Lay Summary
A review of the medical literature since the completion of the Human Genome Project in 2003 revealed that race, a social construct created to justify slavery, continues to be used as a genetic category, due to a lack of understanding of the continuous nature of human genetic variation.
Keywords: African ancestry, African American, black, clinical trials, genetic variation, evolutionary biology
Abstract
Social scientists have long understood race to be a social category invented to justify slavery and evolutionary biologists know the socially constructed racial categories do not align with our biological understanding of genetic variation. The completion of the Human Genome Project in 2003 confirmed humans are 99.9% identical at the DNA level and there is no genetic basis for race. A systematic review of the PubMed medical literature published since 2003 was conducted to assess the use of African ancestry to denote study populations in genetic studies categorized as clinical trials, to examine the stated rationale for its use and to assess the use of evolutionary principles to explain human genetic diversity. We searched for papers that included the terms ‘African’, ‘African American’ or ‘Black’ in studies of behavior (20 papers), physiological responses, the pharmacokinetics of drugs and/or disease associations (62 papers), and as a genetic category in studies, including the examination of genotypes associated with life stress, pain, stuttering and drug clearance (126 papers). Of these, we identified 74 studies in which self-reported race alone or in combination with admixture mapping was used to define the study population. However, none of these studies provided a genetic explanation for the use of the self-identified race as a genetic category and only seven proffered evolutionary explanations of their data. The concept of continuous genetic variation was not clearly articulated in any of these papers, presumably due to the paucity of evolutionary science in the college and medical school curricula.
INTRODUCTION
Social scientists have long understood race to be a social construct used in its most benign form to categorize groups of people according to a small group of phenotypes and cultural differences and in its most insidious form to assign value to a social hierarchy. The concept and significance of race varies around the world with the USA first creating and assigning value to racial categories to justify slavery. Biologists and geneticists have historically been divided as to whether ‘race’ also defines distinct biological and genetic categories. Herein, race without notations is used to denote its use as a social construct, while ‘race’ denotes its use as a biological or genetic entity. Definitions of these latter terms vary widely. Genetic ‘race’ has been viewed as a result of human migration with genetic isolation leading to the development of distinct populations that share DNA as the result of common descent. While the terms genetic ‘race’ and biological ‘race’ have been used interchangeably, Templeton [1] defines biological race as (i) geographically defined populations within a species that have sharp boundaries that separate them from other species or as (ii) distinct evolutionary lineages within a species characterized by a continuous line of descent. He emphasizes that genetic differentiation alone is insufficient to define a subspecies or race under either of these definitions as both require that genetic differentiation exists across sharp boundaries and not as gradual changes.
As a taxonomic term, race defines an informal subdivision of subspecies which are physically and genetically different. However, the species Homo sapiens cannot be further subdivided into subspecies which are physically and genetically different. Thus, for H.sapiens, the species and subspecies are the same—H.sapiens sapiens (Box 1). In 2003, Phase 1 of the Human Genome Project (HGP) demonstrated that humans populating the earth today are on average 99.9% identical at the DNA level, there is no genetic basis for race, and there is more genetic variation within a race than between them [2]. In addition, genetic isolation, sharp boundaries and distinct evolutionary lineages of ‘races’ do not exist. Thus, the idea of ‘race’ as a genetic category was presumably put to rest. The continued acceptance of ‘race’ as an appropriate biological category would have to be predicated on data indicating there are genes distinct to one ‘race’ that are transcribed in one ‘race’, but not another and human genetic variation is not continuous. This is distinct from small differences in allele frequencies due to mutations in a given family’s genetic lineage as will be discussed later in this manuscript.
Box 1. Taxonomy of Homo sapiens sapiens
Kingdom—Animalia
Phylum—Chordata
Class—Mammalia
Subclass—Eutheria
Order—Primates
Family—Hominidae
Genus—Homo
Species—Sapiens
Subspecies—Sapiens
Humans—Homo sapiens sapiens
Evolutionary biologist Joseph L. Graves Jr sought to resolve the misunderstandings and misconceptions with the publication of The Emperor’s New Clothes: Biological Theories of Race at the Millennium 2 years prior to the sequencing of the human genome. It discussed the origins of the race concept, scientific racism, the misapplications of Darwinism, eugenics and the fallacies of the association of race with IQ and disease [3]. This was followed by publication of The Race Myth: Why We Pretend Race Exists in America in 2005, which discussed how the socially constructed racial categories do not align with our biological understanding of genetic variation [4].
At this same time that the human genome was sequenced and the concept of biological ‘race’ and racism were under renewed scrutiny, the FDA and the US Patent Office approved a drug named BiDil® to treat heart failure in ‘self-identified African Americans’ [5, 6]. In brief, the US Patent Office had issued a method patent in 1989 to a company as they had combined two generic anti-hypertensive drugs that had each been in use for 20 years into one medication. Eight years later the company submitted a New Drug Application to the FDA as they had demonstrated the combined drugs were as biopotent as the co-administered drugs. The application was denied as there were too many variables as primary endpoints to interpret two decades of clinical trials data with any certainty. However, in 2000, the company submitted a new patent application for the use of the drug by African Americans. In 2002, the US Patent Office granted the patent for ‘methods for treating and preventing mortality associated with heart failure in an African-American patient’ and in 2005, the FDA approved BiDil® for ‘self-identified African Americans’.
Examination of the rationale for the approval of this drug provides insight into problems of experimental design, but provided no insight into the rationale to view African Americans as physiologically distinct (Table 1). The report of a 2:1 black/white mortality rate due to heart failure at age 35–74 was an erroneous calculation as it does not take into account that 71% of Caucasian American mortality due to heart failure occurred after age 74. When corrected, the ratio was ∼1.1:1. The report of the lack of effectiveness of angiotensin-converting enzyme (ACE) inhibitors in African Americans based on blood pressure measurements was also disputed as other publications reported ACE inhibitors to be equally effective in African Americans compared to Caucasian Americans [7, 8]. The single study of nitric oxide turnover examined umbilical endothelial cells in culture collected from only 13 black women and 12 white women without acknowledging that the endothelial cells were fetal in origin, not maternal [9]. In the two studies of ACE inhibitors and the kinetic study of nitric oxide turnover the rationale for the study of black individuals was epidemiological, rather than biological, that is, the authors cited the greater incidence of hypertension and heart disease in the black population compared to the white population as the rationale for their concept of biological ‘race’.
Table 1.
Rationale for approval of BiDil and assessment of rationale
| Rationale for BiDil approval | Assessment of rationale |
|---|---|
| Disparate burden of heart failure among African Americans | A correct health statistic describing a correlation |
| 2:1 Black/white mortality rate due to heart failure at age 35–74 | Erroneous calculation. The calculation did not take into account that 71% of Caucasian American mortality due to heart failure occurred after age 74 |
| Lack of effectiveness of ACE inhibitors in African Americans | Disputed. Selective citation of the literature. Other studies showed ACE inhibitors to be equally effective in African Americans |
| Ethnic difference in pathophysiology of nitric oxide production and utilization between African Americans and Caucasian Americans | Misuse of the word ethnic. Fewer than 40 African-American participants in each of three studies. Umbilical cells studied are of fetal origin, not maternal |
| 35% decrease in mortality rates of blacks | Unexplained |
A portion of the biomedical community heralded BiDil® as a great discovery, while others voiced strong objections. The immediate response was the publication of articles, editorials, letters to the editor and commentaries in medical journals expounding on issues of ethics, law, commerce, racial categorization and racial profiling raised by the approval of this drug for ‘self-identified African Americans’. Of particular interest was the range of perspectives from the scientific community on the use of ‘race’ as a biological category (Table 2). While the extremes were that ‘race’ was or was not a legitimate biological category, some considered it to be a legitimate proxy for a biological category, while others believed its use, though not ideal, would inevitably continue as it was so entrenched in the biomedical literature. Yet others believed its use would be obviated by the advent of ‘individualized medicine’ at some point in the future, though they did not clarify how population data by race would be applied to individuals. The long-term response has been a growing body of literature addressing the problems associated with the biologization of race and the racialization of medicine [10–21].
Table 2.
Perspectives on use of ‘race’ as a biological category
|
A decade following this controversy the issue of ‘race’ as a genetic category or as a proxy for a genetic category persists. In 2016, the NIH ‘Workshop on the Use of Race and Ethnicity in Genomics and Biomedical Research’ was sponsored by the National Human Genome Research Institute (NHGRI) and the National Institute on Minority Health and Health Disparities (NIMHD) [22]. It was convened to discuss the use of race and ethnicity data in genomics, biomedical and clinical research and their application to minority health and health disparities. The meeting summary emphasized the importance of the discussion to facilitate rigorous scientific study design in order to influence how scientists and the public conceptualize, discuss and react to human differences. It warns that misuse of population descriptors in biomedical research has the potential to perpetuate misinformation, stigmatize certain groups and simplify the complex relationships between individual identity, genetics and health. This warning was never more urgent than it is now during the COVID-19 pandemic, which is underlining the centuries of health disparities burdening African Americans.
EXAMINATION OF THE USE OF AFRICAN ANCESTRY IN GENETIC STUDIES: SYSTEMATIC REVIEW METHODS
We undertook a systematic review of the medical literature to better understand the use of race in biomedical research. To do this, we searched the literature published since 2003 to assess the use of African ancestry and categorization of subjects into African/African American/black populations. We focused on genetic studies categorized as clinical trials since completion of Phase I of the HGP in 2003.
At the outset a National Library of Medicine PubMed database search of the biomedical literature was used to identify papers of interest where Africa, African Americans or blacks were participants in genetics clinical trials. Study of PubMed MeSH terms in consultation with a University of Wisconsin Health Sciences librarian revealed ‘African Continental Ancestry’ to be the MeSH term which encompassed African ancestry, black, African and African American. The terms ‘genetics’ and ‘clinical trials’ were intentionally selected for their breadth to not restrict the breadth of the papers cataloged under ‘African Continental Ancestry’.
A PubMed search of the biomedical literature published from January 2003 to December 2019 was then conducted using the following specific search terms: African continental ancestry, genetics and clinical trials. In each publication the (i) specific use of African continental ancestry, African, African American or black was examined and (ii) the rationale for the use of ‘race’ as a genetic category was examined. (iii) The use of evolutionary explanations of human genetic variation was also assessed to determine if the respective authors framed the population as a biological ‘race’ (Box 2).
Box 2. Evolutionary processes responsible for human genetic variation
Genetic drift: Variation in the relative frequency of different alleles due to the chance disappearance of particular genes as individuals die or do not reproduce.
Gene flow: Transfer of genetic material from one population to another.
Natural selection Survival of the fittest.
Founder effect: The loss of genetic variation that results when a new population is established by a small number of individuals separate from a larger population.
Mutations: The change in the structure of a gene, resulting in a variant form that may be transmitted to subsequent generations, caused by the alteration of single base units in DNA, or the deletion, insertion or rearrangement of larger sections of genes or chromosomes.
A total of 208 full-length, peer-reviewed papers published during this 17-year period were identified [23–61] (Supplementary Material). Each publication was reviewed to identify studies that used African ancestry, African, African American or black (i) as a social construct, (ii) to correlate race with responses or outcomes or (iii) as a genetic category (Table 3). The publications were evaluated by two authors to determine the stated rationale for study of individuals of African ancestry and to assess evolutionary explanations of genetic differences. In addition, a Portable Document Format of the publications was searched using Preview software for the following keywords related to evolutionary processes, family lineage, genetic lineage and admixture: race, African, African ancestry, African American, black, self-identified, self-reported, ethnic, admixture, ancestry informative marker, ancestry proportion, association, Hardy-Weinberg, family, parent, grandfather, grandmother, grandparent, genetic variation, GWAS, linkage, evolution, genetic drift, gene flow, natural selection, founder effect and mutation. Data were entered into a Filemaker database to facilitate analysis.
Table 3.
Uses of African ancestry
| Use of race | Study participants | Percent studies (# participants) | Reference |
|---|---|---|---|
| Race as a social construct | 9.3 % (20) | Supplementary Material | |
| ‘Race’ to study physiological responses, pharmacokinetics of drugs and/or disease associations | 28.4% (62) | Supplementary Material | |
| ‘Race’ as a genetic category examining genotypes associated with a wide range of symptoms, disorders, and diseases | 58.7% (74/126) | ||
| Black study population only | 38.9% (49/126) | ||
| Black and white study populations | 61.1% (77/126) | ||
| Inclusion of self-identified black race/color | 58.7 % (74/126) | ||
| Self-identified black race and admixture mapping | 8.7% (11/126) | [27–37] | |
| Self-identified with 3–4 black grandparents | 2.4% (3/126) | [24–26] | |
| Exclusion by self-identified black race | 0.5% (1/126) | [23] | |
| Exclusion due to mismatch of self-identification and genetically inferred ethnicity | 0.9% (2/126) | [38, 39] | |
| Parents and 1 child | 0.8% (1/126 | [39] | |
| Parents and 3 offspring | 0.8% (1/126) | [46] | |
| Sibling pairs as well as affected relative pairs (sibling, first cousin, uncle, grandparent) | 0.8% (1/126) | [28] | |
| Two family members with disease | [32] | ||
| 22% family members | [57] | ||
| Families with two alcohol dependent sibling | 0.5% (1/126) | [32] | |
| Study participants unrelated | 0.9% (2/126) | [47, 48] | |
| African-American cohort studies- | |||
| African-American Heart Failure Trial | 0.9% (2/215) | [49, 50] | |
| African-American Study of Kids | 1.4% (3/215) | [51–53] | |
| Strong African-American Families | 0.9% (2/215) | [54–56] | |
| MESA study | [58] | ||
| Heritage study | [59–61] |
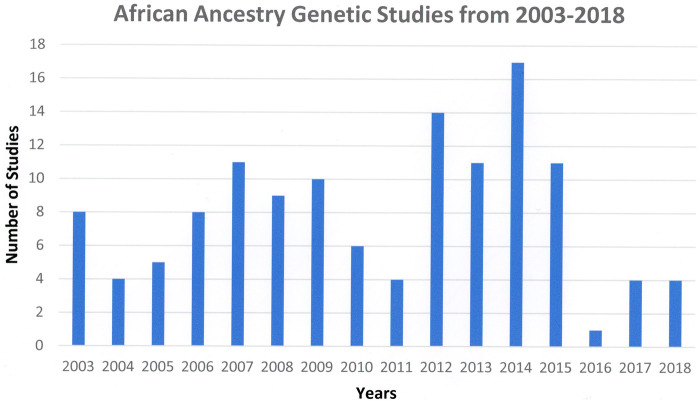
The terms African, African ancestry, African American or black were used as a social construct/category in 9.3% (20) of the 208 papers reviewed [23–61] (Supplementary Material). An additional 28.4% (61) of the studies used these terms to describe studies of physiological responses, the pharmacokinetics of drugs and/or disease associations. In the remaining 58.6% (126 papers) of the studies, African ancestry, African, African American or black were used to define a population in studies including the examination of genotypes associated with life stress, pain, stuttering and drug clearance. This latter group is the subject of further analysis in this review as we wish to determine the author’s conceptions of ‘race’. In these studies, the terms race and ethnicity were often used interchangeably to denote a population of African origin and/or a black study population, though they are not equivalent terms, ethnicity encompassing common ancestry, shared beliefs, cultural traditions, religion and language as well as race. The number of genotyping studies since 2003 does not reflect a specific pattern, the same research group being responsible for multiple papers (Fig. 1). However, it does indicate that there was not a precipitous decline upon completion of Phase I of the HGP.
Figure 1.
Number of African ancestry genetic studies by year since completion of Phase I of the Human Genome Project in 2003
CHARACTERIZATION OF AFRICAN ANCESTRY IN STUDY PARTICIPANTS: SUMMARY FINDINGS
The systematic review assessed (i) the use of African ancestry to denote study populations in genetic studies categorized as clinical trials, (ii) examined the stated rationale for its use and (iii) assessed the use of evolutionary principles to explain human genetic diversity.
African ancestry to denote study populations
Self-reported race
Self-reported race, surrogate-reported race or self-reported skin color were used in 58.7% of the studies to identify and include individuals of African ancestry, while a single study used self-identified race to exclude participants of African ancestry [23] (Table 3). A portion of the studies included individuals who self-identified as African American or black only if they also reported that three or four grandparents self-identified as African American or black [24–26]. Self-identification was also used in combination with admixture mapping using ancestry informative markers [27–37]. Two studies excluded individuals when there was a mismatch between self-identification and genetically inferred ethnicity [38, 39].
Comparison of black and white populations
Black populations only were studied in 38.9%, including large cohort studies, such as the African-American Heart Failure Trial, African-American Study of Kids and Strong African-American Families [50–56]. Black populations were compared to white reference populations in 61.6% (77) of the studies.
Identification of related study participants
To determine whether study participants were related and therefore genetically similar, the studies were also reviewed to determine whether two or more members of the same family were studied. Table 3 shows that different approaches were taken ranging from parents and one to three children or sibling pairs. While it was stated that African-American families were studied, it was not always clear whether two or more members of the same family were studied and whether those two members were direct ancestors, that is, a child, parents and grandparents, but not aunts, uncles and cousins. An example is the study of Musani et al. [57], who indicates the population included 22% family members; however, it is not possible to determine which portion of the data are based on direct ancestors or whether all data were pooled across seven families.
The size of study populations
A subset of 117 of the studies focused on African Americans/American blacks was further examined to assess the size of the study populations. Thirty-one studies had 100 or fewer participants, 87 fewer than 500 participants and 30 over 500 participants (range 502–5047). There was no discussion as to whether the data from these studies could be generalized to any other African American/black population.
Rationale for use of African ancestry as a genetic category
The studies that undertook genotyping of an African/African American/black population did not state their rationales for using ‘race’ as a genetic category with the exception of the study by Horowitz et al. [40], where the authors acknowledged that, while race is a social construct, ancestry has important biological implications; thus, they used ‘race’ as a genetic group. However, biological and genetic are not equivalent terms as biological impact does not necessarily occur through a genetic mechanism. In fact, social determinants of poor health—primarily income, education, occupation—are largely responsible for the majority of biological outcomes that impact black and brown people as the result of poor air, water, food, soil and housing quality. Thus, it is not clear whether the biological implications to which Horowitz et al. referred specifically indicated the need for a genetic approach.
The introductions of the remainder of the publications often cited previous literature which correlated the incidence of a disease with people of African ancestry, suggestive of a genetic basis. Thus, this was an epidemiological, not a causal, justification for a genetic study. Alternatively, papers cited previous genetic studies of people of African ancestry that also did not justify the use of African ancestry as a genetic group.
Assessment of the use of evolutionary principles to explain human genetic diversity
To better understand how the authors viewed ‘race’, we reviewed each genetic study to determine if evolutionary explanations for human genetic variation were offered to determine or infer the author’s position on biological/genetic ‘race’.
In 71 of the 126 genetic studies of African or African-American population, Hardy-Weinberg equilibrium (HWE) was assessed to determine whether or not the gene was evolving. When a population is in HWE for a gene, the population is by definition not evolving. This assumes there is random mating, no mutation, no gene flow, an infinite population size and no selection. If the Hardy-Weinberg value is >0.01, it is concluded that a gene is not evolving. Conversely, if the gene is not in equilibrium (<0.01), the differences are due to mutation, non-random mating, gene flow, finite population size (genetic drift) and/or natural selection. In the 71 studies, polymorphisms were determined to be in equilibrium or excluded if they were not. A portion indicated HWE was assessed, but did not indicate in the results whether the polymorphisms were in equilibrium.
In five studies, evolutionary principles were offered to explain the results or to indicate that evolutionary explanations could not be ruled out. Elhassan et al. [41] described an episode of genetic drift to define the migration of a large east African population out of Africa based on mitochondrial cytochrome C oxidase subunit II (MT-CO2) sequence analysis and genome-wide microsatellite data. East Africans were shown to possess more ancestral lineages in comparison to various other continental populations, concluding that east Africa as the likely spot from which migration toward Asia took place, placing this population at the root of the human evolutionary tree.
Thompson et al. [42] analyzed the contribution of cysteinyl leukotriene 2 receptor gene variation to the development of asthma in the inhabitants of the south Atlantic island of Tristan da Cunha, a population characterized by both a founder effect and a 47% prevalence of atopy, the tendency to develop allergic diseases.
In a study by Sinues et al. [43] of 317 Mestizo Ecuadoreans, Ecuadoreans of combined Spanish and South American Indian descent, revealed CYP3A5*3 allele frequency to be significantly lower in Ecuadorians than in Spaniards and other white populations and higher than in Central Americans, Asians and blacks CYP3A4*1B was more common in Ecuadorians than in the Caucasian or Asian reference populations, but less present compared to a black reference population. The authors presume that the presence of the negative selection factor has been less present in Central and South America. In addition to differential selection, a founder effect and genetic drift could not be excluded.
Ryckman et al. [44] examined genetic polymorphisms in interleukin-1α, -1β, -6 and -8, and tumor necrosis factor-α and their receptors for association with cervical cytokine concentrations in a population of African Americans and European Americans. The goal was to determine if these variants ‘interact’ with polymorphisms in toll-like receptor 4, which was previously shown to associate with pro-inflammatory cervical cytokine concentrations, and to determine if findings are affected by bacterial vaginosis. Several SNPs in IL-1RAP and IL-1R2 were associated with IL-1α or IL-1 β concentrations in African Americans, while only the IL-1RAP SNP was associated with cervical cytokine concentrations in European Americans. They speculate the observed differences in allele frequencies between African Americans and those of European descent may represent convergent evolution and partially explain population disparity in pregnancy-related phenotypes that are cytokine concentration-dependent.
van Zyl et al. [45] identified twenty-five single nucleotide polymorphisms in the low-density lipoprotein receptor (LDLR) gene in a black South African population. One rare variant of the gene (rs17249141) was significantly associated with lower low-density lipoprotein cholesterol levels, while four variants (rs2738447, rs14158, rs2738465 and rs3180023) were significantly associated with elevated low-density lipoprotein cholesterol levels. All of the polymorphisms were in HWE, except the rs6413503 variant of the LDLR gene. However, this was thought to be due to an excess of homozygote mutants as a result of the genotyping assay and the fact that a large number of heterozygotes were not included in the analysis. It was therefore not suspected to be due to genetic drift, non-random mating, selection, or population structure.
INTERPRETATIONS AND RECOMMENDATIONS: THE WAY FORWARD
This systematic review indicated that the completion of Phase I of the Human Genome Project did not uniformly trigger a re-evaluation of the use of ‘race’ in genetic studies. Instead, the papers reviewed cited epidemiological data as the justification for a genetic approach or publication of a prior genetic study that either preceded the HGP or failed to take its findings into consideration. While the burden of poor health or a specific disease to those of African ancestry is of great concern, it is not in and of itself a justification for a genetic approach, given the multitude of social factors that impact biological outcomes.
In the absence of a stated rationale or proffered evolutionary explanations in the papers reviewed, we infer the authors of these studies believe African ancestry denotes a biological ‘race’ of people of common descent who share DNA unique from the rest of mankind. Presumably they do not accept that all people who populate the earth today had common ancestors who migrated out of Africa 30 000–50 000 years ago or that common ancestors did migrate, but subsequently isolated to form distinct evolutionary lineages with distinct gene sequences that differentiate ‘white’ from ‘black’ disease.
We conclude that an understanding of evolutionary biology, specifically the continuous nature of human genetic variation, is missing [62–72] compounded by a misreading of federal directives to include minorities in medical research. Fortunately, both of these misunderstandings can be remedied.
The required understanding of the continuous nature of human genetic variation
To understand the continuous nature of human variation, it is useful for the initiate to consider the concept of a cline, a term proposed by Huxley in 1938 [73]. A cline is a measurable gradient in a single characteristic of a species across its geographical range. Cline is not a term used frequently in biology, but greatly facilitates a basic understanding of the gradient of genetic variation in humans as we migrated out of Africa. In brief, a portion of the African population migrated out of Africa ∼30 000–50 000 years ago, settled at some distance from the parent population, and became a reproductive group. A portion of this second group migrated, settled at some distance and again reproduced. Migration, settling, reproduction and further migration resulted in a gradient of alleles across a geographical range defining clines, hence, the term clinal genetic variation. A cline in what is present day Turkey had a different allele frequency from a cline in present day China or India, even if all of the occupants are considered Asian. Similarly, a cline in what is present day Morocco had a different allele frequency different from what is represented today in Atlanta, Georgia. Thus, it is impossible to specify a specific allele or an allele frequency that would typify ‘African ancestry’ for 1.3 billion Africans on the African continent or 45 million African Americans in the USA.
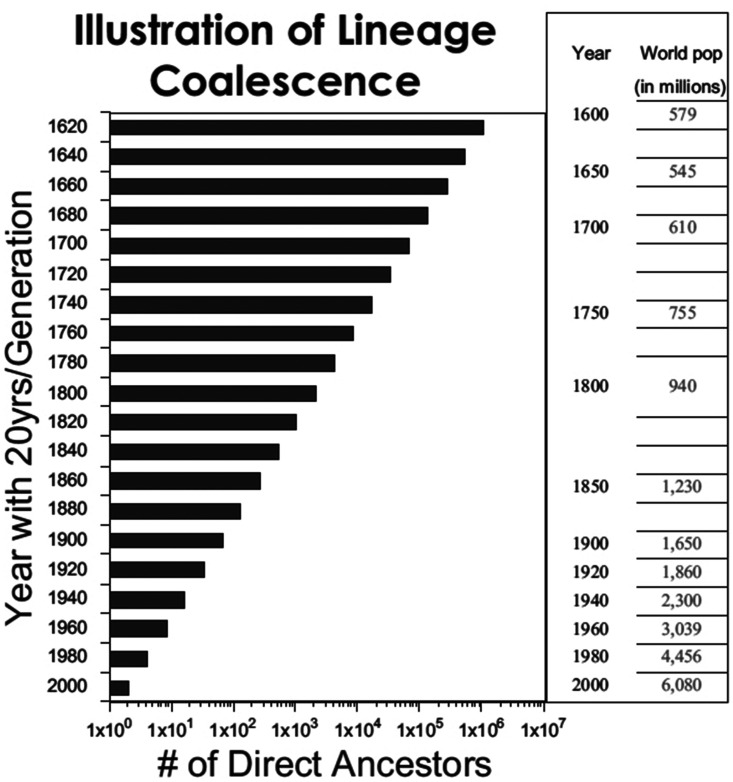
Because allele frequencies vary across geographical space, it follows that people of ‘African ancestry’ do not represent a homogeneous group. Only those who share direct ancestors—parents, grandparents and all generations of great grandparents—are genetically related. Figure 2 demonstrates biological lineage coalescence as described by Jackson [74]. The number of direct ancestors is plotted as a function 20-year generations. As the number of ancestors increases exponentially going back in time, the number of actual humans on the planet decreases. Therefore, it is apparent that biological lineages must converge around shared ancestors, thus increasing the potential for genetic similarity among all modern humans. The genetic contribution of the gametes of all direct ancestors coalesce to contribute to the genome of the individual. Any individual’s genome is the result of reproduction of direct ancestors, direct ancestors who are 99.9% identical at the DNA level. The issue is genetic lineage. Not genealogy. Not skin color. Thus, if authors use African ancestry as a genetic category or as a proxy for a genetic category, it implies that a mutation in a direct ancestor occurred at a specific point in time and this allele was inherited by everyone who shares anywhere from 1% to 99% African ancestry and ‘self-identifies’ as having African heritage. This is the genetic version of the one-drop rule [75].
Figure 2.
Biological lineage coalescence. Note that the number of ancestors increases exponentially going back in time but that the number of actual humans on the planet decreases. Therefore, it is obvious that biological lineages must converge around shared ancestors, thus, increasing the potential for genetic similarity among all modern humans. This figure was reproduced with the permission of Oxford University Press from the work of Dr Fatimah Jackson
Given this heterogeneity in allele frequencies, the only way any specific African ancestry study can be reproduced is if the exact same cohort of ‘self-identified’ Africans or African Americans are studied again. Any different combination of people of African ancestry would result in different findings. Thus, it is not at all surprising that a portion of the studies reported here showed no or very small statistical differences. Even those genetic studies that showed statistically significant differences may not if the study were conducted on any other random cohort of ‘self-identified’ people of African ancestry. Thus, the scientific gold standard of reproducibility is not met when ‘race’ is used as a genetic category in these studies. Even if ‘race’ were used as a proxy, it is not clear how the findings of genetic studies of fewer than 500 participants of ‘self-identified’ African ancestry could be generalized to everyone of African descent. Nor can statistical significance be equated with clinical significance given the host of societal and environmental factors which impact expression of a symptom or disease. None of the papers reviewed address when this hypothetical mutation in direct ancestors took place, such that all black people are genetically similar with the same propensities to certain diseases unique from other ‘races’. There is not a plausible explanation.
As stated above, the majority of the health disparities experienced by African Americans are due to social determinants of health, thus, while genetic discoveries and new technologies offer great promise, the public should not be led to believe that genetic solutions to health disparities are imminent given that 34 million Americans have not completed high school, 38.1 million live below the poverty level, 13.55 million are unemployed and 44 million are without health insurance [75]. Extreme caution must also be exercised in positing that race-based genetics is the path to individualized medicine. In individualized medicine, will the genome scan of an African-American individual only be assessed for alleles believed to reflect African ancestry? Will this be done only if the individual ‘self-identifies’ as such? Will the same mutation not be sought in a ‘white’ person with the same disease? Misconceptions of ‘race’ and errors could result in missed diagnoses, stigmatization of entire populations or remedies proffered to one population over another, errors that would only further divide our nation.
Re-education on guidelines for inclusion of minorities in medical research
There has been insufficient education of scientist, physicians, physician-scientists and the public regarding the rationale for inclusion of minorities in medical research, particularly Office of Management and Budget (OMB) Directive 15 [76], the Guidelines for the Inclusion of Women and Minorities in Medical Research [77, 78] and the Belmont Report to expand human subjects protection [79].
While slavery officially ended with the Emancipation Proclamation in 1862, the Civil Rights Act was not passed for another 104 years. Thereafter, the federal government deemed it necessary to monitor discrimination of minorities in housing, banking and education by collecting demographic data by race and ethnicity. In 1977, the US OMB Directive 15 clarified that these federal classifications were for record keeping, collection and presentation of data on race and ethnicity in Federal program administrative reports and statistical analyses [76]. The Directive specifically states that these classifications should not be interpreted as being scientific or anthropological in nature, though they have been. Then in 1986 the NIH issued guidelines encouraging inclusion of women in clinical research as historically women were underrepresented in studies and clearly differed biologically from white men on whom most medical research had been conducted [77]. In 1989, NIH expanded the guidelines calling for the inclusion of both women and minorities in clinical research, absent an explanation that minorities did not differ biologically from white men [78]. Thus, an assumption among scientists and physicians was perpetuated that people of color, like women, are biologically different from white men.
Federal funding agencies in the USA also require scientists to report the race and ethnicity of their study populations. In social science studies where race is studied as a social construct, racial and ethnic minorities would logically be included in a study at a level to achieve statistical significance. In biological and genetic studies, some scientists and physicians have followed suit, despite race being a social category. Thus, in genetics black people and white people have been included to achieve a level of statistical significance and viewed as different ‘races’. However, all investigators who conduct studies on human beings are required to have taken a human subjects tutorial. These tutorials explain that human subject guidelines initially focused on ‘Do no harm’, but were expanded in 1979 to include respect for persons, beneficence and social justice [79]. Thus, minorities are not to be included because they are innately biologically or genetically different, but because social justice dictates all Americans share the risks and benefits of medical research.
SUMMATION AND REMEDIES
In 2004, the Director of the NHGRI wrote ‘“Race” and “ethnicity” are poorly defined terms that serve as flawed surrogates for multiple environmental and genetic factors in disease causation. Research must move beyond these weak and imperfect proxy relationships to define the more proximate factors that influence health’ [80]. Many of these more proximate factors are not genetic. In 2016, NHGRI and NIMHD co-sponsored the ‘Workshop on the Use of Race and Ethnicity in Genomics and Biomedical Research’ to explore how genomics and biomedical research can describe research participant’s diverse backgrounds and experiences in ways that are scientifically and socially meaningful [22]. Participants included genomic, clinical, epidemiologic and social science researchers in addition to NIH and government stakeholders. The workshop did not aim to create unanimous recommendations or formal consensus, but did produce a summary upon which it was reported there was broad agreement. However, it fell far short of addressing and resolving misconceptions of ‘race’, focusing instead on instructions on collecting and reporting race and ethnicity as to not limit the ways in which a population can be reported. The summary also discouraged the use of race and ethnicity as a proxy for ‘expanded data categories’. Here we would encourage extreme caution against the selection of expanded data categories where the causes, effects and answers are not in the primary DNA sequence.
It is unfortunate it did not clarify that one can self-identify one’s cultural identity, religious identity and sexual identity, but one does not self-identify one’s DNA. We also regret that alternative models were not summarized that are able to simultaneously consider race/ethnicity, culture, genetics, disease incidence and geography as distinct, but very important, categories as demonstrated by the Ethnogenetic Layering Approach of Jackson in 2004 [74]. Her model acknowledges the genetic makeup present at birth as well as environmental factors that impact expressed genotype while taking into consideration the many cultural factors. The model acknowledges the rich contribution of each factor without conflating ‘race’ and self-identification with genetics.
Table 4 lists issues to be remedied by individual investigators as well as issues to be remedied at the systemic level. The first step for individuals in either group is to re-examine and question their own individual beliefs and assumptions as to what race is and is not, the same exercise the entire nation is facing. The next step is to embrace the findings of the HGP and replace misconceptions with an understanding of evolutionary biology and a corrected view of federal directives. Though it is difficult to give up strongly held beliefs, the transition will be greatly facilitated by layering new learning to supplant the incorrect information that ‘race’ is genetic. Table 5 lists the highlights of the new understanding achieved. ‘Race’ is not genetic. ‘Race’ nor ethnicity is suitable proxies for a genetic category. The scientific community and institutions need to discontinue its use in genetics immediately and embrace the findings of the HGP. No substitute term is required to divide us into genetic categories as we are all H.sapiens sapiens. One can self-identify one’s cultural, religious and sexual identity, but not one’s DNA. We are not better people if we attempt to infuse genetics with cultural humility or political correctness. The biomedical community must get its own house in order before it can hope to educate the public and restore the much-needed public trust of science. Only then can the promises of individualized medicine be realized in the future in an equitable and ethical manner.
Table 4.
Remedies to Correct Misconceptions of ‘Race’
| Individual scientist/physician level - | |
| Identify your broad assumptions implicit in the use of ‘race’ in biomedical research and medical practice. | |
|
| |
| Examine the use of ‘race’ in the studies you cite as background to your own study. | |
| Differentiate correlation from causation. Understand that these studies are reproducible only if | |
| the exact same people are studied. Conduct literature reviews using Scopus and Web of | |
| Science, instead of only PubMed which does not include important studies not | |
| included in biomedical journals that focus only on organ systems or diseases. | |
|
| |
| If you have used ‘race’ as a genetic category, re-analyze your data with and without ‘race’ as a | |
| variable, and re-assess your conclusions and their generalizability | |
|
| |
| Invite an evolutionary biologist to meet with your research group to assure your work reflects | |
| the fact that human genetic variation is continuous. | |
|
| |
|
| |
| Invite an ethicist to meet with your research group to identify the risks associated with the use of ‘race’ as a proxy. | |
|
| |
| Take responsibility for the education of the public as the public media is often not qualified to do so. | |
|
| |
|
| |
| Systems level – Funding agencies/scientific societies/publishers/editors/teaching institutions | |
|
| |
| • Discontinue use of ‘race’ and/or ethnicity as a biological/genetic category. | |
|
| |
|
Social science - Race is a social construct. |
|
| Ethnicity is defined as belonging to a common group, | |
| often linked by race, nationality, religion, geographic | |
| area, and/or language, none of which is genetic. | |
| Biological sciences – Need to adhere to the scientific/taxonomical definition of race as a subdivision of subspecies. | |
|
| |
| • Discontinue use of ‘self-identification’ of research participants in biological/genetic studies. | |
|
| |
| There is no genetic basis for ‘self-identification’ > One’s genetic makeup is defined at birth and subsequently modified by mutations resulting from environmental factor. | |
| One cannot pick one’s genetic makeup. Cultural identity, religious identity, gender identity, sexual preference identity are self-identified. Not genetics. | |
|
| |
|
| |
|
• Federal agencies, scientific societies, medical schools, and universities should launch an educational effort to correct scientific misconceptions of ‘race’. | |
|
| |
|
| |
| NIH must stand my its findings that ‘race’ is not a genetic category and take responsibility for the lapse in the education of scientists and health professionals since the completion of the HGP. | |
|
| |
| NIH must clarify to the public that one can choose one’s racial and ethnic identity, but not their genetic makeup. Science is the not the domain of individual opinion or political correctness. | |
|
| |
| Re-educate all scientists about OMB Directive 15. | |
|
| |
| Modify the Federal guidelines for inclusion of women and minorities to clarify that minorities are to be included to address a social justice issue. Not because minorities have a different genetic makeup/physiology. | |
|
| |
| Revise the human subject tutorials required by Institutional Review Boards to clarify that inclusion of minorities is about social justice and not due to biological/genetic differences. | |
|
| |
| Federal funding agencies must clarify that the grant form requiring investigators to identify the study population by race/ethnicity is to monitor inclusion, such that all Americans share the risk and benefits of medical research. Not because minorities are different than the rest of Homo sapiens sapiens. | |
|
| |
| Scientific societies and editorial boards need to re-envision interdisciplinarity to invite scientists who focus on marginalized topics that are controversial because they counter mainstream misconceptions. | |
|
| |
| Scientific societies and medical schools need to design continuing medical education courses to correct misconceptions of ‘race’ and explaining human genetic variation. | |
|
| |
| Medical schools need to require an evolutionary biology course as a prerequisite for medical school and incorporate evolutionary biology into the medical school curricula. | |
|
| |
| Colleges and universities need to incorporate courses in evolutionary biology for all undergraduate science majors | |
|
| |
Table 5.
Corrected conceptions of race
| ‘Race’ is not genetic |
|
|
| Ethnicity—defined as belonging to a common group, often linked by race, nationality, religion, geographic area and/or language—is also not genetic. The terms race and not interchangeable |
|
|
| Self-identification—a gold standard only in the social sciences—cannot be used as a proxy for a genetic category |
|
|
| One can self-identify one’s cultural identity, religious identity and sexual identity, but not one’s DNA |
|
|
| The initial assessment regarding the inappropriate use of ‘race’ must be at the time of experimental design and prior to the awarding of research funds, not at the time of publication |
|
|
| No substitution for ‘race’ is required as we are all Homo sapiens sapiens |
|
|
| ‘Science’ that is not reproducible is not science |
|
|
| If an allele is present in a given number of people, it does not make it true for an entire ‘race’. It may be true for direct ancestors in a family |
|
|
| Declaring that DNA has a ‘race’ or ‘ethnicity’ is not an expression of cultural humility or political correctness as these are not the domains of genetics |
|
|
| Re-education of the biomedical community regarding misconceptions of ‘race’ is required and within reach |
Supplementary data
Supplementary data is available at EMPH online.
Supplementary Material
Acknowledgements
The authors wish to thank Drs Fatimah Jackson, Joseph L. Graves Jr and John Pool for helpful scientific discussions and James Barney and Emanuel Scarbrough for many helpful discussions on lay understandings of race.
Conflict of interest: None declared.
REFERENCES
- 1.Templeton AR. Biological races in humans. Stud Hist Philos Biol Biomed Sci 2013;44:262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins FS, Morgan M, Patrinos A.. The Human Genome Project: lessons from large-scale biology. Science 2003;300:286–90. [DOI] [PubMed] [Google Scholar]
- 3.Graves JL Jr. The Emperor’s New Clothes: biological Theories of Race at the Millenium. New York, NY: Rutgers University Press, 2003. [Google Scholar]
- 4.Graves JL Jr. The Race Myth. New Brunswick, NJ: Plume, 2005. [Google Scholar]
- 5.Nissen SE; Cardiovascular and Renal Drugs Advisory Committee. Report from the Cardiovascular and Renal Drugs Advisory Committee: US Food and Drug Administration; June 15–16, 2005; Gaithersburg, MD. Circulation 2005;112:2043–6. [DOI] [PubMed] [Google Scholar]
- 6.Methods of Treating and Preventing Congestive Heart Failure with Hydralazine Compounds and Isosorbide Dinitrate or Isosorbide Mononitrate. http://patft.uspto.gov/netacgi/nphParser?Sect2=PTO1&Sect2=HITOFF&p=1&u=/netahtml/PTO/searchbool.html&r=1&f=G&l=50&d=PALL&RefSrch=yes&Query=PN/6465463 (29 April 2021, date last accessed).
- 7.Kahn DF, Duffy SJ, Tomasian D. et al. Effects of black race on forearm resistance vessel function. Hypertension 2002;40:195–201. [DOI] [PubMed] [Google Scholar]
- 8.Cardillo C, Kilcoyne CM, Cannon RO, Panza JA.. Racial differences in nitric oxide-mediated vasodilator response to mental stress in the forearm circulation. Hypertension 1998;31:1235–9. [DOI] [PubMed] [Google Scholar]
- 9.Kalinowski L, Dobrucki IT, Malinski T.. Race-specific differences in endothelial function: predisposition of African Americans to vascular disease. Circulation 2004;109:2511–7. [DOI] [PubMed] [Google Scholar]
- 10.Goodman AH. Why genes don't count (for racial differences in health). Am J Public Health 2000;90:1699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long JC, Kittles RA.. Human genetic diversity and the nonexistence of biological race. Hum Biol 2003;75:449–71. [DOI] [PubMed] [Google Scholar]
- 12.Lewontin RC. The fallacy of racial medicine: confusions about human races. Genewatch 2005;18:5–7. [PubMed] [Google Scholar]
- 13.Graves JL Jr, Rose MA.. Against racial prejudice. Patterns Prejudice 2006;40:481–93. [Google Scholar]
- 14.Azoulay KG. Reflections on race and biologization of difference. Patterns Prejudice 2006;40:353–79. [Google Scholar]
- 15.Weigmann K. Racial medicine: here to stay? The success of the International HapMap Project and other initiatives may help to overcome racial profiling in medicine, but old habits die hard. EMBO Rep 2006;7:246–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison GT, Smart A, Tutton R. et al. Racial categories in medicine: a failure of evidence-based practice? PLoS Med 2007;4:e287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long JC, Kittles RA.. Human genetic diversity and the nonexistence of biological race. 2003. Hum Biol 2009;81:777–98. [DOI] [PubMed] [Google Scholar]
- 18.Graves JL. Biological V. Social definitions of race: implications for modern medical research. Rev Black Polit Econ 2010;37:43–60. [Google Scholar]
- 19.Hoquet T. Biologization of race and racialization of human. Vernier, Buffon, Linnaeus. In: Bancel D (ed.). The Invention of Race: Scientific and Popular Representations. New York: Routledge, 2014, 17–32. [Google Scholar]
- 20.Graves JL Jr. Why the nonexistence of biological races does not mean the nonexistence of racism. Am Behav Sci 2015;59:1574–995. [Google Scholar]
- 21.Yudell M, Roberts D, DeSalle R, Tishkoff S.. Taking race out of human genetics. Science 2016;351:564–5. [DOI] [PubMed] [Google Scholar]
- 22.National Human Genome Institute. Workshop on the Use of Race and Ethnicity in Genomics and Biomedical Research. https://www.genome.gov/Pages/About/IRMinorities/2016_Oct_Workshop_Summary_and_Themes.pdf (29 April 2021, date last accessed).
- 23.Paddock S, Laje G, Charney D. et al. Association of GRIK4 with outcome of antidepressant treatment in the STARD cohort. Am J Psychiatry 2007;164:1181–8. [DOI] [PubMed] [Google Scholar]
- 24.Nieves JW, Cosman F, Grubert E. et al. Skeletal effects of vitamin D supplementation in postmenopausal black women. Calcif Tissue Int 2012;91:316–24. [DOI] [PubMed] [Google Scholar]
- 25.Pretorius MM, Gainer JV, Van Guilder GP. et al. The bradykinin type 2 receptor BE1 polymorphism and ethnicity influence systolic blood pressure and vascular resistance. Clin Pharmacol Ther 2008;83:122–9. [DOI] [PubMed] [Google Scholar]
- 26.Langaee TY, Gong Y, Yarandi HN. et al. Association of CYP3A5 polymorphisms with hypertension and antihypertensive response to verapamil. Clin Pharmacol Ther 2007;81:386–91. [DOI] [PubMed] [Google Scholar]
- 27.Frey HA, Stout MJ, Pearson LN. et al. Genetic variation associated with preterm birth in African-American women. Am J Obstet Gynecol 2016;215:235.e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochs-Balcom HM, Sun X, Chen Y. et al. Putative linkage signals identified for breast cancer in African American families. Cancer Epidemiol Biomarkers Prev 2015;24:442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedrich DC, Genro JP, Sortica VA. et al. Distribution of CYP2D6 alleles and phenotypes in the Brazilian population. PLoS One 2014;9:e110691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cozier YC, Palmer JR, Rosenberg L.. Comparison of methods for collection of DNA samples by mail in the Black Women's Health Study. Ann Epidemiol 2004;14:117–22. [DOI] [PubMed] [Google Scholar]
- 31.Zuo L, Luo X, Krystal JH. et al. The pharmacokinetics of codeine and its metabolites in Blacks with sickle cell disease. Eur J Clin Pharmacol 2009;65:651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Gelernter J, Kranzler HR. et al. Ordered subset linkage analysis based on admixture proportion identifies new linkage evidence for alcohol dependence in African-Americans. Hum Genet 2013;132:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kangelaris KN, Sapru A, Calfee CS. et al. The association between a Darc gene polymorphism and clinical outcomes in African American patients with acute lung injury. Chest 2012;141:1160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo L, Luo X, Krystal JH. et al. The efficacies of clozapine and haloperidol in refractory schizophrenia are related to DTNBP1 variation. Pharmacogenet Genomics 2009;19:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerhard T, Gong Y, Beitelshees AL, INVEST Investigators et al. Alpha-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES). Am Heart J 2008;156:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanata CM, Nititham J, Taylor KE. et al. Genetic contributions to lupus nephritis in a multi-ethnic cohort of systemic lupus erythematous patients. PLoS One 2018;13:e0199003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giri A, Hartmann KE, Aldrich MC. et al. Admixture mapping of pelvic organ prolapse in African Americans from the Women's Health Initiative Hormone Therapy trial. PLoS One 2017;12:e0178839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aissani B, Wiener H, Zhang K.. Multiple hits for the association of uterine fibroids on human chromosome 1q43. PLoS One 2013;8:e58399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo L, Wang K, Zhang XY. et al. Association between common alcohol dehydrogenase gene (ADH) variants and schizophrenia and autism. Hum Genet 2013;132:735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horowitz CR, Abul-Husn NS, Ellis S. et al. Determining the effects and challenges of incorporating genetic testing into primary care management of hypertensive patients with African ancestry. Contemp Clin Trials 2016;47:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elhassan N, Gebremeskel EI, Elnour MA. et al. The episode of genetic drift defining the migration of humans out of Africa is derived from a large east African population size. PLoS One 2014;9:e97674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson MD, Storm van's Gravesande K, Galczenski H. et al. A cysteinyl leukotriene 2 receptor variant is associated with atopy in the population of Tristan da Cunha. Pharmacogenetics 2003;13:641–9. [DOI] [PubMed] [Google Scholar]
- 43.Sinues B, Vicente J, Fanlo A. et al. CYP3A5 3, CYP3A4 1B and MDR1 C3435T genotype distributions in Ecuadorians. Dis Markers 2008;24:325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman KK, Williams SM, Krohn MA. et al. Interaction between interleukin-1 receptor 2 and Toll-like receptor 4, and cervical cytokines. J Reprod Immunol 2011;90:220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Zyl T, Jerling JC, Conradie KR. et al. Common and rare single nucleotide polymorphisms in the LDLR gene are present in a black South African population and associate with low-density lipoprotein cholesterol levels. J Hum Genet 2014;59:88–94. [DOI] [PubMed] [Google Scholar]
- 46.Feitosa MF, Borecki IB, Rankinen T. et al. Lack of pleiotropic genetic effects between adiposity and sex hormone-binding globulin concentrations before and after 20 weeks of exercise training: the HERITAGE family study. Metabolism 2003;52:35–41. [DOI] [PubMed] [Google Scholar]
- 47.An P, Borecki IB, Rankinen T. et al. Evidence of major genes for plasma HDL, LDL cholesterol and triglyceride levels at baseline and in response to 20 weeks of endurance training: the HERITAGE Family Study. Int J Sports Med 2005;26:414–9. [DOI] [PubMed] [Google Scholar]
- 48.Xue L, Wang X, Xu J. et al. ISL1 common variant rs1017 is not associated with susceptibility to congenital heart disease in a Chinese population. Genet Test Mol Biomarkers 2012;16:679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNamara DM, Tam SW, Sabolinski ML. et al. Endothelial nitric oxide synthase (NOS3) polymorphisms in African Americans with heart failure: results from the A-HeFT trial. J Card Fail 2009;15:191–8. [DOI] [PubMed] [Google Scholar]
- 50.McNamara DM, Taylor AL, Tam SW. et al. G-protein beta-3 subunit genotype predicts enhanced benefit of fixed-dose isosorbide dinitrate and hydralazine: results of A-HeFT. JACC Heart Fail 2014;2:551–7. [DOI] [PubMed] [Google Scholar]
- 51.Anthony EG, Richard E, Lipkowitz MS. et al. Association of the ADRB2 (rs2053044) polymorphism and angiotensin-converting enzyme-inhibitor blood pressure response in the African American Study of Kidney Disease and Hypertension. Pharmacogenet Genomics 2015;25:444–9. [DOI] [PubMed] [Google Scholar]
- 52.Chen TK, Choi MJ, Kao WH. et al. Examination of potential modifiers of the association of APOL1 alleles with CKD progression. Clin J Am Soc Nephrol 2015;10:2128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhatnagar V, Garcia EP, O’Connor DT. et al. CYP3A4 and CYP3A5 polymorphisms and blood pressure response to amlodipine among African-American men and women with early hypertensive renal disease. Am J Nephrol 2010;31:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beach SR, Brody GH, Lei MK. et al. Differential susceptibility to parenting among African American youths: testing the DRD4 hypothesis. J Fam Psychol 2010;24:513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brody GH, Chen YF, Beach SR. et al. Differential sensitivity to prevention programming: a dopaminergic polymorphism-enhanced prevention effect on protective parenting and adolescent substance use. Health Psychol 2014;33:182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brody GH, Beach SR, Philibert RA. et al. Prevention effects moderate the association of 5-HTTLPR and youth risk behavior initiation: gene x environment hypotheses tested via a randomized prevention design. Child Dev 2009;80:645–61. [DOI] [PubMed] [Google Scholar]
- 57.Musani SK, Fox ER, Kraja A. et al. Genome-wide association analysis of plasma B-type natriuretic peptide in blacks: the Jackson Heart Study. Circ Cardiovasc Genet 2015;8:122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frazier-Wood AC, Manichaikul A, Aslibekyan S. et al. Genetic variants associated with VLDL, LDL and HDL particle size differ with race/ethnicity. Hum Genet 2013;132:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feitosa ME, Rice T, Borecki IB. et al. Pleiotropic QTL on chromosome 12q23-q24 influences triglyceride and high-density lipoprotein cholesterol levels: the HERITAGE family study. Hum Biol 2006;78:317–27. [DOI] [PubMed] [Google Scholar]
- 60.An P, Perusse L, Rankinen T. et al. Familial aggregation of exercise heart rate and blood pressure in response to 20 weeks of endurance training: the HERITAGE family study. Familial aggregation of exercise heart rate and blood pressure in response to 20 weeks of endurance training: the HERITAGE family study. Int J Sports Med 2003;24:57–62. [DOI] [PubMed] [Google Scholar]
- 61.Ukkola O, Rankinen T, Rice T. et al. ; HERITAGE Family Study. Interactions among the beta2- and beta3-adrenergic receptor genes and total body fat and abdominal fat level in the HERITAGE Family Study. Int J Obes Relat Metab Disord 2003;27:389–93. [DOI] [PubMed] [Google Scholar]
- 62.Graves JL. African Americans in evolutionary science: where we have been, and what’s next. Evo Edu Outreach 2019;12:18–27. [Google Scholar]
- 63.Lieberman L, Hampton RE, Littlefield A, Hallead G.. Race in biology and anthropology: a study of college texts and professors. J Res Sci Teach 1992;29:301–21. [Google Scholar]
- 64.Morning A. Reconstructing race in science and society: biology textbooks, 1952-2002. Am J Sociol 2008;114(Suppl):S106–S317. [DOI] [PubMed] [Google Scholar]
- 65.Donovan BM. Reclaiming race as a topic of the U.S. biology textbook curriculum. Sci Edu 2015;99:1092–117. [Google Scholar]
- 66.Nesse RM, Stearns SC.. The great opportunity: evolutionary applications to medicine and public health. Evol Appl 2008;1:28–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nesse RM, Bergstrom CT, Ellison PT. et al. Evolution in health and medicine Sackler colloquium: making evolutionary biology a basic science for medicine. Proc Natl Acad Sci U S A 2010;107(Suppl 1):1800–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nesse RM. Ten questions for evolutionary studies of disease vulnerability. Evol Appl 2011;4:264–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antolin MF, Jenkins KP, Bergstrom CT. et al. Evolution and medicine in undergraduate education: a prescription for all biology students. Evolution 2012;66:1991–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nesse RM, Ganten D, Gregory TR, Omenn GS.. Evolutionary foundations for molecular medicine. J Mol Med (Berl) 2012;90:509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wells JCK, Nesse RM, Sear R. et al. Evolutionary public health: introducing the concept. Lancet 2017;390:500–9. [DOI] [PubMed] [Google Scholar]
- 72.Grunspan DZ, Moeller KT, Nesse RM, Brownell SE.. The state of evolutionary medicine in undergraduate education. Evol Med Public Health 2019;2019:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huxley J. Clines: an auxiliary taxonomic principle. Nature 1938;142:219–20. [Google Scholar]
- 74.Jackson FLC. Human genetic variation and health: new assessment approaches based on ethnogenetic layering. Brit Med Bull 2004;69:215–35. [DOI] [PubMed] [Google Scholar]
- 75.US Census Bureau. census.gov.
- 76.OMB 15. The Standards for Maintaining, Collecting, and Presenting Federal Data on Race and Ethnicity. Federal Register 1997;62:58782.
- 77.Including Women and Minorities in Clinical Research Background. https://orwh.od.nih.gov/womens-health/clinical-research-trials/nih-inclusion-policies/including-women-and-minorities (30 April 2021, date last accessed).
- 78.NIH Policy and Guidelines on the Inclusion of Women and Minorities as Subjects in Clinical Trials. https://grants.nih.gov/policy/inclusion/women-and-minorities/guidelines.htm (29 April 2021, date last accessed).
- 79.The Belmont Report. https://www.hhs.gov/ohrp/regulations-and-policy/belmont-report/index.html.
- 80.Collins FS. What we do and don’t know about ‘race’, ‘ethnicity’, genetics and health at the dawn of the genome era. Nat Genet 2004;36:S13–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.