Abstract
Personal genomic educational testing (PGET) has been suggested as a strategy to improve student learning for pharmacogenomics (PGx), but no randomized studies have evaluated PGET’s educational benefit. We investigated the effect of PGET on student knowledge, comfort, and attitudes related to PGx in a nonblinded, randomized controlled trial. Consenting participants were randomized to receive PGET or no PGET (NPGET) during 4 subsequent years of a PGx course. All participants completed a pre‐survey and post‐survey designed to assess (1) PGx knowledge, (2) comfort with PGx patient education and clinical skills, and (3) attitudes toward PGx. Instructors were blinded to PGET assignment. The Wilcoxon Rank Sum test was used to compare pre‐survey and post‐survey PGx knowledge, comfort, and attitudes. No differences in baseline characteristics were observed between PGET (n = 117) and NPGET (n = 116) participants. Among all participants, significant improvement was observed in PGx knowledge (mean 57% vs. 39% correct responses; p < 0.001) with similar results for student comfort and attitudes. Change in pre/post‐PGx knowledge, comfort, and attitudes were not significantly different between PGET and NPGET groups (mean 19.5% vs. 16.7% knowledge improvement, respectively; p = 0.41). Similar results were observed for PGET participants carrying a highly actionable PGx variant versus PGET participants without an actionable variant. Significant improvement in Likert scale responses were observed in PGET versus NPGET for questions that assessed student engagement (p = 0.020) and reinforcement of course concepts (p = 0.006). Although some evidence of improved engagement and participation was observed, the results of this study suggest that PGET does not directly improve student PGx knowledge, comfort, and attitudes.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Multiple studies have supported the use of personal genomic educational testing (PGET) in the Doctor of Pharmacy (PharmD) curriculum. However, these studies have major limitations, including limited sample sizes, lack of randomization, and self‐selection of students into PGET intervention groups.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study investigates the effect of PGET on PharmD student knowledge and attitudes related to pharmacogenomics (PGx). The study is designed to address limitations in prior studies.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study contradicts existing literature and indicates that PGET provides no significant benefits in terms of improved knowledge or attitudes for PharmD students regarding PGx.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Our results indicate that implementation of PGET in PharmD education is not an effective method to meet educational goals related to PGx.
INTRODUCTION
Despite the growth and evolution of pharmacogenomics (PGx) as a field of biomedical research, healthcare providers have historically been slow to adopt PGx testing in the clinical setting. A major reason is a lack of knowledge among healthcare professionals regarding interpretation and clinical utility of PGx tests. 1 , 2 Pharmacists are poised to bridge this gap in knowledge due to their training in the requisite fields of pharmacokinetics, pharmacodynamics, and therapeutics. Accrediting organizations now require that pharmacy schools include PGx in their Doctor of Pharmacy (PharmD) curriculum. 3 , 4 However, pharmacy students have reported that their pharmacy school education has not prepared them for a career in PGx. 5 Personal genomic educational testing (PGET), the testing of student’s own genetic variants, is an innovative teaching tool to facilitate learning. PGET constitutes a potentially powerful strategy to meet educational goals through the improved engagement and motivation characteristic of utilizing one’s own genetic results. Despite PGET’s potential, rigorous and well‐powered studies that evaluate the efficacy of this intervention are not available.
Although the potential benefits of PGET are well‐recognized, only a handful of studies have been performed evaluating educational outcomes associated with PGET. Rigorous evidence around PGET is particularly important considering the financial costs and potential ethical issues associated with extensive genotyping of students in the PharmD curriculum. Evidence around PGET has broadly supported the improvement in knowledge and attitudes of PGx genotyping, but multiple major limitations exist for these studies. 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 No study has included a randomization to genotyped and non‐genotyped groups. Bias was further introduced into these studies because control groups generally choose not to be genotyped or genotyping is performed based on self‐selection into elective courses. The small sample size of these studies prevented sufficient power to evaluate changes in attitudes or educational performance. Furthermore, these studies did not include analysis of the influence of PGx testing as well as carriage of clinically actionable PGx variants on objective outcomes, including course grades. In order to ensure that an evidence‐based teaching standard is applied to PGET, rigorous and well‐powered studies are needed to assess the benefit of PGET in PharmD classrooms.
The present study was designed to address these limitations in the existing literature. Here, we describe a nonblinded, randomized controlled trial that includes prospective randomization of students to PGx testing and no PGx testing groups. A large number of students were recruited from 4 consecutive years of a three credit, required PharmD course at the University of Arizona (UArizona) College of Pharmacy (Applied Pharmacogenetics and Precision Medicine [PHPR 887]). Our primary objective was to assess the effect of PGET on student PGx knowledge. Secondary objectives included assessment of the effect of PGET on comfort and attitudes regarding PGx and assessment of carriage of actionable PGx variants on student knowledge, comfort, and attitudes regarding PGx.
METHODS
Study design
The PharmD curriculum at UArizona includes a required three‐credit hour course titled “Applied Pharmacogenetics and Precision Medicine” (PHPR 887). This course is offered to third year PharmD students to provide foundational knowledge regarding PGx and skills to guide clinical decision making in the context of PGx results. As part of this course in the spring semesters of 2017, 2018, 2019, and 2020, students were given the opportunity to participate in a study that provided a student’s own PGx results at no cost to the student (Supplementary Tables S1–S3). No major curricular changes were implemented either in PHPR 887 or in the PharmD curriculum between the years when participation in this study was offered. Participation in this study was entirely voluntary and did not influence course grades in any way. This study is registered on clinicaltrials.gov (NCT04889014). Students could only participate if they provided written informed consent for the study. Consultation with a genetic counselor was available to all participants in this study. This study received ethics approval from the UArizona Institutional Review Board, including a human subjects’ research oversight exemption from the under 45 CFR 46.101(b).
All consenting participants were asked to complete a pre‐survey, which included an assessment of PGx knowledge and a self‐assessment including three domains: comfort with PGx clinical skills, comfort with PGx patient education, and attitudes toward PGx. Following the pre‐survey, participants were randomized in a one‐to‐one ratio to two groups (Figure 1). PGET group participants received their own PGx testing results prior to the course modules covering material tested in the knowledge assessment and no PGET (NPGET) group participants did not receive their own PGx testing results until after study completion. During course modules, which included traditional lecture format as well as clinical case discussions related to content covered in the knowledge‐based PGx questions, participants randomized to PGET were encouraged to use their own PGx test results during in‐class exercises. Participants randomized to NPGET were encouraged to use PGx test results for a hypothetical patient from the same panel used to test participants in the PGET group. Instructors and all study personnel were blinded to participant PGET assignment, PGx results, and survey results. Blinding was accomplished using a unique six‐digit study ID, which allowed for deidentification of PGx test results, survey data, and final grades. Following presentation of course modules related to content covered in the knowledge assessment, all participants were asked to complete a post‐survey and retrospective survey, described in detail below. Following completion of the study, all participants randomized to NPGET were given an opportunity to receive their own PGx test results at no cost.
FIGURE 1.
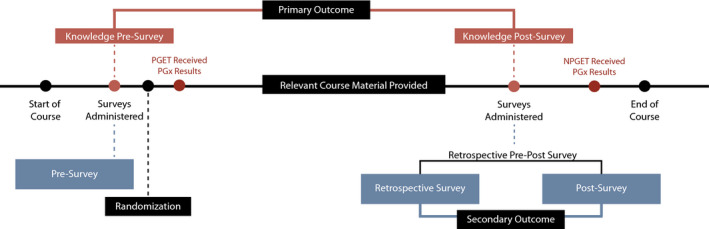
Timeline for participants enrolled in the study. Assessments of PGx knowledge were administered before randomization during the pre‐survey and after all relevant course material was presented during the retrospective pre‐post survey. The primary analysis assessed the improvement in PGx knowledge based on correct responses to questions in the pre‐survey and post‐survey. Assessments of level of comfort with PGx clinical skills, PGx patient education, and attitudes toward PGx were taken during pre‐survey, retrospective survey, and post‐survey. Secondary analyses included differences in Likert scale responses to the retrospective survey and post‐survey. PGx indicates pharmacogenomics; PGET, personal genomic educational testing; NGET, no personal genomic educational testing
Data collection and outcomes
Survey instruments
All participants completed a pre‐survey designed to assess demographic characteristics, PGx knowledge, self‐assessment of three domains: comfort with PGx clinical skills, comfort with PGx patient education, and attitudes toward PGx. Correct responses in the knowledge assessment were used to determine differences in performance between the PGET and NPGET groups, which was the primary outcome of this study. The knowledge assessment included 10 multiple choice questions and one multi‐select (“select all that apply") question related to application of PGx knowledge (Supplementary Table S4). The content of the knowledge survey was created by PHPR 887 course concept mapping and questions included material for the following drug‐gene pairs: clopidogrel‐CYP2C19, simvastatin‐SLCO1B1, warfarin‐CYP2C9, warfarin‐VKORC1, selective serotonin reuptake inhibitors (SSRIs)‐CYP2C19, SSRIs‐CYP2D6, codeine‐CYP2D6, tricyclic antidepressants (TCAs)‐CYP2D6, mercaptopurine‐TPMT, abacavir‐HLA‐B, and carbamazepine‐HLA‐B. Student responses to knowledge‐based survey questions were graded as correct/incorrect and used to make comparisons between groups related to performance on individual questions as well as overall performance. The multi‐select ("select all that apply") question was graded as correct or incorrect without providing credit for a partially correct answer. The original survey included 12 knowledge assessment questions, but one question was withdrawn from analysis as correct answers were different between years due to emerging literature and updated recommendations in Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines. The pre‐survey also included 27 Likert scale response questions for student self‐assessment of three domains: comfort with PGx clinical skills (12 items), comfort with PGx patient education (6 items), and attitudes toward PGx (9 items; Supplementary Tables S5 and S6). Participants responded to survey questions using a Likert scale, with answers ranging from strongly disagree to strongly agree, or extremely uncomfortable to extremely comfortable. All surveys were administered online via Qualtrics.
Following presentation of course modules, all participants were asked to complete a retrospective pre‐post survey, which included retaking the knowledge assessment as well as a post‐survey and a retrospective survey using the same Likert scale questions described above (Figure 1). For the post‐survey, participants were asked to assess their comfort and attitudes at the time of the post‐survey. For the retrospective survey, participants were asked to assess their comfort and attitudes at the time of the pre‐survey (prior to randomization). The retrospective survey was conducted because prior evidence suggests that survey respondents tend to overestimate their comfort prior to an intervention and this technique has been shown to have greater accuracy over pre‐survey responses. 20 , 21 , 22 This response‐shift bias occurs when the student’s frame of reference for the self‐assessment changes between the pre‐survey and the retrospective survey due to the influence of the educational program.
Genotyping
All participants’ DNA were provided via a mouthwash or cheek swab (buccal cell) collection kit. The PGx panel used to test participants varied during the 4‐year study. The genes and variants included on each panel are listed in the Supplementary Materials (Supplementary Tables S1–S3). Genotyping was accomplished using the IDgenetix(R) (AltheaDx) PGx test platform, which has been previously described, in the 2017 cohort. 23 , 24 Genotyping was accomplished using in‐house genotyping with TaqMan(R) (Applied Biosystems) in the 2018 cohort, as described in the Supplementary Methods. Finally, genotyping was accomplished using the Oneome Rightmed(R) (Oneome) in the 2019 and 2020 cohorts. As described in the statistical analyses, sensitivity analyses were performed based on the PGx platform used in order to evaluate consistency of results in light of differences in PGx reports. De‐identified genotyping results were collected by study personnel to investigate the influence of the presence of actionable PGx variants on participant knowledge, comfort, attitudes, and course performance.
Participant genotype data were used to derive clinically actionable PGx genotypes in order to assess the influence of clinically actionable PGx results on PGx knowledge and Likert scale question responses (Supplementary Table S7). Clinically actionable PGx genotypes were determined based on CPIC guidelines and constituted any diplotype or genotype with a strong therapeutic recommendation for a change in therapy (e.g., drug avoidance, dose adjustment, or drug substitution). 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 Strong recommendations in CPIC guidelines indicate that “The evidence is high quality and the desirable effects clearly outweigh the undesirable effects.” Therapeutic recommendations that were classified as optional or moderate or recommended no change in drug therapy were not considered actionable for the purpose of this study. Therapeutic recommendations from CPIC guidelines that were present for a variant regardless of genotype (e.g., any VKORC1 genotype influencing warfarin treatment) were also excluded. In CPIC guidelines, moderate recommendations indicate that “There is a close or uncertain balance as to whether the evidence is high quality and the desirable clearly outweigh the undesirable effects” and optional recommendations indicate that “The desirable effects are closely balanced with undesirable effects, or the evidence is weak or based on extrapolations. There is room for differences in opinion as to the need for the recommended course of action.”
Course performance
De‐identified final numeric grades in the PHPR 887 course were collected to determine if course performance varied based upon the PGET intervention or presence of actionable PGx variants. Study personnel were able to access numeric final grade data but were unable to match this data to individual participants.
Statistical analysis
A complete case analysis approach was taken such that statistical analysis only included participants for which no missing data were present. Participants with any missing data were excluded from all analyses. Demographic data collected on the pre‐survey were compared in participants randomized to PGET and NPGET using t‐tests for continuous data, Wilcoxon Rank Sum test for ordinal variables, and χ2 for dichotomous variables. Randomization of participants was performed using “proc surveyselect” in SAS version 9.4 (SAS) to randomize into two groups (simple random selection). The predefined primary outcome of the study was change in participant PGx knowledge. Change in participant PGx knowledge was assessed using the difference in total number of correct responses to the 11 PGx knowledge questions on the pre‐survey and the post‐survey. The primary analysis assessed this improvement in PGx knowledge in participants randomized to PGET compared to NPGET using a Wilcoxon Rank Sum test. For the primary analysis, a p value less than 0.05 by two‐tailed test was considered significant. All other analyses were considered secondary and hypothesis generating. Using the O’Brien‐Castelloe approximation implemented in “proc power” (SAS version 9.4) with two‐sided alpha = 0.05 and n = 232, this study has 83.2% power to detect a difference in knowledge improvement of a mean 0.6 correct responses between PGET and NPGET groups. 49 , 50
In order to characterize improvement of knowledge during the course of the study, McNemar’s test was used to compare performance on PGx knowledge questions between pre‐survey and post‐survey responses regardless of group. McNemar’s test was also used to compare performance on PGx knowledge questions in PGET and NPGET groups on the pre‐survey and post‐survey separately. For all other individual survey questions, Likert scale responses were compared in PGET and NPGET groups using the Wilcoxon Rank Sum test. In order to compare improvement in Likert scale responses, the difference between the retrospective survey and post‐survey response in numeric scores was calculated and compared overall and between PGET and NPGET groups using the Wilcoxon Rank Sum test.
Multivariable linear regressions were performed to identify variables that associated with improvement in PGx knowledge, improvement in both comfort and attitudes combined, and final grades. Improvement in attitudes was calculated as difference between retrospective survey and post‐survey in cumulative Likert score responses across all attitudes survey questions. Poisson regression models included demographic characteristics, PGET assignment, and study year. Poisson regressions were assessed for fit using the Pearson Chi Square dispersion statistic and re‐evaluated using a negative binomial model as dictated by fit. Several sensitivity analyses were also conducted. First, we compared PGx attitudes survey responses on the pre‐survey versus the retrospective survey to confirm participant overestimation of comfort prior to the intervention. Second, the primary analysis was performed by year and by platform to account for regularly updated PGx recommendations and the change in PGx platform across study years. Finally, presence of a highly actionable PGx variant and the number of highly actionable variants in each participant was evaluated for effect on improvement in knowledge using a Wilcoxon Rank Sum test and Pearson Correlation test, respectively. The highly actionable PGx variant analysis was restricted to those participants assigned to PGET because NPGET participants had not received their PGx results prior to both pre‐ and post‐surveys.
A Rasch analysis was performed on the primary outcome and survey data to ensure utility of count data and Likert scale response data as a continuous outcome (1–6 points for strongly disagree to strongly agree or extremely uncomfortable to extremely comfortable). The survey responses fit and met all assumptions of the Rasch model, indicating that ordinal raw scores could be transformed into interval‐scaled measures for statistical testing. Any p values less than 0.001 are reported as p less than 0.001. Analyses were performed using R software version 4.0.0 (R) and SAS version 9.4. A de‐identified dataset containing the study results and the R code associated with our analysis can be found at https://github.com/karneslab/PGET.
RESULTS
Over 4 years, a total of 453 students were enrolled in the required PGx course and 318 (70%) consented to participation in the study (Supplementary Table S8). A total of 78 consented participants were removed from the analysis for missing genotype data (n = 16), missing survey data (n = 25), or both (n = 44) due to participants not providing their DNA sample and not responding or incompletely responding to surveys. A total of 43 participants excluded due to missing data had been randomized to PGET and 42 to NGET groups. A total of 233 participants were included in the final study analysis, including 117 study participants randomized to the PGET group and 116 participants to the NPGET group (Table 1). This represented 51.4% of total students enrolled in the PGx course from 2017 to 2020. Demographic characteristics for study participants were similar to all enrolled students, although the study tended to have a higher rate of female participants (Table 1; Supplementary Table S8). The demographic survey data indicated that the most common age group was 20–29 years (83.7%) and the majority of participants were female (67.8%). Overall, more than half of participants reported a grade point average (GPA) above 3.5 (55.4%) and participants indicated they had little prior experience in formal undergraduate coursework in genetics (24.5%). Randomization to PGET and NPGET groups did not result in significantly different characteristics.
TABLE 1.
Characteristics of study participants randomized to PGET and NPGET
| Participant characteristic |
PGET a (n = 117) |
NPGET a (n = 116) | p value b |
|---|---|---|---|
| Sex (male) | 39 (33.33) | 36 (31.03) | 0.814 |
| Age {years [mean (SD)]} | 0.4903 | ||
| <20 | 0 (0) | 0 (0) | |
| 20–29 | 96 (82.05) | 99 (85.34) | |
| 30–39 | 18 (15.38) | 15 (12.93) | |
| >40 | 3 (2.56) | 2 (1.72) | |
| Current GPA | 0.9376 | ||
| <2.5 | 0 (0) | 0 (0) | |
| 2.5–2.9 | 2 (1.71) | 4 (3.45) | |
| 3.0–3.5 | 51 (43.59) | 47 (40.52) | |
| 3.6–4.0 | 64 (54.7) | 65 (56.03) | |
| Pre‐pharmacy education | 0.5419 | ||
| Prerequisites | 45 (38.46) | 41 (35.34) | |
| B.A. Degree | 3 (2.56) | 9 (7.76) | |
| B.S. Degree | 65 (55.56) | 54 (46.55) | |
| Master’s Degree | 3 (2.56) | 8 (6.9) | |
| Doctorate | 1 (0.85) | 4 (3.45) | |
| Genetics experience | 0.9446 | ||
| None | 24 (20.51) | 26 (22.41) | |
| Prerequisites | 65 (55.56) | 61 (52.59) | |
| Genetics Course | 21 (17.95) | 17 (14.66) | |
| >1 Course | 7 (5.98) | 10 (8.62) | |
| Major/degree focus | 0 (0) | 2 (1.72) | |
| Personal experience with genotyping | 16 (13.68) | 20 (17.24) | 0.5675 |
| Campus location | 0.7414 | ||
| Tucson | 97 (82.91) | 99 (85.34) | |
| Phoenix | 20 (17.09) | 17 (14.66) | |
| Genotyping method | 0.23189 | ||
| Althea (2017) | 31 (26.5) | 20 (17.24) | |
| In‐house (2018) | 21 (17.95) | 23 (19.83) | |
| OneOme (2019, 2020) | 65 (55.56) | 73 (62.93) | |
| Actionable Variant c | 61 (52.14) | 57 (49.14) | 0.9493 |
Abbreviations: GPA, grade point average; NGET, no personal genomic educational testing; PGET, personal genomic educational testing; PGx, pharmacogenetics.
Values are reported as number (percentage) unless otherwise noted.
Student’s t‐test was used to compare continuous variables, Wilcoxon Rank Sum test was used to compare ordinal variables, and χ2 test was used to compare nominal variables between PGET (n = 117) and NPGET groups (n = 116).
Clinically actionable PGx genotypes were determined based on Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines and constituted any diplotype or genotype with a strong therapeutic recommendation for a change in therapy (e.g., drug avoidance, dose adjustment, or drug substitution). Therapeutic recommendations that were optional or moderate or recommended no change in drug therapy were not considered actionable. Therapeutic recommendations from CPIC guidelines that were present for a variant regardless of genotype (e.g., any VKORC1 genotype influencing warfarin treatment) were also excluded.
Among all participants, a significant improvement was observed in PGx knowledge over the course of the study. For instance, the number of correct answers to PGx knowledge questions improved in post‐surveys compared to pre‐surveys (mean 59% vs. 40% correct, p < 0.001 [Table 2; Supplementary Table S9]). Significant improvement was also observed in surveys assessing comfort with PGx clinical skills, PGx patient education, and attitudes toward PGx (Table 3; Supplemental Tables S10, S11). All participants demonstrated a mean positive change in knowledge and attitudes scores over the course of the study.
TABLE 2.
Comparison of knowledge‐based pharmacogenomics questions at time of pre‐ and post‐survey, among all study participants
| Knowledge question | Pre‐survey a | Post‐survey a | p value b , c |
|---|---|---|---|
| Q1 | 145 (62.23) | 197 (84.55) | <0.001 |
| Q2 | 123 (52.79) | 203 (87.12) | <0.001 |
| Q3 | 81 (34.76) | 153 (65.67) | <0.001 |
| Q4 | 97 (41.63) | 133 (57.08) | <0.001 |
| Q5 | 81 (34.76) | 123 (52.79) | <0.001 |
| Q6 | 35 (15.02) | 75 (32.19) | <0.001 |
| Q7 | 58 (24.89) | 92 (39.48) | <0.001 |
| Q8 | 61 (26.18) | 87 (37.34) | <0.001 |
| Q9 | 90 (38.63) | 133 (57.08) | <0.001 |
| Q10 | 142 (60.94) | 181 (77.68) | <0.001 |
| Q11 | 116 (49.79) | 124 (53.22) | 0.4655 |
| Total | 1029 (40.15) | 1501 (58.56) | <0.001 |
Abbreviation: PGx, pharmacogenomics.
Values are reported as number of correct answers (%).
McNemars Test was to compare the number of correct answers between pre‐survey and post‐survey responses (n = 233).
For analysis of total correct answers in pre‐survey versus post‐survey, Wilcoxon Rank Sum Test was used to compare the total score between pre‐ and post‐surveys (n = 233).
TABLE 3.
Change in PGx knowledge and attitudes over the course of the study for all participants and by randomized group
| Outcome | Improvement during study | Improvement by randomized group | ||||
|---|---|---|---|---|---|---|
| Retrospective survey a | Post‐survey a | p value b | PGET a | NPGET a | p value c | |
| Knowledge improvement | 4.42 (2.24) | 6.44 (2.49) | <0.001 | 2.14 (2.88) | 1.84 (2.52) | 0.405 |
| Comfort with PGx clinical skills and patient education | 50.89 (20.72) | 80.45 (12.17) | <0.001 | 36.39 (19.72) | 34.72 (20.31) | 0.569 |
| Attitudes toward PGx | 41.54 (6.33) | 47.14 (6.45) | <0.001 | 5.98 (5.97) | 5.22 (6.17) | 0.342 |
Abbreviations: NGET, no personal genomic educational testing; PGET, personal genomic educational testing; PGx, pharmacogenomics.
Values reported as mean change, defined as the mean difference in total score between pre‐survey and post‐survey knowledge‐based PGx questions, and retrospective survey and post‐survey Likert scale response questions.
Wilcoxon Signed Rank test was used to compare mean scores on knowledge‐, comfort‐, and attitudes‐based questions among all participants (n = 233).
Wilcoxon Rank Sum test was used to compare mean changes between PGET (n = 117) and NPGET groups (n = 116).
In our primary analysis, improvement in PGx knowledge was not different in PGET and NPGET groups (mean 2.14 vs. 1.84 increase in correct answers, respectively [p = 0.405]). The proportional improvement in correct responses was 19.5% in the PGET group and 16.7% in the NPGET group (Figure 2; Supplementary Table S9). Similarly, cumulative scores for change in retrospective versus post‐survey Likert scale responses were not significantly different (Table 3; Supplementary Tables S10, S11). Although not statistically significant, the average difference was higher in the PGET group for all categories, including improvement in PGx knowledge, comfort, and attitudes. In sensitivity analyses, similar results were observed by study year and by PGx platform. No significant improvement in PGx knowledge was observed in participants with an actionable PGx variant versus no actionable variant (mean 2.33 vs. 1.98 increase in correct answers, respectively [p = 0.64], Supplementary Tables S12, S13).
FIGURE 2.
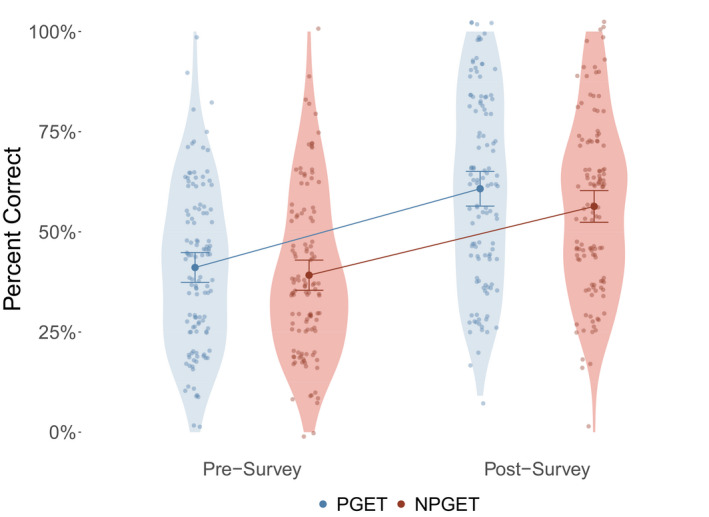
Cumulative performance on PGx knowledge‐based questions. Change in PGx knowledge was determined by finding the difference between total pre‐survey and post‐survey knowledge assessment scores. The bold circle and error bars indicate the mean and 95% confidence intervals for the mean. This change in knowledge was used to compare performance between participants randomized to PGET and NPGET, using a Wilcoxon Rank Sum test. The proportional improvement in correct responses was larger in the PGET group (19.5%) than in the NPGET group (16.7%), which did not result in statistical significance (p = 0.405). PGx indicates pharmacogenomics; PGET, personal genomic educational testing; NGET, no personal genomic educational testing
Of the 27 Likert scale questions administered, two questions from the attitudes toward PGx domain were found to have nominally significantly different responses between the PGET and NPGET groups. Participants in the PGET group were more likely to agree with the statements: “I feel more personally engaged because I am participating in the pharmacogenetics study” (p = 0.020) and “My participation in this pharmacogenetics study will reinforce the concepts taught in class” (p = 0.006 [Figure 3; Supplementary Table S11]). All Likert scale questions showed significantly higher scores on pre‐survey versus retrospective survey responses, suggesting that participants overestimated their comfort levels with PGx prior to the study intervention. Similar results were achieved when improvement in survey responses was measured based on the pre‐survey and post‐survey difference rather than the retrospective and post‐survey difference. Poisson regression models used to evaluate demographic and experiential variables on major outcomes showed no variables significantly predicted improvement in knowledge (Supplementary Table S14). Student GPA was strongly associated with final course grade (beta = 0.05 [0.01], p = 0.001), indicating that higher student‐reported GPA predicted a higher final course grade. In negative binomial regression, no variables significantly predicted improvement in attitudes toward PGx.
FIGURE 3.
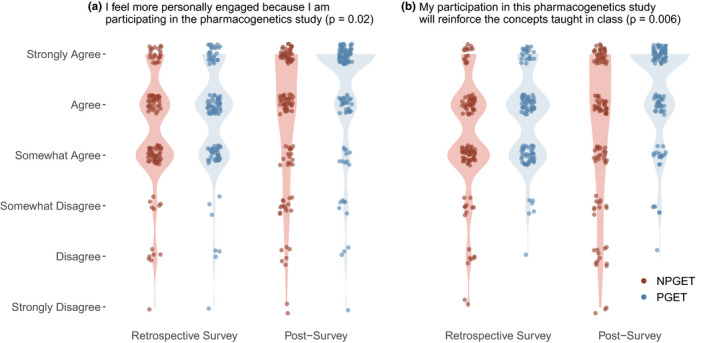
Violin plots showing density of student responses to Likert scale questions by randomized group on the retrospective survey and post‐survey. Likert scale responses varied from 1 (strongly disagree) to 6 (strongly agree). Panel (a) shows responses to the question “I feel more personally engaged because I am participating in the pharmacogenetics study.” Participants randomized to PGET showed a significantly larger increase in attitudes responses between the retrospective and post‐survey using a Wilcoxon Rank Sum (p = 0.02). Panel (b) shows responses to the question “My participation in this pharmacogenetics study will reinforce the concepts taught in class.” Participants randomized to PGET showed a significantly larger improvement in attitudes between the retrospective and post‐survey using a Wilcoxon Rank Sum test (p = 0.006). PGx indicates pharmacogenomics; PGET, personal genomic educational testing; NGET, no personal genomic educational testing
DISCUSSION
We present here the first randomized study aimed at assessing the efficacy of PGET in the PharmD classroom. This study addresses many of the biases and limitations in the existing literature by enrolling a large number of participants across multiple years, by randomizing participants from the same course to PGET and NPGET groups, and by incorporating detailed surveys, an objective knowledge assessment, PGx test results, and final grade data. Our results indicate that panel‐based PGET during a PharmD course has no impact on improvement in student knowledge regarding PGx. We consider our results to have important implications for implementation of PGET in PharmD programs in light of the high cost of PGx genotyping and the potential ethical, legal, and regulatory issues around genotyping of students.
Our findings also indicate that knowledge, comfort, and attitudes related to PGx are not significantly improved with panel‐based PGET. Whereas there was evidence of improved engagement by participants randomized to PGET, this did not translate into improvement in objective outcomes. Although our study was powered for our a priori effect size estimate, the nonsignificant increase in improvement in knowledge and attitudes for the PGET group suggests that improved learning might be observed in larger studies. Nevertheless, our results indicate that the effect of PGET on PGx‐related educational improvement and attitudes toward PGx is likely to be small. Although no significant difference was observed for the primary outcome, we observed nominally significantly improved Likert scale responses for questions evaluating student engagement and reinforcement of concepts covered in class. These results should be interpreted with caution, as no multiple comparisons adjustment was used. However, these results suggest a modest benefit for PGET and, combined with our observation of overall improvement in performance on knowledge questions throughout the course, indicate the validity of the data collected.
Our results contradict the majority of studies evaluating PGET as a learning and engagement strategy in clinical classrooms. Previous evidence evaluating PGET has broadly supported that PGET improves students’ knowledge and attitudes in a variety of settings. 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 A recent systematic review and meta‐analysis has summarized the cumulative results of these studies quite effectively. 14 The authors observed that PGET had a positive effect on student survey responses regarding attitudes and perceptions in studies without a control group (p = 0.009) and in studies with a control group (p = 0.025). However, the authors point to a number of limitations across these studies, including small sample sizes, lack of a control group, and self‐selection of students into PGET intervention groups. The authors also acknowledge the potential value of future studies that utilize randomization and more consistent “placebo” approaches for the control group. In this body of literature, self‐selection of students into PGET intervention groups has likely biased prior studies because students with positive perceptions of PGx are more likely to undergo PGET as well as have improved PGx knowledge. The use of a randomization to PGET and NPGET groups in the present study reduces the potential for this source of bias and is a potential explanation for the stark differences observed between the present study and previously published investigations of PGET.
Although the current study was designed to address the above limitations in the previous literature, there are several limitations worthy of mention in the current study. Because our genotyping platform used in the first year of the study became unavailable during the 2018 Spring semester, our study implemented different genotyping platforms across different years and these genotyping platforms contained different numbers of variants. Whereas the number of genes and variants tested varied between years, genotyping in all 4 years included variants in CYP2C9, CYP2C19, CYP4F2, and VKORC1 and sensitivity analyses suggested that results were consistent regardless of the genotyping platform used. CPIC guidelines also changed throughout the study years, affecting the correct answers to our original knowledge questions, and causing one question to be eliminated. However, we did not observe differences in our results when sensitivity analyses were performed by year. Our study also did not evaluate long‐term outcomes of PGET on student performance or professional engagement following graduation from a PharmD program, which might be expected based on the relevance of PGx test results throughout one’s lifetime. Our actionable variant analyses should be interpreted with caution as participants were tested for different PGx variants in different years. 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 Although some evidence of improved engagement and participation was observed, these results must be interpreted with caution considering the low statistical power to observe differences for these outcomes due to multiple testing. Finally, students with unfavorable opinions of PGx and/or a lack of PGx knowledge may have elected not to participate in this study, resulting in a majority of participants with favorable PGx opinions and/or high PGx knowledge. In this case, our results may have been biased toward a null result, because participants had high PGx knowledge or favorable opinions at baseline with limited room for improvement across the study. In our study, more than 60% of participants had at least a Bachelor’s degree, over 20% had taken a genetics course, and over 15% had prior personal experience with genotyping. Although we expect our results to be broadly generalizable to other PharmD programs, there may be a greater potential for PGx knowledge and attitudes improvement in students with less prior genetics education and experience.
Because the present study includes a relatively large sample size and a formal randomization to PGET and NPGET intervention groups, this study constitutes the most rigorous evaluation of educational outcomes with PGET to date. Although some evidence of improved engagement and participation was observed, our results indicate that PGET has no effect or at least a very limited effect on student’s ability to apply PGx‐related knowledge nor on students’ attitudes toward clinical utility of PGx testing. We consider our results to have important implications for implementation of PGET in PharmD programs in light of the high cost of PGx genotyping and the potential ethical, legal, and regulatory issues around genotyping of students. We believe that this randomized prospective study evaluating educational outcomes with PGET constitutes a significant contribution to the literature related to PGET and will ultimately support more evidence‐based teaching interventions.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
C.G., M.M.L., H.E.S., and J.H.K. wrote the manuscript. M.M.L., P.J.C., C.C., D.Q., W.K., D.N., and J.H.K. designed the research. M.M.L., H.E.S., H.P., D.Q., D.N., T.W., and J.H.K. performed the research. C.G., H.E.S., S.M., P.J.C., H.P., and T.W. analyzed the data.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors would especially like to thank the participants in the University of Arizona Doctor of Pharmacy curriculum who participated in this study. We would like to thank the staff at AltheaDx, including Ali Cullors, Stephani Stewart, and Andrew Lukowiak. We would like to thank the staff at Oneome, including Kristen Bauer, Lilly Drew, and Ross Higgins. Finally, we would like to thank Bruce Johnson, John Murphy, Barb Collins, Nkiru Nwachukwu, Caitlin Cameron, and Jeannie Lee at the University of Arizona College of Pharmacy. The pharmacogenomic testing companies utilized, including Oneome (Minneapolis, MN) and AltheaDx (San Diego, CA), had no role in the design, analysis, and writing of this manuscript.
Grace C, Larriva MM, Steiner HE, et al. Efficacy of personal pharmacogenomic testing as an educational tool in the pharmacy curriculum: A nonblinded, randomized controlled trial. Clin Transl Sci. 2021;14:2532–2543. 10.1111/cts.13121
Chloe Grace and Marti M. Larriva contributed equally to this work.
Funding information
This work was funded in part by the University of Arizona College of Pharmacy. J.H.K. is supported by the National Institutes of Health’s National Heart, Lung, and Blood Institute (NHLBI) under grant K01HL143137. The sources of funding had no role in the design, analysis, and writing of this manuscript.
REFERENCES
- 1. Stanek EJ, Sanders CL, Taber KAJ, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91:450‐458. [DOI] [PubMed] [Google Scholar]
- 2. Calabro GE, Tognetto A, Mazzaccara A, et al. Capacity building of health professionals on genetics and genomics practice: evaluation of the effectiveness of a distance learning training course for Italian physicians. Front Genet. 2021;12:626685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy JE, Green JS, Adams LA, Squire RB, Grace GM, McKay A. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am J Pharm Educ. 2010;74:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Empey PE, Stevenson JM, Tuteja S, et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype‐guided antiplatelet therapy. Clin Pharmacol Ther. 2018;104:664‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaksman N, Barnett M, Hakobyan L, Kutcher I, Louie MC. The impact of incorporating of pharmacogenomics into the pharmacy curriculum on student interest. Pharmacy Education. 2012;12:31‐36. [Google Scholar]
- 6. Salari K, Karczewski KJ, Hudgins L, Ormond KE. Evidence that personal genome testing enhances student learning in a course on genomics and personalized medicine. PLoS One. 2013;8:e68853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vernez SL, Salari K, Ormond KE, Lee SS. Personal genome testing in medical education: student experiences with genotyping in the classroom. Genome Med. 2013;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krynetskiy E, Lee Calligaro I. Introducing pharmacy students to pharmacogenomic analysis. Am J Pharm Educ. 2009;73:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Knoell DL, Johnston JS, Bao S, Kelley KA. A genotyping exercise for pharmacogenetics in pharmacy practice. Am J Pharm Educ. 2009;73:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frick A, Benton C, Suzuki O, et al. Implementing clinical pharmacogenomics in the classroom: student pharmacist impressions of an educational intervention including personal genotyping. Pharmacy (Basel). 2018;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arwood MJ, McDonough CW, Cavallari LH, et al. Evaluating an interactive teaching approach with personal genotyping to provide pharmacy students with a knowledge base for clinical pharmacogenetics. JACCP. 2021;4:343‐351. [Google Scholar]
- 12. Adams SM, Anderson KB, Coons JC, et al. Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am J Pharm Educ. 2016;80:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weitzel KW, McDonough CW, Elsey AR, Burkley B, Cavallari LH, Johnson JA. Effects of using personal genotype data on student learning and attitudes in a pharmacogenomics course. Am J Pharm Educ. 2016;80:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burghardt KJ, Ward KM, Howlett BH, Burghardt PR. Personal genotyping and student outcomes in genetic and pharmacogenetic teaching: a systematic review and meta‐analysis. Pharmacogenomics. 2021;22(7):423‐433. [DOI] [PubMed] [Google Scholar]
- 15. O’Brien TJ, LeLacheur S, Caitlin C, et al. Impact of a personal CYP2D6 testing workshop on physician assistant student attitudes toward pharmacogenetics. Pharmacogenomics. 2016;17:341‐352. [DOI] [PubMed] [Google Scholar]
- 16. Nickola TJ, Munson AM. Pharmacogenomics primer course for first professional year pharmacy students. Pharmacogenomics. 2014;15:39‐48. [DOI] [PubMed] [Google Scholar]
- 17. Remsberg CM, Bray BS, Susan KW, et al. Design, implementation, and assessment approaches within a pharmacogenomics course. Am J Pharm Educ. 2017;81:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber KS, Jensen JL, Johnson SM. Anticipation of personal genomics data enhances interest and learning environment in genomics and molecular biology undergraduate courses. PLoS One. 2015;10:e0133486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang X, Hartman MR, Harrington KT, et al. Using next‐generation sequencing to explore genetics and race in the high school classroom. CBE Life Sci Educ. 2017;16(2):ar22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Howard GS. Response‐shift bias: a problem in evaluating interventions with pre/post self‐reports. Evaluation Review. 1980;4:93‐106. [Google Scholar]
- 21. Testa S, Di Cuonzo D, Ritorto G, et al. Response shift in health‐related quality of life measures in the presence of formative indicators. Health Qual Life Outcomes. 2021;19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McClimans L, Bickenbach J, Westerman M, Carlson L, Wasserman D, Schwartz C. Philosophical perspectives on response shift. Qual Life Res. 2013;22:1871‐1878. [DOI] [PubMed] [Google Scholar]
- 23. Sugarman EA, Cullors A, Centeno J, Taylor D. Contribution of pharmacogenetic testing to modeled medication change recommendations in a long‐term care population with polypharmacy. Drugs Aging. 2016;33:929‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bradley P, Shiekh M, Mehra V, et al. Improved efficacy with targeted pharmacogenetic‐guided treatment of patients with depression and anxiety: a randomized clinical trial demonstrating clinical utility. J Psychiatr Res. 2018;96:100‐107. [DOI] [PubMed] [Google Scholar]
- 25. Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharmacol Ther. 2018;103:210‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bell GC, Caudle KE, Whirl‐Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther. 2017;102:213‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birdwell KA, Decker B, Barbarino JM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther. 2015;98:19‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown JT, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin Pharmacol Ther. 2019;106:94‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clancy JP, Johnson SG, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clin Pharmacol Ther. 2014;95:592‐597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy [published online ahead of print January 2, 2021]. Clin Pharmacol Ther. 10.1002/cpt.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Desta Z, Gammal RS, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz‐containing antiretroviral therapy. Clin Pharmacol Ther. 2019;106:726‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gammal RS, Court MH, Haidar CE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin Pharmacol Ther. 2016;99:363‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goetz MP, Sangkuhl K, Guchelaar HJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther. 2018;103:770‐777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonsalves SG, Dirksen RT, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for the use of potent volatile anesthetic agents and succinylcholine in the context of RYR1 or CACNA1S genotypes. Clin Pharmacol Ther. 2019;105:1338‐1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther. 2015;98:127‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson JA, Caudle KE, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics‐Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017;102:397‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karnes JH, Rettie AE, Somogyi AA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA‐B Genotypes and Phenytoin Dosing: 2020 Update. Clin Pharmacol Ther. 2021;109:302‐309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lima JJ, Thomas CD, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther. 2021;109(6):1417‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin MA, Hoffman JM, Freimuth RR, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA‐B Genotype and Abacavir Dosing: 2014 update. Clin Pharmacol Ther. 2014;95:499‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moriyama B, Owusu Obeng A, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin Pharmacol Ther. 2017;102:45‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muir AJ, Gong L, Johnson SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon‐alpha‐based regimens. Clin Pharmacol Ther. 2014;95:141‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Phillips EJ, Sukasem C, Whirl‐Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for HLA genotype and use of carbamazepine and oxcarbazepine: 2017 update. Clin Pharmacol Ther. 2018;103:574‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin‐induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96:423‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Relling MV, McDonagh EM, Chang T, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for rasburicase therapy in the context of G6PD deficiency genotype. Clin Pharmacol Ther. 2014;96:169‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Relling MV, Schwab M, Whirl‐Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther. 2019;105:1095‐1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saito Y, Stamp LK, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA‐B) genotype and allopurinol dosing: 2015 update. Clin Pharmacol Ther. 2016;99:36‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scott SA, Sangkuhl K, Stein CM, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Theken KN, Lee CR, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and nonsteroidal anti‐inflammatory drugs. Clin Pharmacol Ther. 2020;108:191‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang Y. Size and power estimation for the Wilcoxon‐Mann‐Whitney test for ordered categorical data. Stat Med. 2011;30:3461‐3470. [DOI] [PubMed] [Google Scholar]
- 50. Mollan KR, Trumble IM, Reifeis SA, et al. Precise and accurate power of the rank‐sum test for a continuous outcome. J Biopharm Stat. 2020;30:639‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


