Abstract
Members of the family Herpesviridae have enveloped, spherical virions with characteristic complex structures consisting of symmetrical and non-symmetrical components. The linear, double-stranded DNA genomes of 125–241 kbp contain 70–170 genes, of which 43 have been inherited from an ancestral herpesvirus. In general, herpesviruses have coevolved with and are highly adapted to their hosts, which comprise many mammalian, avian and reptilian species. Following primary infection, they are able to establish lifelong latent infection, during which there is limited viral gene expression. Severe disease is usually observed only in the foetus, the very young, the immunocompromised or following infection of an alternative host. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Herpesviridae, which is available at ictv.global/report/herpesviridae.
Keywords: Herpesviridae, ICTV Report, taxonomy
Virion
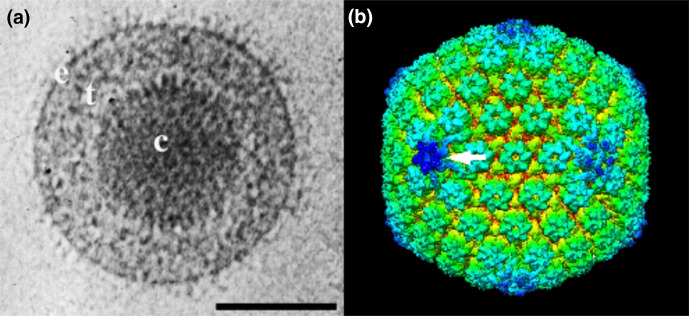
Virions consist of a core, capsid, tegument and envelope (Table 1, Fig. 1) [1]. The core comprises the viral genome packaged into the capsid as a linear, dsDNA molecule. The capsid is a T=16 icosahedron containing 162 capsomers arranged as 150 hexons, 11 pentons and one portal. The tegument consists of inner and outer layers. The lipid envelope contains integral viral glycoproteins forming a network of spikes.
Table 1.
Characteristics of members of the family Herpesviridae
|
Example: |
herpes simplex virus type 1 (JN555585), species Human alphaherpesvirus 1, genus Simplexvirus, subfamily Alphaherpesvirinae |
|---|---|
|
Virion |
Spherical (150–200 nm) particles with condensed DNA core, icosahedral capsid, tegument and a lipid envelope containing glycoproteins |
|
Genome |
125–241 kbp of linear dsDNA |
|
Replication |
Infection has lytic and latent phases; transcription occurs in the nucleus by a kinetic cascade; DNA replicates by a rolling-circle mechanism to generate concatemers, from which genomes are cleaved and packaged into preformed capsids; virions mature in the cytoplasm |
|
Translation |
Occurs from capped, polyadenylated mRNAs, some of which are spliced |
|
Host range |
Mammals, birds and reptiles |
|
Taxonomy |
Realm Duplodnaviria, kingdom Heunggongvirae, phylum Peploviricota, class Herviviricetes, order Herpesvirales; 3 subfamilies, >10 genera and >100 species |
Fig. 1.
Herpes simplex virus type 1 virion and capsid structure. (a) Electron cryo-microscopic image of a virion showing the capsid (c), tegument (t) and envelope (e). Scale bar, 100 nm. From [2] with permission. (b) Three-dimensional image reconstruction of a capsid showing hexons, pentons and the portal (arrow). From [5].
Genome
The genome is 125–241 kbp. The arrangement of direct or inverted repeats at the termini or internally results in several classes of genome architecture. The genome contains 70–170 genes encoding proteins, 43 of which are shared across the family, suggesting a common replication strategy [2]. Additional genes encoding nontranslated RNAs may be present.
Replication
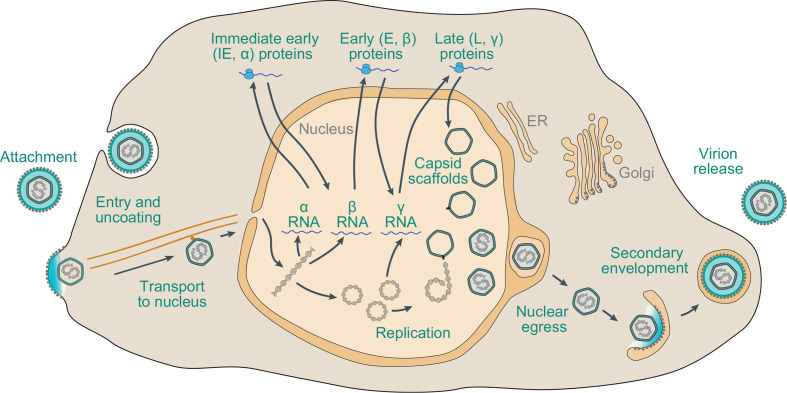
Herpesviruses have been discovered in a wide range of vertebrates (reptiles, birds and mammals). The most extensively studied animals are host to members of several species. Most herpesviruses have coevolved and sometimes cospeciated within a single host lineage, although foundational cross-species transmission events appear to have occurred. In general, lytic infection involves attachment and penetration by the interaction of virion envelope proteins with cell surface receptors, followed by entry via membrane fusion at the cell surface or after endocytosis (Fig. 2) [3, 4]. The capsid uncoats and is transported to a nuclear pore, and the genome enters the nucleus. Transcription occurs in a kinetic cascade: immediate early genes encode regulatory functions, early genes encode the DNA replication complex and a variety of proteins involved in modifying host cell metabolism or immune responses, and late genes primarily encode virion proteins. Viral DNA synthesis occurs by a rolling-circle replication mechanism to generate concatemers, from which genomes are cleaved and packaged into capsids. Capsids bud through the inner nuclear membrane and are then de-enveloped by fusion with the outer nuclear membrane and released into the cytoplasm. Assembly of tegument proteins and secondary envelopment to generate mature virions occur in a Golgi or post-Golgi compartment. Virions exit the cell by exocytosis or cell-to-cell spread.
Fig. 2.
Schematic representation of the lytic replication cycle of a representative herpesvirus in permissive cells.
Herpesviruses are restricted in their natural host range and highly adapted to their hosts, with severe infection usually observed only in the foetus, the very young, the immunocompromised or in an alternative host. Typically, a primary, systemic infection is established via a cell-associated viraemia, followed by a latent phase in which dormant virus occasionally reactivates. Herpesviruses operate a range of modulation mechanisms to manage host immunity.
Taxonomy
Current taxonomy: ictv.global/taxonomy. The family Herpesviridae includes three subfamilies, and belongs to the order Herpesvirales along with the families Alloherpesviridae and Malacoherpesviridae. This order is in turn classified alongside the order Caudovirales in the kingdom Heunggongvirae.
Resources
Current ICTV Report on the family Herpesviridae: ictv.global/report/herpesviridae.
Funding information
Production of this Profile, the ICTV Report and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
The authors are the ICTV Herpesvirales Study Group. The ICTV Report Consortium is Stuart G. Siddell, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, F. Murilo Zerbini, Donald B. Smith and Arvind Varsani.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 2.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Krug LT, Pellett PE. In: Fields Virology. Howley PM, Knipe DM, editors. Wolter Kluwer; 2021. The Family Herpesviridae: A Brief Introduction; pp. 212–234. [Google Scholar]
- 4.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, et al. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. [PubMed] [Google Scholar]
- 5.McElwee M, Vijayakrishnan S, Rixon F, Bhella B. Structure of the herpes simplex virus portal-vertex. PLoS Biol. 2018;16:e2006191. doi: 10.1371/journal.pbio.2006191. [DOI] [PMC free article] [PubMed] [Google Scholar]