ABSTRACT
The Pseudomonas aeruginosa lipoprotein LbcA was discovered because it copurified with and promoted the activity of CtpA, a carboxyl-terminal processing protease (CTP) required for type III secretion system function and virulence in a mouse model of acute pneumonia. In this study, we explored the role of LbcA by determining its effect on the proteome and its participation in protein complexes. lbcA- and ctpA-null mutations had strikingly similar effects on the proteome, suggesting that assisting CtpA might be the most impactful role of LbcA in the bacterial cell. Independent complexes containing LbcA and CtpA, or LbcA and a substrate, were isolated from P. aeruginosa cells, indicating that LbcA facilitates proteolysis by recruiting the protease and its substrates independently. An unbiased examination of proteins that copurified with LbcA revealed an enrichment for proteins associated with the cell wall. One of these copurification partners was found to be a new CtpA substrate and the first substrate that is not a peptidoglycan hydrolase. Many of the other LbcA copurification partners are known or predicted peptidoglycan hydrolases. However, some of these LbcA copurification partners were not cleaved by CtpA, and an in vitro assay revealed that while CtpA and all of its substrates bound to LbcA directly, these nonsubstrates did not. Subsequent experiments suggested that the nonsubstrates might copurify with LbcA by participating in multienzyme complexes containing LbcA-binding CtpA substrates.
IMPORTANCE Carboxyl-terminal processing proteases (CTPs) are widely conserved and associated with the virulence of several bacteria, including CtpA in Pseudomonas aeruginosa. CtpA copurifies with the uncharacterized lipoprotein LbcA. This study shows that the most impactful role of LbcA might be to promote CtpA-dependent proteolysis and that it achieves this as a scaffold for CtpA and its substrates. It also reveals that LbcA copurification partners are enriched for cell wall-associated proteins, one of which is a novel CtpA substrate. Some of the LbcA copurification partners are not cleaved by CtpA but might copurify with LbcA because they participate in multienzyme complexes containing CtpA substrates. These findings are important because CTPs and their associated proteins affect peptidoglycan remodeling and virulence in multiple species.
KEYWORDS: Pseudomonas aeruginosa, cell envelope, proteases
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative bacterium that is widespread in the environment and responsible for serious opportunistic infections, especially in health care settings (1). As with any bacterial pathogen, the cell envelope of P. aeruginosa is the focal point for its interaction with a host. Many critical virulence factors are assembled or exported through the cell envelope, including the type III secretion system (T3SS) and various polysaccharides, which play important roles in acute and chronic infections, respectively (2, 3). The peptidoglycan cell wall is a layer of the cell envelope that helps maintain bacterial cell integrity, has to be remodeled during growth and virulence factor assembly/export, and is one of the most important targets for antibiotics (4–6). Therefore, a comprehensive understanding of proteins involved in cell wall assembly, remodeling, and associated functions has obvious significance.
The cleavage of proteins and peptides in the cell envelope plays roles in protein export, protein quality control and turnover, signal transduction, and the integrity and functions of the cell wall (7–10). One widely conserved family of serine proteases that function within the bacterial cell envelope is the carboxyl-terminal processing proteases (CTPs). Their name arose from early findings that CTPs can process protein substrates to a functional form by removing a small segment from their C-terminal end (11–13). However, more recent findings have demonstrated that at least one CTP removes an N-terminal segment from its substrate (14). It has also emerged that bacterial CTPs can completely degrade some substrates, including peptidoglycan cross-link hydrolases and at least one lytic transglycosylase, in Escherichia coli and P. aeruginosa (15–18). Therefore, CTPs appear to be one of the mechanisms used by bacteria to control cell wall hydrolase activity.
Another feature that has been uncovered in the last few years, for two quite different CTPs, is that they can function in partnership with another protein. The E. coli Prc protease interacts with the tetratricopeptide repeat (TPR)-containing outer membrane (OM) lipoprotein NlpI to degrade the NlpC/P60 endopeptidase MepS, which is a peptidoglycan cross-link hydrolase (16). Structural analysis suggests that NlpI recruits MepS and helps to feed it into the Prc active site for destruction (19). However, Prc can also cleave at least three substrates without the help of NlpI (15, 18, 20). In P. aeruginosa, the CtpA protease interacts with the TPR-containing lipoprotein LbcA to degrade four different predicted peptidoglycan cross-link hydrolases, the NlpC/P60 endopeptidases PA1198 and PA1199 and the LytM/M23 endopeptidases MepM and PA4404 (17). Despite the obvious similarities between the NlpI-Prc and LbcA-CtpA systems, it is not at all clear if they function by the same molecular mechanisms. E. coli Prc and P. aeruginosa CtpA are not orthologs, being quite different in size and belonging to different CTP subfamilies (21–23). In fact, P. aeruginosa has a second CTP that is the apparent ortholog of E. coli Prc (24–26). In addition to the differences between the E. coli Prc and P. aeruginosa CtpA proteases, although their NlpI and LbcA lipoprotein partners both have the short degenerate TPR motifs, they are very different in size and share no obvious primary sequence homology when aligned (17).
In this study, we have investigated the role of LbcA in P. aeruginosa. We found that lbcA- and ctpA-null mutations have remarkably similar effects on the proteome, suggesting that the most impactful role of LbcA might be to facilitate CtpA-dependent proteolysis. LbcA does this, at least in part, by binding to CtpA and its substrates directly, but independently, to bring them together for proteolysis. We also found that the most abundant LbcA copurification partners are enriched for several cell wall-associated proteins and that one of these proteins is a newly discovered CtpA substrate. Some other LbcA copurification partners are not CtpA substrates, and unlike the substrates, our data suggest that they might associate with LbcA indirectly by participating in cell wall enzyme complexes.
RESULTS
lbcA- and ctpA-null mutations have overlapping effects on the proteome.
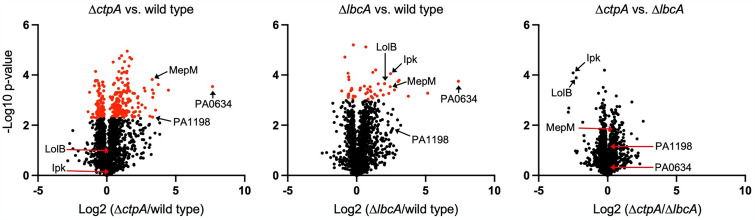
To begin to investigate the role of LbcA, we used label-free quantitative proteomics (LFQP) to compare protein levels in wild-type, ΔctpA, and ΔlbcA strains (see Tables S1 to S3 in the supplemental material). When protein levels were compared between strains, it was apparent that the ΔctpA and ΔlbcA mutations had overlapping effects on the proteome compared to the wild type, whereas there were fewer differences in protein levels when the two mutants were compared to each other (Fig. 1). A closer examination of the data supported the conclusion of the overlap between the effects of the ΔctpA and ΔlbcA mutations. For example, when ranked by fold increases in abundance compared to the wild type, the top 50 proteins in the ΔctpA mutant all had increased levels in the ΔlbcA mutant as well (Tables S1 and S2). Two CtpA substrates, MepM and PA1198, could be quantified in all three strains and were within the top 25 proteins ranked by fold increases in abundance when either the ΔctpA or ΔlbcA mutant was compared to the wild type (Fig. 1; Tables S1 and S2). Other changes in the mutants were also similar, and many might be caused by effects on gene expression. For example, PA0634 had the largest increase in abundance in the ΔctpA and ΔlbcA mutants, and several other proteins encoded by genes closely linked to PA0634 were also increased in both mutants (Fig. 1; Tables S1 and S2). This region encodes the R- and F-type pyocins, which are induced by stresses, including DNA damage and the overproduction of the extracytoplasmic function sigma factor PA4896 (27, 28). Stress from the accumulating substrates of CtpA might induce pyocin gene expression indirectly. There was also overlap in the most-reduced-abundance proteins in the ΔctpA and ΔlbcA mutants, which included some regulators, components, and effectors of the type III secretion system (T3SS) (Tables S1 and S2). Although our experiments were not done under T3SS-inducing conditions, this is consistent with the ΔctpA mutant compromising T3SS function (23). There were two striking differences between the ΔctpA and ΔlbcA mutants, which were increased abundances of LolB and Ipk in the ΔlbcA mutant only (Fig. 1). However, this is a trivial consequence of lbcA being replaced by the gentamicin resistance gene aacC1 in the same orientation as that of lbcA in order to maintain sufficient expression of the essential downstream genes lolB and ipk (17). Therefore, these LFQP data support the hypothesis that a major role of LbcA is to enable CtpA-dependent proteolysis.
FIG 1.
ΔlbcA and ΔctpA mutations have overlapping effects on the proteome. Each chart shows the relative protein levels (x axis) between the pair of strains indicated above and the statistical significance of each difference (y axis). Circles represent individual proteins. Data for the CtpA and LbcA proteins themselves are not included in the plots. Symbols in red indicate measurements considered significant after a 5% Benjamini-Hochberg-based false discovery rate cutoff was applied.
LbcA can form independent complexes with CtpA and its substrates.
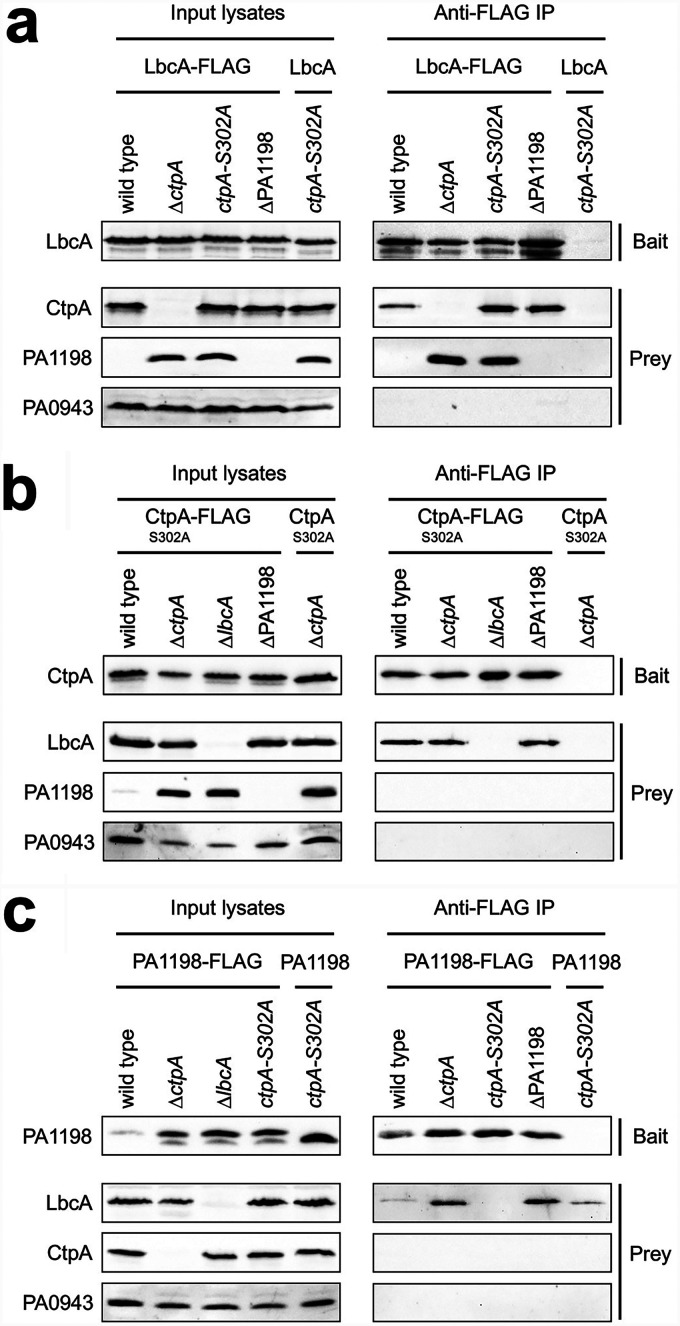
LbcA has several tetratricopeptide repeat (TPR) motifs, which mediate protein-protein interactions and the formation of multiprotein complexes (17, 29). Therefore, we hypothesized that LbcA enables CtpA-dependent proteolysis by acting as a scaffold protein to recruit CtpA and its substrates independently. To test this, we used coimmunoprecipitation (co-IP) assays to investigate in vivo complexes containing LbcA, CtpA, and the representative CtpA substrate, PA1198.
The first bait was LbcA with a C-terminal FLAG tag (LbcA-FLAG), which was produced in the wild-type, ΔctpA, ctpA-S302A, or ΔPA1198 strain. The ctpA-S302A mutation destroys the proteolytic activity of CtpA, allowing the analysis of complexes that contain an intact substrate (17). Analysis of anti-FLAG monoclonal antibody immunoprecipitates revealed that both CtpA and PA1198 copurified with LbcA-FLAG in the ctpA-S302A strain (Fig. 2a). PA1198 still copurified with LbcA-FLAG in a ΔctpA mutant, and CtpA still copurified with LbcA-FLAG in a ΔPA1198 mutant. Therefore, these data suggest that LbcA can form independent complexes with CtpA and its substrate PA1198 in vivo. As a specificity control, the unrelated periplasmic PA0943 protein (30) was not detected by immunoblot analysis of LbcA-FLAG immunoprecipitates from any strain. Furthermore, when the LbcA bait did not have a FLAG tag, none of the proteins tested were detected above trace levels in anti-FLAG immunoprecipitates (Fig. 2a).
FIG 2.
LbcA can form independent complexes with CtpA and its substrates. Each panel shows data from immunoblot analyses of input lysates and immunoprecipitates (Anti-FLAG IP). Strain genotypes are shown above each lane. Polyclonal antisera used for detection are shown at the left. (a) Baits were LbcA-FLAG or the LbcA negative control. (b) Baits were CtpA-S302A–FLAG–His6 or the CtpA-S302A–His6 negative control. (c) Baits were PA1198-FLAG or the PA1198 negative control. Immunoblots are single representatives of results from several independent replicate experiments.
Next, we used CtpA-S302A with a C-terminal FLAG-His6 tag as the bait protein (17). Analysis of the anti-FLAG immunoprecipitates revealed that LbcA copurified with CtpA, but the PA1198 substrate did not (Fig. 2b). This further supports the presence of an LbcA-CtpA complex, but it suggests that CtpA cannot form a stable complex with its substrate alone. Indeed, when we went on to use the PA1198-FLAG substrate as the bait, LbcA copurified with PA1198, but CtpA did not (Fig. 2c). Therefore, all of these co-IPs suggest that LbcA can form independent complexes with CtpA or its substrate, but CtpA cannot form a stable binary complex with its substrate alone. This supports the hypothesis that LbcA functions, at least in part, by binding to CtpA and its substrate independently in order to bring them together for proteolysis.
LbcA copurification partners are enriched for cell wall enzymes and binding proteins.
The above-described experiments suggested that a major role of LbcA is to facilitate CtpA-dependent proteolysis, which it achieves by acting as a scaffolding protein. Therefore, we reasoned that another unbiased approach to learn more about the targets of the LbcA-CtpA complex, and perhaps any additional role(s) of LbcA, would be to identify LbcA binding partners. For this, two independent purifications of LbcA-FLAG from a ctpA-S302A strain were analyzed by mass spectrometry (Table S4). To control for specificity, we also analyzed two independent purifications in which the LbcA bait protein was not FLAG tagged (Table S5). Sixty-seven known or predicted envelope proteins were enriched in the LbcA-FLAG purification (Table S6). CtpA was the most abundant of these proteins based on peptide spectral matches (PSMs), and three of the known CtpA substrates, MepM, PA1198, and PA1199, were also detected. A striking observation was that half of the 18 most abundant proteins that copurified with LbcA-FLAG were known or predicted cell wall-associated proteins (Table S6). This is in contrast to only 5 of the remaining 49 lower-abundance proteins. Therefore, the most abundant LbcA copurification partners were enriched for cell wall enzymes and cell wall-binding proteins. These data suggest that LbcA is associated with multiple cell wall-related proteins, beyond the previously known proteolytic substrates of CtpA.
Proteins in complex with LbcA in vivo include CtpA substrates and nonsubstrates.
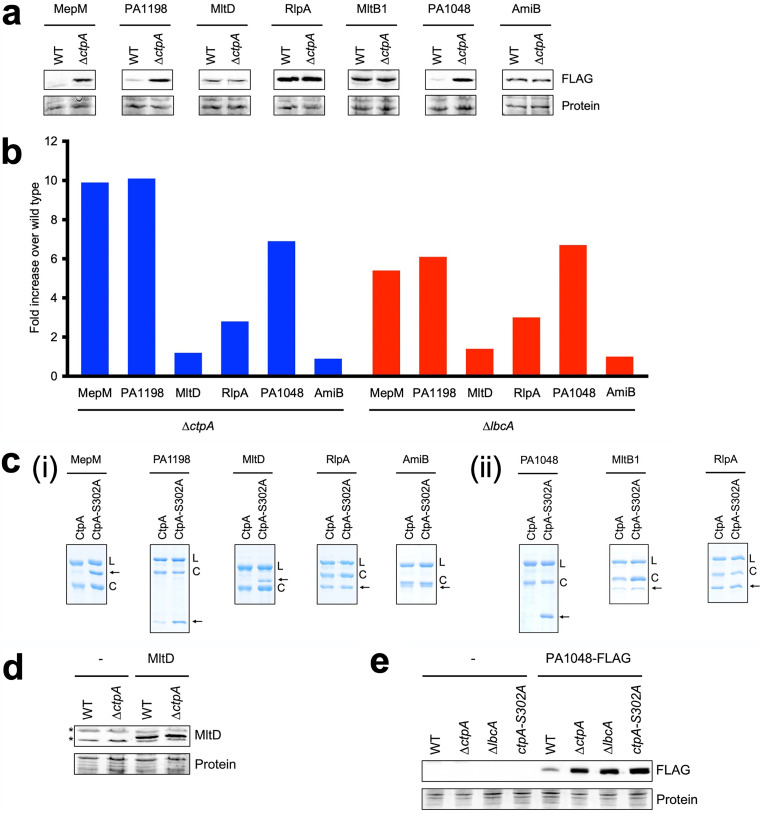
We could not follow up on all 67 envelope proteins that were enriched in the LbcA-FLAG immunoprecipitation (Table S6). However, one of our motivations was to identify new substrates of the LbcA-CtpA complex. In a previous study, the CtpA substrates MepM and PA1198 were discovered because they were the most abundant proteins trapped by an inactive LbcA-CtpA-S302A complex after in vivo cross-linking (17). Many other proteins were trapped exclusively by the inactive LbcA-CtpA-S302A complex, but again, they were too numerous for individual follow-up. Therefore, for this study, out of the 67 proteins that copurified with LbcA-FLAG, we decided to focus on those that had also been trapped by the inactive LbcA-CtpA-S302A complex exclusively in the previous work (17). The addition of this criterion reduced the number of LbcA-FLAG copurification partners for follow-up to seven proteins, all of which are predicted cell wall hydrolases or peptidoglycan-binding proteins (Table 1). These included the two known CtpA substrates MepM and PA1198 but also three proteins, PA1048, MltB1, and AmiB, that had not been investigated previously for the possibility of a relationship with LbcA and/or CtpA. The remaining two proteins, the lytic transglycosylases RlpA and MltD, were assessed as possible CtpA substrates in the previous study. However, RlpA-FLAG and MltD-FLAG did not accumulate in a ΔctpA mutant, suggesting that they are not CtpA substrates (17). Nevertheless, we included RlpA and MltD in our follow-up here because the previous analysis was not exhaustive.
TABLE 1.
Envelope proteins that copurified with LbcA and were also isolated by an LbcA-CtpA-S302A protease trapa
| Protein | Predicted feature or function | Abundance rank with LbcAb | Abundance rank with LbcA-CtpA trapc | CtpA substrate of Srivastava et al.d |
|---|---|---|---|---|
| PA1198 | Peptidoglycan NlpC/P60 endopeptidase | 3 | 2 | Yes |
| PA1048 | Peptidoglycan-binding OmpA C-terminal domain | 4 | 7 | NT |
| RlpA | Peptidoglycan lytic transglycosylase | 5 | 4 | No |
| MltD | Peptidoglycan lytic transglycosylase | 10 | 3 | No |
| MltB1 | Peptidoglycan lytic transglycosylase | 13 | 6 | NT |
| MepM | Peptidoglycan LytM/M23 endopeptidase | 18 | 1 | Yes |
| AmiB | Peptidoglycan amidase | 65 | 23 | NT |
Cell envelope proteins that copurified with LbcA-CtpA-S302A, but not with LbcA-CtpA, following in vivo cross-linking (17) and also copurified with LbcA-FLAG from a ctpA-S302A strain in this study with average peptide spectral matches ≥5-fold higher than those in an LbcA no-FLAG-tag negative-control purification (see Tables S4 to S6 in the supplemental material).
Rank based on the average number of peptide spectral matches among 67 cell envelope proteins that copurified with LbcA-FLAG (Table S6).
Rank based on the average number of peptide spectral matches among 25 cell envelope proteins that copurified exclusively with the LbcA-CtpA-S302A trap (17).
In the study by Srivastava et al., a protein was classified as a CtpA substrate if a FLAG-tagged version accumulated in a ΔctpA mutant in vivo and the protein was degraded by CtpA in vitro (17). NT, not tested by Srivastava et al.
We tested rigorously if these proteins might be CtpA substrates, with the previously characterized substrates MepM and PA1198 serving as positive controls. To screen in vivo, we constructed plasmids encoding C-terminally FLAG-tagged versions, expressed from a nonnative promoter, and determined if they accumulated in a ΔctpA mutant (Fig. 3a). As reported previously, the CtpA substrates MepM and PA1198 accumulated in the ΔctpA mutant, whereas RlpA and MltD did not (17). MltB1 and AmiB did not accumulate in the ΔctpA mutant, suggesting that like RlpA and MltD, they are not CtpA substrates. However, PA1048 behaved like the known CtpA substrates. It was barely detectable in the wild type but accumulated robustly in the ΔctpA mutant (Fig. 3a). We also examined the data for these proteins from our previous LFQP analysis and found that the endogenous proteins had a pattern similar to that of the plasmid-encoded FLAG-tagged versions. MepM, PA1198, and PA1048 levels were approximately 6- to 10-fold higher in the ΔctpA and ΔlbcA mutants, whereas MltD, RlpA, and AmiB had more modest changes (Fig. 3b) (note that in the LFQP analysis, MltB1 was not detected well in multiple samples and so could not be quantified).
FIG 3.
Analysis of LbcA copurification partners. (a) Immunoblots of whole-cell lysates of wild-type (WT) and ΔctpA strains containing a plasmid encoding C-terminally FLAG-tagged versions of the indicated proteins. Proteins were detected with anti-FLAG monoclonal antibodies (FLAG). (b) LFQP analysis of endogenous proteins in ΔctpA and ΔlbcA mutants. (c) In vitro proteolysis assay. Natively purified (i) or renatured (ii) N-terminally His6-tagged versions of the proteins indicated at the top were incubated with LbcA-His6 and either CtpA-His6 or CtpA-S302A–His6 for 3 h at 37°C. Samples were separated on SDS-PAGE gels and stained with Coomassie brilliant blue. L, LbcA-His6; C, CtpA-His6 or CtpA-S302A–His6. Arrows indicate the N-terminally His6-tagged protein being tested. (d) Immunoblot analysis of whole-cell lysates of wild-type and ΔctpA strains containing a plasmid encoding untagged MltD or the empty vector (−). MltD was detected with a polyclonal antiserum. All strains had a ΔmltD mutation. Asterisks indicate cross-reactive proteins. (e) Immunoblot analysis of whole-cell lysates of wild-type, ΔctpA, ΔlbcA, and ctpA-S302A strains containing a plasmid encoding PA1048-FLAG or the empty vector (−). For panels a, d, and e, loading was determined by Ponceau S total protein staining of the nitrocellulose membrane used for detection (Protein). Immunoblots are single representatives of results from several independent replicate experiments.
Next, we tested if N-terminally His6-tagged versions of these proteins could be cleaved by CtpA in vitro, as described previously (17). In agreement with the in vivo data, MepM and PA1198 were degraded, whereas RlpA and AmiB were not, which confirmed that RlpA and AmiB are not CtpA substrates (Fig. 3c). However, MltD was degraded in vitro, even though MltD-FLAG and endogenous MltD did not accumulate in ΔctpA mutants in vivo. This left open the possibility that MltD is a CtpA substrate (see below). PA1048 and MltB1 were insoluble when overproduced in E. coli prior to purification. To circumvent this, we solubilized PA1048 and MltB1 by denaturation and then renatured them on a nickel-agarose column prior to elution. It was possible that this renatured PA1048 would be misfolded, rendering it a substrate of CtpA. We could not eliminate that possibility, but to ensure that this procedure would not render any protein a CtpA substrate, we also repurified RlpA in the same way. Analysis of the renatured proteins showed that PA1048 was degraded by CtpA, whereas MltB and the RlpA control were not (Fig. 3c). Therefore, together, all of these analyses showed that the LbcA copurification partners RlpA, AmiB, MltB1, and possibly MltD are not CtpA substrates.
PA1048 is a novel substrate of CtpA.
We investigated PA1048 and MltD further because the previous analyses suggested that PA1048 is a new CtpA substrate but were inconclusive for MltD, which was degraded in vitro but did not accumulate in vivo in a ΔctpA mutant (Fig. 3a to c). It was possible that endogenous MltD production was downregulated in a ΔctpA mutant, which could obscure the detection of decreased degradation in LFQP analyses. This was not a concern for the plasmid-encoded MltD-FLAG, but in this case, the C-terminal FLAG tag could have interfered with MltD degradation. To address these issues, we raised a polyclonal MltD antiserum and used it to detect untagged MltD encoded on a plasmid with a nonnative promoter. This confirmed that MltD does not accumulate in a ΔctpA mutant (Fig. 3d). Therefore, MltD is not a CtpA substrate in vivo, at least under the growth conditions used in this study (see Discussion).
Our analysis had suggested that PA1048 is a newly discovered CtpA substrate because PA1048-FLAG accumulated in a ΔctpA strain, the level of endogenous PA1048 was much higher in ΔctpA and ΔlbcA strains, and His6-PA1048 was degraded by LbcA-CtpA in vitro (Fig. 3a to c). However, to ensure that PA1048 behaves like all previously known substrates, we also tested the effects of ΔlbcA and ctpA-S302A mutations on constitutively produced PA1048 in vivo. PA1048-FLAG accumulated to the same extent in the ΔctpA, ΔlbcA, and ctpA-S302A strains (Fig. 3e). Therefore, PA1048 is the fifth substrate of CtpA to be discovered because it accumulates in all CtpA protease-defective strains in vivo and is degraded by the LbcA-CtpA complex in vitro. Unlike the other four CtpA substrates, MepM, PA1198, PA1199, and PA4404, PA1048 is not a predicted peptidoglycan cross-link hydrolase. It is a predicted outer membrane (OM) lipoprotein, with an N-terminal domain of unknown function and an OmpA-like C-terminal domain that binds to peptidoglycan noncovalently (see Discussion).
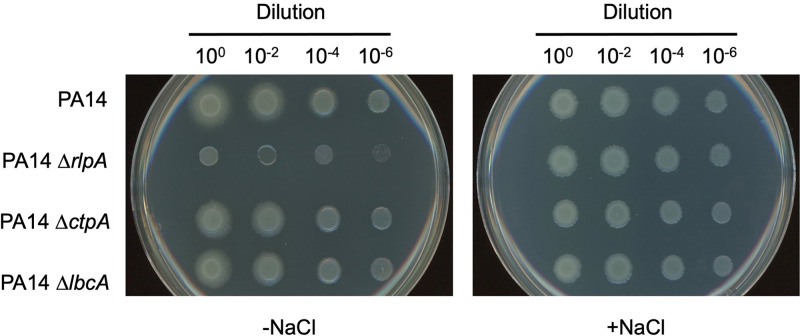
LbcA is not essential for RlpA function.
One of the proteins that copurified with LbcA, but is not a CtpA substrate, is RlpA (Table 2 and Fig. 3). We considered the possibility that a complex containing LbcA and RlpA might occur because LbcA plays a role in facilitating RlpA function. A P. aeruginosa rlpA-null mutant was reported to have a viability defect when plated on Luria-Bertani (LB) agar lacking NaCl (31). Therefore, we reasoned that we could use that phenotype to test if LbcA is required for RlpA function. However, when we introduced an rlpA in-frame deletion mutation into the PAK strain used in our studies, the mutant grew normally on LB agar lacking NaCl (data not shown). We suspected that this might be due to the strain background because the original ΔrlpA phenotype was reported in strain PA14 only (31). Therefore, we introduced rlpA, lbcA, and ctpA deletion mutations into strain PA14. In this strain background, the ΔrlpA mutant had the previously reported viability defect when plated onto LB agar without NaCl (Fig. 4). However, ΔlbcA or ΔctpA mutants did not have the same phenotype, which suggests that any interaction between LbcA and RlpA is not essential for RlpA function (Fig. 4). We also considered the reverse possibility, that RlpA does play a role in the functioning of the LbcA-CtpA proteolytic complex. However, a ΔrlpA mutation had no effect on the levels of CtpA substrates in vivo, suggesting that RlpA does not promote or inhibit LbcA-CtpA proteolytic function (data not shown).
TABLE 2.
Strains and plasmids
| P. aeruginosa strain or plasmid | Genotype or feature(s) | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAK | Wild type | 46 |
| PA14 | Wild type | 47 |
| AJDP730 | PAK ΔctpA | 17 |
| AJDP1091 | PAK ΔlbcA::aacC1 | 17 |
| AJDP1140 | PAK ctpA-S302A | 17 |
| AJDP1202 | PAK ΔmltD | This study |
| AJDP1331 | PAK ΔPA1198 | This study |
| AJDP1332 | PAK ΔctpA ΔPA1198 | This study |
| AJDP1420 | PAK ΔrlpA | This study |
| AJDP1493 | PAK ΔctpA ΔmltD | This study |
| AJDP1494 | PAK ΔctpA ΔrlpA | This study |
| AJDP1515 | PA14 ΔrlpA | This study |
| AJDP1534 | PA14 ΔctpA | This study |
| AJDP1535 | PA14 ΔlbcA::aacC1 | This study |
| Plasmids | ||
| pHERD20T | Ampr; pMB1 ori, araBp expression vector | 48 |
| pET-24b(+) | Kanr; pMB1 ori, T7p expression vector | Novagen |
| pEX18Ap | Ampr; pMB1 ori, oriT sacB+ | 49 |
| pQE-30 | Ampr; Col E1 ori, T5p expression vector | Qiagen |
| pAJD2227 | araBp-ctpA-S302A-His6 in pHERD20T | 23 |
| pAJD2290 | T7p-′ctpA-His6 in pET-24b(+) | 23 |
| pAJD2378 | araBp-ctpA-S302A-FLAG-His6 in pHERD20T | This study |
| pAJD2417 | araBp-lbcA-FLAG in pHERD20T | This study |
| pAJD2653 | T7p-′lbcA-his6 in pET-24b(+) | 17 |
| pAJD2655 | T7p-′ctpA-S302A-his6 in pET-24b(+) | 17 |
| pAJD2799 | araBp-mepM-FLAG in pHERD20T | 17 |
| pAJD2816 | araBp-mltD-FLAG in pHERD20T | 17 |
| pAJD2820 | araBp-PA1198-FLAG in pHERD20T | 17 |
| pAJD2876 | araBp-rlpA-FLAG in pHERD20T | 17 |
| pAJD2916 | araBp-PA1198 in pHERD20T | This study |
| pAJD2946 | T5p-his6-′mepM in pQE-30 | 45 |
| pAJD2948 | T5p-his6-′PA1198 in pQE-30 | This study |
| pAJD2967 | T5p-his6-′rlpA in pQE-30 | This study |
| pAJD2968 | T5p-his6-′mltD in pQE-30 | This study |
| pAJD2986 | araBp-rlpA in pHERD20T | This study |
| pAJD2987 | araBp-mltD in pHERD20T | This study |
| pAJD2994 | T5p-his6-′PA1048 in pQE-30 | This study |
| pAJD3000 | T5p-his6-′mltB1 in pQE-30 | This study |
| pAJD3002 | T5p-his6-′amiB in pQE-30 | This study |
| pAJD3005 | araBp-lbcA in pHERD20T | This study |
| pAJD3006 | araBp-mltB1-FLAG in pHERD20T | This study |
| pAJD3008 | araBp-PA1048-FLAG in pHERD20T | This study |
| pAJD3021 | araBp-amiB-FLAG in pHERD20T | This study |
| pAJD3119 | T7p-FLAG-′lbcA in pET-24b(+) | This study |
FIG 4.
Evidence that LbcA or CtpA is not essential for RlpA function. Serial dilutions of normalized saturated cultures were spotted onto LB agar without NaCl (−NaCl) or containing 1% (wt/vol) NaCl (+NaCl) and incubated at 37°C for approximately 16 h.
Only CtpA and its substrates bind to LbcA directly.
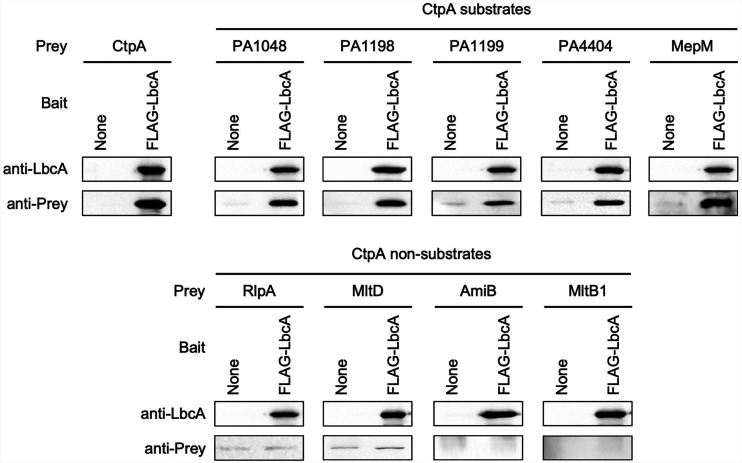
Our experiments had revealed that LbcA can form a complex with CtpA and its substrates independently in vivo but that some proteins that are not CtpA substrates also copurify with LbcA. However, the co-IP experiments that led to those conclusions cannot distinguish between direct and indirect interactions. If LbcA acts as a scaffold to facilitate CtpA-dependent proteolysis, we hypothesized that CtpA and its substrates should bind to LbcA directly and independently. Some non-CtpA substrates might also bind to LbcA directly, perhaps as part of an undiscovered LbcA function. Therefore, we used an in vitro LbcA binding assay to uncover which proteins can or cannot bind to LbcA directly.
A purified FLAG-LbcA protein was tested for its ability to interact with individually purified prey proteins. Binding reactions and washes were done using buffer containing 0.5 M NaCl and 0.1% (wt/vol) Triton X-100 to ensure high stringency. These experiments confirmed that LbcA and CtpA interact directly and that all five known CtpA substrates bound to LbcA directly as well, including the new PA1048 substrate discovered in this study (Fig. 5). In striking contrast, all four non-CtpA substrates, which we had identified as LbcA complex members in vivo and focused on in this study, were unable to bind to LbcA directly (Fig. 5). Lowering stringency by reducing the salt concentration, or using LbcA bait protein with the FLAG tag on the C terminus rather than the N terminus, did not alter any of these conclusions (data not shown). These results support the idea that a major role of LbcA is to act as a scaffolding protein that binds to CtpA and its substrates directly, bringing them together for proteolysis. However, they suggest that some non-CtpA substrates that were cross-linked to an inactive LbcA-CtpA-S302A complex in a previous study, and that copurified natively with LbcA in this study, do not bind to LbcA directly. Therefore, the in vivo association of these non-CtpA substrates with LbcA might be indirect, perhaps due to their participation in multiprotein complexes that include one or more proteins that can bind to LbcA. We investigated this possibility in our final series of experiments.
FIG 5.
Only CtpA and its substrates bind to LbcA directly in vitro. Anti-FLAG M2 affinity resin was used to purify protein from E. coli lysates containing the empty pET-24b(+) vector (None) or a derivative encoding FLAG-LbcA. The purified anti-FLAG M2 agarose immunocomplexes were then incubated with approximately 1.5 μg of a purified prey protein (Prey) for 1 h, washed extensively in high-stringency buffer containing 0.5 M NaCl and 0.1% (wt/vol) Triton X-100, and analyzed by immunoblotting. Immunoblots are single representatives of results from several independent replicate experiments.
LbcA-independent complexes between CtpA substrates and nonsubstrates in vivo.
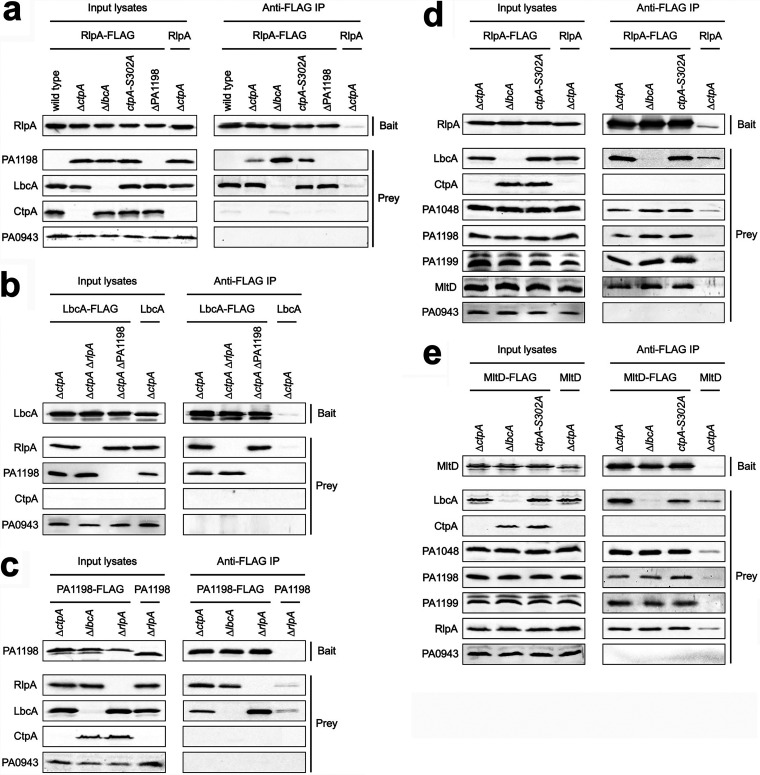
We used co-IP assays to probe the relationship between LbcA and its copurification partners. We chose PA1198 as a representative CtpA substrate and RlpA as a representative non-CtpA substrate. When RlpA-FLAG was the bait, LbcA and PA1198 copurified with it, as expected from the results of our previous experiments (Fig. 6a) (detectible amounts of PA1198 copurified only in the strains where it could not be degraded by CtpA). However, PA1198 still copurified with RlpA-FLAG in a ΔlbcA mutant. Therefore, scaffolding by LbcA is not the explanation for RlpA and PA1198 copurification, and this is consistent with the inability of LbcA and RlpA to interact directly (Fig. 5). It also raised the possibility that interactions between CtpA substrates and nonsubstrates, such as PA1198 and RlpA, bridge the interaction between nonsubstrates and LbcA. This could explain why proteins such as RlpA and MltD were enriched in the LbcA-CtpA-S302A trap in our previous study (17). However, LbcA still copurified with RlpA-FLAG in a ΔPA1198 strain, showing that bridging by PA1198 cannot be the only explanation (Fig. 6a). These conclusions were corroborated when LbcA-FLAG was the bait. RlpA copurified with LbcA-FLAG in a ΔPA1198 strain, and PA1198 copurified with LbcA-FLAG in a ΔrlpA strain (Fig. 6b) (all strains had a ΔctpA mutation to ensure similar levels of the endogenous PA1198 prey in all comparisons). The conclusions were also supported with a PA1198-FLAG bait, which copurified with RlpA and LbcA, even in strains that contained only one of them (Fig. 6c).
FIG 6.
Coimmunoprecipitation analysis suggests the formation of LbcA-independent complexes between CtpA substrates and nonsubstrates. Each panel shows immunoblot analysis of input lysates and immunoprecipitates (Anti-FLAG IP). Strain genotypes are shown above each lane. Polyclonal antisera used for detection are shown at the left. (a) Baits were RlpA-FLAG or the RlpA negative control. (b) Baits were LbcA-FLAG or the LbcA negative control. (c) Baits were PA1198-FLAG or the PA1198 negative control. (d) Baits were RlpA-FLAG or the RlpA negative control. (e) Baits were MltD-FLAG or the MltD negative control. Immunoblots are single representatives of results from several independent replicate experiments.
The above-described experiments suggested that LbcA-independent associations between CtpA substrates and nonsubstrates occur in vivo. This could explain, at least in part, why some nonsubstrates copurified with LbcA and were enriched in an inactive LbcA-CtpA-S302A complex. However, bridging by PA1198 cannot be the sole explanation. Therefore, we expanded the experiments to test if other CtpA substrates and nonsubstrates copurified with RlpA-FLAG. In addition to PA1198, the CtpA substrates PA1048 and PA1199 and the nonsubstrate MltD also copurified with RlpA-FLAG (Fig. 6d) (our polyclonal antisera for the CtpA substrates MepM and PA4404 have low avidity, and so we could not reach a conclusion for them). All of these complexes occurred in both lbcA+ and ΔlbcA strains, demonstrating that scaffolding by LbcA was not the explanation. Finally, a similar set of experiments using MltD-FLAG as the bait showed that it copurified with the CtpA substrates PA1048, PA1198, and PA1199 and the nonsubstrate RlpA (Fig. 6e). Once again, all of these complexes were independent of LbcA. Together, all of these experiments suggest that complexes between peptidoglycan hydrolases and other peptidoglycan-binding proteins occur in P. aeruginosa, and this might explain how some non-CtpA substrates are found in LbcA-containing complexes.
DISCUSSION
LbcA was discovered because it copurified with CtpA, and it was then found to be required for CtpA-dependent proteolysis of four substrates (17). In this study, we combined unbiased approaches with focused experiments to explore the role of LbcA in P. aeruginosa and to gain insight into how it promotes CtpA activity. Our findings suggest that facilitating CtpA-dependent proteolysis might be the most impactful role of LbcA, which it achieves, at least in part, by acting as a scaffold for CtpA and its substrates. These conclusions are supported by our findings of overlapping effects of ΔctpA and ΔlbcA mutations on the proteome, the occurrence of independent complexes containing LbcA and CtpA or LbcA and a substrate in vivo, and the direct interaction between purified LbcA and CtpA proteins and between LbcA and each of the CtpA substrates in vitro. The most abundant LbcA copurification partners were enriched for cell wall enzymes and other cell wall-associated proteins. However, some of these LbcA copurification partners are not CtpA substrates, and in contrast to the substrates, those that we tested here did not bind to LbcA directly. Their copurification might be explained by an indirect association with LbcA due to their participation in multiprotein complexes containing CtpA substrates. Finally, we have also discovered a novel substrate of the LbcA-CtpA complex, the uncharacterized PA1048 protein.
The LbcA-CtpA system is analogous to the E. coli NlpI-Prc system. Both are composed of a TPR-containing outer membrane lipoprotein in complex with a CTP, and both degrade peptidoglycan hydrolases (15–18). Their similarity is further supported by our new finding that LbcA acts as a scaffold because in vivo pulldown experiments suggested that NlpI might bind to the Prc protease and its substrates independently as well (16). However, while these two systems are analogous, they are not orthologous. LbcA is approximately twice the size of NlpI, and their primary sequences are not homologous. Furthermore, Prc is a member of the CTP-1 subfamily, whereas CtpA is in the CTP-3 subfamily. In fact, P. aeruginosa has a second CTP that is the ortholog of E. coli Prc, although little is known about its function. Finally, in other ongoing work, we have discovered that the LbcA-CtpA and NlpI-Prc systems have significant differences in their stoichiometries and structures (H. Hsu, M. Wang, A. Kovach, A. J. Darwin, and H. Li, unpublished data). Therefore, it is intriguing that these two systems have evolved to use quite different components, functioning in different ways, to achieve the common goal of degrading peptidoglycan hydrolases.
We discovered that PA1048 is a substrate of the LbcA-CtpA complex (Fig. 3). In contrast to the four substrates that were already known, PA1048 is not a predicted peptidoglycan hydrolase. However, it does have a link to peptidoglycan because its C-terminal domain, representing approximately 50% of the mature protein, is predicted to adopt an OmpA C-terminal domain fold, which interacts with peptidoglycan noncovalently. This domain occurs in several P. aeruginosa proteins, some of which have been demonstrated to interact with peptidoglycan (32–34). Proteins with the OmpA C-terminal domain are widespread in Gram-negative bacteria, but their N-terminal domains vary. Some have an N-terminal OM porin domain like OmpA itself, whereas others, including PA1048, are lipoproteins that attach to the OM by their acylated N termini. The attachment of these proteins to the OM and the cell wall explains how they can affect envelope integrity. For example, P. aeruginosa OprF and PA1041 have been linked to cell envelope maintenance (33, 35). A PA1048-null mutant was reported to be sensitive to multiple antibiotics (36). Also, we have preliminary data that a ΔPA1048 mutant has increased sensitivity to sodium dodecyl sulfate (SDS)-EDTA, which suggests a compromised OM (M. Wang and A. J. Darwin, unpublished data). Global studies suggested that PA1048 gene expression is likely to be under the direct control of the AlgR virulence regulator, indicating that PA1048 might be especially important under AlgR-inducing conditions (37, 38). Finally, the four peptidoglycan hydrolase substrates of CtpA are potentially dangerous if their activities are not constrained. Perhaps PA1048 might also be dangerous if its levels are too high at the wrong time. For example, even though PA1048 is not a peptidoglycan hydrolase itself, perhaps it can affect peptidoglycan hydrolase activity indirectly.
In order to conclude that a protein is a protease substrate, it must accumulate in a protease-defective mutant in vivo and be degraded by the protease in vitro. Either one alone is not sufficient because accumulation in vivo could be an indirect consequence of absent protease activity, and many proteases can degrade nonphysiological substrates in vitro. Therefore, even though MltD was degraded by CtpA in vitro, it is apparently not a physiological substrate because MltD did not accumulate in a ΔctpA mutant (Fig. 3). Similarly, only 1 of 10 proteins degraded by E. coli Prc in vitro was found to accumulate in a Prc-deficient mutant (15). This was not surprising because some of the other nine proteins were cytoplasmic. Similarly, it is possible that MltD is not degraded by the LbcA-CtpA complex in vivo because the two are physically separated from each other. For example, MltD and the LbcA-CtpA complex might localize to different regions of the OM or perhaps be in different membranes altogether. When the sorting signal from P. aeruginosa MltD was added to a fluorescent reporter protein, it localized to the inner membrane of P. aeruginosa (39). Therefore, P. aeruginosa MltD was proposed to be anchored to the inner membrane, which would separate it from the LbcA-CtpA complex. However, these findings have not been corroborated by determining the location of MltD itself, and our attempts to do so have been inconclusive, possibly due to the low abundance of MltD and the low avidity of our antiserum. Of course, we cannot rule out the possibility that MltD would be degraded by CtpA in vivo under conditions that we are unaware of. However, we have seen no evidence of MltD accumulation in a ΔctpA mutant, including at the different phases of growth.
When LbcA was isolated from P. aeruginosa lysates, the most abundant envelope proteins that copurified with it were enriched for cell wall-associated functions (see Table S6 in the supplemental material). We followed up on seven of those proteins and found that four of them were not CtpA substrates, the lytic transglycosylases RlpA, MltD, and MltB1 and the amidase AmiB. These nonsubstrates had a striking and unequivocal difference from the CtpA substrates when we tested their ability to bind to LbcA directly in vitro. All five known CtpA substrates bound to LbcA, whereas all four of the nonsubstrates that we tested did not (Fig. 5). A definitive conclusion cannot be made from a negative result, but the uniform ability of all CtpA substrates to bind to LbcA directly, coupled with the failure of all four nonsubstrates to do so, strongly suggests that these nonsubstrates copurified with LbcA due to indirect interactions. This was supported by co-IP experiments that suggested the occurrence of LbcA-independent complexes in vivo, which included CtpA substrates and nonsubstrates (Fig. 6). It is also consistent with the known and hypothesized occurrence of multienzyme complexes involved in peptidoglycan remodeling during cell growth and division (5, 40). We could not determine which CtpA substrate(s) might bridge the interactions between LbcA and the nonsubstrates. Individual substrate deletion mutations did not prevent RlpA from copurifying with LbcA (Fig. 6 and data not shown), and analysis of multiple substrate deletion mutants has not been possible due to severe growth deficiencies. We also tested if purified RlpA was captured by LbcA in vitro if CtpA substrates were also added, but this was not successful. Of course, it is also possible that undiscovered CtpA substrates were involved. Finally, an alternative explanation is that some proteins copurified with LbcA due to an association with peptidoglycan rather than participating in LbcA-independent protein complexes. However, we found that treatment of lysates with lysozyme prior to the co-IP procedure did not alter the results (data not shown).
Does LbcA bind to any proteins directly other than CtpA and its substrates, and does LbcA have any role(s) besides facilitating CtpA-dependent proteolysis? The strikingly similar effects of ΔctpA and ΔlbcA mutations on global protein levels suggest that the most impactful role of LbcA is to support proteolysis by CtpA. In addition, global phenotype analysis has shown that ctpA- and lbcA-null mutants have very similar phenotypes in several other Pseudomonas species (41). However, these analyses do not rule out other roles for LbcA. Furthermore, it is important to note a bias in our study because the LbcA copurification partners that we focused on here were those that were also cross-linked to an inactive LbcA-CtpA-S302A complex, but not to an active LbcA-CtpA complex, in a previous study (17). This should enrich for protease substrates and proteins bound to those substrates, and our follow-up experiments have supported that. Therefore, it remains possible that LbcA has other roles and other direct binding partners beyond CtpA and its substrates. In fact, in the analogous NlpI-Prc system of E. coli, evidence suggests that NlpI might have roles beyond facilitating Prc-dependent proteolysis (42). Those authors proposed that NlpI is a general scaffold for peptidoglycan hydrolases, binding to non-Prc substrates and influencing their activity. NlpI and LbcA are not homologous, but both contain TPRs that facilitate protein-protein interactions, and so LbcA has the potential to do something similar to NlpI. In fact, NlpI binds to E. coli MepS and MepM (although it promotes the degradation of only the former), and P. aeruginosa LbcA also binds to the MepS homologs PA1198 and PA1199 and to MepM (16, 17, 42). Therefore, NlpI and LbcA have homologous direct binding partners. If LbcA does have direct binding partners that are not CtpA substrates, some obvious candidates would be the cell wall-associated proteins that copurified with LbcA but were not investigated as part of this study (Table S6). This will be an interesting topic for future studies. Regardless, even if LbcA only binds to CtpA and its substrates directly, it could still have a role beyond promoting degradation. For example, if there is a population of LbcA in the cell that is not bound to CtpA but is bound to the substrate(s), it could influence its activity or location and that of any other proteins in complex with those substrates.
In summary, we have shown that LbcA and CtpA have similar impacts on the proteome and that LbcA facilitates CtpA-dependent proteolysis by acting as a scaffold for the protease and its substrates, discovered the first CtpA substrate that is not a peptidoglycan hydrolase, and found evidence of complexes between CtpA substrates and nonsubstrates in vivo. Goals for the future include investigating the significance of complexes containing CtpA substrates and nonsubstrates and whether or not LbcA can influence the function of these proteins beyond promoting their degradation and a focused investigation into the possibility that LbcA has undiscovered direct binding partners beyond CtpA and its proteolytic substrates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth.
Strains and plasmids are listed in Table 2. Bacteria were grown in Luria-Bertani (LB) broth, composed of 1% (wt/vol) tryptone, 0.5% yeast extract, and 1% (wt/vol) NaCl, or on LB agar, at 30°C or 37°C. To select for P. aeruginosa exconjugants after mating with E. coli donor strains, bacteria were recovered on Vogel-Bonner minimal agar (43).
Plasmid and strain constructions.
The construction of strains with ΔctpA, ΔlbcA::aacC1, and ctpA-S302A mutations was described previously (17). To construct strains with ΔPA1198, ΔmltD, or ΔrlpA in-frame deletion mutations, two fragments of ∼0.55 kb each corresponding to regions flanking the deletion site were amplified by PCR and cloned into pEX18Ap. The plasmids were integrated into the P. aeruginosa chromosome after conjugation from E. coli, and sucrose-resistant, carbenicillin-sensitive segregants were then isolated on LB agar containing 10% sucrose (44). Deletions were verified by PCR analysis of genomic DNA.
Plasmids encoding C-terminally FLAG- and/or His6-tagged proteins were constructed by amplifying the genes from P. aeruginosa PAK DNA using a downstream primer that included a region encoding the tag, followed by a stop codon. For plasmids encoding proteins without any added tags, the downstream primer annealed immediately downstream of the stop codon. The amplified fragments were cloned into pHERD20T using restriction sites incorporated by the PCR primers. Plasmids encoding proteins with N-terminal His6 tags for protein purification were constructed by amplifying the genes encoding the mature proteins (no N-terminal signal sequence) from P. aeruginosa PAK DNA, using a downstream primer that annealed immediately after the stop codon. These fragments were cloned into pQE-30 using restriction sites incorporated by the PCR primers. The plasmid encoding FLAG-′LbcA was constructed by amplifying the region encoding the mature part of LbcA from P. aeruginosa PAK DNA, using a forward primer that included a region encoding the FLAG tag and a downstream primer that annealed immediately after the stop codon. This fragment was cloned into pET-24b(+) using NdeI and HindIII restriction sites incorporated by the PCR primers.
Label-free quantitative proteomics.
Bacteria were inoculated into 100 ml of LB broth in a 500-ml flask to an optical density at 600 nm (OD600) of 0.05 and grown at 37°C with aeration for 5 h. Cells from the equivalent of 100 ml of culture at an OD600 of 1 were resuspended in 10 ml of a solution containing 8 M urea, 3% (wt/vol) sodium dodecyl sulfate (SDS), 100 mM Tris-HCl, and 1 mM EDTA (pH 8.0) and sonicated. One milliliter was transferred to a microcentrifuge tube, and insoluble debris was collected by centrifugation in a microcentrifuge at maximum speed for 20 min. The protein concentration of the clarified lysates, determined with the Bio-Rad DC protein assay kit, was approximately 1.2 to 1.5 mg/ml. Triplicate samples were generated for each strain, each from an independent culture. SDS was eliminated with an S-trap column, and proteins were digested with trypsin and then identified and quantified by the NYU School of Medicine Proteomics Laboratory using a Thermo Scientific Orbitrap Elite hybrid ion trap-orbitrap mass spectrometer. Label-free quantification (LFQ) values were normalized and filtered for proteins with at least two or more peptides identified, intensity values were log2 transformed, samples were grouped based on identity (wild type, ΔctpA, or ΔlbcA) and filtered for proteins that were identified in all triplicate samples of at least one strain, missing values were replaced from a normal distribution, and Student’s t test was then applied, followed by a 5% Benjamini-Hochberg-based false discovery rate cutoff.
Anti-FLAG coimmunoprecipitation assay.
Bacteria were inoculated into 100 ml of LB broth in a 250-ml flask to an OD600 of 0.05 and grown at 37°C with aeration for 5 h. Equivalent amounts of bacterial cells from all strains were collected by centrifugation, washed in a solution containing 10 mM Tris-HCl and 10% (wt/vol) glycerol (pH 7.5), and resuspended in 2 ml nondenaturing lysis buffer (NDLB) (50 mM Tris-HCl, 300 mM NaCl, 5 mM EDTA, 10% [wt/vol] glycerol [pH 7.5]). Roche complete protease inhibitors were added, cells were disrupted by sonication, and 1% (wt/vol) lauryldimethylamine N-oxide (LDAO) was then added, followed by incubation with rotation for 1 h at 4°C. Insoluble material was removed by centrifugation at 16,000 × g for 30 min at 4°C. Thirty microliters of anti-FLAG M2 affinity resin (Sigma-Aldrich) in NDLB was added to the supernatant, and the mixture was incubated for 2 h at 4°C with rotation. A 1-ml spin column (catalog number 69725; Pierce) was used to wash the resin 10 times with 500 μl NDLB containing 0.1% (wt/vol) Triton X-100. Proteins were eluted by the addition of 100 μl of 400 μg/ml 3× FLAG peptide in NDLB containing 0.1% (wt/vol) Triton X-10 and incubation with rotation at 4°C for 30 min. In some cases, proteins present in these samples were identified by liquid chromatography-mass spectrometry (NYU School of Medicine Proteomics Laboratory).
Polyclonal antiserum production and immunoblotting.
E. coli strain M15[pREP4] (Qiagen) containing a pQE-30 derivative encoding His6-PA1048, His6-RlpA, or His6-MltD was grown in LB broth to mid-log phase at 37°C with aeration. Protein production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Proteins were purified under denaturing conditions by nickel-nitrilotriacetic acid (NTA)-agarose affinity chromatography as described by the manufacturer (Qiagen). Polyclonal rabbit antisera were raised by Covance Research Products Inc. Specificity was verified by the detection of proteins in whole-cell lysates of P. aeruginosa strains that either lacked the target protein (deletion mutant) or had it present at endogenous levels (chromosomally encoded) or overproduced from a plasmid. Polyclonal antisera for CtpA, LbcA, MepM, PA0943, PA1198, and PA1199 were described previously (17, 23, 30).
Samples separated by SDS-PAGE were transferred to nitrocellulose by semidry electroblotting. Chemiluminescence detection followed incubation with one of the diluted polyclonal antisera described above or anti-FLAG M2 monoclonal antibody (Sigma) and then goat anti-rabbit IgG (Sigma)– or goat anti-mouse IgG (Sigma)–horseradish peroxidase conjugates, used at the manufacturer’s recommended dilution.
Determination of protein abundance in vivo.
Saturated cultures were diluted into 5 ml of LB broth, containing 150 μg/ml carbenicillin and 0.02% (wt/vol) arabinose, in 18-mm-diameter test tubes so that the OD600 was 0.05. The cultures were grown on a roller drum at 37°C for 5 h. Cells were harvested by centrifugation, resuspended in SDS-PAGE sample buffer at equal concentrations (based on the culture OD600), and analyzed by immunoblotting as described above.
Protein purification.
LbcA-His6, CtpA-His6, and CtpA-S302A–His6 were purified exactly as described previously (45). For His6-PA1198, His6-MepM, His6-MltD, His6-RlpA, and His6-AmiB, E. coli strain M15[pREP4] containing a pQE-30 plasmid derivative was grown in 500 ml LB broth at 37°C with aeration to an OD600 of 0.6 to 1.0. Protein production was induced by adding 1 mM IPTG, and incubation at 37°C was continued for 3 h, after which cells were harvested by centrifugation. Proteins were purified under native conditions by NTA-agarose affinity chromatography in buffer containing 50 mM NaH2PO4 and 300 mM NaCl, as recommended by the manufacturer (Qiagen). Proteins were eluted in 1-ml fractions using 50 mM NaH2PO4–300 mM NaCl (pH 8) buffer containing 50, 100, 150, 200, or 250 mM imidazole.
His6-PA1048 and His6-MltB1 encoded by pQE-30 plasmid derivatives were purified differently because they were insoluble when overproduced in E. coli. Proteins were denatured and solubilized by resuspending the harvested cells in a solution containing 6 M guanidine hydrochloride, 100 mM NaH2PO4, 10 mM Tris-HCl, 5 mM imidazole, and 1% (wt/vol) Triton X-100 (pH 7.5) and stirring at room temperature for 1 h. The suspension was sonicated, and insoluble material was removed by centrifugation at 10,000 × g for 30 min. Ni-NTA-agarose was added to the supernatant, and it was stirred for 1 h at room temperature. The Ni-NTA-agarose resin was collected in a drip column and washed with 30 ml of a solution containing 100 mM NaH2PO4, 10 mM Tris-HCl, 1% (wt/vol) Triton X-100, 20 mM imidazole, and 8 M urea (pH 7.5). Proteins were renatured by washing the resin with 50 ml of a solution containing 50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole (pH 8). Proteins were eluted in 1.5-ml fractions using 50 mM NaH2PO4–300 mM NaCl (pH 8) buffer containing 50, 100, 150, 200, or 250 mM imidazole. His6-RlpA was also repurified in this way to demonstrate that the denaturation/renaturation process would not render a non-CtpA substrate susceptible to cleavage.
In vitro proteolysis assay.
In vitro proteolysis assays were done similarly to those described previously (17, 45). N-terminally His6-tagged test proteins were mixed with CtpA-His6 or CtpA-S302A–His6, together with LbcA-His6, all purified as described above, and incubated at 37°C for 3 h. Reactions were terminated by adding SDS-PAGE sample buffer and incubating the mixture at 90°C for 10 min. The samples were separated by SDS-PAGE and stained with ProtoBlue Safe (National Diagnostics).
LbcA direct binding assay.
E. coli strain ER2566 (New England BioLabs) containing a pET-24b(+) derivative encoding FLAG-LbcA was grown in 250 ml LB broth at 37°C with aeration to an OD600 of approximately 0.6. The production of FLAG-LbcA in the E. coli cytoplasm was induced by adding 1 mM IPTG, and incubation at 37°C was continued for 3 h, after which cells were harvested by centrifugation. Cells were washed in a solution containing 10 mM Tris-HCl and 10% (wt/vol) glycerol (pH 7.5) and resuspended in 5 ml NDLB. Roche complete protease inhibitors were added, cells were disrupted by sonication, and 1% (wt/vol) LDAO was then added, followed by incubation with rotation for 1 h at 4°C. Insoluble material was removed by centrifugation at 16,000 × g for 30 min at 4°C. A final concentration of 5% (wt/vol) glycerol was added to the supernatant, which was stored in aliquots at −70°C (this was the FLAG-LbcA soluble lysate). A negative-control lysate was generated in the same way using E. coli ER2566 containing the empty vector plasmid pET-24b(+).
For direct binding assays, purified baits were prepared by adding 10 μl of EZview red anti-FLAG M2 affinity resin (Sigma-Aldrich) in NDLB to 50 μl of either the FLAG-LbcA soluble lysate or the negative-control soluble lysate, and the total volume was adjusted to 0.5 ml with NDLB. This mixture was incubated with rotation at 4°C for 2 h, and the resin was collected by centrifugation and then washed five times with 0.5 ml high-salt NDLB (NDLB containing 0.5 M NaCl) containing 0.1% (wt/vol) Triton X-100. The purified anti-FLAG M2 agarose immunocomplex was blocked by resuspending the mixture in 0.5 ml high-salt NDLB containing 5% (wt/vol) bovine serum albumin, incubating the mixture with rotation at 4°C overnight, and then washing the resin five times with 0.5 ml high-salt NDLB containing 0.1% (wt/vol) Triton X-100. To test for direct binding, the resin was resuspended in 0.5 ml high-salt NDLB containing approximately 1.5 μg of a His6-tagged prey protein, which had been purified as described above. Binding was allowed to occur by incubation with rotation at 4°C for 1 h, and the resin was collected by centrifugation, washed five times with 0.5 ml high-salt NDLB containing 0.1% (wt/vol) Triton X-100, resuspended in 100 μl SDS-PAGE sample buffer, and boiled for 10 min to elute proteins. Ten microliters of these samples was loaded into each gel lane for SDS-PAGE and immunoblot analyses.
ACKNOWLEDGMENTS
Research was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award number R01AI136901. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Mass spectrometric protein identification done by the NYU Grossman School of Medicine’s Proteomics Laboratory was partly supported by the NYU Grossman School of Medicine.
We thank Alexis Sommerfield for providing critical comments on a draft version of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Andrew J. Darwin, Email: andrew.darwin@med.nyu.edu.
Joseph Bondy-Denomy, University of California, San Francisco.
REFERENCES
- 1.Moore NM, Flaws ML. 2011. Epidemiology and pathogenesis of Pseudomonas aeruginosa infections. Clin Lab Sci 24:43–46. 10.29074/ascls.24.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sibinelli-Sousa S, Hespanhol JT, Bayer-Santos E. 2021. Targeting the Achilles’ heel of bacteria: different mechanisms to break down the peptidoglycan cell wall during bacterial warfare. J Bacteriol 203:e00478-20. 10.1128/JB.00478-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 7.Barchinger SE, Ades SE. 2013. Regulated proteolysis: control of the Escherichia coli sigma(E)-dependent cell envelope stress response. Subcell Biochem 66:129–160. 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- 8.De Geyter J, Tsirigotaki A, Orfanoudaki G, Zorzini V, Economou A, Karamanou S. 2016. Protein folding in the cell envelope of Escherichia coli. Nat Microbiol 1:16107. 10.1038/nmicrobiol.2016.107. [DOI] [PubMed] [Google Scholar]
- 9.Do T, Page JE, Walker S. 2020. Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. J Biol Chem 295:3347–3361. 10.1074/jbc.REV119.010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paetzel M. 2019. Bacterial signal peptidases. Subcell Biochem 92:187–219. 10.1007/978-3-030-18768-2_7. [DOI] [PubMed] [Google Scholar]
- 11.Che Y, Fu A, Hou X, McDonald K, Buchanan BB, Huang W, Luan S. 2013. C-terminal processing of reaction center protein D1 is essential for the function and assembly of photosystem II in Arabidopsis. Proc Natl Acad Sci USA 110:16247–16252. 10.1073/pnas.1313894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh K, Yamamoto Y. 2007. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth Res 94:203–215. 10.1007/s11120-007-9191-z. [DOI] [PubMed] [Google Scholar]
- 14.Deng CY, Zhang H, Wu Y, Ding LL, Pan Y, Sun ST, Li YJ, Wang L, Qian W. 2018. Proteolysis of histidine kinase VgrS inhibits its autophosphorylation and promotes osmostress resistance in Xanthomonas campestris. Nat Commun 9:4791. 10.1038/s41467-018-07228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PC, Chen CS, Wang S, Hashimoto M, Huang WC, Teng CH. 2020. Identification of MltG as a Prc protease substrate whose dysregulation contributes to the conditional growth defect of Prc-deficient Escherichia coli. Front Microbiol 11:2000. 10.3389/fmicb.2020.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci USA 112:10956–10961. 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, Darwin AJ. 2018. A proteolytic complex targets multiple cell wall hydrolases in Pseudomonas aeruginosa. mBio 9:e00972-18. 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YJ, Choi BJ, Park SH, Lee HB, Son JE, Choi U, Chi WJ, Lee CR. 2021. Distinct amino acid availability-dependent regulatory mechanisms of MepS and MepM levels in Escherichia coli. Front Microbiol 12:677739. 10.3389/fmicb.2021.677739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su MY, Som N, Wu CY, Su SC, Kuo YT, Ke LC, Ho MR, Tzeng SR, Teng CH, Mengin-Lecreulx D, Reddy M, Chang CI. 2017. Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8:1516. 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chueh CK, Som N, Ke LC, Ho MR, Reddy M, Chang CI. 2019. Structural basis for the differential regulatory roles of the PDZ domain in C-terminal processing proteases. mBio 10:e01129-19. 10.1128/mBio.01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoge R, Laschinski M, Jaeger KE, Wilhelm S, Rosenau F. 2011. The subcellular localization of a C-terminal processing protease in Pseudomonas aeruginosa. FEMS Microbiol Lett 316:23–30. 10.1111/j.1574-6968.2010.02181.x. [DOI] [PubMed] [Google Scholar]
- 22.Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res 38:D227–D233. 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology (Reading) 151:2251–2261. 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- 25.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene 498:242–253. 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of σ22 (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62:412–426. 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 27.Llamas MA, Mooij MJ, Sparrius M, Vandenbroucke-Grauls CM, Ratledge C, Bitter W. 2008. Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol Microbiol 67:458–472. 10.1111/j.1365-2958.2007.06061.x. [DOI] [PubMed] [Google Scholar]
- 28.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 29.Zeytuni N, Zarivach R. 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20:397–405. 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Seo J, Brencic A, Darwin AJ. 2009. Analysis of secretin-induced stress in Pseudomonas aeruginosa suggests prevention rather than response and identifies a novel protein involved in secretin function. J Bacteriol 191:898–908. 10.1128/JB.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgenson MA, Chen Y, Yahashiri A, Popham DL, Weiss DS. 2014. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol Microbiol 93:113–128. 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X, Ye F, Lin S, Yang F, Chen Z, Cao Y, Chen Z, Gu J, Lu G. 2019. Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket. Biochem Biophys Res Commun 511:875–881. 10.1016/j.bbrc.2019.02.104. [DOI] [PubMed] [Google Scholar]
- 33.Paulsson M, Kragh KN, Su YC, Sandblad L, Singh B, Bjarnsholt T, Riesbeck K. 2021. Peptidoglycan-binding anchor is a Pseudomonas aeruginosa OmpA family lipoprotein with importance for outer membrane vesicles, biofilms, and the periplasmic shape. Front Microbiol 12:639582. 10.3389/fmicb.2021.639582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock RE, Irvin RT, Costerton JW, Carey AM. 1981. Pseudomonas aeruginosa outer membrane: peptidoglycan-associated proteins. J Bacteriol 145:628–631. 10.1128/jb.145.1.628-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawling EG, Brinkman FS, Hancock RE. 1998. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J Bacteriol 180:3556–3562. 10.1128/JB.180.14.3556-3562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fajardo A, Martinez-Martin N, Mercadillo M, Galan JC, Ghysels B, Matthijs S, Cornelis P, Wiehlmann L, Tummler B, Baquero F, Martinez JL. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS One 3:e1619. 10.1371/journal.pone.0001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong W, Zhao J, Kang H, Zhu M, Zhou T, Deng X, Liang H. 2015. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43:8268–8282. 10.1093/nar/gkv747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lizewski SE, Schurr JR, Jackson DW, Frisk A, Carterson AJ, Schurr MJ. 2004. Identification of AlgR-regulated genes in Pseudomonas aeruginosa by use of microarray analysis. J Bacteriol 186:5672–5684. 10.1128/JB.186.17.5672-5684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewenza S, Mhlanga MM, Pugsley AP. 2008. Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol 190:6119–6125. 10.1128/JB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331. 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, Carlson HK, Esquivel Z, Sadeeshkumar H, Chakraborty R, Zane GM, Rubin BE, Wall JD, Visel A, Bristow J, Blow MJ, Arkin AP, Deutschbauer AM. 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557:503–509. 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- 42.Banzhaf M, Yau HC, Verheul J, Lodge A, Kritikos G, Mateus A, Cordier B, Hov AK, Stein F, Wartel M, Pazos M, Solovyova AS, Breukink E, van Teeffelen S, Savitski MM, den Blaauwen T, Typas A, Vollmer W. 2020. Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J 39:e102246. 10.15252/embj.2019102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. 10.1016/S0021-9258(18)65874-0. [DOI] [PubMed] [Google Scholar]
- 44.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung S, Darwin AJ. 2020. The C terminus of substrates is critical but not sufficient for their degradation by the Pseudomonas aeruginosa CtpA protease. J Bacteriol 202:e00174-20. 10.1128/JB.00174-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strom MS, Lory S. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol 165:367–372. 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 48.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download jb.00393-21-s0001.xlsx, XLSX file, 1.4 MB (1.4MB, xlsx)
Tables S4 to S6. Download jb.00393-21-s0002.xlsx, XLSX file, 0.03 MB (35.4KB, xlsx)