then analyzed in a multivariate logistic regression model. Data analysis was performed with Stata version 4.0 (Stata Corp., College Station, Tex.).
RESULTS
Study population.
Between June 1996 and January 1997, 740 endocervical swabs were collected and DNA extracted, 722 (97.7%) of which were found to be positive by β-globin PCR. The 722 women from whom these swabs were collected constituted the study population. Most of these women were Senegalese (76.2%), their ages ranged from 22 to 58 years (mean ± standard deviation = 35.6 ± 7.4), and they had worked an average of 7.8 years as registered FCSW (7.8 ± 7.0) with an average of 7.3 clients per week (7.3 ± 7.8).
C. trachomatis prevalence and distribution of risk factors.
Of 722 samples, 206 (28.5%) were positive by diagnostic PCR and confirmed by Southern blot hybridization. Syphilis infection was also common, with 132 cases in 659 subjects tested (20.0%). N. gonorrhoeae was detected in 44 (6.7%) women. G. vaginalis was also commonly identified, with 25.4% positivity, while yeast infections were identified among 39 (5.9%) subjects (Table 1). Upon physical examination, 114 (16.5%) women were found to have cervical inflammation at the time of the visit.
TABLE 1.
Study population characteristics
| Variable | % Prevalence (no. positive/total no.) | % Prevalence (C. trachomatis status)
|
|
|---|---|---|---|
| Positive | Negative | ||
| C. trachomatis | 28.5 (206/722) | 206 (n) | 516 (n) |
| Laboratory diagnoses | |||
| N. gonorrhoeae | 6.7 (44/660) | 5.8 (11/191) | 7.0 (33/469) |
| T. vaginalis | 12.6 (83/660) | 13.5 (26/192) | 12.2 (57/468) |
| G. vaginalis | 25.4 (168/661) | 23.6 (45/191) | 26.2 (123/470) |
| Yeast infection | 5.9 (39/658) | 2.1 (4/191) | 7.5 (35/467) |
| Syphilis | 20.0 (132/659) | 20.3 (39/192) | 19.9 (93/467) |
| Cervical inflammation | 16.5 (114/693) | 14.7 (29/198) | 17.2 (85/495) |
| Serostatus | |||
| HIV-1 | 9.4 (68/722) | 8.7 (18/206) | 9.7 (50/516) |
| HIV-2 | 9.4 (68/722) | 9.2 (19/206) | 9.5 (49/516) |
| HIV-D | 1.5 (11/722) | 1.0 (2/206) | 1.7 (9/516) |
| Nationality | |||
| Senegalese | 76.2 (550/722) | 76.2 (157/206) | 76.2 (393/516) |
| Other | 23.8 (172/722) | 23.8 (49/206) | 23.8 (123/516) |
| Age (yr) | |||
| ≤30 | 26.4 (188/713) | 24.1 (49/203) | 27.3 (139/510) |
| 31–36 | 28.8 (205/713) | 22.7 (46/203) | 31.2 (159/510) |
| 37–41 | 21.5 (153/713) | 25.6 (52/203) | 19.8 (101/510) |
| >41 | 23.4 (167/713) | 27.6 (56/203) | 21.8 (111/510) |
| Duration of registered prostitution (yr) | |||
| ≤1 | 27.0 (195/722) | 22.8 (47/206) | 28.7 (148/516) |
| 2–6 | 25.5 (184/722) | 26.2 (54/206) | 25.2 (130/516) |
| 7–13 | 24.4 (176/722) | 26.2 (54/206) | 23.6 (122/516) |
| >13 | 23.1 (167/722) | 26.2 (51/206) | 22.5 (116/516) |
| No. of clients per weeka | |||
| ≤3 | 33.3 (196/589) | 26.5 (45/170) | 36.0 (151/419) |
| 4–5 | 17.8 (105/589) | 15.9 (27/170) | 18.6 (78/419) |
| 6–10 | 29.7 (175/589) | 33.5 (57/170) | 28.2 (118/419) |
| >10 | 19.2 (113/589) | 24.1 (41/170) | 17.2 (72/419) |
| Condom usea | |||
| Never | 4.2 (28/665) | 3.1 (6/194) | 4.7 (22/471) |
| Sometimes | 46.5 (309/665) | 43.3 (84/194) | 47.8 (225/471) |
| Always | 49.3 (328/665) | 53.6 (104/194) | 47.6 (224/471) |
Women were given the option not to answer.
There were 68 HIV-1-positive (9.4%), 68 HIV-2-positive (9.4%), and 11 dually infected (1.5%) women in the study, with the remainder being HIV seronegative (n = 579; 79.6%). The majority of the HIV-infected women were healthy and asymptomatic, with only 3.5% of the study population fitting criteria for CDC stage III and 2.2% in CDC stage IV or above (7). Few women reported never using condoms (n = 28; 4.2%), while 309 (46.5%) reported using them sometimes, and most reported always using them (n = 328; 49.3%).
Relationship between chlamydial infection and risk factors.
In the univariate analysis, an increased number of partners per week was found to be a significant risk factor for C. trachomatis infection (Table 2). In contrast, the presence of a yeast infection and younger age at registration (25 to 27 years old) were inversely associated with C. trachomatis infection.
TABLE 2.
Relationship between C. trachomatis infection and risk factorsa
| Variable | Crude ORb (95% CI) | Adjusted ORc (95% CI) |
|---|---|---|
| Laboratory diagnoses | ||
| N. gonorrhoeae | 0.81 (0.40–1.63) | 0.81 (0.38–1.70) |
| T. vaginalis | 1.13 (0.69–1.86) | 1.28 (0.73–2.25) |
| Syphilis | 1.03 (0.67–1.56) | 1.10 (0.68–1.78) |
| HIVd | 0.88 (0.59–1.33) | 0.75 (0.44–1.29) |
| Any STDe | 1.01 (0.73–1.40) | 0.97 (0.67–1.42) |
| G. vaginalis | 0.87 (0.59–1.29) | 1.02 (0.65–1.59) |
| Yeast infection | 0.26 (0.92–0.75) | 0.28 (0.10–0.83) |
| Condom use | ||
| Always | 1.00 | 1.00 |
| Sometimes | 0.80 (0.57–1.13) | 0.87 (0.59–1.31) |
| Never | 0.59 (0.23–1.49) | 0.74 (0.26–2.16) |
| Clients (per 10 clients/wk) | 1.30 (1.04–1.62) | 1.37 (1.06–1.77) |
| Age at registration (yr) | ||
| ≤24 | 1.00 | 1.00 |
| 25–27 | 0.59 (0.36–0.97) | 0.69 (0.39–1.21) |
| 28–32 | 1.14 (0.75–1.74) | 1.25 (0.76–2.05) |
| >32 | 1.08 (0.69–1.67) | 1.01 (0.58–1.77) |
| Registered prostitution (yr) | ||
| ≤1 | 1.00 | 1.00 |
| 2–6 | 1.31 (0.83–2.06) | 1.18 (0.70–2.00) |
| 7–13 | 1.39 (0.88–2.20) | 1.48 (0.85–2.58) |
| >13 | 1.38 (0.87–2.20) | 1.44 (0.80–2.60) |
| Nationality | ||
| Senegalese | 1.00 | 1.00 |
| Other | 1.00 (0.68–1.46) | 1.10 (0.65–1.84) |
OR and AOR values in boldface are statistically significant (P < 0.05).
Univariate analysis by χ2 test.
Multivariate analysis by logistic regression including all variables listed in this table was performed on data from 563 subjects.
HIV includes HIV-1, HIV-2, and dual infections.
Any STD includes any infection with N. gonorrhoeae, T. vaginalis, syphilis, or HIV.
The multivariate model included other STD infections (N. gonorrhoeae, T. vaginalis, syphilis, and HIV), the presence of yeast or of G. vaginalis, condom use, the number of partners per week, the number of years of prostitution, and the age at registration. An increased number of partners per week was found to be significantly associated with chlamydial infection (odds ratio [OR] = 1.37 [expressed per 10 clients/week]; 95% confidence interval [CI] = 1.06 to 1.77). When stratified per quartiles, women with over 10 partners per week had the highest risk of infection (OR = 2.01; 95% CI = 1.14 to 3.54), and women with 5 to 10 partners per week were at a higher risk (OR = 1.68; 95% CI = 1.02 to 2.77) compared to women with fewer than 3 partners per week. The presence of a yeast infection was significantly associated with lowered risk for chlamydial infection (OR = 0.28; 95% CI = 0.10 to 0.83). Surprisingly, no statistically significant association with other concurrent STDs was found, and no association was found between HIV seropositivity and chlamydial infection.
Genotyping and phylogenetic analysis.
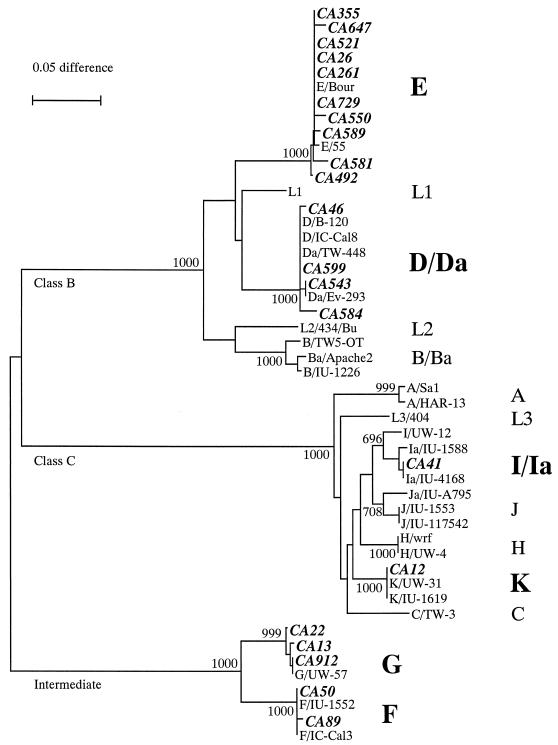
A total of 21 samples positive for C. trachomatis by diagnostic PCR were subjected to omp1 amplification and then sequenced. The phylogenetic tree showed that the genotyped strains were quite diverse, with representatives from all three genetic groups and most strains (14 of 21; 66.7%) belonging to class B (Fig. 1). The most common C. trachomatis genotype identified was E (n = 10; 47.6%), followed by D/Da (n = 4; 19.0%) and G (n = 3; 14.3%). Two strains (9.5%) of genotype F and only 1 strain (4.8%) each of genotypes Ia and K were found. No strains from genotypes J and H were identified, although these genotypes are also associated with urogenital infections.
FIG. 1.
Phylogenetic tree based on the nucleotide sequence of the omp1 gene variable domains from 49 strains. The neighbor-joining tree was prepared by using the Kimura's two-parameter method and by removing positions at which gaps were created. Taxa in boldface represent the sequences from the 21 strains from this study and are denoted by their identification number. Other taxa are sequences obtained from previously published studies (1, 8, 11, 16, 18, 28, 29, 32, 34–36, 47, 48). Branch lengths are proportional to the sequence divergence between taxa in the tree, as illustrated by the scale bar. Numbers at the nodes indicate bootstrap values, based on 1,000 bootstrap replicates. DNA sequence homology groups are labeled as classes B, C, and intermediate.
Relationship between C. trachomatis infection, infecting genotype, and presence of cervicitis.
Of the 206 women identified as C. trachomatis positive, 198 (96.1%) had observations available on the physical aspect of their cervix at the time of visit, and only 29 of these (14.6%) showed visible signs of cervical inflammation or friability. A comparative analysis was performed between symptomatic and asymptomatic C. trachomatis-infected women to ensure that any difference in inflammatory response was not due to other concurrent STD infection or to genital tract syndrome. No significant difference was found between the two groups (data not shown).
All 10 women infected with genotype E had a healthy cervix, while 5 of 11 of those with the non-E genotype had visible signs of cervical inflammation on clinical examination (P < 0.05, Fisher's exact test).
DISCUSSION
This study was performed to determine the prevalence of C. trachomatis in a cohort of regularly monitored FCSW in Dakar, Senegal, to identify the potential risk factors for chlamydial genital infection, to assess the genotypic diversity of the microorganism in this population, and to identify any association between genotypes and cervical inflammation. This was one of the first studies to use a sensitive PCR assay to screen a large number of women not selected for disease and thus allowed us to better estimate the real magnitude of the reservoir of asymptomatic C. trachomatis infections in this high-risk population. The study was warranted since most chlamydial infections are asymptomatic in women and because such infections may increase the risk of HIV infection or facilitate its spread. Also, little is known about the genotype distribution of C. trachomatis in largely asymptomatic populations and about their respective biological and clinical implications.
Recently, there have been considerable advances in developing nucleic acid amplification tests for C. trachomatis diagnosis and genotyping. Overall, nucleic acid amplification assays are far more sensitive than earlier tests (33); they now bypass culture of the organism and allow direct genotyping. Most recent population-based studies favor the use of ligase chain reaction on urine samples to detect C. trachomatis, since this diagnostic test is noninvasive and the sampling can be performed even outside of traditional health care facilities. However, this commercial assay is too expensive to be used routinely in countries with limited resources for health care. Considering that the FCSW in our study were coming to the clinic for their routine gynecological examination, the use of an endocervical swab was considered to be easily implemented and offered the advantage of directly testing the primary tissue targeted in a genital chlamydial infection. Therefore, a PCR-based assay with such swabs was considered appropriate for this study.
The PCR with primers CTP1 and CTP2 was previously found to have a detection limit of 1 to 0.1 inclusion forming units (IFU) by gel electrophoresis and 0.1 to 0.01 IFU after hybridization with internal probe CTP3 (25). This highly sensitive PCR was therefore capable of detecting infections with low copy numbers of C. trachomatis, which are often associated with asymptomatic infections; such samples would have been found to be falsely negative by other microbiological assays. However, the high sensitivity of this PCR assay may also make it more susceptible to contamination. Various measures to minimize the risk of sample contamination were implemented from the start of this study: a limited number of samples were manipulated together, negative controls were included in every amplification and hybridization reaction and, most importantly, DNA extraction and PCR amplification took place in separate buildings.
It has earlier been shown that polymerases are particularly sensitive to inhibitors that can be found at significant levels in complex secretions such as cervicovaginal fluids or urine (3). To prevent loss of sensitivity of the PCR assay, all samples were subjected to a standard phenol-chloroform extraction and alcohol precipitation. The β-globin PCR not only allowed us to assess whether the DNA extraction had been properly performed but also served as internal control to assess PCR reaction inhibition. The high rate of positivity for β-globin PCR (97.7%) indicates that DNA extraction was largely successful and that the samples were mostly free of potential polymerase inhibitors.
Many studies conducted in clinical settings perform STD diagnosis based on the presence of symptoms and/or clinical signs to confirm the identity of the suspected agents. Since C. trachomatis-infected women are frequently asymptomatic, the true prevalence of C. trachomatis is difficult to measure without routine screening tests. When such screening studies are actually undertaken, high rates of chlamydial infections can be found, particularly when nucleic acid amplification assays are used. A study of women attending a family planning clinic in Papua New Guinea determined the prevalence of C. trachomatis to be 17.3% by using a combination of PCR and direct immunofluorescence assays (37). Even recent studies performed in developed countries show relatively high prevalence rates in asymptomatic populations. In a study performed among 13,000 female Army recruits from Fort Jackson, SC, 9.2% were found positive for C. trachomatis DNA in their urine samples by ligase chain reaction, while the prevalence was more than 15% among recruits from the southern states (12). A study performed with the same diagnostic C. trachomatis PCR assay used in the present study found a prevalence of 9.2% in a group of asymptomatic young women volunteers and 11.8% in a population of randomly selected asymptomatic patients attending an inner-city gynecology outpatient clinic in Amsterdam (23). All the rates reported above were unexpectedly high, particularly for populations which were considered low risk. In the present study, we found a C. trachomatis prevalence in Dakar commercial sex workers of 28.5%, which is higher than the 14.3% previously reported in the same population (43). This is not surprising, considering that the earlier study used less-sensitive detection methods such as culture and enzyme-linked immunosorbent assay (33), and the authors themselves stated that this rate was probably an underestimate (43). Thus, we feel that the combination of the higher sensitivity of the assay used and the high-risk behavior of the study population can explain such a high prevalence of C. trachomatis genital infections.
Previous epidemiological studies on chlamydial infection have identified a variety of risk factors, including the number of partners, an age under 25 years, cervical ectopy, concurrent gonoccal infection, a history of sexually transmitted diseases, HIV seropositivity and seroconversion, the duration of prostitution, and the lack of condom use (5, 12, 42). In our study, risk determinant analysis showed that increased number of partners was a risk for C. trachomatis infection; however, no significant association was found with other risk factors, including other STDs or HIV.
One possible explanation for the lack of association between chlamydial infection and previously described risk factors were the differences in STD risk and infection in our study population, as evidenced by differences in HIV rates. Brunham and colleagues described a study of commercial sex workers in Nairobi, Kenya, where chlamydial infection (relative risk [RR] = 1.6; 95% CI = 1.2 to 2.2) and the number of reinfections (RR = 14.5; 95% CI = 3.1 to 258) were both higher among HIV-positive than among HIV-negative women (4, 5). However, that study described a high rate of HIV with 162 of 302 women (54%) starting out as HIV-1 positive and 20% more seroconverting during the follow-up period of 17 months. In the present study, there were 68 HIV-1-positive (9.4%), 68 HIV-2-positive (9.4%), and 11 dually infected (1.5%) women, and our study population as a whole was largely asymptomatic (94.3% in CDC stages I or II). In our analysis of risk factors for C. trachomatis infection, neither HIV serostatus nor CDC stage were significant in the univariate level, and thus the clinical stage was not included in the multivariate analysis. We also did not find an increased susceptibility to other STD infections frequently observed in high-risk populations with high rates of HIV (5, 19, 21), perhaps due to the lack of disease or immune suppression. In addition to a lower prevalence of HIV in our population, regular STD surveillance is part of the Senegalese government-run program for prostitution. The women are seen every 2 to 3 months; thus, STDs are detected early and treated immediately. Before resuming work, women are tested again to ensure clearance of the infection. Thus, the women in our study maintain high-risk behavior, but the clinic provides regular and prompt health care which may distinguish this cohort from others. Therefore, the lack of association between C. trachomatis infection and HIV in this study compared to previously published work (22, 30) may be in part due to the lower rates of HIV among the Dakar commercial sex workers and their overall health status.
Another possible explanation for the lack of association between chlamydial infection and other risk factors (including STDs), is the high sensitivity of the diagnostic C. trachomatis PCR assay employed. This particular assay is capable of detecting infections with very low copy numbers of C. trachomatis and thus would detect a higher rate of asymptomatic infections. These infections would have gone undetected by other microbiological assays, like those used in earlier epidemiological studies. To date, most of the studies that report a significant association between C. trachomatis infection and STDs and HIV used non-PCR-based assays (5, 13, 22, 30). The association may only exist in the case of an infection with high copy numbers of C. trachomatis and/or with symptomatic infections, but further work will be needed to confirm this hypothesis.
We found that the presence of yeast on a genital secretion smear was associated with significantly lower risk of C. trachomatis infection both in the univariate and multivariate analysis (adjusted OR = 0.34; 95% CI = 0.11 to 0.99). There are multiple explanations for such an association. The observation of yeast on a smear indicates an overgrowth of the organism in the genital tract, and this may inhibit infection with C. trachomatis or indirectly hinder the growth of C. trachomatis by rendering the genital environment inhospitable. It should be noted that the increased presence of yeast may also indicate the current or recent use of antibiotics which could affect the balance of vaginal flora, thereby limiting chlamydial infections or perhaps even clearing them. We were unable to address this important question, since data were not gathered on concomitant antibiotic use, and self-prescribed antibiotic use is a frequent practice.
To our knowledge, this is the first study to describe the genotypic diversity of C. trachomatis genital infections in West Africa. It is also the first molecular epidemiology study of C. trachomatis performed in a largely healthy, clinically monitored high-risk population. Since only a subset (10.2%) of samples positive by diagnostic PCR were genotyped, we performed a comparative analysis on the C. trachomatis-positive women with genotyped specimens and women whose specimen was not genotyped. No significant differences in sociodemographic characteristics were observed in a multivariate analysis (data not shown); we therefore feel that the identified genotypes may be considered representative of the C. trachomatis infections in this population. Despite the small number of samples genotyped, a heterogeneous population of C. trachomatis was found in the cohort, including genotypes D, E, F, G, Ia, and K. As in other studies on genotypic diversity in cervical infections (2, 5, 40, 41, 46), D and E were the predominant genotypes identified in cervical samples (66.7%). The predominance of these class B genotypes in different studies and at different times may suggest a real biological advantage over other genotypes.
In the present study, most of the C. trachomatis infections seemed to be asymptomatic, with only 29 of the 206 (14.1%) C. trachomatis-positive women showing signs of cervical inflammation. There is currently no clear consensus on the association between C. trachomatis strains and the severity of disease in a genital infection, since identical genotypes or serovars have been associated with asymptomatic or symptomatic infections in different studies (2, 9, 40, 41, 46). Interestingly, genotype E was found in about half of the characterized samples, and none (0 of 10) of the E strains were associated with visible signs of cervical inflammation, compared to 5 of 11 non-E genotypes (P < 0.02; Fisher's exact test). These results are suggestive of differences in pathogenicity by genotype, with genotype E being less pathogenic. In support of this finding, van Duynhoven et al. (41), in a study performed at University Hospital in Rotterdam, observed that women infected with genotypes E, F, and G less frequently had ≥10 detectable leukocytes per microscopic field on cervical smears than C-complex or D/D− genotypes, indicating a lower degree of cervical inflammation. In a histopathological study performed in San Francisco, Dean et al. (9) showed that genotype E infections were largely asymptomatic (92%) and less often associated with plasma cells and neutrophils on endometrial biopsy than infections with genotypes F, F variants, I, D, H, K, and G.
A high prevalence of genotype E (10 of 21, 47.6%) and its lack of associated clinical symptoms may suggest this genotype is more successful than other, less-prevalent genotypes in our study population. Indeed, a successful genotype would be one that goes undetected for a longer period of time, enhancing dissemination. Variability of the main C. trachomatis antigen MOMP is presumably the result of host selection and bacterial adaptation. Thus, the MOMP sequence that elicits a milder immune response in the infected host could be an adaptive mode of evolution to escape immune pressure and might therefore confer a transmission advantage over other MOMP genotypes. The high rate of asymptomatic C. trachomatis infection and the high rate of genotype E detected in our cohort support this hypothesis. Further studies are required to test this hypothesis and to assess its generalizability for other populations.
The presence of a large asymptomatic pool of C. trachomatis infection in a high-risk population such as FCSW may be of significant concern for any HIV prevention program. Indeed, the high frequency of sexually transmitted diseases in areas such as sub-Saharan Africa has been raised as a possible explanation for the ravaging HIV epidemic (13, 15, 30). It would therefore be quite important to assess if symptomatic and asymptomatic chlamydial infections are equal in their risk for HIV infection and/or if C. trachomatis genotypes differ in their ability to influence HIV infection.
In conclusion, we have detected a high prevalence of C. trachomatis genital infection among healthy FCSW in Dakar, Senegal, by using a highly sensitive diagnostic PCR. An increased number of sexual partners was associated with risk of infection, while the presence of a yeast infection was negatively associated with chlamydial infection. Most of the infections were asymptomatic and were caused by a diverse group of chlamydial genotypes. Interestingly, the most commonly detected genotype (E) was not associated with signs of cervical inflammation compared to non-E genotypes. Future studies will be needed to further investigate the role of C. trachomatis genotypes and disease manifestations and its implication for facilitation of the HIV epidemic.
ACKNOWLEDGMENTS
We thank Boris Renjifo for providing us with β-globin primers P55 and P57, Ruth Ravkin (Department of Medical Microbiology, Brigham and Women's Hospital, Boston, Mass.) for kindly providing us with C. trachomatis L2 serovar cultures, and Fredrik Vannberg and Pulin Shah for technical assistance. We also thank the personnel of the Laboratoire de Bactériologie-Virologie, Hôpital Le Dantec, and at the Institut d'Hygiène Sociale, Dakar, Senegal, for their invaluable help.
This work was supported by grants from the U.S. Army Medical Research and Material Command (17-96-C-5005) and from the National Institutes of Health (N01-AI-35173-123). I.T. and J.L.S. are Fogarty International Fellows (5 D43 TW00004).
REFERENCES
- 1.Baehr W, Zhang Y X, Joseph T, Su H, Nano F E, Everett K D, Caldwell H D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci USA. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batteiger B E, Lennington W, Newhall W J, Katz B P, Morrison H T, Jones R B. Correlation of infecting serovar and local inflammation in genital chlamydial infections. J Infect Dis. 1989;160:332–336. doi: 10.1093/infdis/160.2.332. [DOI] [PubMed] [Google Scholar]
- 3.Bauwens J E, Clark A M, Stamm W E. Diagnosis of Chlamydia trachomatis endocervical infections by a commercial polymerase chain reaction assay. J Clin Microbiol. 1993;31:3023–3027. doi: 10.1128/jcm.31.11.3023-3027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham R, Yang C L, Maclean I, Kimani J, Maitha G, Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (Omp1) gene. J Clin Investig. 1994;94:458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunham R C, Kimani J, Bwayo J, Maitha G, Maclean I, Yang C L, Shen C X, Roman S, Nagelkerke N J D, Cheang M, Plummer F A. The epidemiology of chlamydia trachomatis within a sexually transmitted diseases core group. J Infect Dis. 1996;173:950–956. doi: 10.1093/infdis/173.4.950. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. Morbid Mortal Weekly Rep. 1987;36:1S–15S. [PubMed] [Google Scholar]
- 8.Dean D, Millman K. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J Clin Investig. 1997;99:475–483. doi: 10.1172/JCI119182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean D, Oudens E, Bolan G, Padian N, Schachter J. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J Infect Dis. 1995;172:1013–1022. doi: 10.1093/infdis/172.4.1013. [DOI] [PubMed] [Google Scholar]
- 10.Dean D, Stephens R S. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J Clin Microbiol. 1994;32:1506–1510. doi: 10.1128/jcm.32.6.1506-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fielder T J, Peterson E M, de la Maza L M. Nucleotide sequence of DNA encoding the major outer membrane protein of Chlamydia trachomatis serovar L3. Gene. 1991;101:159–160. doi: 10.1016/0378-1119(91)90240-c. [DOI] [PubMed] [Google Scholar]
- 12.Gaydos C A, Howell M R, Pare B, Clark K L, Ellis D A, Hendrix R M, Gaydos J C, McKee K T, Jr, Quinn T C. Chlamydia trachomatis infections in female military recruits. New Engl J Med. 1998;339:739–744. doi: 10.1056/NEJM199809103391105. [DOI] [PubMed] [Google Scholar]
- 13.Ghys P D, Fransen K, Diallo M O, Ettiegne-Traore V, Coulibaly I M, Yeboue K M, Kalish M L, Maurice C, Whitaker J P, Greenberg A E, Laga M. The associations between cervicovaginal HIV shedding, sexually transmitted diseases and immunosuppression in female sex workers in Abidjan, Cote d'Ivoire. AIDS. 1997;11:F85–F93. doi: 10.1097/00002030-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Grayston J T, Wang S. New knowledge of chlamydiae and the diseases they cause. J Infect Dis. 1975;132:87–105. doi: 10.1093/infdis/132.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Grosskurth H, Mosha F, Todd J, Mwijarubi E, Klokke A, Senkoro K, Mayaud P, Changalucha J, Nicoll A, ka-Gina G, et al. Impact of improved treatment of sexually transmitted diseases on HIV infection in rural Tanzania: randomised controlled trial. Lancet. 1995;346:530–536. doi: 10.1016/s0140-6736(95)91380-7. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton P T, Malinowski D P. Nucleotide sequence of the major outer membrane protein gene from Chlamydia trachomatis serovar H. Nucleic Acids Res. 1989;17:8366. doi: 10.1093/nar/17.20.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay P E, Ghaem-Maghami S. Chlamydia and non-gonococcal urethritis. Curr Opin Infect Dis. 1997;10:44–49. [Google Scholar]
- 18.Hayes L J, Clarke I N. Nucleotide sequence of the major outer membrane protein gene of Chlamydia trachomatis strain A/SA1/OT. Nucleic Acids Res. 1990;18:6136. doi: 10.1093/nar/18.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamenga M C, De Cock K M, St. Louis M E, Toure C K, Zakaria S, N'Gbichi J M, Ghys P D, Holmes K K, Eschenbach D A, Gayle H D, et al. The impact of human immunodeficiency virus infection on pelvic inflammatory disease: a case-control study in Abidjan, Ivory Coast. Am J Obstet Gynecol. 1995;172:919–925. doi: 10.1016/0002-9378(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 20.Kanki P, M'Boup S, Marlink R, Travers K, Hsieh C C, Gueye A, Boye C, Sankale J L, Donnelly C, Leisenring W, et al. Prevalence and risk determinants of human immunodeficiency virus type 2 (HIV-2) and human immunodeficiency virus type 1 (HIV-1) in west African female prostitutes. Am J Epidemiol. 1992;136:895–907. doi: 10.1093/aje/136.7.895. [DOI] [PubMed] [Google Scholar]
- 21.Korn A P, Landers D V. Gynecologic disease in women infected with human immunodeficiency virus type 1. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;9:361–370. [PubMed] [Google Scholar]
- 22.Laga M, Manoka A, Kivuvu M, Malele B, Tuliza M, Nzila N, Goeman J, Behets F, Batter V, Alary M, et al. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS. 1993;7:95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- 23.Lan J, Melgers I, Meijer C J, Walboomers J M, Roosendaal R, Burger C, Bleker O P, van den Brule A J. Prevalence and serovar distribution of asymptomatic cervical Chlamydia trachomatis infections as determined by highly sensitive PCR. J Clin Microbiol. 1995;33:3194–3197. doi: 10.1128/jcm.33.12.3194-3197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan J, Ossewaarde J M, Walboomers J M, Meijer C J, van den Brule A J. Improved PCR sensitivity for direct genotyping of Chlamydia trachomatis serovars by using a nested PCR. J Clin Microbiol. 1994;32:528–530. doi: 10.1128/jcm.32.2.528-530.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan J, Walboomers J M, Roosendaal R, van Doornum G J, MacLaren D M, Meijer C J, van den Brule A J. Direct deletion and genotyping of Chlamydia trachomatis in cervical scrapes by using polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1060–1065. doi: 10.1128/jcm.31.5.1060-1065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison R P, Manning D S, Caldwell H D, editors. Immunology of Chlamydia trachomatis infections: immunoprotective and immunopathogenic responses. New York, N.Y: Raven Press; 1992. [Google Scholar]
- 27.Palmer L, Falkow S. A common plasmid of Chlamydia trachomatis. Plasmid. 1986;16:52–62. doi: 10.1016/0147-619x(86)90079-x. [DOI] [PubMed] [Google Scholar]
- 28.Peterson E M, Markoff B A, de la Maza L M. The major outer membrane protein nucleotide sequence of Chlamydia trachomatis, serovar E. Nucleic Acids Res. 1990;18:3414. doi: 10.1093/nar/18.11.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickett M A, Ward M E, Clarke I N. Complete nucleotide sequence of the major outer membrane protein gene from Chlamydia trachomatis serovar L1. FEMS Microbiol Lett. 1987;42:185–190. [Google Scholar]
- 30.Plummer F A, Simonsen J N, Cameron D W, Ndinya-Achola J O, Kreiss J K, Gakinya M N, Waiyaki P, Cheang M, Piot P, Ronald A R, et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 31.Sarr A D, Hamel D J, Thior I, Kokkotou E, Sankale J L, Marlink R G, Coll-Seck E M, Essex M E, Siby T, Ndoye I, Mboup S, Kanki P J. HIV-1 and HIV-2 dual infection: lack of HIV-2 provirus correlates with low CD4+ lymphocyte counts. AIDS. 1998;12:131–137. doi: 10.1097/00002030-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Sayada C, Denamur E, Elion J. Complete sequence of the major outer membrane protein-encoding gene of Chlamydia trachomatis serovar Da. Gene. 1992;120:129–130. doi: 10.1016/0378-1119(92)90022-h. [DOI] [PubMed] [Google Scholar]
- 33.Schachter J. Which test is best for chlamydia? Curr Opin Infect Dis. 1999;12:41–45. doi: 10.1097/00001432-199902000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Stephens R S, Mullenbach G, Sanchez-Pescador R, Agabian N. Sequence analysis of the major outer membrane protein gene from Chlamydia trachomatis serovar L2. J Bacteriol. 1986;168(3):1277–1282. doi: 10.1128/jb.168.3.1277-1282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens R S, Sanchez-Pescador R, Wagar E A, Inouye C, Urdea M S. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169:3879–3785. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stothard D R, Boguslawski G, Jones R B. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theunissen J J, Kariwiga G, Ossewaarde J M, van Rijsoort-Vos J H, Stolz E, van der Meijden W I. Prevalence of Chlamydia trachomatis in women attending a family planning clinic in Papua New Guinea. Genitourin Med. 1995;715:295–298. doi: 10.1136/sti.71.5.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers K, Mboup S, Marlink R, Gueye-Nidaye A, Siby T, Thior I, Traore I, Dieng-Sarr A, Sankale J L, Mullins C, et al. Natural protection against HIV-1 infection provided by HIV-2. Science. 1995;268:1612–1615. doi: 10.1126/science.7539936. [DOI] [PubMed] [Google Scholar]
- 40.van de Laar M J, van Duynhoven Y T, Fennema J S, Ossewaarde J M, van den Brule A J, van Doornum G J, Coutinho R A, van den Hoek J A. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin Med. 1996;72:261–265. [PMC free article] [PubMed] [Google Scholar]
- 41.van Duynhoven Y T, Ossewaarde J M, Derksen-Nawrocki R P, van der Meijden W I, van de Laar M J. Chlamydia trachomatis genotypes: correlation with clinical manifestations of infection and patients' characteristics. Clin Infect Dis. 1998;26:314–322. doi: 10.1086/516291. [DOI] [PubMed] [Google Scholar]
- 42.Van Duynhoven Y T, van de Laar M J, Schop W A, Mouton J W, van der Meijden W I, Sprenger M J. Different demographic and sexual correlates for chlamydial infection and gonorrhoea in Rotterdam. Int J Epidemiol. 1997;26:1373–1385. doi: 10.1093/ije/26.6.1373. [DOI] [PubMed] [Google Scholar]
- 43.Van Dyck E, Samb N, Sarr A D, Van de Velden L, Moran J, Mboup S, Ndoye I, Lamboray J L, Meheus A, Piot P. Accuracy of two enzyme immunoassays and cell culture in the detection of Chlamydia trachomatis in low and high risk populations in Senegal. Eur J Clin Microbiol Infect Dis. 1992;11:527–534. doi: 10.1007/BF01960808. [DOI] [PubMed] [Google Scholar]
- 44.Wang S P, Grayston J T. Three new serovars of Chlamydia trachomatis: Da, Ia and L2a. J Infect Dis. 1991;163:403–405. doi: 10.1093/infdis/163.2.403. [DOI] [PubMed] [Google Scholar]
- 45.Wang S P, Kuo C C, Barnes R C, Stephens R S, Grayston J T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis. 1985;152:791–800. doi: 10.1093/infdis/152.4.791. [DOI] [PubMed] [Google Scholar]
- 46.Workowski K A, Stevens C E, Suchland R J, Holmes K K, Eschenbach D A, Pettinger M B, Stamm W E. Clinical manifestations of genital infection due to Chlamydia trachomatis in women: differences related to serovar. Clin Infect Dis. 1994;19:756–760. doi: 10.1093/clinids/19.4.756. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Y, Zhang Y X, Watkins N G, Caldwell H D. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect Immun. 1989;57:1040–1049. doi: 10.1128/iai.57.4.1040-1049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y X, Morrison S G, Caldwell H D. The nucleotide sequence of major outer membrane protein gene of Chlamydia trachomatis serovar F. Nucleic Acids Res. 1990;18:1061. doi: 10.1093/nar/18.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]