Abstract
This multicenter clinical study was aimed at conducting a targeted pharmacogenomic association analysis of residual on‐clopidogrel platelet reactivity in 474 Caribbean Hispanic patients. Platelet reactivity was measured using the VerifyNow P2Y12 assay and clopidogrel resistance was defined as P2Y12 reaction units (PRUs) greater than or equal to 208. Genotyping was performed using the whole‐genome Infinium MEGA BeadChip array. An ancestry‐adjusted, weighted polygenic risk score (wPGxRS) was developed to account for the effect of multiple variants on PRU and compared between clopidogrel responders and nonresponders. The mean PRU across the study cohort was 173.8 ± 68.5 and 33.5% of patients were defined as clopidogrel resistant. Multivariate linear regression showed that 19% of PRU variability was attributed to nine independent predictors, with CYP2C19*2 (rs4244285) accounting for ~ 7% of observed PRU variation (p < 0.001). PON1 rs662, ABCB1/MDR1 rs2032582, PEAR1 rs12041331 carrier status, and the interaction between African ancestry and rs12041331 carriers also predicted PRU among the participants (p ≤ 0.05). A clear gene‐dose effect was detected between PRU and CYP2C19*2 genotype, consistent with previous studies in European patient populations, as well as rs12777823. Importantly, a significant positive correlation was detected between our novel wPGxRS (4 variants) and PRU among the Hispanic patient population (r p = 0.35, p < 0.001). Moreover, the wPGxRS discriminated between nonresponders and responders (p = 0.003), indicating that this multigene‐based score is a useful predictor of clopidogrel resistance among Caribbean Hispanics. Taken together, these results help close the gap of knowledge on clopidogrel pharmacogenomics and its potential clinical implementation in this under‐represented population.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
CYP2C19*2 is significantly associated with an impaired response to clopidogrel in patients of mainly European ancestry, but this genetic variant accounts for only a small fraction of observed variability in clopidogrel resistance.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study answers the question of whether there is an association of relevant candidate pharmacogenes involved in clopidogrel absorption, metabolism, and pharmacodynamics with antiplatelet response among Caribbean Hispanics using a multigene‐based score approach.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study closes the existing gap of knowledge about clopidogrel pharmacogenomics in Caribbean Hispanics by both confirming and identifying relevant genetic determinants of clopidogrel responsiveness in this underrepresented population after developing an ancestry‐based polygenic risk score model.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
By gaining a better understanding of underlying determinants of poor clopidogrel response among marginally represented groups, such as Caribbean Hispanics, worldwide efforts to translate such findings into a global precision medicine paradigm of true benefit for everyone can finally bring cardiovascular health equity to the population at large.
INTRODUCTION
Clopidogrel, a P2Y12 receptor inhibitor, is widely recommended for secondary prevention of coronary artery disease (CAD) to reduce major adverse cardiovascular events (MACEs) among patients with acute coronary syndrome (ACS) or stable CAD undergoing percutaneous coronary intervention (PCI). Despite its proven effectiveness as part of dual antiplatelet therapy (DAPT), some patients do not attain adequate antiplatelet effects. 1 , 2 Studies have shown that certain genetic variants in clinically relevant pharmacogenes (e.g., CYP2C19*2) are significantly associated with an impaired response to clopidogrel. 1 , 3 , 4 Moreover, carrying at least one nonfunctional CYP2C19 allele results in reduced formation of the active clopidogrel metabolite when compared with patients with a wild‐type genotype. 5 However, CYP2C19 accounts for only a small fraction of observed variability in response to clopidogrel. In addition, poor responders to clopidogrel (~ 30%) can also be identified among patients who are noncarriers of any known reduced‐function CYP2C19 allele, suggesting that other genetic markers for clopidogrel response prediction remain to be discovered. Notably, most of these studies have been conducted among individuals of European ancestry with Hispanics and other minorities being under‐represented. 6 , 7
Although routine implementation of CYP2C19 testing to guide antiplatelet therapy selection has remained controversial due to a lack of large randomized controlled trials evaluating this approach, the American College of Cardiology Foundation/American Heart Association (ACC/AHA) ACS guidelines noted that genetic testing might be considered on a case‐by‐case basis to identify whether a high‐risk patient is predisposed to inadequate platelet inhibition with clopidogrel. 8 Likewise, the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines recommend an alternative antiplatelet therapy (if not contraindicated) for carriers of CYP2C19 risk alleles, particularly for those with a poor metabolizer (PM) status, when CYP2C19 genotype status is known. 9 A CYP2C19 genotype‐guided strategy for optimal selection of antiplatelet therapy has been proven beneficial in several clinical utility studies; however, none of these trials were performed with Caribbean Hispanic patients. 10 , 11
To date, there is a gap of knowledge about clopidogrel pharmacogenomics in Caribbean Hispanics, as they make up less than 1% of participants in previous studies. This lack of representation tends to exacerbate already existing healthcare disparities. 12 , 13 Our study assessed the association of relevant candidate pharmacogenes involved in clopidogrel pharmacokinetics and pharmacodynamics with antiplatelet response among Caribbean Hispanics to identify genetic determinants of clopidogrel responsiveness using a multigene‐based score approach in this under‐represented population. Because genetic variants on these candidate pharmacogenes were chosen from published reports of significant association with clopidogrel response in other populations, it provides justification of their use to predict risk scores in our study sample rather than applying to an independent cohort.
METHODS
This is a multicenter, cross‐sectional, clinical protocol to perform a combined targeted and untargeted pharmacogenomic association analysis of clopidogrel resistance in Caribbean Hispanics (institutional review board [IRB] approval #A4070417). To this purpose, 549 patients of Hispanic origin who reside in the Commonwealth of Puerto Rico and received either a 600 or 300 mg loading dose plus a daily 75 mg maintenance dose or only a 75mg daily maintenance dose of clopidogrel for more than 7 days (i.e., alone or as a component of DAPT in mainly post‐PCI patients with ACS or stable CAD) were enrolled in this study between January 2018 and June 2020 (Figure S1). Inclusion and exclusion criteria are described as Supplementary Table S1.
Blood samples (20 ml) for genetic testing and rapid ex vivo residual platelet function analysis were collected at a single time point on the day of recruitment while under clopidogrel maintenance dosing. Platelet reactivity was measured within 4 h after blood withdrawal using the VerifyNow P2Y12 assay (i.e., PRUTest; Accumetrics) following the manufacturer’s instructions. The results of residual platelet function tests were expressed as PRU and used as the primary end point to determine platelet inhibition by clopidogrel in each participant. High on‐treatment platelet reactivity (HTPR) was defined as PRU greater than or equal to 208, 14 , 15 which indicates poor clopidogrel response (i.e., resistant status). Additionally, MACEs were evaluated in a subset of patients (n = 305) in whom clinical outcomes data were available. Further details are provided as Supplementary Material.
Genotyping
Genotyping was performed using a HiScan system with the whole‐genome Infinium Multi‐Ethnic Hispanic AMR/AFR MEGA BeadChip array, according to the manufacturer’s instructions (Illumina). Single‐nucleotide polymorphisms (SNPs) occurring on candidate genes of clinical relevance were chosen to perform the corresponding targeted analysis based on prior literature and preliminary unpublished data from our studies in Caribbean Hispanics. 6 , 15 Only SNPs at these loci that were present on the array with a minor allele frequency (MAF) greater than 1% were included in our analyses. The mean genotype call rate of these ~ 1.4 M SNPs was greater than 99%. All SNPs within the regions of interest on preselected pharmacogenes were in Hardy‐Weinberg equilibrium (p ˃ 0.05). Genotype results of selected sites in candidate genes were confirmed by TaqMan SNP genotyping assay methodology using a StepOne Plus Real‐Time polymerase chain reaction (PCR) system, according to standard protocols (Applied Biosystems). Genotype concordance was greater than 97% in a subset of duplicate samples. Standard quality control measures were conducted using PLINK version 1.07. Ancestral proportions were estimated using ADMIXTURE and used as covariates in statistical analyses to account for the potential confounder effect of such a varying degree of ancestry on the expected pharmacogenetic associations of clopidogrel resistance in Caribbean Hispanics.
Statistical analysis
Summary statistics (e.g., means [SDs]) and frequencies for the study population as well as measures of Hardy–Weinberg equilibrium and linkage disequilibrium (LD) metrics were calculated using PLINK version 1.07. A linear regression analysis under an additive genetic model without conditioning was conducted to assess whether relevant covariates (see in Supplementary Material) significantly predicted PRU in Caribbean Hispanic patients. A binary logistic regression was also performed to examine whether carrier status at certain loci had a significant effect on the odds of observing clopidogrel resistance (i.e., HTPR). Kruskal‐Wallis rank sum test was conducted to assess if there were significant differences in PRU between genotypes at preselected loci. Pairwise post hoc comparisons were also examined. Additionally, a two‐tailed Mann‐Whitney U test was used to examine such differences in PRU by carrier status at some loci. All statistical tests were two‐sided.
A weighted polygenic pharmacogenomic risk score (wPGxRS) was applied to determine the cumulative effects of significantly associated candidate SNPs on PRU measures (see Supplementary Material for details). The wPGxRS of responders and nonresponders were compared by Wilcoxon signed‐rank test with continuity correction. Furthermore, a Pearson’s correlation analysis was conducted between the derived wPGxRS and PRU values, with Cohen’s standard used to evaluate the strength of the relationship.
RESULTS
Table 1 describes the baseline characteristics of the participants from the study population with full genetic and clinical data available (n = 474; Figure S1). The average age of all participants in this study was ~ 68 years old, with 55% identified as men. Overall, 78.3% of participants are greater than or equal to 60 years old and 18.6% were within the 45–59 years old range. Largely, most were middle‐aged men, with high prevalence of conventional risk factors (i.e., overweight [28.4 kg/m2]; hypertension [83.8%]; hypercholesterolemia/ dyslipidemias [71.9%], and type‐2 diabetes [54.8%], among others). Furthermore, 20% of participants were on proton pump inhibitors (PPIs; mainly pantoprazole); whereas statins and calcium channel blockers (CCBs) were prescribed in 79.1% and 26.8% of patients, respectively. Patients who were taking aspirin administered as part of DAPT represented 63.3% of the total cohort. In 72.6% of participants, clopidogrel was given for a stable CAD/ ACS indication. Figure 1 depicts the distribution of PRU values in all individuals of the study cohort (mean = 173.75 ± 68.5; range: 2–328). Among all enrolled patients, 33.5% were deemed to be resistant to clopidogrel, based on a PRU cutoff value of 208. PRU values among women were significantly higher than men (i.e., 186 vs. 163, p < 0.001).
TABLE 1.
Characteristics of participants in the study cohort
| Variable | Mean | SD | SEM | Min | Max | Median |
|---|---|---|---|---|---|---|
| Age (years) | 68.01 | 10.95 | 0.51 | 27.00 | 94.00 | 69.00 |
| BMI (kg/m2) | 28.40 | 5.71 | 0.27 | 11.48 | 52.67 | 27.67 |
| Ancestrya proportions | ||||||
| European (Iberians) | 0.70 | 0.13 | 0.01 | 0.23 | 0.93 | 0.72 |
| Native American | 0.11 | 0.04 | 0.00 | 0.03 | 0.50 | 0.11 |
| African (Yoruba) | 0.19 | 0.13 | 0.01 | 0.03 | 0.72 | 0.15 |
| Variable | n | % |
|---|---|---|
| Diabetes mellitus | 260 | 54.85 |
| Hypertension | 397 | 83.75 |
| Dyslipidemias | 341 | 71.94 |
| Smoking | 64 | 13.50 |
| MACEb | 42 | 13.77 |
| Myocardial infarctions (i.e., STEMI and NSTEMI)b | 19 | 6.23 |
| Stent thrombosisb | 15 | 4.92 |
| Strokes/TIAb | 14 | 4.59 |
| Deathsb | 4 | 1.31 |
| Bleedings eventsc | 77 | 16.24 |
| Aspirin users | 300 | 63.29 |
| Statins users | 375 | 79.11 |
| CCBs users | 127 | 26.79 |
| PPIs users | 95 | 20.04 |
| LVEF ≤40% | 39 | 8.22 |
| ACS and stable CAD | 344 | 72.57 |
| Coronary artery stentingd | 177 | 37.34 |
| Peripheral artery disease | 114 | 24.05 |
| Gender | ||
| Female | 212 | 44.73 |
| Male | 262 | 55.27 |
Due to rounding errors, percentages may not equal 100%.
Abbreviations: ACS, acute coronary syndrome; BMI, body mass index; CAD, coronary artery disease; CCB, calcium channel blocker; LVEF, left ventricular ejection fraction; MACE, major adverse cardiovascular event; NSTEMI, non‐ST‐elevation myocardial infarction; PPI, proton pump inhibitor; STEMI, ST‐elevation myocardial infarction; TIA, transient ischemic attack.
aAverage European, Native American and African ancestral genomic proportions at population level were estimated by ADMIXTURE software using the whole‐genome Infinium Multi‐Ethnic Hispanic AMR/AFR MEGA BeadChip array from Illumina.
bMACE reports available only in a subset of 305 patients.
cBleeding events is a combination of major and minor events.
dAll patients with ACS and some with CAD underwent PCI (i.e., percutaneous coronary intervention) or coronary artery stenting.
FIGURE 1.
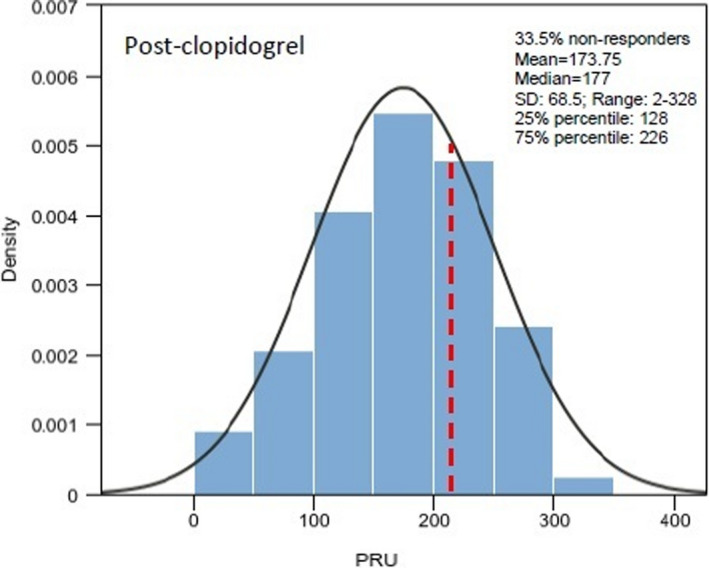
Histogram and probability density function (PDF) to show distribution of P2Y12 reaction unit (PRU) values in all individuals of the study cohort who received clopidogrel treatment. Class intervals include data greater than the lower limit and equal to the upper limit of each interval
Figure 2 illustrates the genotype frequency distribution at 12 of the 17 relevant SNP sites within the predefined pharmacogenes of clinical relevance for clopidogrel responsiveness. 6 No deviations from expected Hardy–Weinberg proportions were found at any of these loci (p > 0.05). The overall MAFs of all tested SNPs in the entire study cohort are shown in Table 2. This table also shows the corresponding MAFs at all these loci for participants with resistant (PRU ≥ 208) or sensitive (PRU < 208) phenotypes and African‐enriched ancestry (≥0.25), as well as the odds ratios (ORs; 95% confidence intervals [CIs]) for the association with resistance and the p values for the MAF comparisons among relevant subgroups. Notably, only CYP2C19*2 (plus rs12777823, which is in LD) were significantly associated with the resistant phenotype in this subgroup.
FIGURE 2.
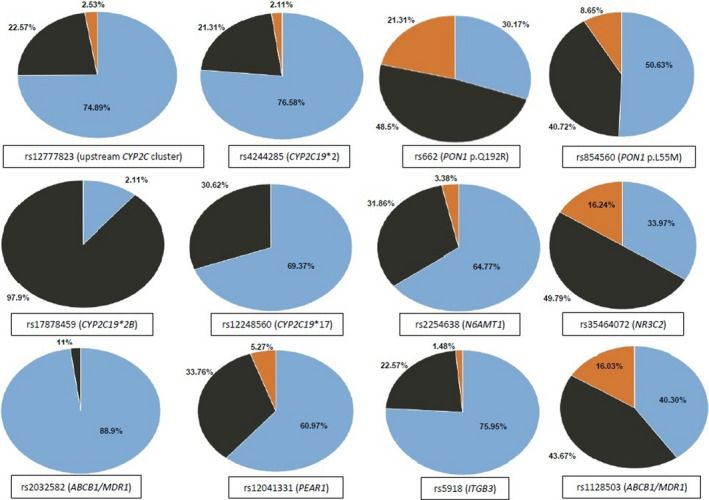
Frequency distributions of genotypes at different tested genetic loci of clinical relevance. Homozygous for major allele are shown in blue color, heterozygous in black, and homozygous for minor allele in orange
TABLE 2.
Allelic frequencies of SNPs on candidate genes tested for the association with platelet function (i.e., PRU) in Caribbean Hispanic patients on clopidogrel (all participants and by African ancestry or resistant status)
| SNP (rs numbera/allele) | Chr. | Position | Gene | MAF | 95% CI | African‐enriched ancestryb (n = 129) | p value c | PRU ≥ 208 (Resistant, n = 184) | PRU < 208 (Sensitive, n = 365) | p valued | ORs [95% CI]e |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4244285 (CYP2C19*2)1 | 10q23.33 | 94,781,859 | CYP2C19 | 0.13 | 0.09–0.16 | 0.14≈ | 0.77 | 0.18↑ | 0.10↓ | 0.013 | 1.94 [1.17–3.22] |
| rs662 (PON1 p.Q192R)f | 7q21.3 | 95,308,134 | PON1 | 0.45 | 0.40–0.49 | 0.50↑ | 0.31 | 0.51↑ | 0.43↓ | 0.08 | 1.38 [0.97–1.97] |
| rs2032582 | 7q21.12 | 87,531,302 | ABCB1/MDR1 | 0.44 | 0.39–0.48 | 0.44≈ | 0.85 | 0.47↑ | 0.43≈ | 0.37 | 1.16 [0.81–1.66] |
| rs12248560 (CYP2C19*17) | 10q23.33 | 94,761,900 | CYP2C19 | 0.15 | 0.12–0.18 | 0.20↑ | 0.19 | 0.12↓ | 0.17↑ | 0.11 | 0.66 [0.39–1.12] |
| rs4986893 (CYP2C19*3) | 10q23.33 | 94,780,653 | CYP2C19 | ND | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ |
| rs28399504 (CYP2C19*4) | 10q23.33 | 94,762,706 | CYP2C19 | 0.002 | 0.0001–0.012 | 0.002≈ | 0.98 | ND | 0.002≈ | 0.39 | ‐‐ |
| rs41291556 (CYP2C19*8) | 10q23.33 | 94,775,416 | CYP2C19 | ND | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ | ‐‐ |
| rs12777823f | 10 | 94,645,745 | CYP2C cluster | 0.14 | 0.11–0.17 | 0.16↑ | 0.57 | 0.18↑ | 0.11↓ | 0.032 | 1.78 [1.08–2.93] |
| rs1128503 | 7q21.12 | 87,550,285 | ABCB1/MDR1 | 0.38 | 0.33–0.42 | 0.31↓ | 0.13 | 0.41↑ | 0.36↓ | 0.26 | 1.23 [0.85–1.77] |
| rs854560 (PON1 p.L55 M)f | 7q21.3 | 95,316,772 | PON1 | 0.29 | 0.25–0.33 | 0.30≈ | 0.98 | 0.27↓ | 0.30≈ | 0.46 | 0.76 [0.51–1.14] |
| rs2254638 | 21q21.3 | 28,883,961 | N6AMT1 | 0.19 | 0.15–0.23 | 0.23↑ | 0.33 | 0.18≈ | 0.20≈ | 0.57 | 0.87 [0.55–1.38] |
| rs17878459 (CYP2C19*2B) | 10q23.33 | 94,775,165 | CYP2C19 | 0.01 | 0.003–0.02 | 0.01≈ | 0.99 | <0.01≈ | 0.01≈ | 0.91 | 0.99 [0.18–5.47] |
| rs12041331 | 1q23.1 | 156,899,922 | PEAR1 | 0.22 | 0.18–0.26 | 0.34↑ | 0.009 | 0.19↓ | 0.23≈ | 0.17 | 0.79 [0.51–1.22] |
| rs35464072 | 4q31 | 149,326,236 | NR3C2 | 0.41 | 0.36–0.45 | 0.31↓ | 0.032 | 0.41≈ | 0.41≈ | 0.99 | 0.98 [0.69–1.41] |
| rs1057910 (CYP2C9*3) | 10q23.33 | 94,981,296 | CYP2C9 | 0.06 | 0.04–0.08 | 0.03↓ | 0.10 | 0.06≈ | 0.06≈ | 0.99 | 0.99 [0.47–2.09] |
| rs762551 (CYP1A2*1F) | 15q24.1 | 74,749,576 | CYP1A2 | 0.29 | 0.25–0.33 | 0.36↑ | 0.14 | 0.26↓ | 0.29≈ | 0.45 | 0.85 [0.57–1.27] |
| rs5918 (A2 allele) | 17q21.32 | 47,283,364 | ITGB3 | 0.13 | 0.10–0.16 | 0.17↑ | 0.27 | 0.14≈ | 0.12≈ | 0.51 | 1.20 [0.71–2.02] |
Abbreviations: Chr., chromosome; CI, confidence interval; LD, linkage disequilibrium; MAF, minor allele frequency; ND, non detected; ORs, odds ratios; PRU, P2Y12 reaction unit; SNP, single‐nucleotide polymorphism.
aThe rs numbers are the accession numbers in the National Center for Biotechnology Information (NCBI) SNP database, dbSNP. Chr: chromosome location; Position denoted by using the human genome assembly/reference GRCh38 coordinate system.
bMAF among individuals with YRI proportion ≥0.25.
cThe p values for the comparison between overall and African‐related MAFs.
dThe p values for the comparison between resistant and sensitive MAFs. The p values were calculated with the use of two proportions z‐test analysis. ↑, ↓, or ≈ denotes increased, decreased or similar MAFs as compared to the corresponding overall MAFs in the entire study cohort.
eThe ORs and their corresponding 95% CIs for the association with resistance to clopidogrel (PRU ≥ 208) using a Yates‐corrected χ2 test.
fThese two pair of SNPs are in strong LD with each other at their corresponding loci (i.e., CYP2C in chr10 and PON1 in chr7, respectively). In phase alleles are TT/CA on PON1 and AA/GG on the CYP2C cluster (see Supplementary Material for details on these LD analyses).
Associations with platelet reactivity
The stepwise multivariate linear regression model was significant (F(10,198) = 4.79, p < 0.001, R 2 = 0.19), indicating that 19% of PRU variability is accounted for by these predictors (Table 3). Gender was found to be a variable that significantly predicted PRU values in Caribbean Hispanics (p = 0.04). In our findings, men had a decreased mean value of PRU by 17.9 units on average versus women. As expected, the rs4244285 SNP (CYP2C19*2) was found to be a highly significant predictor of platelet reactivity to clopidogrel (p < 0.001). Carriers of a single copy of the rs4244285‐A variant increased the mean PRU value by 40.5 units on average when compared to wild‐type individuals; whereas, homozygous carriers of this allele had a mean PRU increase of 89.3 units. In our study cohort, this variant accounted for ~ 7% of observed PRU variation. In addition, the PON1 rs662, ABCB1/MDR1 rs2032582, and PEAR1 rs12041331 carrier status significantly predicted PRU values in participants (p ≤ 0.05). Compared with wild type patients, carriers of rs662 and rs2032582 increased the corresponding PRU value by 19.4 and 31.9 units on average, respectively. In contrast, carriers of the PEAR1 rs12041331 variant had a decrease in mean PRU by 19.3 units. Surprisingly, African ancestry was a slight but still significant predictor of PRU (p = 0.047). An interaction component (i.e., African ancestry to rs12041331 carrier) was also added to the final model (B = −83.67; p = 0.05), suggesting that the possible effect of this PEAR1 variant on platelet function is likely driven by African heritage. In fact, a significantly higher allele frequency was observed among patients with a relatively large African contribution as compared to the overall MAF (i.e., 0.34 vs. 0.22; p = 0.009; Table 2). Finally, as expected, diabetes also predicted platelet function in participants (p = 0.03), increasing the mean value of PRU by 19.8 units on average.
TABLE 3.
Results for multivariate linear regression with different variables predicting PRU values in Caribbean Hispanics on clopidogrel
| Variable | B | SE | 95% CI | β | t | p value |
|---|---|---|---|---|---|---|
| (Intercept) | 118.70 | 14.60 | [89.90, 147.50] | 0.00 | 8.13 | <0.001 |
| Diabetes | 19.81 | 9.10 | [1.86, 37.76] | 0.15 | 2.18 | 0.031 |
| Gender (M) | −17.87 | 8.63 | [−34.88, −0.85] | −0.13 | −2.07 | 0.040 |
| African ancestry (YRI) | −68.54 | 34.27 | [−136.11, −0.97] | −0.14 | −2.00 | 0.047 |
| rs662 carriers | 19.37 | 9.41 | [0.82, 37.92] | 0.13 | 2.06 | 0.041 |
| rs2032582 carriers | 31.90 | 14.25 | [3.79, 60.01] | 0.15 | 2.24 | 0.026 |
| rs12041331 carriers | −19.37 | 9.87 | [−38.83, 0.09] | −0.14 | −1.96 | 0.051 |
| rs4244285 single carrier | 40.54 | 11.05 | [18.76, 62.33] | 0.24 | 3.67 | <0.001 |
| rs4244285 double carrier | 89.27 | 36.63 | [17.03, 161.52] | 0.16 | 2.44 | 0.016 |
| African ancestry: rs12041331 | −83.67 | 42.28 | [−167.04, −0.30] | −0.19 | −1.98 | 0.049 |
F(10,198) = 4.79, p < 0.001, R 2 = 0.19.
Unstandardized Regression Equation: PRU = 118.70 + 19.81 * [Diabetes] − 17.87 * [Gender: males] − 68.54 * [African Ancestry status] + 19.37 * [rs662 carrier] + 31.90 * [rs2032582 carrier] − 19.37 * [rs12041331 carrier] + 40.54 * [rs4244285 single carrier] + 89.27 * [rs4244285 double carrier] − 83.67 * [African Ancestry: rs12041331 interaction].
Abbreviations: CI, confidence interval; PRU, P2Y12 reaction unit.
Figure 3 shows the results of both the Kruskal‐Wallis and the two‐tailed Mann‐Whitney U tests for assessing the relationship between median PRUs and either the number of risk alleles at CYP2C19 rs4244285 and rs12777823 or the carrier status at PEAR1 rs12041331, PON1 rs662, and ABCB1 rs2032582, respectively. Statistically significant associations were found in the performed analyses (p < 0.01), suggesting that significant differences in PRU distribution were found across all occurring genotypes at each loci regardless of the direction of the relationship (Supplementary Material). No significant difference in median PRU values between wild‐type and carriers of the other tested SNPs were observed (p > 0.05). Furthermore, the rs4244285 and rs662 carrier status, patient age, gender (male), and diabetes were also found to be significant predictors of clopidogrel resistance (PRU > 208) in a binary logistic regression analysis (Supplementary Material).
FIGURE 3.
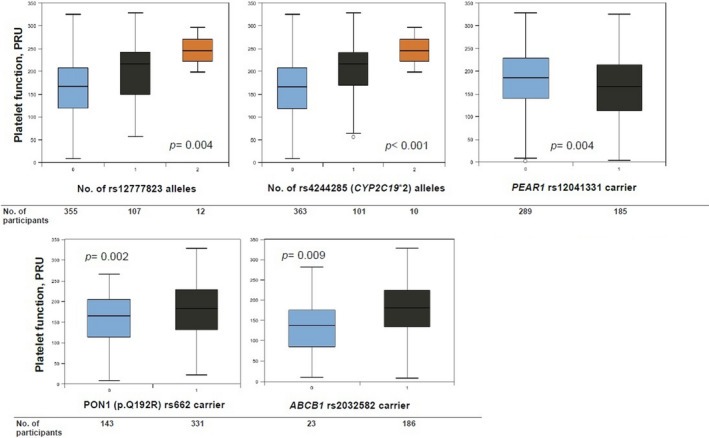
Boxplots of P2Y12 reaction unit (PRU) values (VerifyNow) by genotyping or carrier status to illustrate the association of different tested genetic variants with platelet function measures post clopidogrel administration in participants of the study. Due to their low minor allele frequencies (MAFs), no homozygous for the rs12041331, rs662, and rs2032582 single‐nucleotide polymorphisms (SNPs) were identified
The wPGxRS was calculated by the sum of each individual effect size (i.e., β coefficients) of associated genetic signals in the linear regression model (i.e., PON1 rs662; ABCB1 rs2032582; PEAR1 rs12041331; and CYP2C19*2 rs4244285 variants) multiplied by the number of risk alleles (0, 1, and 2) at such loci, and adjusted by the corresponding ancestral proportions. A significant positive correlation was observed between wPGxRS and PRUs (r p = 0.35, p < 0.001, 95% CI: 0.22–0.46). The correlation coefficient between wPGxRS and PRUs was 0.35, indicating a moderate effect size (Figure 4, left panel). We also assessed the impact of carrying multiple risk alleles on PRU (Figure 4, right panel). Specifically, we tested the four SNPs that were found to be significantly associated with platelet reactivity in the regression analysis to derive the wPGxRS, after adjustment for covariates, plus the rs12777823 variant. Each allele that correlated with PRU was coded as a “risk” allele. Collectively, these SNPs accounted for ~ 12% of the variation in platelet reactivity (F(5,205) = 5.79, p < 0.001, R 2 = 0.12).
FIGURE 4.
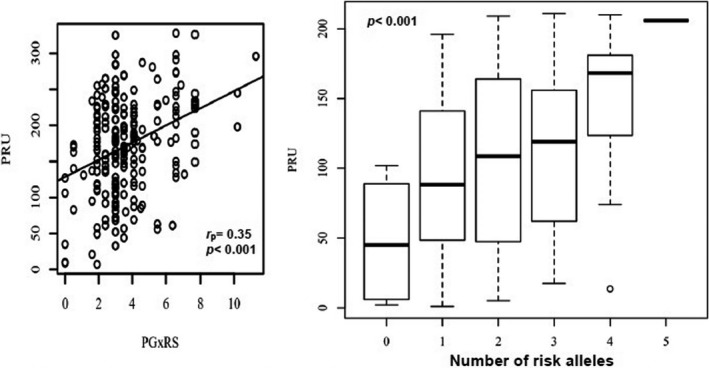
Scatterplot between the weighted polygenic risk score (wPG × Rs) and P2Y12 reaction units (PRUs) with the regression line added for the Pearson’s correlation analysis (left panel) and PRU changes based on increasing number of risk alleles at genetic loci of predictive value for clopidogrel responsiveness in Caribbean Hispanics (right panel). Note: For each box plot, the horizontal line within each box indicates the median; the top and bottom borders of each box indicate the interquartile range. The whiskers extending from each box indicate the 95% confidence interval (CI) and the points beyond the whiskers indicate outliers beyond ±2.5% CI. PG × RS, weighted polygenic risk score
The two‐tailed Wilcoxon signed rank test was statistically significant (V = 166.00, z = −2.97, p = 0.003) for the comparison of the wPGxRS between nonresponders and responders. The median wPGxRS among responders (μ = 3.00) was significantly lower than the corresponding median value among nonresponders (μ = 3.50), underscoring the potential utility of the multigene‐based wPGxRS as a predictor of clopidogrel resistance among Caribbean Hispanics.
Associations with MACEs
Finally, we also assessed the effect of carrying multiple risk alleles on the occurrence of cardiovascular events in a subset of our study cohort (n = 305). Results in Table 4 were significant for the association between an increasing number of risk alleles and MACEs (OR: 8.17, p = 0.04). No significant effects were observed for the odds of having a single cardiovascular event in the study cohort.
TABLE 4.
Effect of cumulative number of risk alleles associated with platelet reactivity (PRU) on the occurrence of cardiovascular end points in the study cohort
| Outcomes | adjusted ORs | 95% CI | p value |
|---|---|---|---|
| MACE (n = 42/305) | 8.17 | [1.30, 51.4] | 0.041 |
| MI (n = 19/305) | 1.27 | [0.70, 2.28] | 0.431 |
| Stent thrombosis (n = 15/305) | 1.09 | [0.62, 2.77] | 0.551 |
| Bleeding (n = 77/474) | 0.37 | [0.069, 1.91] | 0.297 |
The ORs were calculated using Fisher’s exact probability test by comparing patients who carried three or more risk alleles associated with increased platelet reactivity with patients who carried two or fewer of these alleles. ORs were adjusted by critical covariates (e.g., sex, coronary artery stenting, and diabetes). Analyses were conducted in the subset of participants in which clinical event data were available.
Abbreviations: CI, confidence interval; MI, myocardial infarction; MACE, major adverse cardiovascular events; OR, odds ratio; PRU, P2Y12 reaction unit.
Italicized p‐values indicate statistical significance.
DISCUSSION
Our study sought to assess the role of candidate genes and variants implicated in clopidogrel response among Caribbean Hispanic patients with ACS/CAD with or without PCI, as antiplatelet therapy with clopidogrel is still widely used in this under‐represented population. Importantly, our results are consistent with previous association studies with largely European patient populations but extend these findings by identifying novel associations and effect sizes with on‐treatment platelet reactivity in Caribbean Hispanics. Moreover, our candidate gene approach also identified a novel pharmacogenomic polygenic risk score that can effectively distinguish clopidogrel responders and nonresponders in this historically under‐represented patient population.
Participants in our study cohort were on‐average older than those in an International Clopidogrel Pharmacogenomics Consortium (ICPC)‐led genomewide association study (GWAS) of platelet reactivity and cardiovascular response to clopidogrel (i.e., 68.01 vs. 64.6 years old, respectively). 6 However, our patients were also more admixed and genetically heterogeneous than those enrolled in the ICPC study (all self‐reported Whites of European ancestry). Although the relative proportion of men in both studies was, as expected, greater than women (i.e., 77% in the ICPC cohort and 55% in our cohort), gender was more balanced in our study (Table 1). Compared to the ICPC study, a larger proportion of patients from our study had comorbidities that are known to be risk factors for MACEs (i.e., type‐2 diabetes: 55% vs. 24.7% and hypercholesterolemia: 71.9% vs. 64.5%, respectively). 6 Finally, indications for clopidogrel use were similar between both studies, with ACS plus CAD accounting for prescriptions in most participants (72% vs. 97%). Overall, our study sample was smaller but a more diverse group than that from the ICPC study.
The estimated allele frequencies of CYP2C19*2 (12.8%), CYP2C19*17 (15.3%), and PON1 p.Q192R (45.6%) were comparable to those earlier reported by our group in both a previous association study in Puerto Ricans 15 and a secondary analysis of prevalence in 1863 Puerto Ricans from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) cohort, 16 although a slightly lower than expected prevalence of the CYP2C19*2 variant was observed in the current cohort. Approximately 25% of participants carried at least one copy of both the rs12777823 A and CYP2C19*2 (rs4244285 A) alleles, which were the two variants with the largest effect on PRU. This is because these two SNPs on chromosome 10q24 are inherited together and, therefore, in strong LD within our study population (D’: 0.91; R 2: 0.79; χ2: 165.5; p < 0.0001). Notably, about 97% of participants had at least one allelic variant at any of these seven loci, and ~ 42% were carriers of three or more alleles, highlighting the rich repertoire of unique allelic combinations within admixed Caribbean Hispanics.
We found a strong association between rs4244285 and platelet reactivity in Caribbean Hispanics (Tables 2 and 3, Figure 3). The rs4244285 SNP in the CYP2C19 gene, which defines the *2 haplotype, is a synonymous transition in exon 5 (NM_000769.1: c.681G>A, p. Pro227=) that creates an aberrant splice site. This splicing defect alters the mRNA reading frame, by causing a premature stop codon downstream that results in a truncated, nonfunctional protein responsible for its null enzymatic activity. 17 More important, the presence of this nonfunctioning CYP2C19*2 variant correlates with a PM status for several commonly prescribed drugs, including the prodrug clopidogrel. Consequently, a resistant phenotype to antiplatelet therapy with clopidogrel is genetically inferred in those patients who are carriers of the rs4244285‐A risk allele. Notably, this SNP has been found to be significantly associated with lower exposure to the active thiol metabolite in patients treated with clopidogrel, with a diminished platelet responsiveness to clopidogrel ex vivo and a higher rate of MACEs in different studies worldwide. 5 , 6 Furthermore, GWAS for platelet reactivity revealed a strong association for the CYP2C19*2 signal, accounting for most of all the relationship with diminished responsiveness to clopidogrel in both healthy volunteers and patients (i.e., p = 1.5·10−13 in the PAPI study and p = 1.7·10−33 in the ICPC study, respectively). 2 , 6
The vast majority of these prior reports were based on studies conducted in individuals of mostly European ancestry, and Asians to a lesser extent. This study confirmed our preliminary report of a strong association between CYP2C19*2 and ADP‐stimulated platelet reactivity in a cohort of admixed Caribbean Hispanic patients on clopidogrel. 15 Accordingly, our findings provide further evidence in support of a global pharmacogenetic‐guided antiplatelet approach to tailoring DAPT that is based on CYP2C19 genotyping. However, a polygenic risk score‐driven approach improved the predictability of responsiveness to clopidogrel and risk of resistance that may ultimately facilitate DAPT optimization in Caribbean Hispanics.
Carriers of ABCB1 rs2032582 were also correlated with decreased response to clopidogrel due to a higher‐than‐expected platelet function (p < 0.05 vs. noncarriers). This is a nonsynonymous SNP located in exon 21 of the ABCB1 gene, also known as G2677T (p. Ala893Thr), which has been reported as a predictor of lipid‐lowering response. 18 This polymorphism seems to correlate with the well‐known and more commonly reported rs1045642 variant, also known as ABCB1 C3435T. According to previous report, the ABCB1 C3435T plays an important role in intestinal absorption of clopidogrel that further affects the exposure to the active metabolite of clopidogrel and platelet aggregation. 19
The PON1 rs662 (p.Q129R) variant was also found to be associated with resistant status in our study cohort. This PON1 variant has been previously found to be significantly associated with clopidogrel resistance in our study population. 15 The failure by Verma et al. 6 to see signals for the ABCB1 and PON1 SNPs in their recent large GWAS on clopidogrel response seems at odds with what is reported here. However, we believe it is a direct consequence of the differences in genetic backgrounds and individual admixture proportions between our study cohort of Caribbean Hispanics with multiple ethno‐geographic architecture and that in the ICPC study (Whites and Europeans). It is clear that the effect sizes of certain SNPs of predictive value differ across different ethnicities. 20
There were no obvious effects of other studied genes on platelet reactivity to clopidogrel (see Supplementary Material), including the SNP defining the CYP2C19*17 increased function allele. This last finding provides further confirmatory evidence that even in Caribbean Hispanics CYP2C19*17 is not a true independent predictor of on‐treatment platelet reactivity, but rather dependent on rs4244285 genotype. Because rs12248560‐T allele seems to occur only when the rs4244285‐G allele is present, we might speculate that any potential association of rs12248560‐T allele (CYP2C19*17) with PRU could significantly be attenuated in patients who were also either homozygous or heterozygous for the major allele at rs4244285.
In our study cohort, we found that the minor “A” allele of rs12041331 was independently associated with significantly lower platelet aggregation (p = 0.004; Figure 3). PEAR1 (platelet endothelial aggregation receptor 1 gene, chromosome 1q23.1) encodes a platelet cell surface receptor that, upon activation, results in the amplification of the glycoprotein IIb/IIIa pathway and sustained platelet aggregation. 21 Prior studies have postulated that SNP rs12041331, located in the first intron of PEAR1, regulates expression of PEAR1 protein level in a dose‐dependent manner. The major G allele at rs12041331 has been linked to greater PEAR1 protein expression in human platelets.
The PEAR1 rs12041331 A‐allele has been previously found to be significantly associated with decreased agonist‐induced platelet aggregation in both European and African Americans, accounting for ~ 15% of total phenotypic variation in platelet function and providing a plausible biologic mechanism to explain the observed pharmacogenetic association (Figure 3). 22 , 23 , 24 A GWAS of platelet aggregation in African Americans confirmed the association of the rs12041331 intronic variant in PEAR1 with multi‐agonist‐induced aggregation. 22 However, this specific PEAR1 rs12041331 SNP was only replicated in European Americans for ADP‐induced aggregation and its impact on platelet response in Caribbean Hispanics receiving DAPT is yet to be determined. Accordingly, the observed association with PRU (i.e., ADP‐induced aggregation) in our study cohort of admixed Caribbean Hispanics, as well as its interaction with the African ancestry proportion of participants, is novel. Platelet hyperaggregability ex vivo is associated with greater risk of MACEs in cardiovascular patients receiving antiplatelet therapy for secondary prevention. 25 Likewise, it also correlates with lower CAD survival rates, mainly among African Americans, after adjusting for confounders. 26 Furthermore, platelet aggregation seems to show higher heritability in African than European descendants. Therefore, the genetics of this physiologically relevant biochemical process can be a key factor accounting for this racial difference.
For PEAR1, the literature also suggests a role in aspirin response. 27 , 28 , 29 Because a subset of participants were on aspirin as part of DAPT, an additional analysis was performed to examine whether aspirin is also playing a role in this association with on‐treatment platelet function (Supplementary Material). After filtering by aspirin use, the association analysis showed that the PRUs were significantly different between rs12041331‐A variant carriers and noncarriers, but in aspirin users only (p = 0.004). No significant differences were observed among patients who do not use aspirin (p = 0.42). Based on this finding, it seems that the effect of rs12041331 on platelet function was linked in part to the use of aspirin. However, all aspirin users in our study cohort were also receiving clopidogrel. Therefore, an impact of this variant on clopidogrel response cannot be completely discarded. 30 Indeed, carriers of the rs12041331‐A allele had an altered response to clopidogrel in a previous clinical trial. 31 Besides, the interaction between aspirin use and rs12041331 carrier status did not have a significant effect on PRU (p = 0.47; see Supplementary Material). Accordingly, the use of aspirin does not seem to significantly alter the relationship between rs12041331 SNP and PRU.
Our novel polygenic risk score (wPGxRS) revealed that patients who carry an increasing number of risk alleles that are independently associated with PRU measures were more likely to experience HTPR compared to those who do not (p < 0.001). Consistent with these findings, when comparing patients by the number of risk alleles, patients who carried three or more alleles associated with platelet reactivity were more likely to experience higher PRU compared with patients who carried two or fewer of these alleles. We also found that carrying multiple risk alleles significantly increased the odds of experiencing a composite of cardiovascular events (OR: 8.17, 95% CI: 1.30, 51.4; p = 0.04) in our study subcohort (Table 4). Similarly, a group of investigators from the ICPC also found no significant association of any individual risk allele across 31 candidate loci with cardiovascular events in a cohort of patients from 13 countries (n = 2134, all of European descent), but the generation of a pharmacogenomic polygenic response score by this group revealed that patients who carried a greater number of alleles previously found associated with increased on‐treatment platelet reactivity were more likely to experience both cardiovascular events (p = 0.01) and cardiovascular‐related death (p = 0.007). 32 The clinical implications of such enhanced risk of MACEs in patients who carried multiple risk alleles in our cohort is noteworthy, suggesting that a polygenic risk approach for the clinical management of these Caribbean Hispanic patients on DAPT not only improve predictability of responsiveness to clopidogrel but also of the risk of MACEs and, therefore, should be a critical component of any future strategies for prevention of poor outcomes at clinical settings.
Overall, we were able to find significant pharmacogenetic associations with on‐clopidogrel platelet reactivity in Caribbean Hispanics among the predefined candidate loci. The effect sizes of these significantly associated loci could likely be a direct consequence of the relative percentage of heritability explained by them combined in this population and the well‐defined biological nature of the PRU phenotype. In fact, Shuldiner et al. have previously established that platelet response to clopidogrel is highly heritable (h(2) = 0.73; p < 0.001). 2
In conclusion, the results of this study represent the largest pharmacogenomic investigation to date of Caribbean Hispanic patients treated with clopidogrel, which is notable as there is an urgent need to increase representation of diverse populations in PGx studies. 33 , 34 , 35 , 36 Such a European‐centric bias of earlier genetic studies, if not properly mitigated, will limit our understanding of underlying determinants of poor clopidogrel response among Caribbean Hispanics. We strongly believe pharmacogenomics can bring cardiovascular health equity to our medically underserved populations by fostering efforts to achieve proper representation of the true human diversity in clinical protocols. The current paucity of data from individuals of non‐European ancestry will preclude any further extrapolation of derived prediction models to the population at large. Consequently, findings from this work are expected to help in part by addressing such an unmet need and, hence, avoiding the potential harm of extrapolating genetic results from one population to another.
CONFLICTS OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
J.D., E.S., D.F.H.‐S., K.M., S.A.S., and G.R. wrote the manuscript. J.D., D.F.H.‐S., K.M., S.A.S., and G.R. designed the research. J.D., E.S., M.M., A.L.‐R., M.R., J.Y.R., P.G., L.I.F.‐M., L.A.V.‐F., O.A., F.M.‐M., and H.N. performed the research. J.D., E.S., M.M., and P.G. analyzed the data.
DISCLAIMER
The authors are solely responsible for the design and conduct of this study, all study analyses, the drafting and editing of the paper, and its final contents. The contents of this manuscript do not represent the views of the National Institutes of Health (NIH) or the United States Government. No funded writing assistance was utilized in the production of this manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the patients for voluntarily participating in this study protocol. A special acknowledgement to the Research Design and Biostatistics Core service of The Alliance, formerly the Puerto Rico Clinical and Translational Research Consortium (PRCTRC), supported by the NIMHD, and the National Institute of Allergy and Infectious Diseases (NIAID) under award #U54MD007587, for helping us with study design and sample size calculations. Additionally, we also want to thank Drs. Abiel Roche‐Lima and Kelvin Carrasquillo from the Bioinformatics Core of the CCRHD‐RCMI Program at the UPR‐MSC, for their assistance with admixture and ancestry measurements. Mr. Mohan Kocherla, MS, MIS, MBA, and his staff at the CLIA‐certified laboratory DNA MedTuning (Center for DNA‐Guided Medicine; Hartford, CT) for running whole‐genome assays of specimens from participants.
Duconge J, Santiago E, Hernandez‐Suarez DF, et al. Pharmacogenomic polygenic risk score for clopidogrel responsiveness among Caribbean Hispanics: A candidate gene approach. Clin Transl Sci. 2021;14:2254–2266. 10.1111/cts.13124
Funding information
This work was supported in part by CCRHD‐RCMI grant #2U54 MD007600‐34 from the National Institute on Minority Health and Health Disparities (NIMHD) of the National Institutes of Health and by the National Institute of General Medical Sciences (NIGMS)‐Research Training Initiative for Student Enhancement (RISE) Program grant R25 GM061838.
Footnotes
Italicized p‐values indicate statistical significance.
REFERENCES
- 1. Simon T, Verstuyft C, Mary‐Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363‐375. 10.1056/NEJMoa0808227 [DOI] [PubMed] [Google Scholar]
- 2. Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2C19 loss‐of‐function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108(7):2244‐2247. [DOI] [PubMed] [Google Scholar]
- 4. Mega JL, Simon T, Anderson JL, et al. CYP2C19 genetic variants and clinical outcomes with clopidogrel: a collaborative meta‐analysis. Circulation. 2009;120(18):S598‐S599. [Google Scholar]
- 5. Mega JL, Close SL, Wiviott SD, et al. Cytochrome p‐450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354‐362. [DOI] [PubMed] [Google Scholar]
- 6. Verma SS, Bergmeijer TO, Gong LI, et al. Genomewide association study of platelet reactivity and cardiovascular response in patients treated with clopidogrel: a study by the international clopidogrel pharmacogenomics consortium. Clin Pharmacol Ther. 2020;108(5):1067‐1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177(1):26‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levine GN, Bates ER, Blankenship JC, et al. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation. 2011;124(23):e574‐e651. [DOI] [PubMed] [Google Scholar]
- 9. Scott SA, Sangkuhl K, Stein CM, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Claassens DMF, Vos GJA, Bergmeijer TO, et al. A genotype‐guided strategy for oral P2Y12 inhibitors in primary PCI. N Engl J Med. 2019;381(17):1621‐1631. [DOI] [PubMed] [Google Scholar]
- 11. Klein MD, Williams AK, Lee CR, Stouffer GA. Clinical utility of CYP2C19 genotyping to guide antiplatelet therapy in patients with an acute coronary syndrome or undergoing percutaneous coronary intervention. Arterioscler Thromb Vasc Biol. 2019;39(4):647‐652. [DOI] [PubMed] [Google Scholar]
- 12. Duconge J, Ruaño G. Admixture and ethno‐specific alleles: missing links for global pharmacogenomics. Pharmacogenomics. 2016;17(14):1479‐1482. [DOI] [PubMed] [Google Scholar]
- 13. Duconge J, Ruaño G. Preventing the exacerbation of health disparities by iatrogenic pharmacogenomic applications: lessons from warfarin. Pharmacogenomics. 2018;19(11):875‐881. [DOI] [PubMed] [Google Scholar]
- 14. Aradi D, Kirtane A, Bonello L, et al. Bleeding and stent thrombosis on P2Y12‐inhibitors: collaborative analysis on the role of platelet reactivity for risk stratification after percutaneous coronary intervention. Eur Heart J. 2015;36(27):1762‐1771. [DOI] [PubMed] [Google Scholar]
- 15. Hernandez‐Suarez DF, Mariana MR, Scott SA, et al. Pharmacogenetic association study on clopidogrel response in Puerto Rican Hispanics with cardiovascular disease: a novel characterization of a Caribbean population. Pharmgenomics Pers Med. 2018;11:95‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Melin K, Moon J‐Y, Qi Q, et al. Prevalence of pharmacogenomic variants affecting the efficacy of clopidogrel therapy in the Hispanic Community Health Study/Study of Latinos cohort. Pharmacogenomics. 2019;20(2):75‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S‐mephenytoin metabolism in humans. J Biol Chem. 1994;269(22):15419‐15422. [PubMed] [Google Scholar]
- 18. Thompson JF, Man M, Johnson KJ, et al. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 2005;5(6):352‐358. [DOI] [PubMed] [Google Scholar]
- 19. Wang XQ, Shen CL, Wang BN, Huang XH, Hu ZL, Li J. Genetic polymorphisms of CYP2C19*2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene. 2015;558(2):200‐207. [DOI] [PubMed] [Google Scholar]
- 20. Shah RR, Gaedigk A. Precision medicine: does ethnicity information complement genotype‐based prescribing decisions? Ther Adv Drug Saf. 2018;9(1):45‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nanda N, Bao M, Lin H, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat‐containing transmembrane receptor, participates in platelet contact‐induced activation. J Biol Chem. 2005;280(26):24680‐24689. [DOI] [PubMed] [Google Scholar]
- 22. Qayyum R, Becker LC, Becker DM, et al. Genome‐wide association study of platelet aggregation in African Americans. BMC Genet. 2015;16:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson AD, Yanek LR, Chen M‐H, et al. Genome‐wide meta‐analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42(7):608‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faraday N, Yanek LR, Yang XP, et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood. 2011;118(12):3367‐3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Breet NJ, van Werkum JW, Bouman HJ, et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA. 2010;303(8):754‐762. [DOI] [PubMed] [Google Scholar]
- 26. Thomas KL, Honeycutt E, Shaw LK, Peterson ED. Racial differences in long‐term survival among patients with coronary artery disease. Am Heart J. 2010;160(4):744‐751. [DOI] [PubMed] [Google Scholar]
- 27. Würtz M, Nissen PH, Grove EL, et al. Genetic determinants of on‐aspirin platelet reactivity: focus on the influence of PEAR1. PLoS One. 2014;9(10):e111816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewis JP, Riaz M, Xie S, et al. Genetic variation in PEAR1, cardiovascular outcomes and effect of aspirin in a healthy elderly population. Clin Pharmacol Ther. 2020;108(6):1289‐1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herrera‐Galeano JE, Becker DM, Wilson AF, et al. A novel variant in the platelet endothelial aggregation receptor‐1 gene is associated with increased platelet agreeability. Arterioscler Thromb Vasc Biol. 2008;28:1484‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis JP, Backman JD, Reny J‐L, et al. Pharmacogenomic polygenic response score predicts ischemic events and cardiovascular mortality in clopidogrel‐treated patients. Eur Heart J Cardiovasc Pharmacother. 2020;6(4):203‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu K, Ye S, Zhang S, et al. Impact of platelet endothelial aggregation receptor‐1 genotypes on platelet reactivity and early cardiovascular outcomes in patients undergoing percutaneous coronary intervention and treated with aspirin and clopidogrel. Circ Cardiovasc Interv. 2019;12:e007019. [DOI] [PubMed] [Google Scholar]
- 32. Lewis JP, Backman JD, Reny J‐L, et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel‐treated patients. Eur Heart J Cardiovasc Pharmacother. 2020;6(4):203‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pendyala LK, Torguson R, Loh JP, et al. Racial disparity with on‐treatment platelet reactivity in patients undergoing percutaneous coronary intervention. Am Heart J. 2013;166(2):266‐272. [DOI] [PubMed] [Google Scholar]
- 34. Gad MM, Elgendy IY, Mahmoud AN, et al. Disparities in cardiovascular disease outcomes among pregnant and post‐partum women. J Am Heart Assoc. 2021;10(1):e017832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breathett K, Spatz ES, Kramer DB, et al. The groundwater of racial and ethnic disparities research. Circ Cardiovasc Qual Outcomes. 2021;14(2):135‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Borrell LN, Elhawary JR, Fuentes‐Afflick E, et al. Race and genetic ancestry in medicine ‐ a time for reckoning with racism. N Engl J Med. 2021;384(5):474‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morisky DE, Green LW, Levine DM. Concurrent and predictive validity of a self‐reported measure of medication adherence. Med Care. 1986;24:67‐74. [DOI] [PubMed] [Google Scholar]
- 38. Thygesen K, Alpert JS, Jaffe AS, et al. Third universal definition of myocardial infarction. Circulation. 2012;126:2020‐2035. [DOI] [PubMed] [Google Scholar]
- 39. Cutlip DE, Windecker S, Mehran R, et al. Clinical end points in coronary stent trials. Circulation. 2007;115:2344‐2351. [DOI] [PubMed] [Google Scholar]
- 40. Hernandez‐Suarez DF, Melin K, Marin‐Maldonado F, et al. Implementing a pharmacogenetic‐driven algorithm to guide dual antiplatelet therapy (DAPT) in Caribbean Hispanics: protocol for a non‐randomised clinical trial. BMJ Open. 2020;10:e038936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li HE, Zhang Y‐J, Li M‐P, et al. Association of N6AMT1 rs2254638 polymorphism with clopidogrel response in Chinese patients with coronary artery disease. Front. Pharmacol. 2018;9:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duconge J, Duconge J, Santiago‐Cartagena E, Monero‐Paredes M, Martienz‐Jimenez JE. Pharmacogenomic study of clopidogrel in Caribbean Hispanics. (2021), Mendeley Data, version 1. 10.17632/5t3d5wtvm3.1 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


