Abstract
The discovery and development of new medicines is expensive, time-consuming, and often inefficient, with many failures along the way. Powered by artificial intelligence (AI), language models (LMs) have changed the landscape of natural language processing (NLP), offering possibilities to transform treatment development more effectively. Here, we summarize advances in AI-powered LMs and their potential to aid drug discovery and development. We highlight opportunities for AI-powered LMs in target identification, clinical design, regulatory decision-making, and pharmacovigilance. We specifically emphasize the potential role of AI-powered LMs for developing new treatments for Coronavirus 2019 (COVID-19) strategies, including drug repurposing, which can be extrapolated to other infectious diseases that have the potential to cause pandemics. Finally, we set out the remaining challenges and propose possible solutions for improvement.
Keywords: Artificial intelligence, Language models, Natural language processing, Drug discovery, Drug development, COVID-19
Introduction
A LM aims to determine the probability of a given sequence of words occurring in a sentence by using different statistical and probabilistic techniques. Powered by AI, a LM acts as a human-like learning process, not only to predict words, but also to understand languages. Moreover, the knowledge gained by the LM can be transferred to other tasks, as humans do when they routinely learn from one task and transfer the knowledge to another. This innovative revolution has massively empowered NLP. Consequently, AI-based LMs have proved their use in a variety of real-world applications, such as chatbots, automated translations, customer experience, sentiment-based news aggregation, and language identification.1 Here, the AI-powered LMs we describe are mainly focused on LM based on a neural architecture.
Innovations in emerging biological technologies have made enormous improvements to our understanding of disease etiology and pathogenesis.2, 3 However, drug discovery and development remain a time-consuming and costly process, beset by high failure rates and uncertainty.4 Significant efforts are being invested in refining, revising, and reforming the de novo drug discovery and development process, with particular emphasis on data-driven approaches to new treatments, improved patient outcomes, and lower costs.5 With a rapid increase in the quantity of biomedical data, a better understanding of the characteristics of the data generated and the types of analysis that can be carried out will be valuable in understanding the potential of data resources.6
Although attention tends to focus on ‘omics data and bioassays generated from large technology platforms, text-based data remains a valuable information resource in the drug discovery and development process. Thus, conventional NLP-based approaches and tools have been developed to uncover hidden information embedded in such documents.7 However, more advanced strategies are urgently needed to harness a growing wealth of available data and to stay abreast of the latest accumulated text-based documents. Notably, AI-powered LMs have the potential to unlock new possibilities for drug development and usher in an era of faster, cheaper, and more effective drug discovery and development.8
Several text-based documents and applications of AI-powered LMs have been shown to be useful in different stages of drug discovery and development (Fig. 1 ). During the preclinical stages, the incomplete understanding of the pathophysiology of complex diseases is one of the significant hurdles for target identification. Furthermore, animal models might not reflect the underlying mechanisms of human disorders. During the clinical phases, patient selection, recruitment, and monitoring pose a strategic challenge. During the post-marketing phase, there are shortfalls in the ability of the current system to effectively and efficiently detect, interpret, and analyze safety signals. In addition, complexity in the regulatory submission process could hinder harmonized communication between pharmaceutical companies and regulatory agencies. In this review, we provide an overview of tangible opportunities for AI-powered LMs in drug discovery and development, and offer potential solutions for key remaining challenges.
Figure 1.
Artificial intelligence (AI)-powered language models in the context of drug discovery and development. The overall stages of the development process are illustrated in the top layer (green), and the objectives from this process are captured in the layer below (blue). The text documents related to each stage are listed, and the opportunities of AI-powered language models are summarized in the following two layers (yellow and pink). Abbreviations: PD, pharmacodynamics; PK, pharmacokinetics.
AI-powered language models
Rapidly evolving LMs have enormously increased our ability to uncover actionable insights from text (Box 1 ). Powered by AI, many intriguing LM infrastructures, particularly transformer-based LMs, have been developed and have shown potential in information retrieval, text classification, text summarization, and sentiment analysis. The heart of transformer-based LMs is sequence-to-sequence learning (Seq2Seq) through self-attention and positional encoding, which has changed the way we work with text data, from processing language to learning language9 (Box 2 ).
Box 1. Evolution of language models.
LMs enable a computer to make sense of the human language by estimating the probability distribution of various linguistic units (e.g., words, sentences, paragraphs, etc.). LMs are mainly divided into two categories: count-based and continuous-space LMs. The classic example of count-based LM is n-gram, which aims to construct the joint probability distribution of sentences to predict the words.103 However, several drawbacks of n-gram LM have limited its real-world applications: (i) the n-gram LM is not capable of inferring new word sequence combinations that were not encountered in the training corpus; and (ii) the n-gram model is not able to take into consideration the semantic relationship among words. The shortcoming of count-based LMs led to the idea of continuous-space LM by applying neural network or dimension reduction techniques to extract syntactic and semantic features of languages.104 Most continuous-space LMs are neural probabilistic-based models.105 Mikolov et al.106 proposed Word2Vec to generate vector representation of words (i.e., word embeddings) by training a shallow neural network to learn the similarities between words. Word2Vec is one of the most widely used neural-based LM and takes breakthroughs to the whole field. Along with other word representation models, (e.g., GloVe107 and FastText108), these word-embedding techniques require less memory and decreased compute time, and have been shown to improve downstream model performance drastically. However, word embeddings provide a one-to-one relationship between word and vector representation that does not solve the problem of polysemous words. Subsequently, RNNs and LSTM were proposed to handle the processing of textual sequence data.12 However, the two algorithms suffer from a vanishing gradient problem and have a difficulty in dealing with long sequence sentences.98 The most innovative groundbreaking NLP framework is transform-based LMs, and the BERT model from Google is an outstanding example of such an approach.16
Box 2. Architectures of transformer-based language models.
Two kinds of transformer developed with distinct learning scenarios currently dominate the field: BERT and Generative Pretrained Transformer (GPT) models.
BERT and its derivatives are built using a complete encoder-decoder transformer, which is fine-tuned for downstream NLP tasks.16 The BERT-based model architecture is task agnostic. There is a need for task-specific data sets and task-specific fine-tuning to achieve optimized model performance. However, it could be challenging to collect task-specific labeled data, especially in the biomedical domain. Furthermore, the generalization made under this paradigm can be inadequate because the model is overly specific to the training distribution and does not generalize well outside it. Two kinds of strategy were developed to overcome these shortcomings. First, the training material was increased in size and diversity to enhance model generalizability for different NLP tasks. One example is the Robustly Optimized BERT Pretraining Approach (RoBERTa).109 Besides the original BERT training corpus, RoBERTa extends the training corpus with news and stories corpora from Common Crawl.110 Subsequently, the model outperformed by 2–20% both BERT and XLNet on GLUE benchmark results with a dynamic masking training strategy. Second, adjusted training strategies were developed to improve model performance. Examples, including ARBERT,27 ELECTRA,26 and DistillBERT,25 provided lite model architectures without losing predictive model performances.
GPT models are autoregressive transformer-based LMs. The models have a task-specific learning architecture without an intensive fine-tuning process. GPT models used the ‘in-context learning’ concept, in which models develop a broad set of skills and pattern recognition abilities at training time and then use those abilities at inference time to adapt to or recognize the desired task rapidly. GPT-3 was released recently, comprising a huge transformer model with 175 billion parameters trained with 45 Tb of compressed plaintext from Common Crawl, plus high-quality reference corpora, such as Wikipedia.111 The GPT-3 model was demonstrated to achieve better state-of-the-art results for various NLP tasks with few-shot learners in task-specific data sets.
A Seq2seq model comprises a combination of an Encoder and a Decoder, aiming to convert sequences from one domain (e.g., a sentence in English) to another (e.g., the exact meaning of the sentence in French).10 Deep-learning model architectures, such as recurring neural network (RNN) or long short-term memory (LSTM), could be used for Encoder and Decoder development. The Encoder takes the sequence as an input and maps the sequence into internal state vectors or context vectors, where the output of the Encoder is then discarded. The generated context vector could encapsulate input sequence information to facilitate the prediction of the Decoder. The training process of the Decoder is referred to as ‘teach forcing’. Specifically, the Decoder takes the extracted context vector of the Encoder as the initial state to generate the output sequence. These outputs are also taken into consideration for future outputs. Seq2seq models have been used to solve complex NLP tasks, such as Machine Translation, Question & Answering (Q&A), Chatbots, Text Summarization, and so on. LSTMs function through a cell, an input gate, an output gate, and a forget gate, avoiding the problem of a vanishing gradient seen, for example, with RNN.11, 12 The main revolutionary part of transformer models is the possibility of directly accessing all positions in the sequence, equivalent to having full random-access memory of the sequence during encoding/decoding.
Transformer-based LMs can mimic some human-like characteristics of constant acquisition, fine-tuning, and transfer of knowledge and skills (Fig. 2 ). First, transformer-based LMs can offer a transfer learning framework.13 For this, the learned knowledge is stored in a pretrained model, allowing users to be further trained with incrementally available information or domain-specific knowledge. One example is BioBERT, which is a pretrained language representation model derived by training the original BERT model with biomedical corpora in PubMed.14 BioBERT outperformed the original BERT base model in most biomedical text-mining tasks, including biomedical named entity recognition, biomedical relation extraction, and biomedical question answering. The same learning strategy was also adopted by ClinicalBERT, which trained the BERT-based models with electronic medical records (EHRs) data to enhance its clinical application.15 Second, transformer-based LMs could be fine-tuned based on the downstream task. A human can apply the right knowledge to solve related questions; this function also appears to be possible with transformer-based LMs. For different tasks, the pretrained language models could be fine-tuned with just one additional output layer to create state-of-the-art models for a range of NLP tasks. For example, the same pretrained BERT model, with one fine-tuning layer, yielded better model performance for 11 state-of-the-art NLP tasks with large margins compared with XXXX. These NLP tasks fall primarily into three categories: text classification, textual entailment, and question answering. More encouragingly, BERT is the first to outperform the human-level performance for two tasks: the Stanford Question Answering Dataset (SQuAD), and the Situations With Adversarial Generations (SWAG).16 Third, transformer-based LMs are capable of summarizing the knowledge embedded in different documents. One of the human learning abilities is to accurately sum up information in documents and turn it into useful knowledge. The long sequence-based transformer model has been demonstrated to generate fluent, coherent multisentence paragraphs; even whole Wikipedia articles could be created this way as a multidocument summarization of source documents.17 However, the vast computational memory requirement of long sequence summarization models has limited their applications. The newly proposed Reformer model of Google shows a tremendously increased capability for handling long sequences gained by adopting a locality-sensitive hashing technique, which will significantly expand the horizon of multidocument summarization.18
Figure 2.
Comparison of artificial intelligence (AI)-powered language models and human intelligence: (1) Transfer learning (green); (2) Apply knowledge (blue); and (3) summarize knowledge (yellow).
Selection of ‘fit-for-purpose’ AI-powered language models
The diversity of transformer-based LMs has massively enhanced capabilities in tackling unstructured text for various real-world applications. However, it is challenging to select and reposition the transformer-based LMs in the context of biomedical applications. The implementation of a ‘fit-for-purpose’ transformer-based LM in drug discovery and drug development is multifactorial; significant factors are the availability of domain-specific training data sets, downstream NLP tasks, and computational power. Crucial steps essential for selecting a ‘fit-for-purpose’ AI-based LM in drug discovery and development are ‘define a purpose’, ‘manage data availability’, and ‘measure scalability’.
Define a purpose
AI-powered LMs have potential at every stage of drug discovery and development, but it is essential to define the purpose before seeking the right AI solution. For example, a scientist at a pharmaceutical company might need to understand the biological role of a protein target and then collate the patents on therapeutic categories to support target identification and validation. For this, an AI-powered Q&A system that can aggregate publicly available literature and medical patent databases might be the right solution. Patient recruiters might be more interested in looking for an automated route to prioritize clinical sites and patients. Accordingly, an AI-based contextual-based patient-matching system might be useful. Drug dossier reviewers might be more interested in a powerful tool to detect safety signals from clinical documents. For this, AI-powered biomedical named-entity recognition (NER) and entity relation extraction approaches could be options. Given the diversity of data and ‘needs’, defining a purpose for any data-driven hypothesis becomes a priority.
Manage data availability
A large body of the text is required to train AI-based language models. The current publically available pretrained LMs are mainly trained based on general knowledge, such as books, news, webpages, social media, and Wikipedia. A few domain-specific LMs, such as BioBERT14 and ClinicalBERT,19 have been proposed to enhance the clinical application by using publicly available biomedical literature or deidentified EHRs. However, labeled data are still required for the fine-tuning process and to enable the model fit for downstream tasks. However, the curation of labeled data is a challenging and time-consuming process, in which significant domain expertise and knowledge are necessary. Furthermore, data generated across drug discovery and development might be company sensitive, posing a challenge around data sharing in LM development.20, 21 Consequently, it is recommended to have a clear view of data availability and the effort required to curate labeled data before selecting a suitable AI-powered LM.
Although data annotation is still the bottleneck in AI-powered LM development, several successful examples exist that can stimulate interest in the community to accelerate further and promote ‘labeled data’ development in the biomedical field. First, crowd-sourced biomedical labeling could be an effective way to curate domain-specific labeling data. The concept of crowd-sourced biomedical labeling aims to outsource biomedical data annotation to a distributed ‘crowd’ of experts worldwide. Some business models, such as Amazon Mechanical Turk, have been developed for this purpose. We recommend establishing a voluntary-based biomedical labeling consortium to facilitate biomedical data annotation. Second, a reorganization of publicly available biomedical corpus would be useful for addressing specific BioNLP tasks. For example, the combined different domain-specific corpus could be a practical approach to creating annotated bioconcept ambiguity data.22 Third, labeling tools could be a solution to facilitate the manual data curation and annotation process. The most common starting point is an Excel/Google spreadsheet to handle common labeling tasks, such as part-of-speech and named entity recognition labeling. However, this could be error-prone because typographic errors in transcription are common and the cells and columns are not the most intuitive way to read a text document. For example, gene name errors are common in scientific literature, with up to 30% of gene names misrepresented.23 Some standard labeling tools, such as Prodigy, LightTag, TagTog, and Datasaur.ai, provide more standardized solutions for offering customizability and handling advanced NLP tasks.24
Measure scalability
The performance improvement of transformer-based LMs results from increased data and model size, computational power, or training procedures. Comparing similarities and differences among popular AI-powered LMs frameworks (Table 1 ) is useful to support model selection. First, speed matters in applying AI-powered LMs in different tasks. For example, suppose the AI-powered LM is targeted toward the patient-monitoring process. In that case, a faster inference speed is set as the highest priority to meet real-time data collection and analysis requirements. Thus, the distilled model architecture, such as DistilBERT,25 ELECTRA,26 and ALBERT,27 might be a reasonable starting point, although a few percentage points might compromise the prediction performance. Second, AI-powered LM development demands vast computation power. Larger documents and higher model parameters lead to better performance, whereas more computational power is a prerequisite to retrain these models with the custom corpus. For example, if the AI-powered LMs aim to identify potential adverse events from clinical notes, pretrained BioBERT or ClinicalBERT are appropriate options to test the necessity of developing the model de novo. Third, an ensemble approach might allow for further performance improvement. For complicated drug discovery and development tasks, such as patient recruitment, a single model might capture one aspect of complexity, whereas a consensus approach might improve patient matching.
Table 1.
Selected examples of transformer-based language models.
| Architectures | BERT | OpenAI GPT | XLNet | ALBERT | RoBERTa | ELECTRA | DistillBERT |
|---|---|---|---|---|---|---|---|
| Pre-training corpus | BooksCorpus and English Wikipedia Size: 16 Gb |
8 million web pages from Common Crawl Size: unknown |
Base: BooksCorpus and English Wikipedia Size: 16 Gb Large: BooksCorpus + English Wikipedia + Giga5 + ClueWeb 2012-B + Common Crawl Size: 113 Gb |
BooksCorpus and English Wikipedia Size: 16 Gb |
BooksCorpus + English Wikipedia + Common Crawl news dataset + Web text corpus + stories from Common Crawl Size: 160 Gb |
BooksCorpus and English Wikipedia Size: 16 Gb |
BooksCorpus and English Wikipedia Size: 16 Gb |
| Model Parameters |
Base: 110M Large: 340M |
GPT-2: 1.5 billion parameters GPT-3: 1.7 billion parameters |
Base: 110M Large: 340M |
Base: 12M Large: 18M |
Base: 125M Large: 355M |
Small: 14M Base: 110M Large: 335M |
Base: 66M |
| Training strategies | Masked Language Model (MLM) and Next Sentence Prediction (NSP) |
process the input text left-to-right, predicting the next word given the previous context | Permutation-based modeling | BERT with reduced parameters for sentence order prediction | BERT with a dynamic masking strategy. Without Next Sentence Prediction (NSP) | ELECTRA models are trained to distinguish “real” input tokens vs. “fake” input tokens generated by another neural network. | BERT base model with a distillation loss function |
| Training time |
Base: 12 days with 8 Nvidia Tesla V100 GPUs Large: 4 days with 64 TPUs or 1 day with 280 Nvidia Tesla V100 GPUs |
Unknown | Large: 2.5 days with 512 TPUs | Large: 1.7 faster than BERT | 1 day with 1024 Nvidia Tesla V100 GPUs | Small: ∼4 days on a v100 GPU | 3.5 days with 8 Nvidia Tesla V100 GPUs |
| Feature dimension |
Base: up to 768 Large: up to 1024 |
GPT-2: Up to 1024 GPT-3: Up to 2048 |
Up to 4096 |
Base: up to 768 Large: up to 1024 |
Base: up to 768 Large: up to 1024 |
Small: up to 256 Base: up to 768 Large: up to 1024 |
Base: up to 768 |
| Performance | Outperformed state-of-the-art in 11 NLP tasks | GP7-2 Achieves state-of-the-art results on 7 out of 8 tested language modeling datasets | 2%∼15% improvement over BERT | Large: 3% improvement over BERT | outperforms 2%-20% both BERT and XLNet on GLUE benchmark results |
Small: perform roughly in between GPT and BERT-Base in terms of GLUE performance Large: ELECTRA achieves state-of-the-art results on the SQuAD dataset |
retaining 97% performance of BERT base model on GLUE benchmark results |
| Weblink | https://github.com/google-research/bert | GPT-2: https://github.com/minimaxir/gpt-2-simple GPT_3: https://github.com/openai/gpt-3 |
https://github.com/zihangdai/xlnet | https://github.com/google-research/ALBERT | https://github.com/pytorch/fairseq/tree/master/examples/roberta | https://github.com/google-research/electra | https://github.com/huggingface/transformers |
| References | 13 | 90, 91 | 92 | 21 | 89 | 20 | 19 |
AI-powered language models in drug discovery
AI offers great potential in drug discovery and development (Table 2 ). Here, we highlight potential opportunities for AI-powered LMs in different drug development stages and suggest possible directions and solutions for further improvement.
Table 2.
Selected examples AI-based NLP applied in drug discovery.
| NLP task | Data set | Algorithm | Notes | Source code | Refs |
|---|---|---|---|---|---|
| Target identification | |||||
| Biomedical Named Entity Recognition | BC2GM; BC5CD; NCBI -Disease; JNLPBA | BioBERT | A multi-ask (MT)-BioNER proposed for biomedical named entity recognition using BioBERT as shared layers and different data sets in task-specific layers | https://github.com/cambridgeltl/MTL-Bioinformatics-2016 | 30 |
| Gene–disease relationship extraction | DisGeNET: database of gene–disease associations | Convolution neural network (CNN) and attention-based BiLSTM | Proposed Deep-GDAE integrates specificities of CNN and an attention-based BiLSTM to classify gene–disease associations | https://github.com/EsmaeilNourani/Deep-GDAE/ | 33 |
| Biomedical text summarization | PubMed | BERT and hierarchical clustering algorithm | Biomedical text summarizer (BERT-based-Summ) proposed by interrogating BERT and hierarchical clustering algorithm to extract biomedical content summarization | https://github.com/BioTextSumm/BERT-based-Summ | 34 |
| Drug properties prediction | 1 million SMILE codes of compounds in ChEMBL database | BiLSTM-based transfer learning | Transfer learning framework, MolPMoFiT, to predict physical and biological endpoints, such as lipophilicity and blood–brain barrier penetration, for given compounds | https://github.com/XinhaoLi74/MolPMoFiT | 40 |
| Virtual screening | SMILES | BERT | MOLBERT model proposed by applying BERT model to SMILES for virtual screening | https://github.com/BenevolentAI/MolBERT | 41 |
| Reshape clinical trails | |||||
| Patient–trial matching | Patient EHR data | ClinicalBERT | Proposed DeepEnroll based on ClinicalBERT jointly encodes enrollment criteria and patient EHRs into shared latent space for patient–trial matching | https://github.com/deepenroll/DeepEnroll | 46 |
| Trial eligibility criteria | Patient EHR data | CrOss-Modal PseudO-SiamEse network (COMPOSE) | COMPOSE aims to address challenges for patient–trial matching; one path of network encodes EC using convolutional highway network | https://github.com/v1xerunt/COMPOSE | 112 |
| Assist the regulatory process | |||||
| Biomedical entity normalization | Clinical notes; PubMed abstract; drug labeling | BERT; BioBERT; ClinicalBERT | Authors proposed entity normalization architecture by fine-tuning pretrained BERT/BioBERT/ClinicalBERT models and applying them to SNOMED-CT coding, MedDRA coding, and Medical Subject Headings (MESH) coding | – | 63 |
| Disease coding | Clinical notes and ICD-10 | BERT | Authors proposed ML model, BERT-XML, for large-scale automated ICD coding from EHR notes | https://github.com/suamin/multilabel-classification-bert-icd10 | 64 |
| Biomedical mention disambiguation | CTDbase and gene2pubmed | CNN with LSTM | Authors developed biomedical corpus for curating biomedical terms ambiguous between one or more concept types; model is used by interrogating LSTM and CNN | – | 22 |
| Advance postmarketing surveillance | |||||
| ADR detection | Bidirectional BiLSTM | Authors proposed RNN model using pretrained word embeddings created from a large, nondomain-specific Twitter data set for ADR extraction | https://github.com/chop-dbhi/twitter-adr-blstm | 113 | |
| Ensemble models of BERT, BioBERT, and ClinicalBERT | Authors proposed ensemble model by integrating BERT, BioBERT, and ClinicalBERT for ADR detection from social media data | – | 75 | ||
Opportunity 1: AI-powered language models to accelerate target identification
Target identification is one of the most crucial steps in the drug discovery pipeline to establish the biological origin of disease and design appropriate interventions.28 Typically, target identification involves various considerations from both scientific and economic perspectives. The project team, with experts from diverse disciplines, need to define the disease area and the expected therapeutic effects. Then, they need to look for potential biochemical, cellular, or pathophysiological mechanisms tailored to the disease. Next, a comprehensive investigation of the targets involving different tools might be conducted to further prioritize targets for development. Importantly, the prioritized targets should be competitive in terms of therapeutic efficacy, safety, and intellectual property perspectives. However, the vast array of information might be widely distributed in the public domain literature, patent documents, and biomedical databases. It is often too great a challenge to curate the data manually using conventional simple search-based approaches.
AI-powered LMs can advance findings and accelerate target identification. Automatic biomedical named entity recognition (BioNER) is a practical approach to uncover the hidden relationship among chemicals, genes, targets, and diseases embedded in free-text documents.29 Khan et al. 30 proposed a multiple-tasking learning architecture for BioNER using the BioBERT. These approaches outperformed state-of-the-art approaches, such as bidirectional LSTM (BiLSTM), conditional random fields (CRF),31 and multitask learning neural network with shared character and word layers (MTM-CW),32 for chemical, disease, and gene entity recognition. Nourani et al. 33 developed a hybrid transfer learning framework (Deep-GDAE) for biomedical association extraction from PubMed literature, which integrates attrition-based BiLSTM and a convolutional neural network (CNN) based on feature information extracted from BERT and BioBERT base models. Deep-GDAE yielded a high performance (79.8% of F-measure) for gene–disease relationship extraction. Another promising application of AI-powered LMs is to summarize the essential information from biomedical literature for accelerating target identification. Moradi et al. 34 applied the BERT base and large models for biomedical text summarization to create a synthetic abstract based on full PubMed articles. The approaches achieved state-of-the-art results; performance could be further improved by using domain-specific contextual embedding from BioBERT.
The concept of transformer-based LM has been leveraged into chemoinformatics to advance drug–target relationship prediction.35 The Simplified Molecular Input Line Entry System (SMILES) is a comprehensive yet straightforward chemical language in which molecules and reactions can be specified using ASCII characters representing atom and bond symbols. Similarly, FASTA is useful in analyzing protein structure and function because it finds regions of local or global similarity between protein or DNA sequences.36 Inspired by transformer-based pretrained LMs, the large body of information in SMILES or FASTA files could be assimilated in the same way that humans do with sentences to grasp the semantics of molecules and their relationship to downstream tasks. Unlike early attempts at chemical representation based on deep-learning frameworks, such as Wod2vec and Variational Autoencoders (VAEs),37, 38 transformer-based chemical representation incorporates the attention mechanism (positional encoding) into the learning process to maximize information extraction. One such example is SMILES transformer, which trained 861 000 SMILES from the ChEMBL database, a chemical bioassay repository. The learned chemical representations were fine-tuned to different chemophysical properties, therapeutic targets, and toxicity predictions. This approach significantly outperformed conventional fingerprint-based strategies.39
Other AI frameworks have also been used for virtual screening based on SMILE sequences. Li et al. 40 proposed a transfer learning framework, named Molecular Prediction Model Fine-Tuning (MolPMoFiT), to predict physical and biological endpoints, such as lipophilicity and blood–brain barrier penetration, for the given compounds. The proposed MolPMoFiT comprised two components. First, the authors developed a weight-dropped LSTM model based on 1 million SMILE sequences in the ChEMBL database to predict the masked atoms in the SMILE sequencing. Second, a trained weight-Dropped LSTM model was transferred and fine-tuned for downstream tasks. Similarity, Fabian et al. 41 also adopted the transfer learning framework by training the SMILES sequences with BERT, which was then applied for virtual screening of compound-binding affinity of 69 individual protein targets.
AI-powered LMs have the potential to assess unmet medical needs and provide prioritized targets for high-throughput screening (HTS). The opportunity to accelerate understanding of the current market and potential gaps could facilitate early drug development planning. However, target identification still relies on the generation of experimental data; AI-powered LMs have the potential to promote understanding of the data and to support target identification and prioritization. Currently, AI-powered LM models provide a more informative way to represent text-based input as an n-dimensional vector or high-level representation. However, to further improve target identification performance, fine-tuned models are vital for different downstream tasks. Some more comparative studies and evaluations appeared to set out the pros and cons of AI-powered models compared with conventional approaches, potentially guiding the fit-for-purpose selections of different AI models.
Opportunity 2: AI-powered language models to reshape clinical trials
Clinical trials are resource intensive, accounting for around half of the cost and time in the drug development life cycle, yet there is a high failure rate.42 Unsuccessful clinical trials are attributed to various reasons, some of which are suboptimal patient cohort selection, ineffective patient recruitment strategies, and unsophisticated patient monitoring systems.43 Diverse text-based data sets, including electronic health records (EHRs), clinical trial databases, trial announcements, eligibility databases, social media, and medical literature, provide a unique and immediate entry point for AI-powered LMs to improve clinical trial outcomes.44
Approximately 80% of clinical trials in the USA fail to meet their timelines on patient recruitment.43 The complexity of inclusion/exclusion criteria in terms of suitability, eligibility, motivation, and empowerment poses a challenge for patient recruitment. Poorly matched disease subtypes might make patients unsuitable, and inconsistencies in medical-history recording could render suitable patients ineligible. Patient information is often inconsistent and recorded in an unstructured format, hampering comprehensive patient screening for a given set of inclusion/exclusion criteria.
AI-powered LMs enable automation of the patient recruitment process, alleviating the manual workload burden through advanced information retrieval and prioritization mechanisms. First, AI-powered LMs can learn medical terms and their synonyms to retrieve useful information from clinical documents that might be free-flowing and unstructured. For example, disease heterogeneity often hinders the determination of patient suitability; recurrent models based on the Bidirectional GRU architecture with contextual embedding could effectively boost multilabel disease extraction from EHRs.45 This approach has the potential to stratify patients precisely based on disease subtypes for patient recruitment. Second, AI-powered LMs could synthesize the eligibility criteria into a standardized contextual query to improve the clinical trial-matching process. One such example described the use of BERT-based contextual embedding to match eligibility criteria for patient selection.46 Powered by a cross-model learning infrastructure, the proposed DeepEnroll could jointly encode enrollment criteria and patient EHRs into a shared latent space for matching inference. Eventually, the model outperformed the rule-based matching strategies, with up to a 12.4% improvement in the F-measure (a measure of the accuracy of a test). Third, AI-powered LMs could be combined with other emerging technologies seamlessly to expedite patient stratification. A combination of EHR data, genomics data, or image data holds significant promise to advance precision medicine.47, 48 AI-powered LMs could be used to boost phenotyping capabilities by deep mining from EHRs, registries, hospital records, and health insurance data alongside biobank, genomic, and digital phenotyping information. Finally, AI-powered LMs enable higher patient enrollment rates and better site identification, leading to efficient recruitment of patients. However, site identification is a multifactorial decision. Factors, such as prior experience of the site for a therapeutic area, connection with health nonprofits and patient advocates, historical data of patient retention, and cost-effectiveness are significant contributors. AI-powered LMs could be leveraged to support clinical decision-making by considering these diverse factors, allowing for a balanced decision and the best possibility of success.
Successful completion of clinical trials justifies the massive investment in patient recruitment, but the average patient dropout rate across all clinical trials is ~ 30%.44 Efforts to overcome challenges in clinical trial recruitment and retention continue. These efforts could safeguard the well-being of trial participants, ensure adherence to trial rules and procedures, enhance compliance and retention, collect reliable and high-quality trial data points, and improve real-world outcome monitoring.49 AI-powered LMs, as a combination of ML and digital technologies, could have an essential role in enhancing patient monitoring toward a lower dropout rate and a more efficient data uptake framework.
Digital health technologies, such as wearables, voice technologies, and computer vision, make remote patient monitoring possible.50 These emerging technologies also enable the collection of longitudinal and real-time biometric data sets to provide unique insights into the long-term, real-world impact of pharmacotherapies and treatment protocols. Meanwhile, the implementation of such technologies could relieve patients of their more burdensome tasks during the clinical trial and increase their adherence. More importantly, AI and ML (particularly deep-learning models) could be used to carry out real-time patient monitoring for detecting and logging relevant information.51, 52 For example, powered by AI, voice assistants have been gradually adopted in clinical trials for various tasks, including reminding patients of appointments, recording patient diaries, fostering collaboration between site investigators and sponsors, and increasing physician awareness.53
Although AI-powered LMs, along with digital technologies, have the potential to transform clinical trials, most of the interventions to date have yet to deliver on that potential. Debate over the adoption of AI and mobile platforms in clinical trials is ongoing.54 Regulatory guidance is urgently needed to leverage these promising tools and technologies to advance clinical trials. To fill the gap, the US Food and Drug Administration (FDA) announced a new strategic framework to promote the use of real-world evidence (RWE) to support the development of drugs and biologicals.55 On the other side, to gain the trust of patients in AI and digital technologies, the benefit of RWE should be verified and communicated. One Community of Patients for Research (ComPaRe) study on patient uptake of wearable devices and AI showed that only 20% of participants considered that the benefits of the technology greatly outweighed the dangers.56 Furthermore, the authors found that 35% of patients would refuse to integrate at least one existing or soon-to-be-available intervention using biometric monitoring devices (BMDs) and AI-based tools into their care.
Opportunity 3: AI-powered language model to assist the regulatory process
Regulatory submissions comprise a dossier of documents sent by pharmaceutical companies to health regulatory agencies as evidence of compliance. The process includes many laws, regulatory requirements, and regulatory guidance, which help define how drug companies manufacture their drugs, design clinical trials, report safety findings, and create promotional material. The FDA promotes regulatory submissions of standardized study data in electronic format for investigational new drugs (INDs), new drug applications (NDAs), and biologics license applications (BLAs). For example, the FDA Data Standards Catalog indicates that these data sets should be formatted following the Clinical Data Interchange Standards Consortium (CDISC) Standard Exchange for Nonclinical Data (SEND).57 These standards currently support single-dose general toxicology, repeat-dose general toxicology, and carcinogenicity studies.
Meanwhile, the FDA has an internal database to maintain and organize submission in a data warehouse, such as its Document Archiving, Reporting, and Regulatory Tracking System (DARRTS).58 Essential regulatory documents are required to proceed through the drug development life cycle. These documents, such as regulatory guidance, IND safety reports, NDA/BLA submissions, patient narratives, drug labeling, and FDA Adverse Event Reporting System (FAERS), are a rich source of information. AI-powered LMs offer an unprecedented opportunity for medical officers and others who ensure that drugs are safe and effective in supporting RWE generation for regulatory decision making and better patient outcomes. The FDA promotes developing knowledge management systems to enable better leveraging of AI to advance NLP in the regulatory process.59 Although NLP-derived clinical evidence has not yet been included in regulatory submission documents, it is time to consider how this could be enabled without disrupting assumptions of data integrity and acceptance in the future.
A standardized medical terminology could accurately represent medical knowledge stored in regulatory documents for efficient, evidence-based decision making, and optimal communication among stakeholders.60 Standardized terminologies have been assigned and regulated by health agencies.61 The suggested coding systems were recommended for different domains, such as the International Classification of Diseases (ICD) for disease, WHO Anatomical Therapeutic Chemical (ATC) Classification for medicine, the Systematized Nomenclature of Medicine–Clinical Terms (SNOMED-CT) for diagnoses, Health Level 7 (HL7) for messaging, and Medical Dictionary for Regulatory Activities (MedDRA) for adverse events.62 However, the uptake of standardized medical terminologies in regulatory-related documents is a time-consuming and labor-intensive task. AI-powered LMs could facilitate the coding of regulatory documents for more efficient and effective review, delivery, and information recall.
Biomedical named-entity normalization aims to identify biomedical entities from documents and further link detected entities to their corresponding concepts in a given knowledge base or ontology. Ji et al. 63 proposed a BERT-based ranking model for biomedical entity normalization for SNOMED-CT coding, MedDRA coding, and Medical Subject Headings (MESH) coding. The model adopted domain-specialized BERT architectures, including BioBERT and ClinicalBERT, and yielded a superior performance compared with state-of-the-art approaches without any prior knowledge of medical terminology. BERT-XML was also developed for large-scale automated ICD coding from EHRs.64 Notably, the authors trained the BERT model de novo on EHR notes with multilabel attention for better clinical vocabulary identification. Thus, the proposed model outperformed reported models and further demonstrated that the domain-specific BERT model could improve the performance of downstream tasks.
Apparent ambiguity between different bioconcept types is a potential obstacle for automated bio-NER method development. This ambiguity exists within the particular domains and across other biological concepts. Abbreviation ambiguity means that one entity could map to multiple bioconcepts. For example, the abbreviation ‘BD’ could not only represent Binswanger’s disease, but also Behçet’s disease; this example is relatively easy to resolve.65 However, some ambiguities across bioconcepts are challenging for automated bio-NER methods. For example, CO2 could mean carbon dioxide in chemicals and cytochrome c oxidase subunit 2 gene. Bio-NER is developed based on the different standardized corpus in various domains. The unified biomedical corpus could be a potential solution for biomedical mention disambiguation. For instance, Wei et al. developed a biomedical corpus for curating biomedical terms that are ambiguous between one or more concept types. Using the ensemble model by interrogating LSTM and CNN, the model could achieve F1-scores of 91.94% (micro-averaged) and 85.42% (macro-averaged) for ambiguous entity identification, outperforming the transformer models, such as BioBERT.22 Thus, we recommend further efforts to standardize bioconcepts for enhancing the performance of automated bio-NER methods.
The decision-making process is tied to the regulatory framework, which yields consensual results by integrating different data sets. Medical officers are required to not only review the submission documents, but also take account of historical data and related documents to generate evidence and support decision-making, which is a complicated and time-consuming process. The current regulatory-related databases are independently indexed and maintained, with no interconnection between each other. More importantly, the indexing strategy is mainly identity based, and there exist no semantic relations of entity–entity and document–document types. Reviewers must move from one database to another to collect the relevant information. Powered by AI, a semantic search engine might be a potential solution to improve the effectiveness of information retrieval for extracting the most relevant documents for reviewer convenience.66
Unlike a lexical search, in which the search engine looks for literal matches of query words or variants of them, semantic search can search and rank the relevance with meaning.67 Some early attempts at biomedical question & answering (Q&A) systems, such as BioBERT, have given rise to a new direction.14 Building on this, a publicly available evaluation infrastructure for biomedical semantic indexing and Q&A was developed to evaluate the performance of the developed semantic search engines.68 To take full advantage of the semantic search engine in the regulatory arena, we strongly suggest that regulators should work with the community to develop a regulatory-based semantic search engine that would assist the review of regulatory dossiers. To facilitate this, we list the regulatory data sets that are open to the public in support of these efforts (Table 3 ).
Table 3.
Publicly available FDA documents for promoting AI-powered LMs in regulatory applications.
| Data sets | Descriptions | Potential use in LMs | URL of data files |
|---|---|---|---|
| Drug labeling | Drug labeling comprises a summary of information for safe and effective use of the drug, which is proposed by manufacturer and approved by FDA | Drug labeling could be a useful resource (>120 000 product labelings) to develop biomedical named-entity recognition/normalization, and relation extraction between drug and AEs, drug–drug interaction, etc. | https://dailymed.nlm.nih.gov/dailymed/spl-resources-all-drug-labels.cfm |
| FAERS | FAERS is a database that contains information on AE and medication error reports submitted to FDA | FAERS is designed to support the post-marketing safety surveillance program for drug and therapeutic biologic products of the FDA. There are more than 19 million case reports in FAERS; AI-powered LMs could be applied to carry out AE detection, causal relationship extraction, etc. | https://fis.fda.gov/extensions/FPD-QDE-FAERS/FPD-QDE-FAERS.html |
| Orange Book | Orange Book identifies drug products approved on basis of safety and effectiveness by FDA under the Federal Food, Drug, and Cosmetic Act and related patent and exclusivity information | Orange book provides crucial regulatory information, such as biological equivalence, reference listed drug (RLD), Reference Standard (RS), and patent status. This information could be included in AI-powered LMs to compare drug product information with RLD and RS to facilitate abbreviated new drug application (ANDA) submissions | www.fda.gov/drugs/drug-approvals-and-databases/orange-book-data-files |
| Drugs@FDA | Drugs@FDA includes most drug products approved since 1939. Most patient information, labels, approval letters, reviews, and other information are available for drug products approved since 1998 | Drugs@FDA provides rich information on drug approval history, which could be used as AI-powered LMs to explore underlying reasons for labeling changes and increase business success | www.fda.gov/drugs/drug-approvals-and-databases/drugsfda-data-files |
| FDA Guidance Documents | Guidance documents describe FDA’s interpretation of policy on a regulatory issue (21 CFR 10.115(b)). These documents usually discuss more specific products or issues that relate to design, production, labeling, promotion, manufacturing, and testing of regulated products | FDA Guidance Documents could be useful to implement AI-powered LMs for standardizing and monitoring crucial steps in drug discovery and development in terms of their consistency and alignment with regulatory requirements | www.fda.gov/regulatory-information/search-fda-guidance-documents |
| FDA Acronyms and Abbreviations | FDA Acronyms and Abbreviations database provides a quick reference to acronyms and abbreviations related to FDA activities | Emphasis of FDA Acronyms and Abbreviations is on scientific, regulatory, government agency, and computer application terms. The database includes some FDA organizational and program acronyms. It is a useful resource to define vocabularies in AI-powered LMs and increase model generalization | www.accessdata.fda.gov/scripts/cder/acronyms/index.cfm |
Opportunity 4: AI-powered model to advance postmarketing surveillance
Postmarket surveillance refers to the process of drug safety monitoring once drugs have reached the market, and is an essential part of the science of pharmacovigilance.69 The primary purpose of postmarket surveillance is to further refine, confirm, or refute the safety of a drug or device after use in the general population with a variety of medical conditions. Postmarket surveillance data are primarily derived from: (i) spontaneous/voluntary reporting of cases (e.g., FAERS, Local or Regional Joint Commission Requirement) and scientific literature publications; (ii) observational studies, including automated healthcare databases/social media, and randomized clinical trials; and (iii) active surveillance, such as the Drug-Induced Liver Injury Network (DILIN)70 and FDA Sentinel initiative.71 These real-world data (RWD) and RWE data sets are having an increasing role in healthcare decisions and are adopted by the FDA to monitor postmarket safety and AEs and to make regulatory decisions.72 The safety data accumulated in the postmarket stage provide an excellent resource for AI to deeply mine safety signals and advance pharmacovigilance.
AI-powered LMs have also been shown to be useful in improving the detection of drug–AE associations and in deciphering the causal relationship between the AE and clinical parameters.73, 74 Social media has gradually become one of the major resources for adverse drug reaction (ADR) monitoring. Breden et al. 75 proposed an ensemble model by integrating the BERT-large model, BioBERT, and ClinicalBERT to generate an enhanced automatic ADR detection within Twitter tweets. Relation extraction from clinical notes is a practical approach to detect the causality relationship between AE and relations. Guan et al. 76 combined the BERT model and Edge sampling to identify ADR and disease relationships from Electronic Health Records 1.0 (MADE) with improved performance. This developed model could be used to extract causal relationships from unstructured documents.
User cases: AI-powered model to combat emerging infections
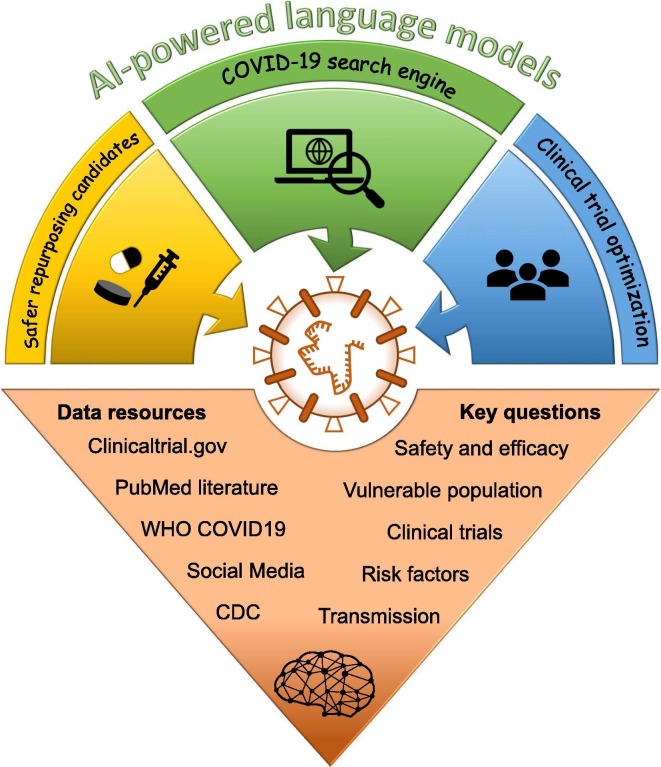
Emerging infectious diseases have been an ever-present threat to public health, and COVID-19 is a recent example.77 At the time of this review, COVID-19 had infected more than 31 million people, killed over 961 000, and resulted in catastrophic social and economic losses. Global efforts have been put into the development of effective treatments to combat this devastating deadly disease. Unfortunately, there are still no approved drugs or vaccines.78 Encouragingly, AI is proving invaluable in the battle against the coronavirus pandemic. Here, we illustrate how AI-powered LMs could aid the development of treatments for COVID-19 (Fig. 3 ).
Figure 3.
Artificial intelligence (AI)-powered language models for accelerating Coronavirus 2019 (COVID-19) treatment development. Potential opportunities, data resources, and key questions are illustrated. Abbreviation: CDC, Centers for Disease Control and Prevention.
COVID-19 search engine
COVID-19 may be the hottest topic in the scientific arena at this moment, resulting in more than 20,000 papers published in 2020 alone. The number is still increasing exponentially, and an average of 300 articles are being published every day. The published literature is a rich resource for promoting the development of COVID-19 treatments. However, there are far too many publications for any researcher to read. Some first efforts (CORD-19 data set) have been made to create the most extensive machine-readable coronavirus literature collection of COVID-19 available for data mining to date.79 The AI-powered COVID-19 search engine is a great solution to help researchers navigate the scientific literature for addressing different questions. More than 50 search and discovery tools have been developed and used for various topics, such as drug repurposing, interaction with other diseases, infection, mortality by demographic, and management policies.80 These AI-powered search engines allow researchers to ask specific questions, such as ‘what approved drug could potentially treat COVID-19?’.
Safer repurposing candidates
Drug repositioning and repurposing is being promoted as a rapid drug development paradigm for COVID-19 therapy.81, 82, 83, 84 Some repurposing candidates, including chloroquine and hydroxychloroquine, were initially authorized by the FDA for hospitalized patients only under careful heart monitoring because of the risk of heart rhythm problems.85, 86 The use of these two drugs for COVID-19 has now been revoked because the evidence suggests that they are unlikely to be an effective treatment. The potential risk of experiencing QTc prolongation with chloroquine or hydroxychloroquine was included in the FDA-approved drug label for their original indications.86 AI-powered LMs could be applied to extract the relationship between repurposed drugs and their potential AEs to prioritize repurposing candidates regarding their safety profiles.
Furthermore, COVID-19 has affected vulnerable populations and patients with pre-existing conditions disproportionately across the world.87 Patients with COVID-19 and pre-existing conditions and older patients have a high probability of encountering drug–drug interactions (DDIs) because they are more likely to take multiple medications.88 Therefore, caution should be considered before prescribing COVID-19 therapy to vulnerable populations. AI-powered models could be used to extract potential DDIs between COVID-19 repurposing candidates and other medicines for prevention.89
Clinical trial optimization
Treating patients with COVID-19 is forcing doctors to make hard decisions between two equally unattractive options: (i) prescribe drugs off-label in the hope that there will be some benefit; or (2) treat patients with standard supportive care for severe respiratory disease. This will continue to be the case until a confirmed randomized controlled trial establishes an effective treatment. Based on statistics from clinicaltrial.gov, there are currently more than 2900 clinical trials related to COVID-19. The number of enrolled patients, age groups, and demographic distributions within the clinical trials are highly variable, with the potential for controversies among trial sponsors. For example, disputes regarding treatment or preventative effects of hydroxychloroquine were reported based on different clinical trials.79, 90, 91, 92 A randomized controlled trial is currently underway in dozens of hospitals around the world proposed by REMAP-CAP using AI to guide researchers toward the most effective treatments for COVID-19.93
Concluding remarks
AI-powered LMs have enormous potential to transform every step of the drug discovery and development pipeline. As such, we expect different stakeholders to implement more investigation and real-world applications. We have illustrated the potential opportunities of AI-powered models in drug discovery and development, focusing on the role of AI-powered LMs for accelerating target identification, optimizing clinical trials, facilitating regulatory decision-making, and enhancing pharmacovigilance. Moreover, we highlighted how AI-powered LMs could promote treatment development in combating the COVID-19 pandemic. However, the implementation of AI-powered LMs in drug discovery is still in its infancy. Furthermore, besides AI-powered LM, other AI-based models been proposed and show promise in tackling different drug discovery and development questions. These are out of the scope of the current review, but we recommended a closer look at other AI-based models, which might combine with AI-power language models to enhance drug discovery and development.94, 95, 96
AI-powered LMs are a fast-evolving field, and many model architectures have been proposed. However, most of the applications in drug discovery and development are based on BERT and its derivates. Other newly developed LMs have claimed superior performance and strength based on evaluation data in the general domain. The utility of these transformer-based LMs in drug discovery and development remains to be established via further investigation and critical evaluation. To carry out a comprehensive assessment of different transformer-based LMs for various tasks in drug discovery and development, more standard benchmark data sets in the biomedical domain, such as BioASQ68 and Biomedical Language Understanding Evaluation (BLUE),97 are urgently needed.
The benefit of learning domain-specific corpus and knowledge has been demonstrated for LMs.14, 19, 98 However, these models have been retrained on top of the BERT-base model. Improved model performance is expected by using the BERT large model. Furthermore, we strongly recommend these advanced transformed-based LMs be retrained by using other regulatory-related documents to enhance their application in the regulatory process. Moreover, novel model architecture, such as GPT-3, showed potential in tackling downstream tasks without task-related fine-tuning data sets. A further investigation of biomedical applications should be conducted if a favorable performance is obtained, which could expand this utility in drug discovery and development.
Being able to explain how AI-powered LMs could be used in drug discovery and development is vital to building trust. Large LMs can produce powerful contextual representations that lead to improvements across many NLP tasks. Our ability to explore the biological relevance of these contextual representations will enhance adoption in the drug discovery and development process. Initial efforts, such as ExBERT, have been proposed to provide insights into the meaning of the contextual representations by matching a human-specified input to similar contexts in a large, annotated data set.99 More in-depth efforts to develop explainable transformers would be beneficial.
AI faces challenges in reproducibility because researchers have difficulty reproducing many vital results, hindering their real-world applications.100 Few efforts have been made to explore the reproducibility of AI-powered LMs.101 Some consortium efforts, such as the Kaggle challenge, might be a suitable platform to organize a comprehensive assessment of the reproducibility of AI-powered LMs with biomedical data, such as EHRs or PubMed literature.
AI-powered LMs have been applied in many different areas of biomedical science. The impact of AI-powered models in all areas of drug discovery and healthcare is already noticeable, especially in transforming clinical trial design. Conventional NLP, along with rule-based matching strategies, has also been extensively applied in drug discovery and development.7, 102 We believe the AI-based LMs could be complementary to conventional approaches to promote drug discovery and development. Here, we have summarized the challenges and opportunities presented by AI-powered LMs to stimulate community efforts for further evaluation and to better position and promote AI-powered LMs in drug discovery and development.
Declaration of interests
R.A.R. is co-founder and co-director of ApconiX, an integrated toxicology and ion channel company that provides expert advice on nonclinical aspects of drug discovery and drug development to academia, industry, and not-for-profit organizations.
Acknowledgments
X.C. and R.A.R. are grateful to the National Center for Toxicological Research (NCTR) for support through the Oak Ridge Institute for Science and Education (ORISE) via an interagency agreement between the Department of Energy and the FDA. R.H. is grateful for participation through part of the Tox21 project. The views presented in this article do not necessarily reflect the opinions of the FDA or the NIH. Any mention of commercial products is for clarification and is not intended as an endorsement. The work was funded and supported by the FDA Medical Countermeasures Initiative (MCMi).
Biographies

Zhichao Liu Zhichao Liu is a technical leader in Artificial Intelligence Research Force (AIRForce), Division of Bioinformatics & Biostatistics, FDA/NCTR. Dr Liu’s background spans the fields of chemistry, biology, and computer science. He has led many cutting-edge projects over the past decade by designing, implementing, and deploying artificial intelligence (AI)/machine-learning (ML) solutions for advanced regulatory sciences. Specifically, Dr Liu developed a standard pipeline for AI-powered drug repositioning to help the industry seek the optimal route to accelerate drug-development efficacy from an advanced regulatory-sciences perspective. Furthermore, Dr Liu developed AI/ML solutions for promoting predictive toxicology, with successful models adopted by the industry and regulators. His achievements have been reflected by Dr Liu being awarded five FDA-wide Awards, nine NCTR-level Awards, two scientific community-level awards, and by more than 100 peer-reviewed publications.

Ruth A. Roberts Ruth A. Roberts is Chair and Director of Drug Discovery at Birmingham University, UK and cofounder of ApconiX, an integrated toxicology and ion channel company. She is a former president of EUROTOX and of the British Toxicology Society (BTS), a Fellow and past president of the Academy of Toxicological Sciences (ATS), a fellow of the Royal College of Pathologists and of the Royal Society of Biology, and Vice Chair of the Health and Environmental Sciences Institute (HESI) Board. Ruth was the recipient of the SOT Achievement award in 2002, the EUROTOX Bo Holmstedt Award in 2009 and the SOT 2018 Founders award, given in recognition of outstanding leadership in fostering the role of toxicological sciences in safety decision making. With more than 150 publications in peer-reviewed journals, she is interested in developing and implementing innovative models in drug discovery and development.

Madhu Lal-Nag Madhu Lal-Nag was awarded a PhD in molecular and cellular oncology from The George Washington University and an MSc in bioscience business from The Keck Graduate Institute of Applied Biosciences, Claremont, CA. She was also awarded an MSc in biochemistry from the University of Mumbai, India. At NCATS, Dr Lal-Nag served as the Director of the Trans NIH RNAi Facility, which runs high-throughput functional genomics screens for the entire NIH intramural program, serving 21 institutes. Dr Lal-Nag moved to the Center for Drug Evaluation and Research at the FDA in December 2018, where she is the Program Director for the Research Governance Council, an advisory council overseeing CDER research. She is active in the microphysiological systems community, continuing to teach workshops and give scientific talks about the role and intersection of alternative animal models and microphysiological systems in evaluating the efficacy and safety of drugs in therapeutic development.

Weida Tong Weida Tong is the Director of the Division of Bioinformatics and Biostatistics at FDA/NCTR. He has published over 300 peer-reviewed papers from his roles in supervising and leading the FDA-led community-wide MicroArray and Sequencing Quality Control (MAQC/SEQC) consortium to analyze technical performance and practical utility of emerging genomic technologies with emphasis on regulatory application and precision medicine; addressing drug safety concerns related to drug-induced liver injury (DILI); developing machine learning (ML) and AI for digital health and drug repositioning; and conducting molecular modeling and QSARs on various toxicological endpoints, such as carcinogenicity.
References
- 1.A.I. breakthroughs in natural-language processing are big for business. www.fortune.com/2020/01/20/natural-language-processing-business/ [Accessed June 24, 2021].
- 2.Liu Z., Zhu L., Roberts R., Tong W. Toward clinical implementation of next-generation sequencing-based genetic testing in rare diseases: where are we? Trends Genet. 2019;35:852–867. doi: 10.1016/j.tig.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider P., Walters W.P., Plowright A.T., Sieroka N., Listgarten J., Goodnow R.A., Jr, et al. Rethinking drug design in the artificial intelligence era. Nat Rev Drug Discov. 2020;19:353–364. doi: 10.1038/s41573-019-0050-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen B., Garmire L., Calvisi D.F., Chua M.-S., Kelley R.K., Chen X. Harnessing big ‘omics’ data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:238–251. doi: 10.1038/s41575-019-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shilo S., Rossman H., Segal E. Axes of a revolution: challenges and promises of big data in healthcare. Nature Medicine. 2020;26(1):29–38. doi: 10.1038/s41591-019-0727-5. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal P., Searls D.B. Literature mining in support of drug discovery. Briefings in Bioinformatics. 2008;9(6):479–492. doi: 10.1093/bib/bbn035. [DOI] [PubMed] [Google Scholar]
- 8.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., et al. Applications of machine learning in drug discovery and development. Nature Reviews Drug Discovery. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. arXiv preprint 2017; arXiv:1706.03762v5.
- 10.Sutskever I, Vinyals O, Le QV. Sequence to sequence learning with neural networks. arXiv preprint 2014: arXiv;1409.3215.
- 11.The fall of RNN/LSTM. www.towardsdatascience.com/the-fall-of-rnn-lstm-2d1594c74ce0. [Accessed June 24, 2021].
- 12.Salehinejad H, Sankar S, Barfett J, Colak E, Valaee S. Recent advances in recurrent neural networks. arXiv preprint 2017; arXiv:1801.01078.
- 13.Parisi G.I., Kemker R., Part J.L., Kanan C., Wermter S. Continual lifelong learning with neural networks: a review. Neural Networks. 2019;113:54–71. doi: 10.1016/j.neunet.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsentzer E, Murphy JR, Boag W, Weng, W-H, Jin D, Naumann T, et al. Publicly available clinical BERT embeddings. arXiv preprint 2019; arXiv:1904.03323.
- 16.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. arXiv preprint 2018; arXiv:1810.04805.
- 17.Liu PJ, Saleh M, Pot E, Goodrich B, Sepassi R, Kaiser L et al. Generating Wikipedia by summarizing long sequences. arXiv preprint 2018; arXiv:1801.10198.
- 18.Kitaev N, Kaiser Ł, Levskaya A. Reformer: the efficient transformer. arXiv preprint 2020; arXiv:2001.04451.
- 19.Huang K, Altosaar J, Ranganath R. Clinicalbert: modeling clinical notes and predicting hospital readmission. arXiv preprint 2019; arXiv:1904.05342.
- 20.Sanz F, Pognan F, Steger-Hartmann T, Díaz C; eTOX, Cases M et al. Legacy data sharing to improve drug safety assessment: the eTOX project. Nature Reviews Drug Discovery 2017; 16(12): 811-812. [DOI] [PubMed]
- 21.Harrison C. GlaxoSmithKline opens the door on clinical data sharing. Nature Reviews Drug Discovery. 2012;11(12):891–892. doi: 10.1038/nrd3907. [DOI] [PubMed] [Google Scholar]
- 22.Wei C-H, Lee K, Leaman R, Lu Z. Biomedical mention disambiguation using a deep learning approach. arXiv preprint 2019; arXiv:1909.10416v1
- 23.Ziemann M., Eren Y., El-Osta A. Gene name errors are widespread in the scientific literature. Genome Biology. 2016;17(1):177. doi: 10.1186/s13059-016-1044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Data labeling for natural language processing. www.topbots.com/data-labeling-for-natural-language-processing/. [Accessed June 24, 2021].
- 25.Sanh V, Debut L, Chaumond J, Wolf T. DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter. arXiv preprint 2019; arXiv:1910.01108.
- 26.Clark K, Luong M-T, Le QV, Manning CD. Electra: pre-training text encoders as discriminators rather than generators. arXiv preprint 2020; arXiv:2003.10555.
- 27.Lan Z, Chen M, Goodman S, Gimpel K, Sharma P, Soricut R. Albert: A lite Bert for self-supervised learning of language representations. arXiv preprint 2019; arXiv:1909.11942.
- 28.Schenone M., Dančík V., Wagner B.K., Clemons P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nature Chemical Biology. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giorgi J.M., Bader G.D. Towards reliable named entity recognition in the biomedical domain. Bioinformatics. 2020;36(1):280–286. doi: 10.1093/bioinformatics/btz504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan MR, Ziyadi M, AbdelHady M. MT-BioNER: multi-task learning for biomedical named entity recognition using deep bidirectional transformers. arXiv preprint 2020; arXiv:2001.08904.
- 31.Liu Z., Fang H., Borlak J., Roberts R., Tong W. In vitro to in vivo extrapolation for drug-induced liver injury using a pair ranking method. ALTEX. 2017;34(3):399–408. doi: 10.14573/altex.1610201. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Zhang Y., Ren X., Zhang Y., Zitnik M., Shang J., et al. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. 2018;35(10):1745–1752. doi: 10.1093/bioinformatics/bty869. [DOI] [PubMed] [Google Scholar]
- 33.Nourani E., Reshadat V. Association extraction from biomedical literature based on representation and transfer learning. Journal of Theoretical Biology. 2020;488 doi: 10.1016/j.jtbi.2019.110112. [DOI] [PubMed] [Google Scholar]
- 34.Moradi M., Dorffner G., Samwald M. Deep contextualized embeddings for quantifying the informative content in biomedical text summarization. Computer Methods and Programs in Biomedicine. 2020;184 doi: 10.1016/j.cmpb.2019.105117. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Guo Y, Wang Y, Sun H, Huang J. SMILES-BERT: large scale unsupervised pre-training for molecular property prediction. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, New York: Association for Computing Machinery; 2019: 429–436
- 36.What is FASTA format? https://zhanglab.dcmb.med.umich.edu/FASTA/ [Accessed June 24, 2021].
- 37.Jaeger S., Fulle S., Turk S. Mol2vec: unsupervised machine learning approach with chemical intuition. Journal of Chemical Information and Modeling. 2018;58(1):27–35. doi: 10.1021/acs.jcim.7b00616. [DOI] [PubMed] [Google Scholar]
- 38.Winter R., Montanari F., Noé F., Clevert D.-A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chemical Science. 2019;10(6):1692–1701. doi: 10.1039/c8sc04175j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda S, Shi S, Ueda HR. SMILES Transformer: pre-trained molecular fingerprint for low data drug discovery. arXiv preprint 2019: arXiv:1911.04738.
- 40.Li X., Fourches D. Inductive transfer learning for molecular activity prediction: next-gen QSAR models with MolPMoFiT. Journal of Cheminformatics. 2020;12(1):27. doi: 10.1186/s13321-020-00430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabian B, Edlich T, Gaspar H, Segler M, Meyers J, Marco Fiscato M, et al. Molecular representation learning with language models and domain-relevant auxiliary tasks. arXiv preprint 2020: arXiv:2011.13230.
- 42.Scannell J.W., Blanckley A., Boldon H., Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nature Reviews Drug Discovery. 2012;11(3):191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 43.Fogel D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp Clin Trials Commun. 2018;11:156–164. doi: 10.1016/j.conctc.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrer S., Shah P., Antony B., Hu J. Artificial intelligence for clinical trial design. Trends in Pharmacological Sciences. 2019;40(8):577–591. doi: 10.1016/j.tips.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Blanco A., Perez-de-Viñaspre O., Pérez A., Casillas A. Boosting ICD multi-label classification of health records with contextual embeddings and label-granularity. Computer Methods and Programs in Biomedicine. 2020;188 doi: 10.1016/j.cmpb.2019.105264. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Xiao C, Glass LM, Sun J. DeepEnroll: patient-trial matching with deep embeddingand entailment prediction. arXiv preprint 2020; arXiv:2001.08179.
- 47.Hall J.L., Ryan J.J., Bray B.E., Brown C., Lanfear D., Newby L.K., et al. Merging electronic health record data and genomics for cardiovascular research: A Science Advisory from the American Heart Association. Circulation: Cardiovascular. Genetics. 2016;9(2):193–202. doi: 10.1161/HCG.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dias R., Torkamani A. Artificial intelligence in clinical and genomic diagnostics. Genome Medicine. 2019;11(1):70. doi: 10.1186/s13073-019-0689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo M. An AI boost for clinical trials. Nature. 2019;573(7775):S100–S102. doi: 10.1038/d41586-019-02871-3. [DOI] [PubMed] [Google Scholar]
- 50.Steinhubl S.R., Wolff-Hughes D.L., Nilsen W., Iturriaga E., Califf R.M. Digital clinical trials: creating a vision for the future. NPJ Digital Medicine. 2019;2(1):126. doi: 10.1038/s41746-019-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Chen P.-H.C., Krause J., Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA. 2019;322(18):1806–1816. doi: 10.1001/jama.2019.16489. [DOI] [PubMed] [Google Scholar]
- 52.Sim I. Mobile devices and health. New England Journal of Medicine. 2019;381(10):956–968. doi: 10.1056/NEJMra1806949. [DOI] [PubMed] [Google Scholar]
- 53.Wilder J.L., Nadar D., Gujral N., Ortiz B., Stevens R., Holder-Niles F., et al. Pediatrician attitudes toward digital voice assistant technology use in clinical practice. Appl Clin Inform. 2019;10(2):286–294. doi: 10.1055/s-0039-1687863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anon. Getting real with wearable data. Nature Biotechnology 2019; 37(4): 331-331. [DOI] [PubMed]
- 55.Statement from FDA Commissioner Scott Gottlieb, M.D., on FDA’s new strategic framework to advance use of real-world evidence to support development of drugs and biologics. www.fda.gov/news-events/press-announcements/statement-fda-commissioner-scott-gottlieb-md-fdas-new-strategic-framework-advance-use-real-world. [Accessed June 24, 2021].
- 56.Tran V.-T., Riveros C., Ravaud P. Patients’ views of wearable devices and AI in healthcare: findings from the ComPaRe e-cohort. NPJ Digital Medicine. 2019;2(1):53. doi: 10.1038/s41746-019-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Study data standards: what you need to know. www.fda.gov/media/98907/download. [Accessed June 24, 2021].
- 58.FDA’s Document Archiving, Reporting, and Regulatory Tracking System (DARRTS). www.fda.gov/media/80214/download. [Accessed June 24, 2021].
- 59.The future of FDA’s electronic safety surveillance. www.fda.gov/news-events/fda-voices/future-fdas-electronic-safety-surveillance. [Accessed June 24, 2021].
- 60.Awaysheh A., Wilcke J., Elvinger F., Rees L., Fan W., Zimmerman K. A review of medical terminology standards and structured reporting. J Vet Diagn Invest. 2018;30(1):17–25. doi: 10.1177/1040638717738276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Centers for Medicare & Medicaid Services, HHS. Medicare and Medicaid programs; electronic health record incentive program. Final rule. Fed Regist 2010; 75(144): 44313-44588. [PubMed]
- 62.Anon. Standards for medical identifiers, codes, and messages needed to create an efficient computer-stored medical record. J. Am. Med. Informatics. 1994;1:1–7. doi: 10.1136/jamia.1994.95236133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji Z., Wei Q., Xu H. Bert-based ranking for biomedical entity normalization. AMIA Summits on Translational Science Proceedings. 2020;2020:269. [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, Liu J, Razavian N. BERT-XML: large scale automated ICD coding using BERT pretraining. arXiv preprint 2020; arXiv:2006.03685.
- 65.Sohn S., Comeau D.C., Kim W., Wilbur W.J. Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics. 2008;9(1):402. doi: 10.1186/1471-2105-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fiorini N., Leaman R., Lipman D.J., Lu Z. How user intelligence is improving PubMed. Nature Biotechnology. 2018;36(10):937–945. doi: 10.1038/nbt.4267. [DOI] [PubMed] [Google Scholar]
- 67.Chen Q., Lee K., Yan S., Kim S., Wei C.-H., Lu Z. BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale. PLOS Computational Biology. 2020;16(4) doi: 10.1371/journal.pcbi.1007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsatsaronis G., Balikas G., Malakasiotis P., Partalas I., Zschunke M., Alvers M.R., et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinformatics. 2015;16(1):138. doi: 10.1186/s12859-015-0564-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beninger P. Pharmacovigilance: an overview. Clinical Therapeutics. 2018;40(12):1991–2004. doi: 10.1016/j.clinthera.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 70.Fontana R.J., Watkins P.B., Bonkovsky H.L., Chalasani N., Davern T., Serrano J., et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32(1):55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.FDA’s Sentinel Initiative. www.fda.gov/safety/fdas-sentinel-initiative. [Accessed June 24, 2021].
- 72.Real-world evidence. www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence. [Accessed June 24, 2021].
- 73.Fan B., Fan W., Smith C., Garner H.S. Adverse drug event detection and extraction from open data: a deep learning approach. Information Processing & Management. 2020;57(1) [Google Scholar]
- 74.Biseda B, Mo K. Enhancing pharmacovigilance with drug reviews and social media. arXiv preprint 2020; arXiv:2004.08731.
- 75.Breden A, Moore L. Detecting adverse drug reactions from Twitter through domain-specific preprocessing and BERT ensembling. arXiv preprint 2020; arXiv:2005.06634.
- 76.Guan H., Devarakonda M. Leveraging contextual information in extracting long distance relations from clinical notes. AMIA Annu Symp Proc. 2020;2019:1051–1060. [PMC free article] [PubMed] [Google Scholar]
- 77.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang W., Cao Z., Han M., Wang Z., Chen J., Sun W., et al. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: open label, randomised controlled trial. BMJ. 2020;369 doi: 10.1136/bmj.m1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brainard J. New tools aim to tame pandemic paper tsunami. Science. 2020;368(6494):924–925. doi: 10.1126/science.368.6494.924. [DOI] [PubMed] [Google Scholar]
- 81.Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368(6493):829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 82.Shaffer L. 15 drugs being tested to treat COVID-19 and how they would work. Nature Medicine. Published online May 15, 2020. http://dx.doi.org/10.1038/d41591-020-00019-9. [DOI] [PubMed]
- 83.Delavan B., Roberts R., Huang R., Bao W., Tong W., Liu Z. Computational drug repositioning for rare diseases in the era of precision medicine. Drug Discovery Today. 2018;23(2):382–394. doi: 10.1016/j.drudis.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. 2020;38(4):379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 85.Mercuro N.J., Yen C.F., Shim D.J., Maher T.R., McCoy C.M., Zimetbaum P.J., et al. Risk of QT interval prolongation associated with use of hydroxychloroquine with or without concomitant azithromycin among hospitalized patients testing positive for Coronavirus Disease 2019 (COVID-19) JAMA Cardiology. 2020;5:1036–1041. doi: 10.1001/jamacardio.2020.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chorin E., Dai M., Shulman E., Wadhwani L., Bar-Cohen R., Barbhaiya C., et al. The QT interval in patients with COVID-19 treated with hydroxychloroquine and azithromycin. Nature Medicine. 2020;26:808–809. doi: 10.1038/s41591-020-0888-2. [DOI] [PubMed] [Google Scholar]
- 87.Selvan M.E. Risk factors for death from COVID-19. Nature Reviews Immunology. 2020;20:407. doi: 10.1038/s41577-020-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roden D.M., Harrington R.A., Poppas A., Russo A.M. Considerations for drug interactions on QTc interval in exploratory COVID-19 treatment. Journal of the American College of Cardiology. 2020;75(20):2623–2624. doi: 10.1016/j.jacc.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang T., Leng J., Liu Y. Deep learning for drug–drug interaction extraction from the literature: a review. Briefings in Bioinformatics. 2019;21:1609–1627. doi: 10.1093/bib/bbz087. [DOI] [PubMed] [Google Scholar]
- 90.Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. New England Journal of Medicine. 2020;383:517–525. doi: 10.1056/NEJMoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Z, Hu J, Zhang Z, Jiang S, Han S, Yan D, et al. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. medRxiv 2020: 2020.2003.2022.20040758.
- 92.Mahévas M., Tran V.T., Roumier M., Chabrol A., Paule R., Guillaud C., et al. Clinical efficacy of hydroxychloroquine in patients with covid-19 pneumonia who require oxygen: observational comparative study using routine care data. BMJ. 2020;369 doi: 10.1136/bmj.m1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.REMAP-CAP response to the COVID-19 pandemic. www.remapcap.org/coronavirus. [Accessed June 24, 20201].
- 94.Paul D., Sanap G., Shenoy S., Kalyane D., Kalia K., Tekade R.K. Artificial intelligence in drug discovery and development. Drug Discovery Today. 2021;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bender A., Cortés-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: ways to make an impact, and why we are not there yet. Drug Discovery Today. 2021;26(2):511–524. doi: 10.1016/j.drudis.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 96.Mak K.-K., Pichika M.R. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today. 2019;24(3):773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 97.Peng Y, Yan S, Lu Z. Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets. arXiv preprint 2019; arXiv:1906.05474.
- 98.Beltagy I, Lo K, Cohan A. SciBERT: a pretrained language model for scientific text. arXiv preprint 2019: arXiv:1903.10676v3.
- 99.Hoover B, Strobelt H, Gehrmann S. exbert: A visual analysis tool to explore learned representations in transformers models. arXiv preprint 2019; arXiv:1910.05276.
- 100.Hutson M. Artificial intelligence faces reproducibility crisis. Science. 2018;359(6377):725–726. doi: 10.1126/science.359.6377.725. [DOI] [PubMed] [Google Scholar]
- 101.Gundersen O.E., Kjensmo S. State of the art: reproducibility in artificial intelligence. AAAI. 2018;2018:1644–1651. [Google Scholar]
- 102.Zhao S., Su C., Lu Z., Wang F. Recent advances in biomedical literature mining. Briefings in Bioinformatics. 2020;22:bbaa057. doi: 10.1093/bib/bbaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Broder A.Z., Glassman S.C., Manasse M.S., Zweig G. Syntactic clustering of the Web. Computer Networks and ISDN Systems. 1997;29(8):1157–1166. [Google Scholar]
- 104.Schwenk H. Continuous space language models. Computer Speech & Language. 2007;21(3):492–518. [Google Scholar]
- 105.Bengio Y., Schwenk H., Senécal J.-S., Morin F., Gauvain J.-L. In: Innovations in machine learning: theory and applications. Holmes D.E., Jain L.C., editors. Springer; Berlin: 2006. Neural probabilistic language models; pp. 137–186. [Google Scholar]
- 106.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. arXiv preprint 2013; arXiv:1310.4546v1.
- 107.Pennington J, Socher R, Manning CD. GloVe: global vectors for word representation. In: Moschitti A, Pang B, Daelemans W, eds. Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP) 2014. Stroudsburg: Association for Computational Linguistics, 2014: 1532–1543.
- 108.Bojanowski P., Grave E., Joulin A., Mikolov T. Enriching word vectors with subword information. Transactions of the Association for Computational Linguistics. 2017;5:135–146. [Google Scholar]
- 109.Liu Y, Ott M, Goyal N, et al. Roberta: A robustly optimized BERT pretraining approach. arXiv preprint 2019: arXiv:1907.11692.
- 110.https: //commoncrawl.org/ [Accessed June 24, 2021].
- 111.Brown TB, Mann B, Ryder N, et al. Language models are few-shot learners. arXiv preprints 2020; arXiv:2005.14165.
- 112.Gao J, Xiao C, Glass LM, Sun J. COMPOSE: cross-modal pseudo-Siamese network for patient trial matching. arXiv preprint 2020; arXiv:2006.08765v1.
- 113.Cocos A., Fiks A.G., Masino A.J. Deep learning for pharmacovigilance: recurrent neural network architectures for labeling adverse drug reactions in Twitter posts. J Am Med Inform Assoc. 2017;24(4):813–821. doi: 10.1093/jamia/ocw180. [DOI] [PMC free article] [PubMed] [Google Scholar]