Abstract
Since the publication of the Human Genome Project, genetic information has been used as an accepted, evidence‐based biomarker to optimize patient care through the delivery of precision health. Pharmacogenetics (PGx) uses information about genes that encode proteins involved in pharmacokinetics, pharmacodynamics, and hypersensitivity reactions to guide clinical decision making to optimize medication therapy selection. Clinical PGx implementation is growing from the dramatic increase in PGx studies over the last decade. However, an overwhelming lack of genetic diversity in current PGx studies is evident. This lack of diverse representation in PGx studies will impede equitable clinical implementation through potentially inappropriate application of gene‐based dosing algorithms, whereas representing a missed opportunity for identification of population specific single nucleotide variants and alleles. In this review, we discuss the challenges of studying PGx in under‐represented populations, highlight two successful PGx studies conducted in non‐European populations, and propose a path forward through community‐based participatory research for equitable PGx research and clinical translation.
INTRODUCTION
Translation of genetic research into clinical practice is currently being implemented as precision health, while the race toward full clinical implementation across practice settings is expanding beyond academic‐based health institutions. When available, genetic information is being used as an accepted, evidence‐based biomarker to optimize care that is quickly gaining support and interest from patients, providers, and payers. When genetic information is not available, race, ethnicity, and family history serve as clinical proxies. In this review, we use the term “European” to represent the biogeographical ancestry group that includes populations primarily of European descent, including European Americans and that could be referenced elsewhere as “White” or “Caucasian.” 1 Due to the inherent bias and complexities of overgeneralization of racial categorization, and the intricacies of using the terms “race,” “ethnicity,” and “genetic ancestry, we use the terms “genetic ancestry” and “genetic ancestry group” to describe the population from which the individual’s recent biological ancestors originated. 1 , 2
Genetics influence an individual’s susceptibility to certain disease states but can also contribute to the wide variability observed with medication response. Pharmacogenetics (PGx) uses information about genes that encode proteins involved in pharmacokinetics, pharmacodynamics, and hypersensitivity reactions to guide clinical decision making to optimize medication therapy selection. Using PGx information to guide clinical decisions parallels the use of other clinical information, similar to how liver and kidney function guides medication therapy decisions. For example, knowing the presence of a CYP2C19 loss of function allele, such as *2 or *3, can help guide antiplatelet therapy decisions as the prodrug clopidogrel requires bioactivation to the active metabolite predominantly by CYP2C19. 3 However, response variability observed from medications due to underlying genetic differences can vary between genetic ancestry groups. Allele frequencies in pharmacogenes differ across genetic ancestry groups and can even differ between subgroups of a specific population, as evidenced by variation in the CYP2C19 *2 allele which varies in frequency from 5.7% to 49.4% within Asian ancestry. 4
With allele frequencies differing across genetic ancestry groups, identification of variants in pharmacogenes that are clinically relevant for that population presents challenges. 4 Despite the relative infancy of the genomic era in healthcare, the outsized influence of European‐based research is apparent. A recent review of genomewide association studies (GWAS) found that 78% of individuals included were of European descent with a small percentage representing Asian, African, and Hispanics, and less than 1% representing all other ethnicities. 5 This review concluded that the bias of European‐based genetic research translated to a non‐European population can result in heterogeneous treatment outcomes. 5
Several reviews have discussed the lack of under‐represented populations in PGx studies, 6 , 7 , 8 , 9 , 10 , 11 all of which note an overwhelming lack of genetic diversity that will impede equitable clinical implementation through inappropriate application of gene‐based dosing algorithms and by missed opportunities for identification of population‐specific single nucleotide variants and alleles. Systematic reviews in Africans, North American Indigenous populations, US Hispanics, Asians, Mexicans, and Brazilians share several common themes, including that there are existing differences in allele frequencies across races in common pharmacogenes, application of European‐based PGx to other races may unintentionally result in heterogeneous clinical outcomes, and that non‐European ethnicities must be represented in PGx studies both for discovery of novel variants and to guide clinical implications through population specific PGx tests and dosing algorithms. 6 , 7 , 8 , 9 , 10 , 11
As precision health is translated from research into clinical practice, the question is no longer if using genetic information will become standard of care, but rather who it will be the standard of care for. As PGx implementation progresses, non‐European populations are being left behind exacerbating existing disparity gaps. This review uses an equity lens, a process to analyze the impact of design on underserved populations to identify and mitigate barriers specific to PGx studies. In doing so, this review explores the challenges of studying PGx in under‐represented populations, highlights successful PGx studies conducted in non‐European populations, and proposes a path forward for equitable PGx research and clinical translation.
CHALLENGES OF PGx IN DIVERSE CLINICAL SETTINGS
Several challenges exist as it pertains to conducting research implementation of PGx in broad populations, including the context of what is considered diversity, the collaborative involvement and participatory research in populations rarely included in clinical research, and using PGx panels developed based on variants/alleles in Europeans in non‐European populations. To assess under‐representation in PGx studies, it is important to scrutinize the definition of under‐representation as it applies to genetic ancestry. The National Institutes of Health (NIH) defines Blacks or African Americans, Hispanics or Latinos, American Indians or Alaska Natives, and Native Hawaiians and other Pacific Islanders as under‐represented in health‐related sciences, while also acknowledging that under‐representation will vary depending on the setting. 12 Under‐representation defined as non‐European populations is insufficient as self‐identified ancestry has questionable reliability and the categories presented to patients and research participants is heterogeneous and inconsistent. Even within the NIH category of “White,” there are more than a dozen subgroup ethnic categories that show comparatively different allele frequencies for certain genes, as described by the Clinical Pharmacogenetics Implementation Consortium. 4
Previously, self‐reported ancestry was thought to be an accurate representation of genetic ancestry; however, it has recently become apparent that there is a discordance between genetic ancestry and self‐reported ancestry. In a previous study of over 3500 participants, 0.14% showed genetic ancestry differing from self‐reported ancestry. 13 However, the four categories used were generic representations of genetic ancestry and the specificity of self‐reported ancestry may be enhanced when participants are faced with a greater number of subgroups to choose from. Additionally, studies have shown that self‐reported African Americans, European Americans, and Latino populations can have different genetic ancestry, especially in an admixed population. 14
In addition to self‐reported ancestry being a controversial marker for genetic ancestry and a proxy for clinical decisions, it is further complicated by the number of choices or categories for genetic ancestry used by researchers and presented to research participants or patients. A recent study by Zhang et al., revealed a dearth of standardization in race and ethnicity categories used in research to categorize race, ethnicity, and genetic ancestry internationally. For example, they found that Malaysia used 24 different categories to classify the category “Asian,” whereas the United States used only three. 4 Different research settings may or may not have standards set by regulatory agencies, which can also compound the complexity of standardizing categorization. For example, in the United States, the NIH defined standards contain five racial categories (American Indian or Alaska Native, Asian, Black or African American, Native Hawaiian or Other Pacific Islander, and White) and two ethnicity categories (Hispanic or Latino and Not Hispanic or Latino) to be used in clinical and medical research. 12
The use of broad categories to capture genetic ancestry could lead to overgeneralization of subgroups resulting in inaccurate translation into clinical care. Similarly, oversimplifying genetic ancestry in studies that show differences in efficacy or adverse events without additional investigation of PGx contributions could be clinically detrimental when translated into practice. If the clinical outcome observed is due to a phenotype (i.e., a poor or ultra‐rapid metabolizer) and that phenotype is found in a subgroup that is driving the difference observed in the outcome, that could be true only for that specific subgroup and the overgeneralization of this outcome to the overall racial category could lead to inappropriate clinical decisions. Clinically, this contributes to the disparity gap as medications are selected, or avoided, based inappropriately on genetic ancestry rather than PGx phenotype. This disparity gap is exacerbated further when we clinically apply findings from a majority European PGx study to a non‐European population as alleles in linkage disequilibrium with alleles causing the clinical impact but not tested for, can also differ across populations. 5 However, in some cases, without broader categorization, substantially increased population sizes are needed to attain power to detect differences. This creates challenges in balancing the need for statistical power and replication studies with accuracy in self‐reported racial categorization.
The second barrier to enhancing inclusion in PGx studies is creating a sustainable, collaborative environment. Challenges in creating a collaborative research environment in any population include participant mistrust, lack of comfort with the research process, lack of information, time and resource constraints, and lack of awareness. 15 A recent study evaluating the reasons for participant enrollment refusal in African Americans revealed that mistrust of genetic research, a commonly cited barrier to research involvement, was only cited about 5% of the time and ranked below the participant not being interested in the research and other convenience factors, such as the time involvement and the site being too far for travel. 16 Another study noted differences in willingness to participate across race but showed that when genetic health‐related or genetic ancestry results were returned and discrimination issues (life and health insurance costs and employment) addressed, those differences were alleviated. 17
Last, a clinical barrier to under‐represented PGx is the application of PGx panels in non‐European populations. Analysis of GWAS studies informing PGx variants showed the majority of studies (52%) were conducted in European populations. 5 A similar analysis of PGx studies showed that the majority (53%, n = 102) were conducted in North America but only five were conducted with American Indian or Alaska Native populations and only six conducted with Hispanic/Latino populations. 18 When using a general PGx panel with the most commonly described variants and alleles in a PGx study in a largely unstudied population differences in allele frequencies can be revealed; however, it is a missed opportunity for identification of novel alleles with clinical impact. A review conducted in an African population on CYP2C9, CYP2C19, CYP2D6, and other CYPs revealed that of 74 PGx studies, only 16% (n = 12) used methodology to detect novel variants. 19
To highlight PGx studies in understudied populations and their methods, we briefly describe two well‐conducted studies in under‐represented populations. Additional PGx studies in under‐represented populations are included as a table in the Supplemental Material.
STUDY HIGHLIGHT: CYP2D6 ELUCIDATION IN THE AMERICAN INDIAN POPULATION
Fohner et al. used a community‐based participatory research (CBPR) approach to develop a partnership with the Confederated Salish and Kootenai Tribes (CSKT) community, and through this prioritized optimization of anticancer agents through PGx testing by focusing on CYP2D6 and tamoxifen. 20 One hundred eighty‐seven CSKT participants underwent CYP2D6 sequencing, resulting in 67 CYP2D6 variants identified, including nine novel variants. Additionally, novel variants are also described in CYP3A4, CYP3A5, and CYP2C9 (Table 1). Allele frequencies were similar to those seen in European‐observed frequencies, with the exception of CYP3A4, and differed from other North American indigenous populations. This study was designed to investigate CYP2D6 variation in an indigenous population previously under‐represented in PGx studies. In doing so, they collaborated with the CSKT community to determine prioritization of a PGx research tract with meaningful impact on tamoxifen optimization within the community. 20
TABLE 1.
Select studies revealing novel PGx variants discovered in diverse populations through sequencing methods
| Citation | Population | Sequencing/genotyping method | Gene | Haplotype | Novel variant rsID | Variant type | Minor allele frequency (%) | Potential medications impacted |
|---|---|---|---|---|---|---|---|---|
| Wright 2010 |
Xhosa n = 15 |
The CYP2D6 gene, upstream sequence and 3′‐UTR was sequenced with bidirectional sequencing with BigDye chemistry version 3.1. Genotyping was done by long‐range PCR, DNA sequencing, and multiplex single nucleotide primer extension analysis. |
CYP2D6 | *73 | rs267608308 | Missense | Sub‐Saharan Africa: 0.48 | TCAs, atomoxetine, opioids, 5‐HT3 antagonists, SSRIs, SNRIs, tamoxifen, antipsychotics |
| *74 | rs267608322 | Missense | Sub‐Saharan Africa: 0.48 | |||||
| Fohner 2013 |
Confederated Salish and Kootenai Tribes n = 187 |
CYP2D6 including exons, introns, upstream, and downstream was resequenced with conventional Sanger sequencing with BigDye chemistry. | CYP2D6 | *2 | rs567431353 | Upstream variant | 0.29 | |
| *1, *127, *141 | rs1269631565 | Intron variant | 0.28 | |||||
| *2 | rs572914357 | Upstream variant | 0.58 |
Abbreviations: PCR, polymerase chain reaction; PGx, pharmacogenetics; SNRI, serotonin‐norepinephrine reuptake inhibitor; SSRI, selective serotonin reuptake inhibitor; TCAs, tricyclic antidepressants.
STUDY HIGHLIGHT: CYP2D6 ELUCIDATION IN THE XHOSA POPULATION
Wright et al. conducted a PGx study in the Xhosa population in South Africa to optimize medication therapy for the treatment of schizophrenia. 21 The Xhosa people are under‐represented in research despite accounting for a large proportion of the South African population. The study recruited individuals of Xhosa ethnicity with written informed consent and institutional approval and was undertaken to elucidate variation in CYP2D6 within the Xhosa people to better guide medications that are CYP2D6 substrates used to treat schizophrenia, such as antipsychotics, including risperidone, aripiprazole, brexpiprazole, clozapine, perphenazine, thioridazine, and paliperidone. This study used two methods for CYP2D6 investigation, including CYP2D6 sequencing in a subgroup and using a CYP2D6 panel in another. CYP2D6 was sequenced in 15 individuals and CYP2D6 was genotyped for over 25 alleles in controls and individuals with schizophrenia using long‐range polymerase chain reaction (PCR), DNA sequencing and single nucleotide primer extension analysis. In total, 56 CYP2D6 variants were identified with allele frequencies unique to the Xhosa population, higher frequencies of *5 and *40, and differing from another South African population. Sequencing revealed two novel alleles in this population, *73 and *74 (Table 1). Notably, 12.5% of participants were either poor or ultrarapid CYP2D6 metabolizers. Overall, this study was designed to detect novel variants and establish allele frequencies in CYP2D6 in a diverse South African population. The clinical impact of this CYP2D6 investigation is important for treatment of schizophrenia within the Xhosa people with CYP2D6 substrates. 21
ELEMENTS OF SUCCESSFUL PGx STUDIES IN UNDER‐REPRESENTED POPULATIONS
The studies showcased above highlight two common themes for successful PGx research in under‐represented populations. One of the most resounding themes is establishing a collaborative environment for research in the population. Both studies worked with under‐represented populations to expand knowledge of a relevant pharmacogene, but importantly sought to optimize therapy for related medications and indications to improve outcomes that were meaningful to the population. Fohner et al. describes eliciting community buy‐in via the development of a Tribal Health and community advisory board that helped facilitate discussions with the community. 20 Although not explicitly described in the study highlighted, more in‐depth references on their collaborative partnership with the CSKT community are available and highlight the community as a stakeholder in the oversight of the project, with research objectives that focus on community health needs, bidirectional learning and communication, and including cultural competency training. 22 , 23 The second common theme for these studies was the sequencing of the pharmacogenes under review. Both studies identified novel variants within CYP2D6 and revealed a unique allele frequency distribution for the population when compared to other populations.
FUTURE DIRECTIONS
Designing PGx studies with an equity lens starts by acknowledging structural inequities and through CBPR efforts with the specific population. CBPR is defined as “a collaborative, action‐oriented research approach that seeks to address health disparities through aligning community members’ insider knowledge of their communities with academic researchers’ methodological expertise,” 24 and should be used to establish meaningful and lasting relationships within communities. With CBPR, mistrust within the community, as well as other reasons for not participating in research, such as general interest, knowledge, and convenience factors, can be addressed and overcome as barriers for participation in PGx research. Engaging a community in research in a positive manner may also inspire those within the community to pursue research as a career. Increased representation of these individuals could result in a positive feedback loop that further strengthens participation in research. The relationship established should also be sustained beyond a single study to continue to provide benefits for both the researcher and the population, with several examples of CBPR being used successfully within American Indian/Alaskan Native communities. 22 , 23 , 24 Carroll et al. provides eight recommendations based on CBPR principles as a framework for increasing American Indian representation in PGx research. 25 Importantly, they highlight building trust within the community, practicing cultural humility, providing resources and support within the community, and finding a balance between the realistic benefits to the community and the knowledge gained in pursuit of research. 25 Although their recommendations focus on the American Indian community, this framework can be adapted for use within other under‐represented and more heterogeneous populations (Figure 1).
FIGURE 1.
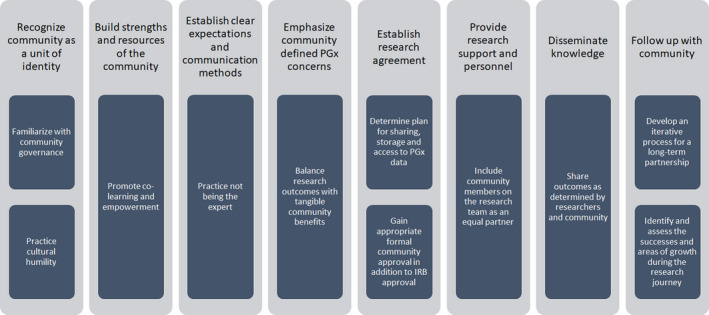
Community‐based participatory research (CBPR) framework for increasing diverse community representation in pharmacogenetics (PGx) studies. IRB, institutional review board.
While establishing a research relationship within a community, resources should be addressed. PGx studies done with predefined genotyped panels, whereas an attractive option due to widespread availability and lesser cost can provide useful information about how common pharmacogene alleles frequencies differ between populations; however, they do not provide insight on new variants unique to that population. The greatest amount of genetic diversity is found outside of European ancestry and using a PGx panel mostly defined by this research is a missed opportunity. Thus, partnerships should be sought with research groups that can provide the technology to sequence. Sequencing allows identification and categorization based on a genetic basis rather than race or ethnicity and may enhance clinical practice by further expanding PGx panels offered. Notably, there are groups that are working to design population‐specific genotyping arrays for under‐represented populations, including Multi‐Ethnic Global Array, Global Screening Array, and the H3Africa Array. 2 The PGx studies highlighted in this review were done with relatively homogenous communities found in a local geographical region. When applying to under‐represented populations that are more heterogeneous and geographically unrestricted, sequencing and use of genetic ancestry groups will be paramount.
As efforts increase to include under‐represented populations in PGx research, it will be important that it is translated into clinical practice. The recent decision that manufacturers of clopidogrel “engaged in unfair and deceptive business practices” resulted in a ruling of over $800 million in penalties as the state of Hawaii claimed that the manufacturers knew that clopidogrel could have diminished or no effect in people of ancestry with higher frequencies of CYP2C19 loss of function alleles. 26 This ruling could have a tremendous impact on the clinical PGx community and it is imperative that translation of PGx into clinical practice is done thoughtfully and equitably.
An additional consideration is standardization of categorizing individuals by race and ethnicity. Zhang et al. showed that race and ethnicity is complex and allele frequencies are heterogeneous across subgroups of ethnicities, thus applying genetic frequency assumptions of a group to a subgroup may be clinically inappropriate. 4 Efforts have been made to address these inconsistencies and include using biogeographical groups based on the geographical distribution of genetic ancestry. 1
Beyond the scope of this review are larger efforts to enroll under‐represented populations in genetic studies. Efforts to increase diversity include RIBEF, 1000 Genomes Project, All of US, and the African Genome Project. In particular, the 1000 Genomes Project sequenced over 2000 people across 26 populations and aims to ensure access and usability of the data while continuing to collect from populations not included in the original project. 27 The All of Us Research Program aims to generate genomic data from their participants across the United States and has a core value devoted to diversity and inclusion. 28
SUMMARY
Genetic variation is linked to medication response variation and is used as an evidence‐based tool in clinical care to optimize medication therapies. Implementation of PGx as it is translated into clinical care from research is increasing; however, the heavy influence of European ancestry genetics in PGx studies is exacerbating the existing healthcare disparity gap, creating a growing need for PGx studies to be done in under‐represented populations so that the promising translation of PGx into clinical care can be implemented equitably. Research within under‐represented populations should begin by addressing structural inequities and social determinants of health with a CBPR approach, as PGx research may not be the highest priority for under‐represented populations.
CONFLICT OF INTEREST
All authors declared no competing interests for this work.
Supporting information
Table S1
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Olihe Okoro, PhD, MPH, MPharm, for her review of this manuscript and expertise and feedback on community based participatory research and social determinants of health.
Luczak T, Stenehjem D, Brown J. Applying an equity lens to pharmacogenetic research and translation to under‐represented populations. Clin Transl Sci. 2021;14:2117–2123. 10.1111/cts.13110
Funding information
No funding was received for this work.
REFERENCES
- 1. Huddart R, Fohner AE, Whirl‐Carrillo M, et al. Standardized biogeographic grouping system for annotating populations in pharmacogenetic research. Clin Pharmacol Ther. 2019;105:1256‐1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peterson RE, Kuchenbaecker K, Walters RK, et al. Genome‐wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CPIC® Guideline for Clopidogrel and CYP2C19, https://cpicpgx.org/guidelines/guideline‐for‐clopidogrel‐and‐cyp2c19/. Accessed April 7, 2021.
- 4. Zhang F, Finkelstein J. Inconsistency in race and ethnic classification in pharmacogenetics studies and its potential clinical implications. Pharmacogenomics Pers Med. 2019;12:107‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Radouani F, Zass L, Hamdi Y, et al. A review of clinical pharmacogenetics studies in African populations. Pers Med. 2020;17:155‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henderson LM, Claw KG, Woodahl EL, et al. P450 pharmacogenetics in indigenous North American populations. J Pers Med. 2018;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Claudio‐Campos K, Duconge J, Cadilla CL, et al. Pharmacogenetics of drug‐metabolizing enzymes in US Hispanics. Drug Metab Pers Ther. 2015;30:87‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lo C, Nguyen S, Yang C, et al. Pharmacogenomics in Asian subpopulations and impacts on commonly prescribed medications. Clin Transl Sci. 2020;13:861‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fricke‐Galindo I, Jung‐Cook H, LLerena A, López‐López M. Interethnic variability of pharmacogenetic biomarkers in Mexican healthy volunteers: a report from the RIBEF (Ibero‐American Network of Pharmacogenetics and Pharmacogenomics). Drug Metab Pers Ther. 2016;31:61‐81. [DOI] [PubMed] [Google Scholar]
- 11. Rodrigues‐Soares F, Kehdy FSG, Sampaio‐Coelho J, et al. Genetic structure of pharmacogenetic biomarkers in Brazil inferred from a systematic review and population‐based cohorts: a RIBEF/EPIGEN‐Brazil initiative. Pharmacogenomics J. 2018;18:749‐759. [DOI] [PubMed] [Google Scholar]
- 12. NOT‐OD‐15‐089: racial and ethnic categories and definitions for NIH diversity programs and for other reporting purposes, https://grants.nih.gov/grants/guide/notice‐files/not‐od‐15‐089.html. Accessed April 7, 2021.
- 13. Tang H, Quertermous T, Rodriguez B, et al. Genetic structure, self‐identified race/ethnicity, and confounding in case‐control association studies. Am J Hum Genet. 2005;76:268‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mersha TB, Abebe T. Self‐reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Human Genomics. 2015;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark LT, Watkins L, Piña IL, et al. Increasing diversity in clinical trials: overcoming critical barriers. Curr Probl Cardiol. 2019;44:148‐172. [DOI] [PubMed] [Google Scholar]
- 16. Shah‐Williams E, Levy KD, Zang Y, et al. Enrollment of diverse populations in the INGENIOUS pharmacogenetics clinical trial. Front Genet. 2020;11:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burnett‐Hartman AN, Blum‐Barnett E, Carroll NM, et al. Return of research‐related genetic test results and genetic discrimination concerns: facilitators and barriers of genetic research participation in diverse groups. Public Health Genomics. 2020;23:59‐68. [DOI] [PubMed] [Google Scholar]
- 18. Popejoy AB. Diversity in precision medicine and pharmacogenetics: methodological and conceptual considerations for broadening participation. Pharmacogenomics Pers Med. 2019;12:257‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajman I, Knapp L, Morgan T, et al. African genetic diversity: implications for cytochrome P450‐mediated drug metabolism and drug development. EBioMedicine. 2017;17:67‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fohner A, Muzquiz LI, Austin MA, et al. Pharmacogenetics in American Indian populations: analysis of CYP2D6, CYP3A4, CYP3A5, and CYP2C9 in the confederated Salish and Kootenai tribes. Pharmacogenet Genomics. 2013;23:403‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wright GEB, Niehaus DJH, Drögemöller BI, et al. Elucidation of CYP2D6 genetic diversity in a unique African population: implications for the future application of pharmacogenetics in the Xhosa population. Ann Hum Genet. 2010;74:340‐350. [DOI] [PubMed] [Google Scholar]
- 22. Boyer BB, Dillard D, Woodahl EL, et al. Ethical issues in developing pharmacogenetic research partnerships with American Indigenous communities. Clin Pharmacol Ther. 2011;89:343‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Woodahl EL, Lesko LJ, Hopkins S, et al. Pharmacogenetic research in partnership with American Indian and Alaska Native communities. Pharmacogenomics. 2014;15:1235‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katigbak C, Foley M, Robert L, et al. Experiences and lessons learned in using community‐based participatory research to recruit Asian American immigrant research participants. J Nurs Scholarsh Off Publ Sigma Theta Tau Int Honor Soc Nurs. 2016;48:210‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carroll DM, Hernandez C, Braaten G, et al. Recommendations to researchers for aiding in increasing American Indian representation in genetic research and personalized medicine. Pers Med. 2021;18:67‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bristol‐Myers, Sanofi ordered to pay Hawaii $834 million over Plavix warning label. US News & World Report, https://www.usnews.com/news/top‐news/articles/2021‐02‐15/bristol‐myers‐sanofi‐ordered‐to‐pay‐hawaii‐834‐million‐over‐plavix‐warning‐label. Accessed April 7, 2021.
- 27. 1000 Genomes Project. Genome.gov, https://www.genome.gov/27528684/1000‐genomes‐ project. Accessed April 7, 2021.
- 28. National Institutes of Health (NIH) . National Institutes of Health (NIH) — All of Us, https://allofus.nih.gov/future‐health‐begins‐all‐us (2020). Accessed April 7, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


