SUMMARY
Sertoli cells are highly polarized testicular supporting cells that simultaneously nurture multiple stages of germ cells during spermatogenesis. Proper localization of polarity protein complexes within Sertoli cells, including those responsible for blood-testis barrier formation, is vital for spermatogenesis. However, the mechanisms and developmental timing that underlie Sertoli cell polarity are poorly understood. We investigate this aspect of testicular function by conditionally deleting Cdc42, encoding a Rho GTPase involved in regulating cell polarity, specifically in Sertoli cells. Sertoli Cdc42 deletion leads to increased apoptosis and disrupted polarity of juvenile and adult testes but does not affect fetal and postnatal testicular development. The onset of the first wave of spermatogenesis occurs normally, but it fails to progress past round spermatid stages, and by young adulthood, conditional knockout males exhibit a complete loss of spermatogenic cells. These findings demonstrate that Cdc42 is essential for Sertoli cell polarity and for maintaining steady-state sperm production.
Graphical abstract
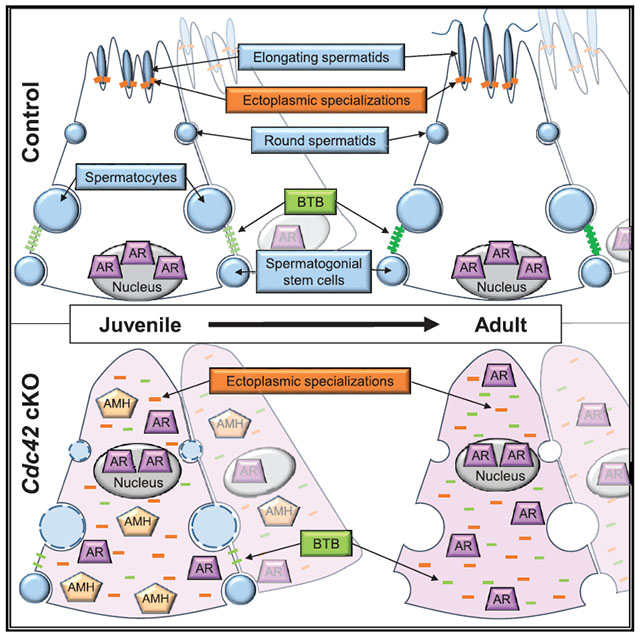
In brief
Sertoli cells of the testicular seminiferous tubule must be highly polarized to simultaneously sustain multiple stages of germ cells during spermatogenesis. Heinrich et al. use a Sertoli-specific conditional deletion mouse model to address the roles of CDC42-mediated apicobasal cell polarity in promoting testis development and spermatogenesis.
INTRODUCTION
Cell polarity, in which different morphologies or molecular components are asymmetrically distributed across a cell, is a critical feature of many cell types. Asymmetries in polarized cells, such as epithelial cells, migratory cells, and stem cells, are required for barrier function, directed cell movement and cell fate decisions, respectively (Campanale et al., 2017). Within epithelial cells, including mature Sertoli cells of the adult mammalian testis, asymmetry across the apical-basal axis is a key cellular characteristic (Gao and Cheng, 2016; Gao et al., 2016). Among the core molecular machinery underlying apicobasal cell polarity are 3 main protein polarity complexes: (1) the Par3/Par6/aPKC (official names PARD3/PARD6A/PRKCZ) complex, (2) the Crumbs (CRB3) complex, and (3) the Scribble (SCRIB) complex. The Par3/Par6/aPKC and CRB3 complexes are typically localized to apical membranes and cell junctions, while the Scribble complex is basolaterally associated (Campanale et al., 2017; Gao et al., 2016); these specific subcellular localizations are vital to maintain unique functions of different cell compartments.
Sertoli cells are the supporting epithelial cells of the testis, whose function is to nurture germ cells in their developmental path from gonocytes (also called prospermatogonia) in fetal and neonatal stages to spermatozoa in the adult. Sertoli cells are the first male-specific cells to differentiate in the fetal gonad and, in conjunction with germ cells, they undergo tubular morphogenesis to form testis cords (Cool et al., 2012), which are the structural and functional units of the testis that give rise to adult seminiferous tubules. As the testis develops, Sertoli cells undergo significant changes in their cellular behavior. From fetal stages until about 10–14 days after birth in rodents, Sertoli cells actively divide (Orth, 1982), increasing the diameter of the seminiferous tubules and the number of potential spermatogonial stem cell (SSC) niches in the testis (Oatley et al., 2011). At the end of this time, Sertoli cells exit the cell cycle and remain quiescent throughout juvenile and adult stages (Kluin etal., 1984; Orth, 1982). As they mature, Sertoli cells also undergo significant changes in the expression and localization of a number of Sertoli-specific proteins that are critical for their supportive roles in the testis: anti-Mullerian hormone (AMH) is downregulated in mature Sertoli cells; GATA1 is initially expressed in immature postnatal Sertoli cells, then upon maturation is upregulated and is expressed only in a subset of seminiferous tubules in a manner dependent on specific stages of germ cells within the tubule (Yomogida etal., 1994); and androgen receptor (AR), the receptor for testosterone produced by interstitial Leydig cells, is translocated from the Sertoli cell cytoplasm to the nucleus upon maturation (Sharpe et al., 2003).
In large part through the guidance of Sertoli cells, steady-state spermatogenesis is sustained by SSCs, the germline stem cell population (sometimes referred to more broadly as a stem/progenitor population called undifferentiated spermatogonia). SSCs proliferate and self-renew to maintain their population, while a subset of SSCs transition into transit-amplifying undifferentiated spermatogonial progenitors (type A undifferentiated spermatogonia), proliferate to form spermatogonial chains, and then proceed through a differentiation program launched in response to retinoic acid (RA). After passing through differentiated spermatogonial stages, spermatogenic cells initiate meiosis and proceed through several spermatocyte stages. After the completion of meiosis, there are several round spermatid and elongating spermatid stages before finally becoming testicular spermatozoa (França et al., 2016; Griswold, 2016; Kanatsu-Shinohara and Shinohara, 2013; Oatley and Brinster, 2012; Yoshida, 2010). In addition to SSC-derived spermatogenesis, there is also a “first wave” of spermatogenesis that occurs in juvenile testes whereby cells directly enter spermatogenesis without passing through a stem cell phase; there are thought to be some unique aspects of the first wave of spermatogenesis relative to the adult steady-state program (Yoshida et al., 2006). During both of these processes, Sertoli cells nurture all stages of germ cells as they proliferate and differentiate to give rise to sperm.
During adult steady-state spermatogenesis, Sertoli cell polarization is essential to support various stages of spermatogenic cells and to compartmentalize its functions, including maintaining germline stem/progenitor cells (as well as differentiating spermatogonia and preleptotene spermatocytes) in the basal compartment of the seminiferous tubule while simultaneously promoting spermiogenesis in the apical adluminal compartment (França et al., 2016; Oatley and Brinster, 2012). A special property of Sertoli cells that allows them to compartmentalize their various cellular interactions is a series of tight junctions that forms the blood-testis barrier (BTB), which sequesters meiotic and post-meiotic cells within the apical compartment and provides an immune-privileged niche for the latter stages of spermatogenesis. Neo-antigens from meiotic and post-meiotic cells arise in developing spermatocytes and spermatids after systemic tolerance is established, which likely renders these germ cells “foreign” by the adaptive immune system. Therefore, the physical sequestration of these antigens is a critical function of Sertoli cells and suggests that Sertoli polarization, including proper basal localization of the BTB, is vital for spermatogenesis (Gao and Cheng, 2016; Gao et al., 2016). Beyond establishing the BTB, Sertoli cells also modulate the immune environment in general via the secretion of cytokines and other factors, as it has been shown that Sertoli cells can significantly enhance the success of xenotransplantation in other organs (Doyle et al., 2012; Dufour et al., 2003; Kaur et al., 2014, 2020).
Although Sertoli cell polarity is vital for successful spermatogenesis, when these cells become polarized and through what mechanisms are poorly understood. A potential player in this process is Rho GTPase signaling, which has been linked to cell polarity and other cellular processes (e.g., cell cycle, cell migration, cytoskeletal structure) in diverse biological contexts (Etienne-Manneville and Hall, 2002; Mack and Georgiou, 2014; Ngok et al., 2014). A central member of the Rho GTPase family is CDC42, which has been implicated in several aspects of cellular biology, including establishing polarity in epithelial cells (Erickson and Cerione, 2001; Kroschewski et al., 1999; Melendez et al., 2011). When activated in its GTP-bound state, CDC42 binds to and activates numerous effector proteins. CDC42 was shown to interact with Par6 of the Par3/Par6/aPKC complex to regulate cell polarity through aPKC signaling (Gao et al., 2016; Garrard et al., 2003; Johansson et al., 2000; Noda et al., 2001). aPKC regulates Sertoli cell F-actin-rich structures called ectoplasmic specializations (ESs), which have a basal and an apical localization, the former of which is the BTB and the latter of which tethers spermatids in place until they are released during spermiation (Russell et al., 1988; Wong et al., 2008). In vitro approaches have implicated CDC42 in the regulation of Sertoli-Sertoli BTB dynamics during the seminiferous epithelial cycle (Wong et al., 2010) and in Sertoli-germ cell interactions at the apical ES mediated by the effector protein IQGAP1 (Lui et al., 2005). However, little is known about the in vivo role of CDC42 within Sertoli cells during testicular differentiation and spermatogenesis.
Here, we show, using Cdc42 conditional knockout (cKO) mice, that Cdc42 activity in Sertoli cells in vivo is not essential for fetal and neonatal testicular differentiation and the postnatal onset of the first wave of spermatogenesis, but it is required for later stages of first-wave spermatogenesis beyond the round spermatid stage. Cdc42 cKO mice exhibited severe defects in testicular structure and function beginning at juvenile stages that continued through adulthood, including the disruption of Sertoli apicobasal polarity and a loss of germ cells by the onset of sexual maturity, leading to a total lack of sperm. Overall, these findings demonstrate that Cdc42 activity in Sertoli cells, likely via its role in mediating apicobasal polarity, is dispensable for early aspects of testicular development but is critical for adult steady-state spermatogenesis.
RESULTS
Cdc42 in Sertoli cells is essential for adult steady-state spermatogenesis
To analyze the role of Cdc42 in Sertoli cells in vivo, cKO mice were generated using Dhh-Cre (referred to as “cKO” hereafter) to delete Cdc42 specifically in Sertoli cells. As a first step, we examined 2-month-old (postnatal day [P] 60) adult cKO male mice, and we found that cKO testis size was severely reduced relative to heterozygous control littermates (Figure 1A). Consistent with this observation, we found that both testis weight and the testis:body weight ratio in cKO males were significantly reduced (75% and 60% reductions, respectively) relative to control littermates (Figures 1B and 1C).
Figure 1. Sertoli-cell-specific ablation of Cdc42 causes complete loss of germ cells in the adult testis.
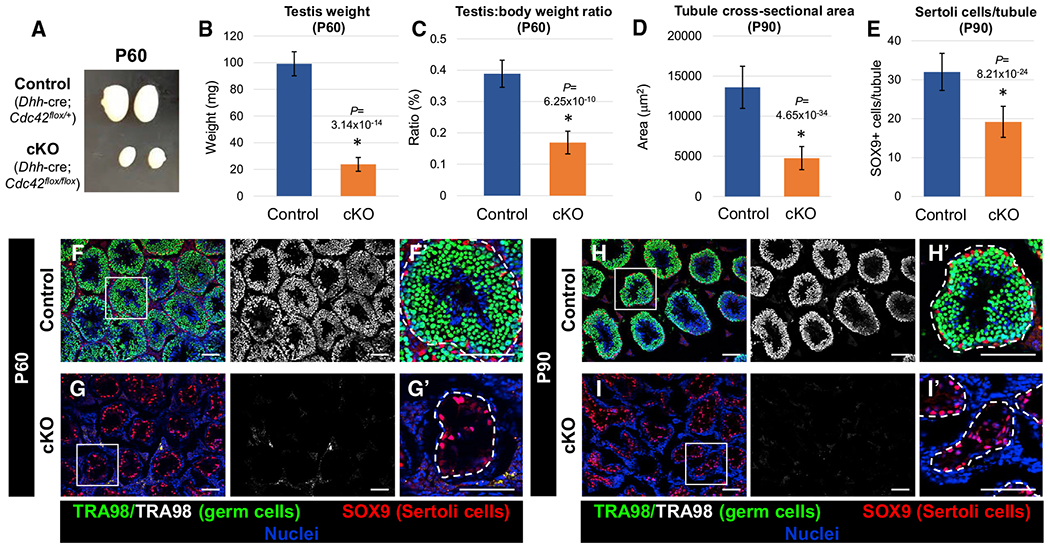
(A) Image showing adult (P60) testis size decrease in cKO (Dhh-Cre;Cdc42flox/flox) testes versus Dhh-Cre;Cdc42flox/+ heterozygous control testes.
(B and C) Graphs showing average testis weight (B) and average testis weight:body weight ratio (C) of P60 control Dhh-Cre;Cdc42flox/+ males versus cKO males (n = 6 control and n = 4 cKO testes, each from independent males).
(D and E) Graphs showing average tubule cross-sectional area (D) and average number of SOX9+ cells per tubule (E) of P90 control (Dhh-Cre;Cdc42flox/+) versus cKO tubules (n = 15 tubules from each of 3 independent males).
(F–I) Immunofluorescence images of P60 (F and G) and P90 (H and I) adult control Dhh-Cre;Cdc42flox/+ (F and H) and cKO (G and I) testes, showing absence of TRA98+ germ cells in cKO testes. (F’)–(I’) are higher-magnification images of the boxed regions in (F)–(I). Dashed lines indicate tubule boundaries. Scale bars, 100 μm.
Data in (B)–(E) are shown as means ± SDs. p values were calculated via a 2-tailed Student’s t test.
To investigate the likely possibility that reduced cKO testis size occurred due to a decrease in germ cell number, we performed immunofluorescence for TRA98, which labels all germ cells within the seminiferous tubules. There was a complete loss of TRA98+ cells in seminiferous tubules of cKO mice as early as P60 (Figures 1F–1I), which resulted in a significant reduction in the cKO seminiferous tubule cross-sectional area (Figure 1D). We examined the expression of SOX9, which labels Sertoli cells, and found that adult cKO mice exhibited a significant reduction (~40% reduction) in the number of Sertoli cells per tubule (Figures 1E–1I).
Lack of Cdc42 disrupts some aspects of adult Sertoli cell development
To address how Cdc42 is required for adult Sertoli cell development, we examined the expression of several Sertoli-specific markers in adult control and cKO testes. GATA1 was expressed in virtually every tubule in cKO testes, in contrast to the expression in a subset of tubules as in control testes (Figures S1A and S1B). AMH expression, which is downregulated in adult control Sertoli cells, was also undetectable in the cKO testis via immunofluorescence or quantitative real-time PCR (data not shown; Figures S1C and S1D). Vimentin (VIM), expressed strongly in the basal cytoplasm near the nucleus in control Sertoli cells, was also strongly expressed in cKO Sertoli cells; however, cKO Sertoli cells displayed an aberrant subcellular localization of VIM in which it was expressed throughout the entire cell (Figures S1E and S1F). In contrast to control Sertoli cells, in which AR was strictly nuclear localized, cKO Sertoli cells exhibited diffuse expression of AR in the cytoplasm, reminiscent of fetal and early postnatal Sertoli cells (Figures S1G and S1H). We also examined the expression of glial cell line-derived neurotrophic factor (GDNF), an essential factor for the maintenance of SSCs, which is expressed in multiple cell types, including Sertoli cells (Chen et al., 2014,2016; Meng et al., 2000). In cKO testes, we observed strong GDNF expression in Sertoli cells, similar to control testes (Figures S1I and S1J). Quantitative real-time PCR analyses of whole adult testes revealed that the Sertoli-specific gene Sox9 was upregulated, likely due to the relative enrichment of Sertoli cells after a significant loss of germ cells in cKO testes, which was confirmed by a 100- to 1,000-fold decrease in the expression of multiple germ cell genes (e.g., Pou5f1 [also known as Oct4] for undifferentiated spermatogonia, Stra8 for differentiating spermatogonia and preleptotene spermatocytes, Ddx4 [also known as Mvh or Vasa] for most germ cells but expressed most strongly in spermatocytes and spermatids) and a lack of sperm in the P60 epididymis (Figures S1K–S1M). Additional normalization to Sox9, to account for this change in the relative proportion of Sertoli cells among all testicular cells in cKO mice, for quantitative real-time PCR analyses of Sertoli-expressed genes (e.g., Gata1, Ar, Cldn11, Ocln, Fshr, Inhbb, Pdgfa, Gdnf), revealed that there were no significant, widespread defects in Sertoli gene expression in cKO testes (Figure S1N). Therefore, some aspects of adult Sertoli cell development were affected in cKO testes, but general Sertoli cell differentiation occurred comparably to controls.
Cdc42 function is essential for maintenance of adult Sertoli cell polarity and BTB function
Given the role of CDC42 in regulating cell polarity, we next sought to determine the effects of Cdc42 deletion on adult Sertoli cell polarity in vivo. In control adult testes, Sertoli cell nuclei are localized basally, adjacent to the basement membrane of the tubules; however, in cKO testes, we often observed Sertoli cell nuclei localized near the lumina of tubules, either as individual nuclei or in small clusters (Figures S2A and S2B). We next assessed the localization of polarity complex proteins within adult cKO Sertoli cells. We examined the localization of ITGB1 (β1-integrin), an integrin protein localized to the apical ES and to the basement membrane in control testes; in contrast, in cKO testes, ITGB1 was localized diffusely throughout the entire Sertoli cell (Figures S2A and S2B). A central protein of the Par polarity complex, PARD3 (Par3), was enriched in the apical compartment and at the BTB in control Sertoli cells, but in cKO cells PARD3 was aberrantly distributed throughout the entire Sertoli cell (Figures S2C and S2D). We then examined the localization of F-actin via phalloidin staining, which was enriched at the actin-rich BTB and ES structures in controls but was inappropriately localized diffusely throughout cKO Sertoli cells (Figures S2C and S2D).
We next examined the expression of basally localized proteins. As expected, CLDN11 (Claudin11), CTNNB1 (β-catenin), and SCRIB were localized specifically at the BTB or in the basal compartment within control tubules, but in cKO testes, all of these proteins were localized throughout the entirety of Sertoli cells (Figures S2E–S2H).
To determine whether the aberrant localization of polarity proteins in cKO testes correlated with defects in BTB function, we assessed its barrier function via a biotin tracer assay. In contrast to controls, in which biotin only penetrated the interstitium and the basal-most layer of germ cells inside the BTB (i.e., spermatogonia and preleptotene spermatocytes), in cKO testes, almost all of the tubules exhibited the presence of biotin deep within tubules near their lumina (Figures S2I and S2J). These analyses confirmed that conditional deletion of Cdc42 resulted not only in disrupted polarity complexes but also in the functional disruption of the BTB in the adult testis.
Testosterone regulates the maintenance of the BTB and dynamics of its assembly and reassembly during spermatogenesis (Meng et al., 2005; Smith and Walker, 2014; Su et al., 2010; Yan et al., 2008); therefore, we addressed whether BTB disruptions were caused by reduced testosterone production due to defective Sertoli-Leydig cell crosstalk by assessing Leydig cell development in adult cKO testes. We observed robust steroidogenic enzyme expression, CYP17A1 and Cyp11a1, in the interstitial compartment of cKO adult testes by immunofluorescence and quantitative real-time PCR (Figures S3A, S3B, and S3K); furthermore, cKO testicular testosterone levels were comparable to controls (Figure S3L). In addition, we examined endothelial cell markers PECAM1 and Cdh5 and saw grossly normal development of blood vessels in the testicular interstitium of cKO testes (Figures S3C, S3D, S3K). However, we did observe occasional ectopic expression of MKI67 (also known as Ki67, a marker of active cell cycle–i.e., not in G0 phase of the cell cycle) in interstitial cells of cKO testes, especially within the peritubular cell layer (Figures S3E and S3F), which are normally negative for MKI67, suggesting some disruption of crosstalk between Sertoli and interstitial cells in cKO testes.
Given that the BTB is disrupted by Cdc42 deletion, we tested the potential consequences of this defect in terms of immune response. The BTB, via sequestration of potentially foreign germ cell neo-antigens, normally prevents the infiltration of immune cells into the lumina of the seminiferous tubules. However, in adult cKO testes, there was an overall increase in testicular CD45+ immune cells, most of which were AIF1+ (IBA1+) macrophages. Macrophages were consistently observed ectopically within cKO tubules and were also increased on the periphery of the peritubular wall in cKO testes relative to controls (Figures S3G–S3J).
Cdc42 expression in Sertoli cells is dispensable for fetal testis differentiation
We next investigated the developmental origins of the defects observed in adult cKO testes. We recently demonstrated that Dhh-Cre is highly specific and effective for targeting Sertoli cells starting as early as embryonic day (E) 12.5 (Heinrich et al., 2020). Furthermore, previously published cell-type-specific and single-cell transcriptome studies indicated that Cdc42 is highly expressed in Sertoli cells (and other cell types) starting at the earliest stages of testicular differentiation until adulthood (Figures S4A–S4D) (Darde et al., 2015, 2019, Green et al., 2018; Jameson et al., 2012; Stévant et al., 2018). Therefore, we wanted to assess whether Cdc42 has essential roles in fetal Sertoli cells that could affect their later development, and Dhh-Cre-mediated deletion would reveal these early roles in Sertoli cells.
As an initial step, we examined several parameters of testicular differentiation in E18.5 cKO fetal testes. Our analyses revealed no differences in general testis cord size, structure, and marker expression in major cell types within cKO testes, including Sertoli cells, germ cells, and Leydig cells. In cKO fetal testes, AMH+ and SOX9+ Sertoli cells were arranged normally along the basal surface of the testis cords, with similar expression of CTNNB1 (β-catenin), an intercellular adhesion molecule with dynamic expression in Sertoli cells (Chang et al., 2008) as compared to control testis cords (Figures 2A–2D). Sertoli cells in cKO testes were in active cell cycle (as defined by MKI67 expression), similar to what was expected for control Sertoli cells (Orth, 1982), and germ cells were centrally localized within the cords and were in cell-cycle arrest (MKI67−), similar to what was expected for control germ cells (Western et al., 2008) (Figures 2E–2J). Leydig cell development and male-specific vascularization of the testis were also normal in cKO samples (Figures 2K and 2L). Finally, we observed no significant difference in CD45+ cells, which were mostly located outside the testis cords, in control versus cKO testes (Figures 2M and 2N). Overall, our results suggest that Cdc42 in Sertoli cells is not essential for fetal testicular somatic and germ cell development.
Figure 2. Cdc42 in Sertoli cells is dispensable for fetal testicular differentiation.
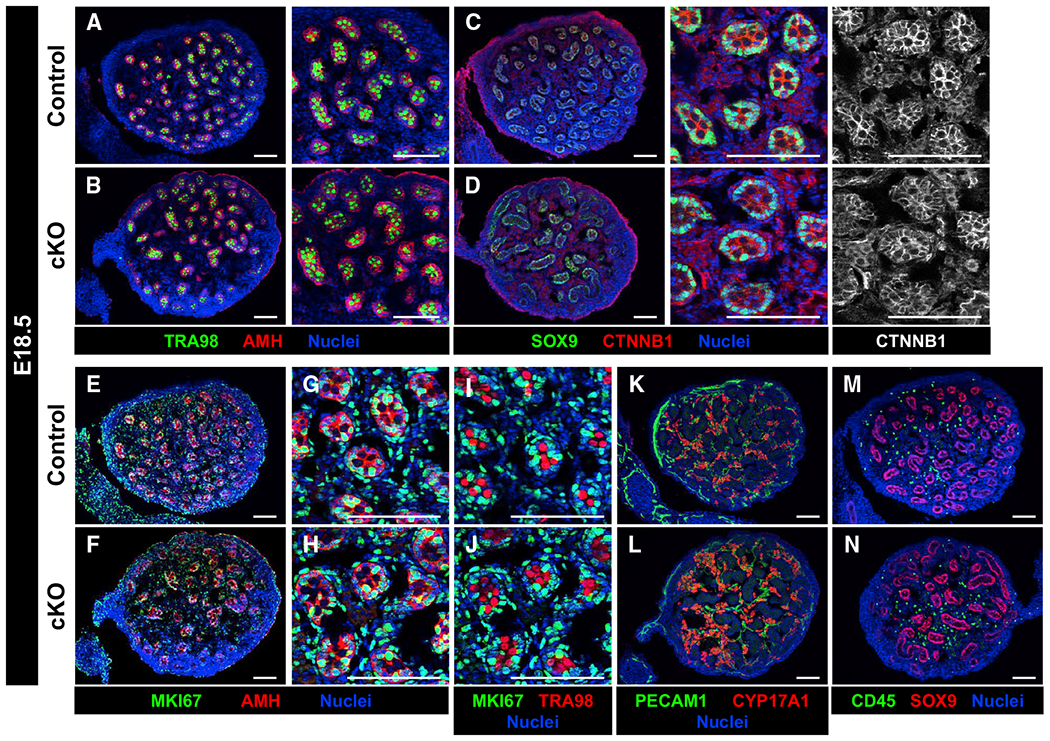
Immunofluorescence images of E18.5 control Dhh-Cre;Cdc42flow/+ (A, C, E, G, I, K, and M) and cKO (B, D, F, H, J, L, and N) testes.
(A–D) Control (A and C) and cKO (B and D) testes contain similar numbers of germ (TRA98+) and Sertoli (AMH+/SOX9+) cells, and they display similar expression of CTNNB1 within tubules.
(E-H) Both control (E and G) and cKO (F and H) Sertoli cells are in active cell cycle (MKI67+).
(I and J) Both control (I) and cKO (J) germ cells are quiescent (MKI67−).
(K–N) Control (K and M) and cKO (L and N) testes show similar numbers of vascular endothelial (PECAM1+), Leydig (CYP17A1+), and immune (CD45+) cells.
Scale bars, 100 μm.
Cdc42 is not required for postnatal testicular differentiation and the onset of the first wave of spermatogenesis
Our findings suggested that spermatogenesis was disrupted in adult cKO testes, but fetal testicular differentiation was unaffected; therefore, we next investigated postnatal developmental stages to determine when testis development and spermatogenesis were affected by Cdc42 conditional deletion. In general, we found that early postnatal spermatogenesis at P7 was unaffected in cKO testes when compared to controls. Undifferentiated spermatogonia, as visualized via GFRA1 and ZBTB16 staining, were present in both control and cKO testes and all of them exhibited proper basal localization within the tubules (Figures 3A–3D). KIT staining revealed that the differentiating spermatogonial population was comparable to controls, and DDX4 and TRA98 staining showed that overall germ cell numbers were comparable between P7 control and cKO testes (Figures 3E–3H). As for Sertoli cells, we found that P7 cKO Sertoli cells generally had similar GATA4, GATA1, AMH, and AR expression as compared to controls (although AMH expression was decreased in some cKO tubules) (Figures 3G–3L). Overall apoptosis levels of Sertoli and germ cells were similar in P7 control versus cKO testes (Figures S5A and S5B); additionally, we saw no differences in vasculature, Leydig cells, or immune cells in P7 cKO testes (Figures S5C–S5H). These data overall suggest that early postnatal testis differentiation and the initiation of the first wave of spermatogenesis was unaffected in cKO testes.
Figure 3. Early postnatal differentiation of Sertoli and germ cells occurs normally in cKO testes.
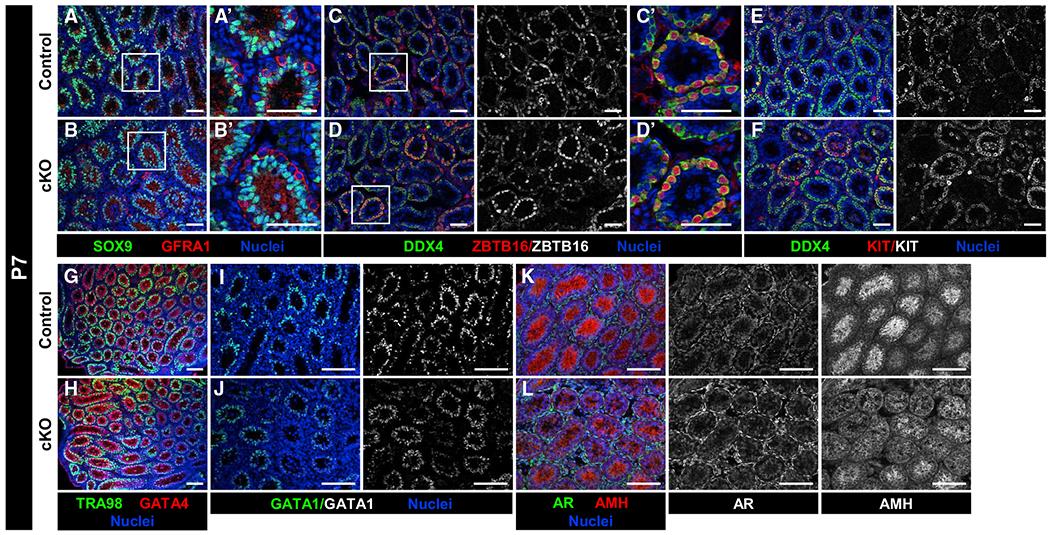
Immunofluorescence images of P7 control Dhh-Cre;Cdc42flox/+ (A, C, E, G, I, and K) and cKO (B, D, F, H, J, and L) testes. (A’)–(D’) are higher-magnification images of the boxed regions in (A)–(D).
(A–H) Compared to P7 controls (A, C, E, and G), P7 cKO testes (B, D, F, and H) have similar numbers and localization of Sertoli cells (SOX9+, GATA4+), undifferentiated spermatogonia (GFRA1+, ZBTB16+), differentiating spermatogonia (KIT+), and overall germ cells (DDX4+, TRA98+).
(I and J) GATA1 expression is detected in both control (I) and cKO (J) Sertoli cells.
(K and L) Both AMH and AR are detected within control (K) and cKO (L) testes, but AMH expression appears slightly decreased in a subset of cKO Sertoli cells.
Scale bars, 100 μm.
Cdc42 is dispensable for onset and maintenance of Sertoli cell quiescence, but is required for juvenile and adult Sertoli cell survival
CDC42 has been implicated in cell-cycle regulation, so we examined whether Cdc42 deletion induced changes in the cell-cycle status of Sertoli cells. By adulthood, normally, Sertoli cells are mitotically quiescent (Orth, 1982), and we observed that adult control Sertoli cells did not express MKI67 (Figure 4A). In adult cKO testes, while Sertoli cells were generally MKI67−, we observed rare Sertoli cells aberrantly expressing MKI67 (Figure 4B); however, cKO Sertoli cells were never observed expressing the mitotic marker phosphohistone H3 (pHH3) (Figures 4C and 4D), suggesting that adult cKO Sertoli cells were not ectopically proliferative.
Figure 4. Cdc42 is not required for onset and maintenance of Sertoli cell quiescence but is required for Sertoli cell survival.
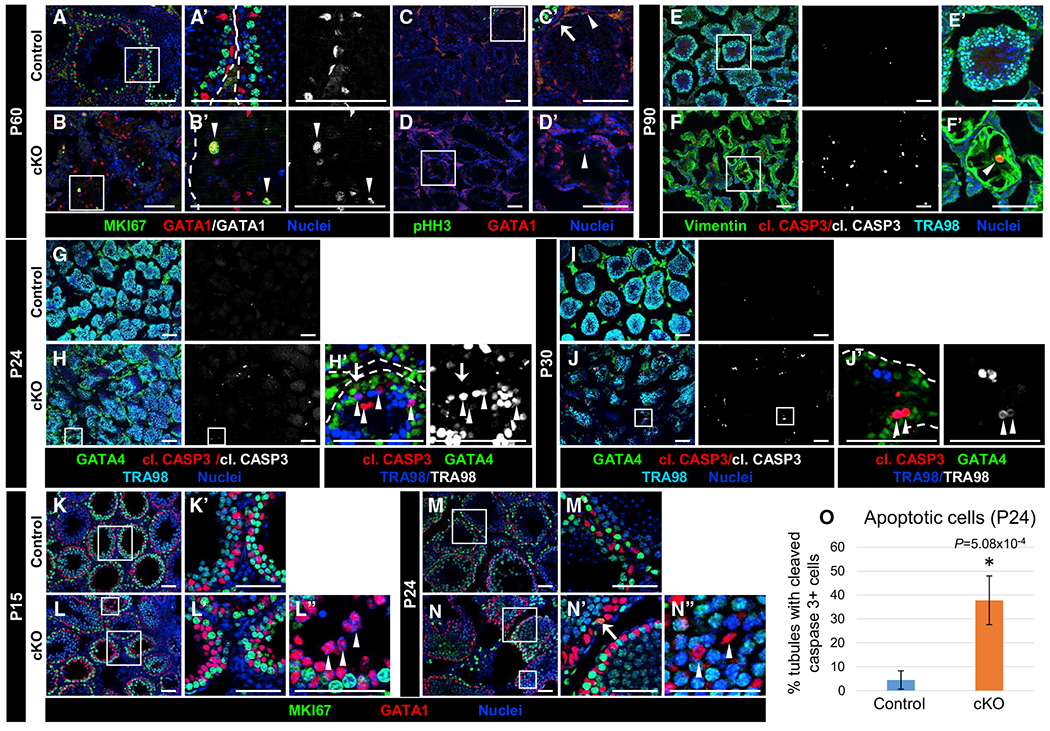
Immunofluorescent images of P60 (A–D), P90 (E and F), P24 (G, H, M, and N), P30 (I and J), and P15 (K and L) control Dhh-Cre;Cdc42flox/+ (A, C, E, G, I, K, and M) and cKO (B, D, F, H, J, L, and N) testes. (A’)–(F’), (H’), (J’), and (K’)–(N”) are higher-magnification images of the boxed regions in (A)–(F), (H), (J), and (K)–(N). Dashed lines throughout the figure indicate tubule boundaries.
(A and B) Both P60 control (A) and cKO (B) Sertoli cells are MKI67−, but rare cKO Sertoli cells expressing MKI67 (arrowheads in B’) are observed.
(C and D) Neither control (C) or cKO (D) Sertoli cells express the mitotic marker pHH3 (arrowheads in C’ and D’); only germ cells in control tubules are pHH3+ (arrow in C’).
(E and F) Cleaved caspase 3+ (apoptotic) Sertoli cells are rarely detected in P90 control (E) testes but are often observed in cKO (F) testes (arrowhead in F’).
(G–J) Compared to control P24 (G) and P30 (I) juvenile testes, cKO juvenile testes (H and J) exhibit increased cleaved caspase 3+ Sertoli cells (GATA1+; arrow in H’) and germ cells (TRA98+; arrowheads in H’ and J’).
(K–N) Relative to control P15 (K) and P24 (M) juvenile Sertoli cells, cKO juvenile Sertoli cells (L and N) show similar immunonegativity for MKI67 (arrowheads in L” and N”), apart from rare MKI67+ cells (arrow in N’). Scale bars, 100 μm.
(O) Graph showing percentage of tubules containing cleaved caspase 3+ cells in P24 control versus cKO testes (n = 15 tubules each from 3 testes each for controls and cKO). Data are shown as means ± SDs. p value was calculated via a 2-tailed Student’s t test.
We next sought to address whether Cdc42 loss affected adult Sertoli cell survival. We observed a significant increase in cleaved caspase 3 staining in adult cKO testes; co-staining revealed that apoptotic cells were Sertoli cells (Figures 4E and 4F), which is consistent with the reduction in Sertoli cell number seen in adult cKO tubules (see Figure 1E). Looking at juvenile stages of development, P24 and P30, we observed an increase in Sertoli cells (GATA4+ cells) expressing cleaved caspase 3; additionally, there was an increased number of germ cells (TRA98+) that were apoptotic in cKO testes (Figures 4G–4J). Quantification of cleaved caspase 3+ cells indicated that apoptosis was significantly increased in cKO tubules at P24 (Figure 4O).
We next assessed whether Cdc42 was required for postnatal Sertoli cells to enter mitotic arrest and become quiescent. We examined postnatal testes at P15 and P24, stages by which Sertoli cells should have entered cell-cycle arrest (Orth, 1982). We observed that Sertoli cells were all MKI67− at P15 and P24, apart from very rare cells, in both controls and cKO testes (Figures 4K–4N); even cKO Sertoli cells that were aberrantly localized in the center of tubules were also MKI67− (Figures 4L and 4N). These data indicate that Cdc42 is not required for the onset and maintenance of quiescence in postnatal Sertoli cells but is required for juvenile and adult Sertoli cell survival.
Disruption of germline and Sertoli cell development occurs in juvenile cKO testes
Our results indicated that fetal and neonatal testis differentiation were unaffected by Cdc42 conditional deletion, so we next examined Sertoli cells in P24 juvenile testes, at which point the BTB is established and other hallmarks of Sertoli cell maturation such as AR nuclear localization and AMH downregulation are observed (Sharpe et al., 2003). At this stage, there were obvious Sertoli cell defects in cKO testes. In contrast to control tubules, in which SOX9+ Sertoli cell nuclei were localized exclusively near the basement membrane, Sertoli cell nuclei in cKO testes were regularly observed near the center of tubules (although most cKO Sertoli cell nuclei were localized near the basement membrane) (Figures 5A and 5B). Consistent with our observations of increased apoptosis (see Figure 4), quantification revealed that there was a significant reduction in Sertoli and germ cells in P24 cKO testes, leading to a reduction in seminiferous tubule cross-sectional areas (Figures 5C–5E).
Figure 5. Juvenile cKO mice show defects in Sertoli cell development.
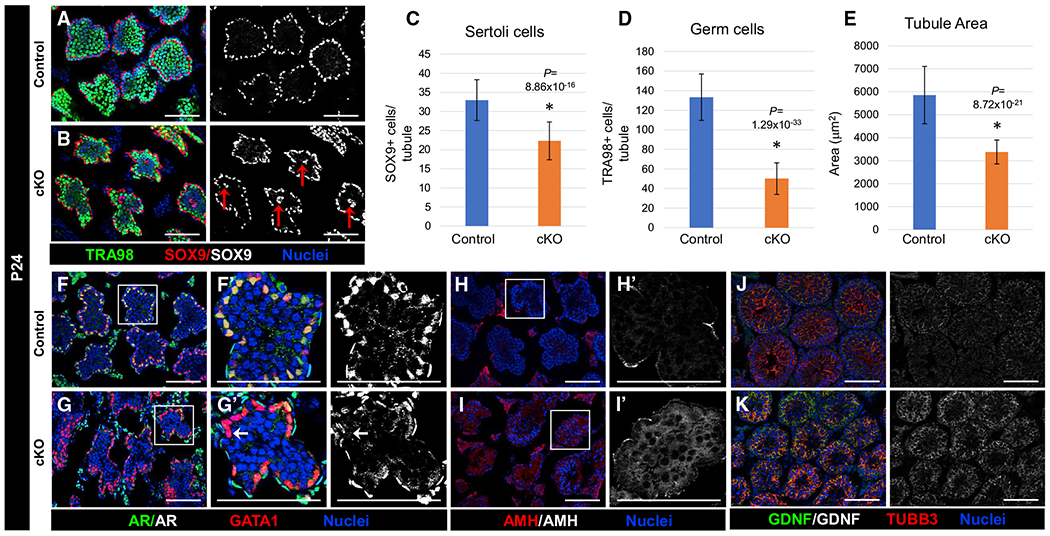
Immunofluorescent images of P24 (A, B, and F–K) control Dhh-Cre;Cdc42flox/+ (A, F, H, and J) and cKO (B, G, I, and K) testes. (F’)–(I’) are higher-magnification images of the boxed regions in (F)–(I).
(A and B) Relative to control testes (A), cKO testes (B) show smallertubules and mislocalization of Sertoli cell nuclei (red arrows in B) near the center of tubules.
(C–E) Graphs showing average number of SOX9+ Sertoli cells per tubule (C), average number of TRA98+ germ cells per tubule (D), and average tubule cross-sectional area (E) in P24 control versus cKO testes (n = 15 tubules each from 3 testes each for controls and cKO). The data in (C)–(E) are shown as means ± SDs. p values were calculated via a 2-tailed Student’s t test.
(F–I) Relative to control (F and H) testes that exhibit nuclear localization of AR and undetectable expression of AMH, cKO testes (G and I) show only weak or sporadic localization of AR in Sertoli cells (arrow in G’) and persistent expression of AMH.
(J and K) Both control (J) and cKO (K) testes exhibit strong expression of GDNF.
Scale bars, 100 μm.
We next examined Sertoli cell phenotypes in cKO testes by examining Sertoli-specific markers. We observed a disruption in the localization of AR in which it was diffusely localized throughout cKO Sertoli cells, similar to our observations in the cKO adult (Figures 5F and 5G). AMH expression remained higher in cKO tubules relative to controls (Figures 5H and 5I), suggestive of a disruption or delay in Sertoli cell maturation. However, we did note that GDNF was still strongly expressed in the Sertoli cells of P24 cKO testes relative to controls (Figures 5J and 5K).
Sertoli cell polarity and BTB function are disrupted in cKO juvenile testes
Given that germ cells were still present in P24 juvenile cKO testes, we next assessed whether cell polarity defects were present at this stage and if they occurred before (and not because of) the severe germ cell loss observed in adult cKO testes. We examined several cell polarity markers and, similar to adult testes, there were widespread, although not universal, defects in polarity protein localization in P24 cKO testes. CLDN11, which was highly enriched in a tight ring-like BTB structure in control testes, was localized in a more diffuse web-like structure in most cKO tubules, suggesting that the BTB was rudimentarily formed, but was also regularly observed throughout the entirety of the tubule in a subset of cKO tubules (Figures 6A and 6B). We also assessed PARD3 expression, and in contrast to basal localization in control tubules, it was diffusely localized throughout cKO tubules (Figures 6C and 6D).
Figure 6. Loss of Cdc42 leads to disruptions in Sertoli cell polarity and BTB function in cKO juvenile testes.
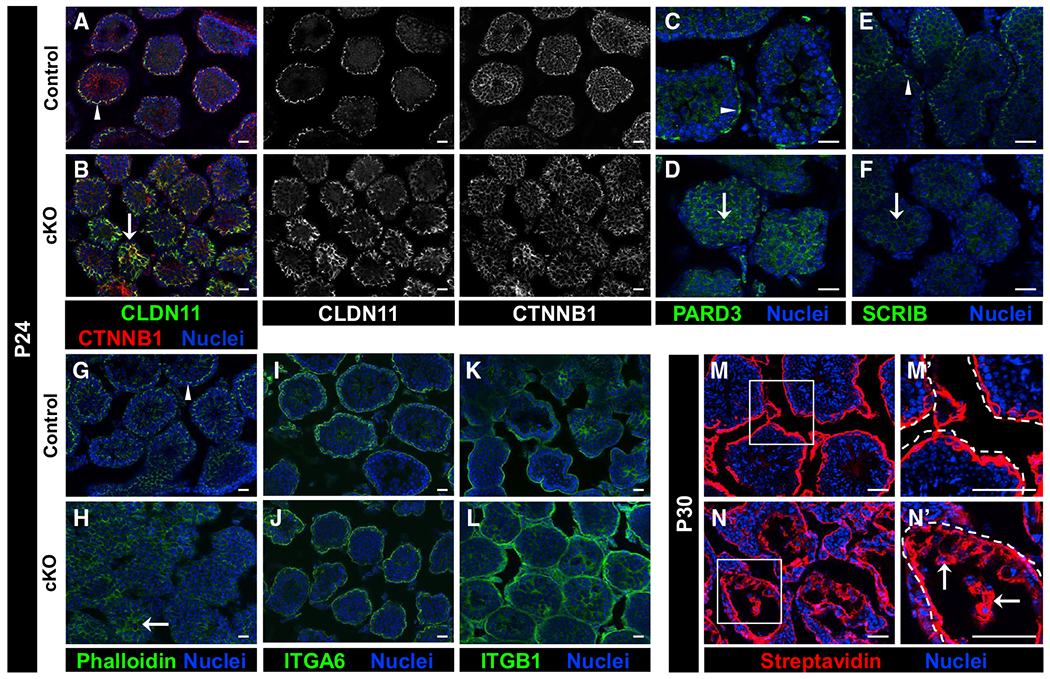
Immunofluorescent images of P24 control Dhh-Cre;Cdc42flox/+ (A, C, E, G, I, and K), P30 control (M), P24 cKO (B, D, F, H, J, and L), and P30 cKO (N) testes. (M’) and (N’) are higher-magnification images of the boxed regions in (M) and (N).
(A–H) While control (A, C, E, and G) testes showed compartment-specific enrichment of CLDN11, CTNNB1, PARD3, SCRIB, and phalloidin staining (arrowheads), cKO testes (B, D, F, and H) exhibited aberrant localization of these factors (arrows).
(I–L) Both control (I and K) and cKO (J and L) testes displayed strong expression of ITGA6 and ITGB1 in the tubule basement membrane; however, cKO testes regularly displayed ectopic expression of ITGB1 in the central portion of tubules.
(M and N) While P30 control (M) testes showed only the interstitial and basal tubular presence of biotin (detected by streptavidin staining) in a tracer assay, cKO testes (N) showed widespread presence of biotin deep within tubules (arrows in N’) (n = 3 testes each for controls and cKO). Dashed lines indicate tubule boundaries.
Scale bars, 50 μm.
In terms of basal markers, we found that SCRIB was not enriched in the basal compartment in cKO testes (Figures 6E and 6F), nor was phalloidin specifically localized to the BTB (Figures 6G and 6H), indicating that there were defects in the basal compartment of juvenile cKO testes. Three-dimensional imaging of phalloidin staining revealed that the actin cytoskeletal structure of cKO tubules was more diffuse and did not possess a clear actin-rich ring-like BTB as compared to controls (Videos S1 and S2).
To address whether Sertoli cell adhesion may have been compromised, we examined the expression of the integrin proteins ITGA6 and ITGB1. While ITGA6 in cKO testes appeared similar to controls, with localization to the basement membrane and basal-most layer of germ cells (Figures 6I and 6J), ITGB1 localization was disrupted in cKO testes, with ectopic localization throughout the entire tubule (Figures 6K and 6L); however, basement membrane expression and the overall expression levels of these integrins did not appear to be reduced.
We next investigated whether there was defective BTB barrier function in juvenile cKO testes. Using a biotin tracer assay, we found that, in contrast to P30 control testes that had effective barrier function, P30 cKO testes exhibited biotin penetration deep within all of the tubules (Figures 6M and 6N), indicating defective barrier function.
To confirm further that cell polarity defects seen in cKO Sertoli cells occur independently of germ cell loss, we examined two other mouse models of germ cell deficiency, KitW/W-v and Dnd1Ter/Ter. We observed that, similar to what we reported in adult testes for KitW/W-v and Dnd1Ter/Ter (Heinrich et al., 2020), several cell polarity complex proteins, but not all, were disrupted in germ cell-deficient juvenile (P28) testes; while CTNNB1 and PARD3 were mislocalized, CLDN11 and SCRIB showed normal localization (Figure S6). Therefore, the loss of germ cells cannot solely account for the cell polarity defects observed (e.g., CLDN11 and SCRIB disruption) in cKO testes, and it is likely that the cell polarity defects in cKO testes are a cause, rather than an effect, of germ cell loss.
Cdc42 in Sertoli cells is essential for progression past the round spermatid stage during first-wave spermatogenesis and for maintenance of germ cells
A degree of germ cell loss was evident by P24 in cKO testes, so we next examined different germ cell types at that age to determine any changes in spermatogenesis that could underlie the observed germ cell loss. GFRA1+ undifferentiated spermatogonia, which represent the most undifferentiated spermatogonial population of which a subset are SSCs, were still observed in their basal location in cKO tubules similarly to controls (Figures 7A and 7B) and in similar numbers (Figure 7C), and GFRA1+ cells were found in 46.0% ± 9.9% of control tubules versus 52.9% ± 4.3% of cKO tubules (p = 0.131, n = 23–104 tubules per testis from 3 testes each for controls and cKO). Furthermore, a similar distribution of GFRA1+ cells per tubule was observed between controls and cKO testes (Figure 7D). These data suggest that SSC maintenance was normal in cKO testes at this stage; to address this issue, we performed quantitative real-time PCR analyses for a number of Sertoli-expressed genes that are implicated in SSC maintenance and found that there was no significant disruption in gene expression specifically related to SSC maintenance (Figures S7A and S7B). In contrast, ZBTB16 expression, which labels a broader population of undifferentiated spermatogonia, revealed a reduction of this spermatogonial population in cKO testes (Figures 7E–7G); however, basal localization of undifferentiated spermatogonia in cKO tubules did not seem to be affected.
Figure 7. Cdc42 in Sertoli cells is essential for progression past the round spermatid stage during first-wave spermatogenesis.
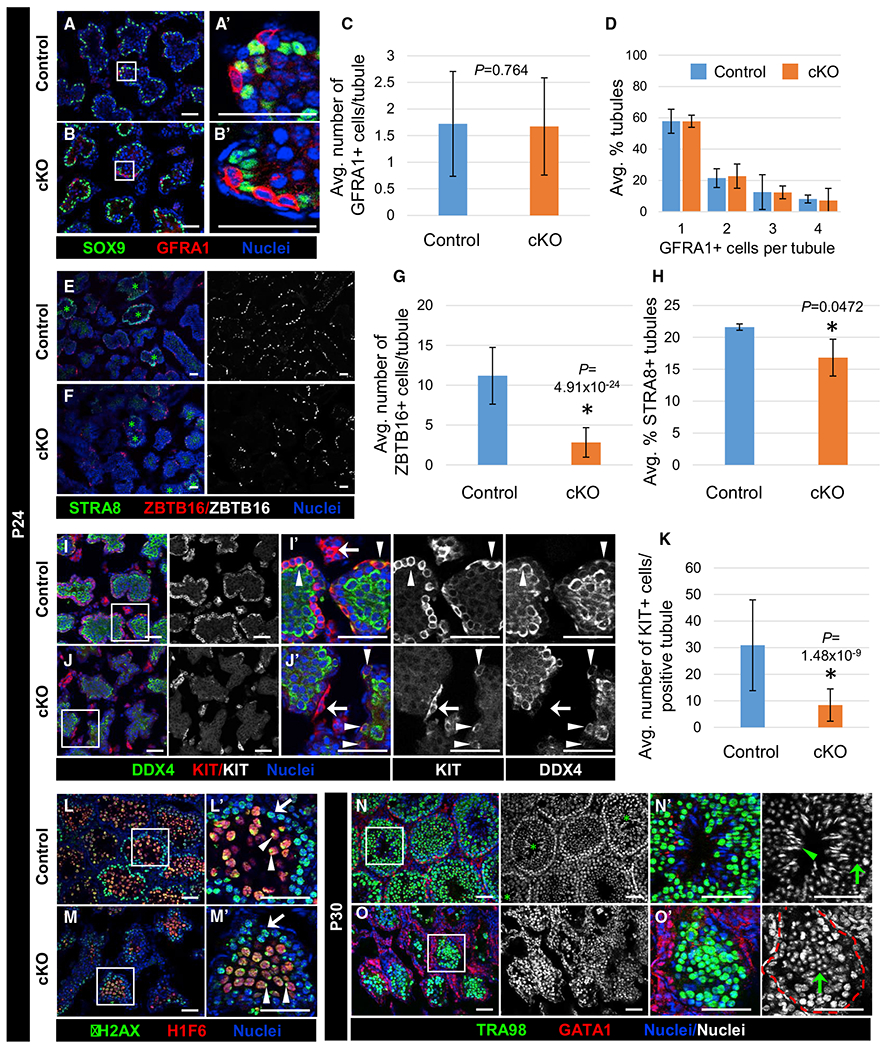
Immunofluorescent images of P24 (A, B, E, F, I, J, L, and M) and P30 (N and O) control Dhh-Cre;Cdc42flox/+ (A, E, I, L, and N) and cKO (B, F, J, M, and O) juvenile testes. (A’), (B’), (I’), (J’), and (L’)–(O’) are higher-magnification images of the boxed regions in (A), (B), (I), (J), and (L)–(O).
(A and B) Both control (A) and cKO (B) testes contain basally localized GFRA1+ undifferentiated spermatogonia.
(C) Graph showing average number of GFRA1+ cells within tubules in P24 control versus cKO testes (n = 23–104 tubules per testis from 3 testes each for controls and cKO).
(D) Graph showing distribution of percentage of tubules containing 1–4 GFRA1+ cells in P24 control versus cKO testes (n = 13–50 tubules per testis from 3 testes each for controls and cKO).
(E and F) Control (E) and cKO (F) testes both contain tubules with STRA8+ preleptotene spermatocytes (green asterisks), but cKO testes contain fewer ZBTB16+ cells.
(G) Graph showing average number of ZBTB16+ cells per tubule in P24 control versus cKO testes (n = 15 tubules each from 3 testes each for controls and cKO).
(H) Graph showing average percentage of tubules containing STRA8+ preleptotene spermatocytes in P24 control versus cKO testes (n = 58–156 tubules per testis from 3 testes each for controls and cKO).
(I and J) Compared to control (I) testes, cKO testes (J) contain fewer KIT+ differentiating spermatogonia (also DDX4+; arrowheads); however, KIT expression in Leydig cells (arrows) is comparable between control and cKO testes.
(K) Graph showing average number of KIT+ cells in P24 control versus cKO tubules (n = 15 tubules each from 3 testes for controls and 2 testes for cKO).
(L and M) γ-H2AX+ leptotene (arrows) and zygotene spermatocytes, as well as H1F6+ (H1T+) pachytene spermatocytes with proper sex-body-enriched γ-H2AX localization (arrowheads), are observed in both control (L) and cKO (M) tubules; however, the number of H1F6+ spermatocytes is decreased in cKO tubules.
(N and O) While P30 control (N) tubules contain round spermatids (green arrow in N’) and elongating spermatids (green arrowhead in N’), P30 cKO tubules only contain round spermatids (green arrow in O’). The red dashed line indicates tubule boundaries.
Scale bars, 50 μm. All graph data are shown as means ± SDs. p values were calculated via a 2-tailed Student’s t test.
Given the loss of ZBTB16+ undifferentiated spermatogonia that we observed in cKO testes, we specifically assessed the differentiating spermatogonial pool via KIT expression and found a dramatic decrease in differentiating spermatogonia at P24 (Figures 7I–7K). To assess whether RA synthesis was reduced, thus potentially affecting spermatogonial differentiation, we performed immunofluorescence and quantitative real-time PCR analyses for RA synthesis enzymes Aldh1a2 (also known as Raldh2) and Aldh1a1 (also known as Raldh2) in P24 cKO testes; immunofluorescence revealed strong expression of both ALDH1A1 and ALDH1A2 in P24 cKO testes, and quantitative real-time PCR assays did not reveal any significantly reduced expression of Aldh1a1 or Aldh1a2 (Figures S7C–S7G). Furthermore, the percentage of tubules containing STRA8+ preleptotene spermatocytes, which depends on RA, was only slightly reduced in P24 cKO testes (Figures 7E, 7F, and 7H). These findings suggest that the loss of differentiating spermatogonia or preleptotene spermatocytes in cKO testes was likely not due to a specific disruption of RA synthesis.
We also found a decrease in spermatocyte and round spermatid populations in P24 cKO testes, as marked by γ-H2AX (official name H2A.X variant histone) and H1F6 (usually known as H1T); however, the morphology of these cell types was unaffected, including the enrichment of γ-H2AX in the XY body of pachytene spermatocytes and a condensed chromocenter within round spermatids (Figures 7L and 7M).
We next determined the extent to which spermatogenesis could proceed in cKO testes. While we were able to confirm the presence of round spermatids via H1F6 staining and nuclear morphology in both control and cKO tubules, elongating spermatids were never observed in any P30 cKO tubules, in contrast to controls (Figures 7N and 7O), indicating that spermatogenesis is arrested at the round spermatid stage in cKO males.
DISCUSSION
In this study, we have obtained insights into the role of CDC42 in Sertoli cells in regard to spermatogenesis and testicular structure and function. Specifically, we provide in vivo evidence that Sertoli cells require Cdc42 function to establish apicobasal polarity critical for supporting adult steady-state spermatogenesis and maintaining germ cells in the testis, but that it is dispensable for embryonic and early postnatal testicular development. These findings are consistent with our recent observations that cell polarity, as visualized by the localization of polarity complexes, is not yet observed in fetal stages. In addition, we have previously shown that Sertoli-specific conditional deletion of Rac1, encoding another Rho GTPase, disrupted cell polarity in the adult testis but did not affect fetal testis development (Heinrich et al., 2020). Although Sertoli cell polarity was not essential for fetal and early postnatal development, onset of the Cdc42 cKO phenotype was detectable as early as P24, and it became more severe with age. Therefore, our data suggest that Cdc42 function, and likely Sertoli cell polarity, is only required starting in juvenile stages, when a greater diversity of spermatogenic stages are present in the testis, which must be simultaneously supported by Sertoli cells.
In addition to this loss of the germline, the structure of adult cKO seminiferous tubules was significantly disrupted, with reduced Sertoli cell number and tubule area, which was likely the result of the increased apoptosis of Sertoli cells. Perhaps most striking was a mislocalization of Sertoli cell nuclei in the seminiferous tubule and universally aberrant localization of polarity protein complexes, such as the Par6/Par3/aPKC and SCRIB complexes, and of factors localized to BTB tight junctions and ES adherens junctions, such as CLDN11, F-actin, ITGB1, ITGA6, and CTNNB1. These results show that CDC42 is an essential factor whose deletion in Sertoli cells causes severe cell polarity defects.
While we hypothesize that these disruptions in protein expression are a result of Cdc42 deletion, it is important to note that germ cell depletion alone causes some similar disruptions, as we have previously reported in adult Kit and Dnd1 mutant testes (Heinrich et al., 2020). Here, in juvenile (P28) Dnd1Ter/Ter and KitW/W-v tubules, we observed the aberrant expression of CTNNB1, ITGB1, ITGA6, and phalloidin, similar to our observations in Cdc42 cKO tubules. However, we observed proper basal expression of SCRIB and CLDN11 in both germ cell-deficient models. While the germ cell population was reduced in P24 cKO tubules, a significant number of TRA98+ cells was still present at this point, so it is unlikely that germ cell loss is the sole cause of the phenotype observed. In addition, Kit mutation results in a failure of undifferentiated spermatogonia to differentiate (thus, only undifferentiated spermatogonia remain in KitW/W-v testes) and also compromises BTB function (Li et al., 2018), but SSCs can be maintained in this environment, indicating that BTB function is not absolutely necessary for a functional SSC niche. Our recent analyses of Rac1 cKO testes also revealed BTB disruption, but with the sustained maintenance of germline stem/progenitor cells; this difference may be due to less severe disruption of apicobasal polarity in Rac1 cKO mice as compared to Cdc42 cKO mice (Heinrich et al., 2020). However, our Cdc42 cKO model appears to exhibit a more severe phenotype than merely a disrupted BTB or apical ES.
In cKO testes, we observed extensive immune cell infiltration into the seminiferous tubules, indicative of a disrupted BTB, which was confirmed by biotin tracer assays at P30 and P90. There are additional aspects of immune function that rely on Sertoli functions beyond BTB regulation, such as the secretion of immunosuppressive molecules (reviewed in (Meinhardt and Hedger, 2011) and (Zhao et al., 2014)) and the acquisition of systemic tolerance via retrograde transcytosis of selective neo-antigens from germ cells phagocytosed by Sertoli cells into the interstitium (Tung et al., 2017). Therefore, defective protein trafficking may be linked to critical functions of Sertoli cells in mediating the unique immune environment of the testis that permits spermatogenesis.
Given that SSCs are absent in cKO testes by adulthood, we examined the expression of Gdnf, which is required for SSC maintenance (Meng et al., 2000). Our analyses revealed the normal expression of Gdnf, as well as other SSC-related factors in cKO Sertoli cells, suggesting that a lack of this and other critical ligands is not the underlying cause of eventual SSC loss in adults. Our analyses also indicated that GFRA1+ cells were not affected in juvenile cKO testes, whereas other cell types, such as differentiating spermatogonia, were significantly reduced in number, indicating that SSCs were not specifically affected by the loss of Sertoli Cdc42. However, the complete loss of germ cells at P60 demonstrates that, ultimately, SSCs cannot be maintained in adult cKO testes. We also assessed the expression of RA-synthesizing enzymes Aldh1a1 and Aldh1a2, since RA is necessary for the initiation of both spermatogonial differentiation and the initiation of meiosis (Griswold, 2016), and potentially also for BTB function (Morales and Cavicchia, 2002). Decreases in RA can cause arrest at both of these stages and could explain the decreasing number of differentiating and differentiated spermatogonia we observed at P24 in cKO testes. However, we observed no significant changes in the expression of RA synthesis enzymes; furthermore, we also detected meiotic cell types in cKO testes. In addition, RA has been proposed to regulate testosterone levels (Appling and Chytil, 1981; Huang et al., 1983), but we observed normal testosterone levels in cKO testes. Therefore, although widespread germ cell loss is evident in our cKO testes, we found no evidence to suggest that this phenotype is the result of any specific effects on SSC maintenance, induction of spermatogonial differentiation, or meiotic entry.
We observed some defects in the differentiation of juvenile cKO Sertoli cells, which was characterized by the aberrant maintenance of AMH expression and the reduced localization of AR to the nucleus (however, there was no period of extended or ectopic proliferation). Therefore, a disruption in Sertoli cell differentiation or maturation cannot be ruled out as a contributor: for example, Sertoli cell-specific AR cKO mice undergo spermatogenic arrest during meiotic stages (Chang et al., 2004; De Gendt et al., 2004). We detected normal testosterone levels and regularly observed round spermatids from the first wave of spermatogenesis within cKO testes, suggesting that androgen-signaling disruptions cannot solely account for the cKO phenotype.
Actin cytoskeletal dynamics are another potential factor to consider in Sertoli cell biology, as the actin cytoskeleton is not only vital for protein vesicular transport but also for filipodia formation and other cellular processes. Since Sertoli cells contact multiple germ cells simultaneously, Sertoli cell cytoarchitecture specifically accommodates physical interactions with germ cells to maintain proper adhesion and intercellular signaling; the intermingling of germ and somatic cells has been carefully described in Drosophila gonads, and Rho GTPases also have been implicated in this phenomenon (Jenkins et al., 2003; Sarkar et al., 2007). Our data do not suggest any defects in cell adhesion, as integrins ITGB1 and ITGA6 were strongly expressed, albeit with slightly altered localization, in cKO testes. CDC42 may be involved in remodeling the actin cytoskeleton to allow its complex architecture to accommodate numerous germ cells at different stages of spermatogenesis. Our 3D imaging of phalloidin staining in cKO tubules supports the idea that the actin cytoskeleton is disrupted.
Finally, it is necessary to consider that the worsening phenotype observed in our cKO testes is likely due in part to the constant communication between Sertoli and germ cells. While Cdc42 ablation may initiate many aspects of the phenotype observed in early postnatal testes, the gradual loss of both Sertoli and germ cells and their subsequent signals to one another are likely to result in a feedforward loop that rapidly advances the phenotype observed in both cell populations. Our analyses of Dnd1 and Kit mutant testes suggest that germ cell loss alone is not sufficient to explain the phenotype in cKO testes, and that deletion of Cdc42 presents additional defects in testicular structure and function. Beyond all of these hypothesized roles for CDC42 that we have discussed, we also concede that, given the strong phenotype we observed, CDC42 could possess one or more unknown roles in Sertoli cell function whose disruption leads to germ cell loss.
In summary, we have found that CDC42 in Sertoli cells is absolutely essential for adult testicular function, particularly through the maintenance of apicobasal cell polarity and establishment of the BTB and apical ES within the seminiferous epithelium. The perturbation of processes regulated by CDC42 affected steady-state spermatogenesis beginning at juvenile stages and resulted in a complete disruption of germ cell maintenance by adulthood. This study demonstrates the broad impact of CDC42 in testicular structure and function, providing insights into the mechanisms underlying the establishment of Sertoli cell polarity and its critical role in ensuring spermatogenesis and fertility.
Limitations of the study
In the present study, we examined the role of CDC42 in testicular development, specifically focusing on Sertoli cells. Our data demonstrated that Cdc42 function is absolutely necessary for apicobasal Sertoli cell polarity and for the maintenance of adult steady-state spermatogenesis, but it is dispensable for embryonic and postnatal testicular differentiation. Given the potentially diverse cellular roles of CDC42, there are several possibilities for the specific function of CDC42 within Sertoli cells. During our analyses, we addressed a number of possible hypotheses regarding the cause of Sertoli cell defects in adult cKO testes, including disruptions in cell cycle, SSC maintenance, hormonal signaling, or, alternatively, indirect effects due to loss of germ cells. Our data largely ruled out these hypotheses, leaving a disruption of apicobasal cell polarity as the most likely cause. However, direct evidence that reduced cell polarity is the primary, specific cause of Sertoli cell disruptions in cKO testes remains to be found; furthermore, there are other possible functions for CDC42 that remain untested. Further investigation into the potential influence of CDC42 on protein trafficking and cytoskeletal remodeling, for example, would be beneficial in exploring its potential role in the establishment of the microenvironment necessary for spermatogenesis and to understand in greater detail its broader role in testicular function.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tony DeFalco (tony.defalco@cchmc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This paper analyzes existing, publicly available microarray and RNA-Seq data. The GEO accession numbers for these datasets are listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact on request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-AIF1 (IBA1) | Wako | Cat#019-19741; RRID: AB_839504 |
| Rabbit polyclonal anti-ALDH1A1 | Abcam | Cat#ab23375; RRID: AB_2224009 |
| Rabbit polyclonal anti-ALDH1A2 | Sigma-Aldrich | Cat#HPA010022; RRID: AB_1844723 |
| Goat polyclonal anti-AMH (MIS) | Santa Cruz Biotechnology | Cat#sc-6886; RRID: AB_649207 |
| Rabbit polyclonal anti-AR | Santa Cruz Biotechnology | Cat#sc-816; RRID: AB_1563391 |
| Rat monoclonal anti-CD45 (clone 30-F11) | BioLegend | Cat#103101; RRID: AB_312966 |
| Rat monoclonal anti-CDH1 (clone ECCD-2) | Thermo Fisher Scientific | Cat#13-1900; RRID: AB_2533005 |
| Rabbit polyclonal anti-CLDN11 | Thermo Fisher Scientific | Cat#36-4500; RRID: AB_2533259 |
| Rabbit polyclonal anti-cleaved Caspase 3 (Asp175) | Cell Signaling Technology | Cat#9661S; RRID: AB_2341188 |
| Goat polyclonal anti-CTNNB1 | Santa Cruz Biotechnology | Cat#sc-1496; RRID: AB_1563968 |
| Goat polyclonal anti-CYP17A1 | Santa Cruz Biotechnology | Cat#sc-46081; RRID: AB_2088659 |
| Rabbit polyclonal anti-DDX4 (MVH) | Abcam | Cat#ab13840; RRID: AB_443012 |
| Rat monoclonal anti-GATA1 (clone N6) | Santa Cruz Biotechnology | Cat#sc-265; RRID: AB_627663 |
| Goat polyclonal anti-GATA4 | Santa Cruz Biotechnology | Cat#sc-1237; RRID: AB_2108747 |
| Rabbit polyclonal anti-GDNF | Santa Cruz Biotechnology | Cat#sc-328; RRID: AB_2247684 |
| Goat polyclonal anti-GFRA1 | Neuromics | Cat#GT15004; RRID: AB_2307379 |
| Rabbit polyclonal anti-Histone H2A.X, phospho (Ser139) | Millipore | Cat#07-164; RRID: AB_310406 |
| Guinea pig polyclonal anti-H1F6 (H1T) | Mary Ann Handel (Inselman et al., 2003) | N/A |
| Rat monoclonal anti-ITGA6 (CD49f) (clone GoH3) | BioLegend | Cat#313602; RRID: AB_345296 |
| Goat polyclonal anti-ITGB1 | Novus/R&D Systems | Cat#AF2405-SP; RRID: AB_416591 |
| Goat polyclonal anti-KIT | R&D Systems | Cat#AF1356; RRID: AB_354750 |
| Rabbit monoclonal anti-MKI67 (Ki67) (clone SP6) | GeneTex | Cat#GTX16667; RRID: AB_422351 |
| Rabbit polyclonal anti-PARD3 (Par3) | Novus | Cat#NBP1-88861; RRID: AB_11056253 |
| Goat polyclonal anti-PECAM1 | R&D Systems | Cat#AF3628; RRID:AB_2161028 |
| Rat monoclonal anti-PECAM1 (clone MEC13.3) | BD Biosciences | Cat#553370; RRID: AB_394816 |
| Rabbit monoclonal anti-phospho-Scribble (Ser1220) (clone D8A2) | Cell Signaling Technology | Cat#12316; RRID: AB_2797883 |
| Rabbit polyclonal anti-SOX9 | Millipore | Cat#AB5535; RRID: AB_2239761 |
| Rabbit polyclonal anti-STRA8 | Abcam | Cat#ab49602; RRID: AB_945678 |
| Rat monoclonal anti-TRA98 (clone TRA98) | Abcam | Cat#ab82527; RRID: AB_1659152 |
| Mouse monoclonal anti-TUBB3 (clone TUJ1) | BioLegend | Cat#801201; RRID: AB_2313773 |
| Chicken polyclonal anti-VIM | BioLegend | Cat#919101; RRID: AB_2565208 |
| Rat monoclonal anti-VIM (clone W16220A) | BioLegend | Cat#699301; RRID: AB_2716136 |
| Mouse monoclonal anti-ZBTB16 (PLZF) (clone 2A9) | Millipore | Cat#OP128-100UG; RRID: AB_213280 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat#712-605-153; RRID:AB_2340694 |
| Alexa Fluor 647 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21447; RRID:AB_2535864 |
| Alexa Fluor 647 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-31573; RRID:AB_2536183 |
| Alexa Fluor 647 Goat anti-Mouse IgG2a | Thermo Fisher Scientific | Cat#A-21241; RRID:AB_2535810 |
| Cy3 AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat#712-165-153; RRID:AB_2340667 |
| Alexa Fluor 555 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21432; RRID:AB_2535853 |
| Alexa Fluor 555 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-31572; RRID:AB_162543 |
| Alexa Fluor 555 Goat anti-Mouse IgG2a | Thermo Fisher Scientific | Cat#A-21137; RRID:AB_2535776 |
| Alexa Fluor 488 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-11055; RRID:AB_2534102 |
| Alexa Fluor 488 Donkey anti-Rat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21208; RRID:AB_141709 |
| Alexa Fluor Plus 488 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A32790; RRID:AB_2762833 |
| Alexa Fluor 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson Immunoresearch | Cat#703-545-155; RRID:AB_2340375 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | Cat#A22287; RRID: AB_2620155 |
| Rhodamine Phalloidin | Thermo Fisher Scientific | Cat#R415; RRID: AB_2572408 |
| Hoechst 33342, Trihydrochloride, Trihydrate | Thermo Fisher Scientific | Cat#H1399 |
| Streptavidin, Alexa Fluor 555 Conjugate | Thermo Fisher Scientific | Cat#S21381; RRID: AB_2307336 |
| EZ-Link Sulfo-NHS-LC-Biotin, No-Weigh Format | Thermo Fisher Scientific | Cat#A39257 |
| Deposited data | ||
| Fetal gonad (E11.5-E13.5) cell-type-specific microarray dataset | (Jameson et al., 2012) | GEO: GSE27715; GDS3995 |
| Fetal testis (E10.5-E16.5) single-cell (NR5A1-eGFP+ cells) RNA-Seq dataset | (Stévant et al., 2018) | GEO: GSE97519 |
| Adult testis single-cell RNA-Seq dataset | (Green et al., 2018) | GEO: GSE112393 |
| Experimental models: organisms/strains | ||
| Mouse: Dhh-Cre: Tg(Dhh-cre)1Mejr | Dies Meijer; (Jaegle et al., 2003; Lindeboom et al., 2003) | RRID: IMSR_JAX:012929 |
| Mouse: Cdc42 fl/fl: Cdc42tm1Yizh/J | Yi Zheng; (Chen et al., 2006) | RRID: IMSR_JAX:027576 |
| Mouse: CD-1 IGS Mouse: Crl:CD1(ICR) | Charles River | Cat#CRL:022; RRID: IMSR_CRL:022 |
| Mouse: C57BL/6J Mouse: JAX:C57BL/6J | The Jackson Laboratory | Cat#JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse: KitW/KitW-v: WBB6F1/J-KitW/KitW-v/J | The Jackson Laboratory | Cat#JAX:100410; RRID: IMSR_JAX:100410 |
| Mouse: Dnd1Ter: Dnd1Ter | Blanche Capel; (Stevens, 1973; Youngren et al., 2005) | MGI:2158668; RRID: MGI:3690452 |
| Oligonucleotides | ||
| Primers for quantitative real-time PCR, see Table S1 | This study | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| The ReproGenomics Viewer | (Darde et al., 2019; Darde et al., 2015) | https://rgv.genouest.org/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice used in this study were housed in the Cincinnati Children’s Hospital Medical Center’s animal care facility, in compliance with institutional and National Institutes of Health guidelines. Institutional ethical approval through the Institutional Animal Care and Use Committee (IACUC) of Cincinnati Children’s Hospital Medical Center was obtained for all animal experiments. Mice were housed in a 12-hour light/12-hour dark cycle and had access to autoclaved rodent Lab Diet 5010 (Purina, St. Louis, MO, USA) and ultraviolet light-sterilized RO/DI constant circulation water ad libitum. Dhh-Cre (Tg(Dhh-cre)1Mejr) mice (Jaegle et al., 2003; Lindeboom et al., 2003) were obtained from Dies Meijer (University of Edinburgh) and Cdc42flox (Cdc42tm1Yizh) mice (Chen et al., 2006; Guo et al., 2013) were obtained from Yi Zheng (Cincinnati Children’s Hospital Medical Center). Both alleles were maintained on a C57BL/6J background. Homozygous Cdc42flox/flox females and hemizygous Dhh-Cre; heterozygous Cdc42flox/+ males were bred, and their offspring were genotyped and used for analysis. Dhh-Cre;Cdc42flox/flox mutant mice begin to exhibit partial hind limb paralysis around 3 weeks but can live a normal lifespan if continuously provided with food on the floor of their cages after weaning (Guo et al., 2013). Dhh-Cre mice were genotyped using primers sense: 5′-ACCCTGTTACGTATAGCCGA-3′ and anti-sense: 5′-CTCCGGTATTGAAACTCCAG-3′; Cre presence was confirmed via production of a 400-bp band. Cdc42 floxed and wild-type alleles were genotyped using primers sense: 5′-TACAGTTGGTACATATTCCGATGGG-3′ and anti-sense: AGACAAAACAAGGTCCAGAAAC-3′; the Cdc42 floxed allele was confirmed via presence of a 465-bp band and the wild-type allele was confirmed via presence of a 361-bp band. For fetal time points, timed matings were arranged. Noon on the day a vaginal plug was observed was designated as E0.5.
Four-week-old KitW/KitW-v compound heterozygous males (WBB6F1/J-KitW/KitW-v/J) and corresponding control mice were purchased from the Jackson Laboratory (stock #100410), on a mixed background of WB/ReJ and C57BL/6J. Dnd1Ter mice (Stevens, 1973; Youngren et al., 2005) were a gift from Blanche Capel (Duke University Medical Center); Dnd1Ter mice were originally on a 129T2/SvEmsJ background, but were backcrossed to C57BL/6J for 3 generations to eliminate the occurrence of testicular teratomas while maintaining complete germ cell depletion during fetal and postnatal stages (Cook et al., 2011). Dnd1Ter/Ter homozygous mice were generated via an intercross of Dnd1Ter/+ heterozygous mice and were genotyped via a Custom TaqMan SNP Genotyping Assay (Applied Biosystems/Thermo Fisher #4332077).
METHOD DETAILS
Immunofluorescence
Testes were dissected in PBS and fixed overnight in 4% paraformaldehyde (PFA) with 0.1% Triton X-100 (PBTx). For adult testes, the capsule was superficially punctured 10-12 times with a 27-gauge needle prior to fixation to ensure penetration of PFA. Following overnight fixation at 4°C, testes were cut in half transversely and placed again in 4% PFA for an additional 2 hours. After several washes in PBTx, fixed testis samples were processed through a sucrose:PBS gradient (10%, 15%, and 20% sucrose) before rocking at 4°C overnight in a 1:1 mixture of 20% sucrose and Optical Cutting Temperature (OCT) medium (Fisher Healthcare, TX; catalog #4585). The following day, testes samples were embedded in OCT medium and stored at −80°C until cryosectioned.
After cryosectioning at 16-20 μm (depending on stage/age; 3D images for Videos S1 and S2 were obtained from 50-μm-thick sections), samples were washed several times in PBTx, then incubated in blocking solution (PBTx + 10% FBS + 10% bovine serum albumin [BSA]) for 1 hour at room temperature. Primary antibodies were diluted in blocking solution and applied to samples overnight at 4°C. Primary antibodies used were (companies/sources and dilutions are listed in parentheses): rabbit anti-AIF1 (Wako Cat# 019-19741, RRID: AB_839504; 1:1,000); rabbit anti-ALDH1A1 (Abcam Cat# ab23375, RRID: AB_2224009; 1:1,000); rabbit anti-ALDH1A2 (Sigma-Aldrich Cat# HPA010022, RRID: AB_1844723; 1:200); goat anti-AMH (Santa Cruz Cat# sc-6886; RRID: AB_649207; 1:500); rabbit anti-AR (Santa Cruz Cat# sc-816, RRID: AB_1563391; 1:300); rat anti-CD45 (BioLegend Cat# 103101, RRID: AB_312966; 1:300); rat anti-CDH1 (Thermo Fisher Cat# 13-1900, RRID: AB_2533005; 1:500); rabbit anti-CLDN11 (Thermo Fisher Cat# 36-4500, RRID: AB_2533259; 1:1,000); rabbit anti-cleaved caspase 3 (Asp175) (Cell Signaling Technology Cat# 9661S, RRID: AB_2341188; 1:250); goat anti-CTNNB1 (Santa Cruz Cat# sc-1496, RRID: AB_1563968; 1:400); goat anti-CYP17A1 (Santa Cruz Cat# sc-46081, RRID: AB_2088659; 1:500); rabbit anti-DDX4 (Abcam Cat# ab13840, RRID: AB_443012; 1:1,000); rat anti-GATA1 (Santa Cruz Cat# sc-265, RRID: AB_627663; 1:1,000); goat anti-GATA4 (Santa Cruz Cat# sc-1237; RRID: AB_2108747; 1:100); rabbit anti-GDNF (Santa Cruz Cat# sc-328, RRID: AB_2247684; 1:750); goat anti-GFRA1 (Neuromics Cat# GT15004, RRID: AB_2307379; 1:100); Rabbit polyclonal anti-Histone H2A.X, phospho (Ser139) (Millipore Cat# 07-164, RRID: AB_310406; 1:1,000); guinea pig anti-H1F6 (H1T) (Mary Ann Handel (Inselman et al., 2003); 1:2,000); rat anti-ITGA6 (BioLegend Cat# 313602, RRID: AB_345296; 1:500); goat anti-ITGB1 (Novus Cat# AF2405-SP, RRID: AB_416591; 1:500); goat anti-KIT (R&D Cat# AF1356, RRID: AB_354750; 1:400); rabbit anti-MKI67 (GeneTex Cat# GTX16667, RRID: AB_422351; 1:300); rabbit anti-PARD3 (Novus Cat# NBP1-88861, RRID: AB_11056253; 1:100); goat anti-PECAM1 (R&D Cat# AF3628, RRID: RRID:AB_2161028; 1:300); rat anti-PECAM1 (BD Biosciences Cat# 553370, RRID: AB_394816; 1:250); rabbit anti-phospho-SCRIB (Ser1220) (Cell Signaling Technology Cat# 12316, RRID: AB_2797883; 1:500); rabbit anti-SOX9 (Millipore Cat# AB5535, RRID: AB_2239761; 1:4,000); rabbit anti-STRA8 (Abcam Cat# ab49602, RRID: AB_945678; 1:250); rat anti-TRA98 (Abcam Cat# ab82527, RRID: AB_1659152; 1:1,000); mouse monoclonal anti-TUBB3 (BioLegend Cat# 801201, RRID: AB_2313773; 1:1,000); chicken anti-VIM (BioLegend Cat# 919101, RRID: AB_2565208; 1:300); rat anti-VIM (BioLegend Cat# 699301, RRID: AB_2716136; 1:300); mouse anti-ZBTB16 (Millipore Cat# OP128-100UG, RRID: AB_213280; 1:250).
After several washes in PBTx, fluorescent secondary antibodies (Alexa-conjugated; from Molecular Probes/Thermo Fisher, all at 1:500 dilution) and nuclear dye (2 μg/ml Hoechst 33342, catalog #H1399, Molecular Probes/Life Technologies/Thermo Fisher) were applied for 1 hour at room temperature. Following several washes in PBTx, samples were mounted on slides in Fluoromount-G (Southern Biotech, AL; catalog #0100-01). Samples were imaged either on a Nikon Eclipse TE2000 microscope (Nikon, Tokyo, Japan) with an Opti-Grid structured illumination imaging system using Volocity software (PerkinElmer, Waltham, MA, USA) or on a Nikon A1 Inverted Confocal Microscope (Nikon, Tokyo, Japan). At least 3 sections from at least n = 3 independent, individual animals (i.e., n = 3 independent biological replicates) were examined for each time point (i.e., fetal stage or age) and/or experimental condition.
Cell counts and tubule area quantification
For all cell counts, at least 15 random tubules within each testis, from at least 3 different sections, were analyzed (n = 3 testes from independent animals). The Fiji/ImageJ (NIH) Cell Counter plug-in was used to manually count positive cells per tubule. For measuring tubule cross-sectional areas, the perimeters of individual tubules were outlined using the Polygon Selection tool, and the area in pixels was calculated with the Measure function. Pixel area was then converted to square microns based on the pixel dimensions of each image. Graph results are shown as mean ± SD. Statistical analyses were performed using a two-tailed Student’s t test, and a P value of p < 0.05 was considered statistically significant.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA was extracted and processed for quantitative real-time PCR. Tissue was homogenized by vortexing in 800μl TRIzol reagent (Invitrogen/Thermo Fisher). RNA extraction was then performed using a TRIzol/isopropanol precipitation method. Briefly, 200 μL of chloroform was added to the Trizol/tissue mixture, shaken by hand, incubated at room temperature for 3 minutes, and centrifuged at 12,000 × g for 10 minutes at 4°C. The upper aqueous layer was carefully recovered and added to 400 μL isopropanol, which was rocked at room temperature for 30 minutes. After centrifugation at 12,000 × g for 10 minutes at 4°C, supernatant was removed, and the pellet was washed with 500 μL of ethanol. After another centrifugation (with same parameters), the RNA pellet was briefly air-dried and diluted in nuclease-free water. RNA quality was assessed by spectrophotometric analysis via absorbance at 260 and 280 nm, in which only RNA samples with a 260/280 ratio greater than or equal to 1.6 was used for quantitative real-time PCR analysis (although sample ratios were usually between 1.7-2.0). An iScript cDNA synthesis kit (BioRad) was used on 500ng of RNA for cDNA synthesis. Quantitative real-time PCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems/Thermo Fisher) on the StepOnePlus Real-Time PCR system (Applied Biosystems/Thermo Fisher). The following parameters were used: 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s, followed by a melt curve run. Primer specificity for a single amplicon was verified by melt curve analysis. Gapdh was used as an internal normalization control. Fold change in mRNA levels was calculated relative to controls using a ΔΔCt method. Specific sample sizes are noted within the corresponding figure legend, but at least n = 3 independent testes were used for each control and cKO condition tested. A two-tailed Student’s t test was performed to calculate P values based on delta Ct values, in which p < 0.05 was considered statistically significant. Sequences of primers used in this study are listed in Table S1.
Hormone measurements
Testosterone (T) measurements were performed by the Ligand Assay and Analysis Core of the University of Virginia Center for Research in Reproduction. For testis homogenate measurements, whole adult testes were mechanically lysed with mortar and pestle in 500 uL PBS, sonicated for 60 s to ensure cell disruption, and centrifuged at 12,000 × g at 4°C for 10 minutes to remove cellular debris. Supernatant was collected and stored at −80°C until analyzed. Measurements were performed on at least n = 3 testes from independent males.
Biotin tracer injections
P30 or P90 male mice (n = 3 males for each age for each control and cKO condition) were anesthetized via inhaled isoflurane before a small incision was made in the lower abdomen, exposing the abdominal cavity. Using forceps to grip the epididymal fat pads, the testis was then gently pulled out of the abdomen. Twenty microliters of either 1mM CaCl2 (in PBS) alone for one testis, or 1mM CaCl2 with 10 mg/mL Biotin (EZ-Link Sulfo-NHS-LC-Biotin, No-Weigh Format; Thermo Fisher #A39257) for the contralateral experimental testis was injected. Testes were then replaced into the abdominal cavity and the incision closed. Mice were kept under anesthesia via isoflurane for an additional 30 minutes before being euthanized by cervical dislocation. Testes were harvested, fixed, and prepared for cryosectioning and immunofluorescence as described above. To detect biotin binding in testicular cryosections, Alexa Fluor-555-conjugated streptavidin (Thermo Fisher #S21381, 2 μg/ml) was used for 1 hour at room temperature, along with Hoechst 33342 dye to stain nuclei.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments, such as the exact value of n, what n represents, precision measures (mean ± SD), and statistical significance can be found in the Figure Legends. Statistical analysis was conducted using Excel (Microsoft). The results are reported as mean ± standard deviation. For cell counts and tubule area quantification, a two-tailed Student’s t test was performed on control versus cKO samples. For quantitative real-time PCR, a two-tailed Student’s t test was performed on delta Ct values (normalized to Gapdh) for each gene for control versus cKO samples. p < 0.05 was considered statistically significant.
Supplementary Material
Highlights.
Cdc42 function in Sertoli cells is required for steady-state spermatogenesis
Sertoli Cdc42 is dispensable for fetal and early postnatal testis development
Loss of Cdc42 disrupts Sertoli cell polarity and blood-testis barrier function
CDC42-mediated Sertoli cell polarity is required starting in juvenile stages
ACKNOWLEDGMENTS
We thank C. Moon for animal husbandry, D. Meijer and Y. Zheng for providing mice, and M. Handel for H1F6 (H1T) antibody. We also thank B. Waller, C. Spinner, and M. White for preliminary experiments. This work was supported by Cincinnati Children’s Hospital Medical Center developmental funds and internal funding and by National Institutes of Health grants R35GM119458 and R01HD094698 to T.D.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109885.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Appling DR, and Chytil F (1981). Evidence of a role for retinoic acid (vitamin A-acid) in the maintenance of testosterone production in male rats. Endocrinology 108, 2120–2124. [DOI] [PubMed] [Google Scholar]
- Campanale JP, Sun TY, and Montell DJ (2017). Development and dynamics of cell polarity at a glance. J. Cell Sci 130, 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H, and Yeh S (2004). Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc. Natl. Acad. Sci. USA 101, 6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Gao F, Guillou F, Taketo MM, Huff V, and Behringer RR (2008). Wt1 negatively regulates beta-catenin signaling during testis development. Development 135, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liao G, Yang L, Campbell K, Nakafuku M, Kuan CY, and Zheng Y (2006). Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc. Natl. Acad. Sci. USA 103, 16520–16525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Brown PR, Willis WB, and Eddy EM (2014). Peritubular myoid cells participate in male mouse spermatogonial stem cell maintenance. Endocrinology 155, 4964–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Willis WD, and Eddy EM (2016). Targeting the Gdnf gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc. Natl. Acad. Sci. USA 113, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MS, Munger SC, Nadeau JH, and Capel B (2011). Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development 138, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool J, DeFalco T, and Capel B (2012). Testis formation in the fetal mouse: dynamic and complex de novo tubulogenesis. Wiley Interdiscip. Rev. Dev. Biol 1, 847–859. [DOI] [PubMed] [Google Scholar]
- Darde TA, Sallou O, Becker E, Evrard B, Monjeaud C, Le Bras Y, Jégou B, Collin O, Rolland AD, and Chalmel F (2015). The ReproGenomics Viewer: an integrative cross-species toolbox for the reproductive science community. Nucleic Acids Res. 43 (W1), W109–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darde TA, Lecluze E, Lardenois A, Stévant I, Alary N, Tüttelmann F, Collin O, Nef S, Jégou B, Rolland AD, and Chalmel F (2019). The ReproGenomics Viewer: a multi-omics and cross-species resource compatible with single-cell studies for the reproductive science community. Bioinformatics 35, 3133–3139. [DOI] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C, et al. (2004). A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc. Natl. Acad. Sci. USA 101, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TJ, Kaur G, Putrevu SM, Dyson EL, Dyson M, McCunniff WT, Pasham MR, Kim KH, and Dufour JM (2012). Immunoprotective properties of primary Sertoli cells in mice: potential functional pathways that confer immune privilege. Biol. Reprod 86, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JM, Rajotte RV, Seeberger K, Kin T, and Korbutt GS (2003). Long-term survival of neonatal porcine Sertoli cells in non-immunosuppressed rats. Xenotransplantation 10, 577–586. [DOI] [PubMed] [Google Scholar]
- Erickson JW, and Cerione RA (2001). Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol 13, 153–157. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, and Hall A (2002). Rho GTPases in cell biology. Nature 420, 629–635. [DOI] [PubMed] [Google Scholar]
- França LR, Hess RA, Dufour JM, Hofmann MC, and Griswold MD (2016). The Sertoli cell: one hundred fifty years of beauty and plasticity. Andrology 4, 189–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, and Cheng CY (2016). Does cell polarity matter during spermatogenesis? Spermatogenesis 6, e1218408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xiao X, Lui WY, Lee WM, Mruk D, and Cheng CY (2016). Cell polarity proteins and spermatogenesis. Semin. Cell Dev. Biol 59, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrard SM, Capaldo CT, Gao L, Rosen MK, Macara IG, and Tomchick DR (2003). Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 22, 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, et al. (2018). A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 46, 651–667.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD (2016). Spermatogenesis: The Commitment to Meiosis. Physiol. Rev 96, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Moon C, Zheng Y, and Ratner N (2013). Cdc42 regulates Schwann cell radial sorting and myelin sheath folding through NF2/merlin-dependent and independent signaling. Glia 61, 1906–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich A, Potter SJ, Guo L, Ratner N, and DeFalco T (2020). Distinct Roles for Rac1 in Sertoli Cell Function during Testicular Development and Spermatogenesis. Cell Rep. 31, 107513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HF, Dyrenfurth I, and Hembree WC (1983). Endocrine changes associated with germ cell loss during vitamin A-induced recovery of spermatogenesis. Endocrinology 112, 1163–1171. [DOI] [PubMed] [Google Scholar]
- Inselman A, Eaker S, and Handel MA (2003). Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet. Genome Res 103, 277–284. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, and Meijer D (2003). The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 17, 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, and Capel B (2012). Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 8, e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins AB, McCaffery JM, and Van Doren M (2003). Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development 130, 4417–4426. [DOI] [PubMed] [Google Scholar]
- Johansson A, Driessens M, and Aspenström P (2000). The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J. Cell Sci 113, 3267–3275. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, and Shinohara T (2013). Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol 29, 163–187. [DOI] [PubMed] [Google Scholar]
- Kaur G, Thompson LA, and Dufour JM (2014). Sertoli cells–immunological sentinels of spermatogenesis. Semin. Cell Dev. Biol 30, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Wright K, Mital P, Hibler T, Miranda JM, Thompson LA, Halley K, and Dufour JM (2020). Neonatal Pig Sertoli Cells Survive Xenotransplantation by Creating an Immune Modulatory Environment Involving CD4 and CD8 Regulatory T Cells. Cell Transplant. 29, 963689720947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin PM, Kramer MF, and de Rooij DG (1984). Proliferation of spermatogonia and Sertoli cells in maturing mice. Anat. Embryol. (Berl.) 169, 73–78. [DOI] [PubMed] [Google Scholar]
- Kroschewski R, Hall A, and Mellman I (1999). Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol 1, 8–13. [DOI] [PubMed] [Google Scholar]
- Li XY, Zhang Y, Wang XX, Jin C, Wang YQ, Sun TC, Li J, Tang JX, Batool A, Deng SL, et al. (2018). Regulation of blood-testis barrier assembly in vivo by germ cells. FASEB J. 32, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom F, Gillemans N, Karis A, Jaegle M, Meijer D, Grosveld F, and Philipsen S (2003). A tissue-specific knockout reveals that Gata1 is not essential for Sertoli cell function in the mouse. Nucleic Acids Res. 31, 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WY, Mruk DD, and Cheng CY (2005). Interactions among IQGAP1, Cdc42, and the cadherin/catenin protein complex regulate Sertoli-germ cell adherens junction dynamics in the testis. J. Cell. Physiol 202, 49–66. [DOI] [PubMed] [Google Scholar]
- Mack NA, and Georgiou M (2014). The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt A, and Hedger MP (2011). Immunological, paracrine and endocrine aspects of testicular immune privilege. Mol. Cell. Endocrinol 335, 60–68. [DOI] [PubMed] [Google Scholar]
- Melendez J, Grogg M, and Zheng Y (2011). Signaling role of Cdc42 in regulating mammalian physiology. J. Biol. Chem 286, 2375–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493. [DOI] [PubMed] [Google Scholar]
- Meng J, Holdcraft RW, Shima JE, Griswold MD, and Braun RE (2005). Androgens regulate the permeability of the blood-testis barrier. Proc. Natl. Acad. Sci. USA 102, 16696–16700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales A, and Cavicchia JC (2002). Spermatogenesis and blood-testis barrier in rats after long-term vitamin A deprivation. Tissue Cell 34, 349–355. [DOI] [PubMed] [Google Scholar]
- Ngok SP, Lin WH, and Anastasiadis PZ (2014). Establishment of epithelial polarity–GEF who’s minding the GAP? J. Cell Sci 127, 3205–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda Y, Takeya R, Ohno S, Naito S, Ito T, and Sumimoto H (2001). Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 6, 107–119. [DOI] [PubMed] [Google Scholar]
- Oatley JM, and Brinster RL (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev 92, 577–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley MJ, Racicot KE, and Oatley JM (2011). Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol. Reprod 84, 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JM (1982). Proliferation of Sertoli cells in fetal and postnatal rats: a quantitative autoradiographic study. Anat. Rec 203, 485–492. [DOI] [PubMed] [Google Scholar]
- Russell LD, Goh JC, Rashed RM, and Vogl AW (1988). The consequences of actin disruption at Sertoli ectoplasmic specialization sites facing spermatids after in vivo exposure of rat testis to cytochalasin D. Biol. Reprod 39, 105–118. [DOI] [PubMed] [Google Scholar]
- Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, and Schulz C (2007). Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr. Biol 17, 1253–1258. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe RM, McKinnell C, Kivlin C, and Fisher JS (2003). Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of test is function in adulthood. Reproduction 125, 769–784. [DOI] [PubMed] [Google Scholar]
- Smith LB, and Walker WH (2014). The regulation of spermatogenesis by androgens. Semin. Cell Dev. Biol 30, 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stévant I, Neirijnck Y, Borel C, Escoffier J, Smith LB, Antonarakis SE, Dermitzakis ET, and Nef S (2018). Deciphering Cell Lineage Specification during Male Sex Determination with Single-Cell RNA Sequencing. Cell Rep. 22, 1589–1599. [DOI] [PubMed] [Google Scholar]
- Stevens LC (1973). A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst 50, 235–242. [DOI] [PubMed] [Google Scholar]
- Su L, Mruk DD, Lee WM, and Cheng CY (2010). Differential effects of testosterone and TGF-β3 on endocytic vesicle-mediated protein trafficking events at the blood-testis barrier. Exp. Cell Res 316, 2945–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung KS, Harakal J, Qiao H, Rival C, Li JC, Paul AG, Wheeler K, Pramoonjago P, Grafer CM, Sun W, et al. (2017). Egress of sperm autoantigen from seminiferous tubules maintains systemic tolerance. J. Clin 127, 1046–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western PS, Miles DC, van den Bergen JA, Burton M, and Sinclair AH (2008). Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26, 339–347. [DOI] [PubMed] [Google Scholar]
- Wong EW, Mruk DD, Lee WM, and Cheng CY (2008). Par3/Par6 polarity complex coordinates apical ectoplasmic specialization and blood-testis barrier restructuring during spermatogenesis. Proc. Natl. Acad. Sci. USA 105, 9657–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EW, Mruk DD, Lee WM, and Cheng CY (2010). Regulation of blood-testis barrier dynamics by TGF-beta3 is a Cdc42-dependent protein trafficking event. Proc. Natl. Acad. Sci. USA 107, 11399–11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HH, Mruk DD, Lee WM, and Cheng CY (2008). Blood-testis barrier dynamics are regulated by testosterone and cytokines via their differential effects on the kinetics of protein endocytosis and recycling in Sertoli cells. FASEB J. 22, 1945–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomogida K, Ohtani H, Harigae H, Ito E, Nishimune Y, Engel JD, and Yamamoto M (1994). Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development 120, 1759–1766. [DOI] [PubMed] [Google Scholar]
- Yoshida S (2010). Stem cells in mammalian spermatogenesis. Dev. Growth Differ 52,311–317. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, and Nabeshima Y (2006). The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 133, 1495–1505. [DOI] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhu W, Xue S, and Han D (2014). Testicular defense systems: immune privilege and innate immunity. Cell. Mol 11, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper analyzes existing, publicly available microarray and RNA-Seq data. The GEO accession numbers for these datasets are listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact on request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-AIF1 (IBA1) | Wako | Cat#019-19741; RRID: AB_839504 |
| Rabbit polyclonal anti-ALDH1A1 | Abcam | Cat#ab23375; RRID: AB_2224009 |
| Rabbit polyclonal anti-ALDH1A2 | Sigma-Aldrich | Cat#HPA010022; RRID: AB_1844723 |
| Goat polyclonal anti-AMH (MIS) | Santa Cruz Biotechnology | Cat#sc-6886; RRID: AB_649207 |
| Rabbit polyclonal anti-AR | Santa Cruz Biotechnology | Cat#sc-816; RRID: AB_1563391 |
| Rat monoclonal anti-CD45 (clone 30-F11) | BioLegend | Cat#103101; RRID: AB_312966 |
| Rat monoclonal anti-CDH1 (clone ECCD-2) | Thermo Fisher Scientific | Cat#13-1900; RRID: AB_2533005 |
| Rabbit polyclonal anti-CLDN11 | Thermo Fisher Scientific | Cat#36-4500; RRID: AB_2533259 |
| Rabbit polyclonal anti-cleaved Caspase 3 (Asp175) | Cell Signaling Technology | Cat#9661S; RRID: AB_2341188 |
| Goat polyclonal anti-CTNNB1 | Santa Cruz Biotechnology | Cat#sc-1496; RRID: AB_1563968 |
| Goat polyclonal anti-CYP17A1 | Santa Cruz Biotechnology | Cat#sc-46081; RRID: AB_2088659 |
| Rabbit polyclonal anti-DDX4 (MVH) | Abcam | Cat#ab13840; RRID: AB_443012 |
| Rat monoclonal anti-GATA1 (clone N6) | Santa Cruz Biotechnology | Cat#sc-265; RRID: AB_627663 |
| Goat polyclonal anti-GATA4 | Santa Cruz Biotechnology | Cat#sc-1237; RRID: AB_2108747 |
| Rabbit polyclonal anti-GDNF | Santa Cruz Biotechnology | Cat#sc-328; RRID: AB_2247684 |
| Goat polyclonal anti-GFRA1 | Neuromics | Cat#GT15004; RRID: AB_2307379 |
| Rabbit polyclonal anti-Histone H2A.X, phospho (Ser139) | Millipore | Cat#07-164; RRID: AB_310406 |
| Guinea pig polyclonal anti-H1F6 (H1T) | Mary Ann Handel (Inselman et al., 2003) | N/A |
| Rat monoclonal anti-ITGA6 (CD49f) (clone GoH3) | BioLegend | Cat#313602; RRID: AB_345296 |
| Goat polyclonal anti-ITGB1 | Novus/R&D Systems | Cat#AF2405-SP; RRID: AB_416591 |
| Goat polyclonal anti-KIT | R&D Systems | Cat#AF1356; RRID: AB_354750 |
| Rabbit monoclonal anti-MKI67 (Ki67) (clone SP6) | GeneTex | Cat#GTX16667; RRID: AB_422351 |
| Rabbit polyclonal anti-PARD3 (Par3) | Novus | Cat#NBP1-88861; RRID: AB_11056253 |
| Goat polyclonal anti-PECAM1 | R&D Systems | Cat#AF3628; RRID:AB_2161028 |
| Rat monoclonal anti-PECAM1 (clone MEC13.3) | BD Biosciences | Cat#553370; RRID: AB_394816 |
| Rabbit monoclonal anti-phospho-Scribble (Ser1220) (clone D8A2) | Cell Signaling Technology | Cat#12316; RRID: AB_2797883 |
| Rabbit polyclonal anti-SOX9 | Millipore | Cat#AB5535; RRID: AB_2239761 |
| Rabbit polyclonal anti-STRA8 | Abcam | Cat#ab49602; RRID: AB_945678 |
| Rat monoclonal anti-TRA98 (clone TRA98) | Abcam | Cat#ab82527; RRID: AB_1659152 |
| Mouse monoclonal anti-TUBB3 (clone TUJ1) | BioLegend | Cat#801201; RRID: AB_2313773 |
| Chicken polyclonal anti-VIM | BioLegend | Cat#919101; RRID: AB_2565208 |
| Rat monoclonal anti-VIM (clone W16220A) | BioLegend | Cat#699301; RRID: AB_2716136 |
| Mouse monoclonal anti-ZBTB16 (PLZF) (clone 2A9) | Millipore | Cat#OP128-100UG; RRID: AB_213280 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat#712-605-153; RRID:AB_2340694 |
| Alexa Fluor 647 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21447; RRID:AB_2535864 |
| Alexa Fluor 647 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-31573; RRID:AB_2536183 |
| Alexa Fluor 647 Goat anti-Mouse IgG2a | Thermo Fisher Scientific | Cat#A-21241; RRID:AB_2535810 |
| Cy3 AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch | Cat#712-165-153; RRID:AB_2340667 |
| Alexa Fluor 555 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21432; RRID:AB_2535853 |
| Alexa Fluor 555 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A-31572; RRID:AB_162543 |
| Alexa Fluor 555 Goat anti-Mouse IgG2a | Thermo Fisher Scientific | Cat#A-21137; RRID:AB_2535776 |
| Alexa Fluor 488 Donkey anti-Goat IgG (H+L) | Thermo Fisher Scientific | Cat#A-11055; RRID:AB_2534102 |
| Alexa Fluor 488 Donkey anti-Rat IgG (H+L) | Thermo Fisher Scientific | Cat#A-21208; RRID:AB_141709 |
| Alexa Fluor Plus 488 Donkey anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#A32790; RRID:AB_2762833 |
| Alexa Fluor 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson Immunoresearch | Cat#703-545-155; RRID:AB_2340375 |
| Chemicals, peptides, and recombinant proteins | ||
| Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | Cat#A22287; RRID: AB_2620155 |
| Rhodamine Phalloidin | Thermo Fisher Scientific | Cat#R415; RRID: AB_2572408 |
| Hoechst 33342, Trihydrochloride, Trihydrate | Thermo Fisher Scientific | Cat#H1399 |
| Streptavidin, Alexa Fluor 555 Conjugate | Thermo Fisher Scientific | Cat#S21381; RRID: AB_2307336 |
| EZ-Link Sulfo-NHS-LC-Biotin, No-Weigh Format | Thermo Fisher Scientific | Cat#A39257 |
| Deposited data | ||
| Fetal gonad (E11.5-E13.5) cell-type-specific microarray dataset | (Jameson et al., 2012) | GEO: GSE27715; GDS3995 |
| Fetal testis (E10.5-E16.5) single-cell (NR5A1-eGFP+ cells) RNA-Seq dataset | (Stévant et al., 2018) | GEO: GSE97519 |
| Adult testis single-cell RNA-Seq dataset | (Green et al., 2018) | GEO: GSE112393 |
| Experimental models: organisms/strains | ||
| Mouse: Dhh-Cre: Tg(Dhh-cre)1Mejr | Dies Meijer; (Jaegle et al., 2003; Lindeboom et al., 2003) | RRID: IMSR_JAX:012929 |
| Mouse: Cdc42 fl/fl: Cdc42tm1Yizh/J | Yi Zheng; (Chen et al., 2006) | RRID: IMSR_JAX:027576 |
| Mouse: CD-1 IGS Mouse: Crl:CD1(ICR) | Charles River | Cat#CRL:022; RRID: IMSR_CRL:022 |
| Mouse: C57BL/6J Mouse: JAX:C57BL/6J | The Jackson Laboratory | Cat#JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse: KitW/KitW-v: WBB6F1/J-KitW/KitW-v/J | The Jackson Laboratory | Cat#JAX:100410; RRID: IMSR_JAX:100410 |
| Mouse: Dnd1Ter: Dnd1Ter | Blanche Capel; (Stevens, 1973; Youngren et al., 2005) | MGI:2158668; RRID: MGI:3690452 |
| Oligonucleotides | ||
| Primers for quantitative real-time PCR, see Table S1 | This study | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| The ReproGenomics Viewer | (Darde et al., 2019; Darde et al., 2015) | https://rgv.genouest.org/ |


