Abstract
Recent studies have shown phenotypic and metabolic heterogeneity in related species including Streptococcus oralis, a typical oral commensal bacterium, Streptococcus mutans, a cariogenic bacterium, and Streptococcus gordonii, which functions as an accessory pathogen in periodontopathic biofilm. In this study, metabolites characteristically contained in the saliva of individuals with good oral hygiene were determined, after which the effects of an identified prebiotic candidate, D-tagatose, on phenotype, gene expression, and metabolic profiles of those three key bacterial species were investigated. Examinations of the saliva metabolome of 18 systemically healthy volunteers identified salivary D-tagatose as associated with lower dental biofilm abundance in the oral cavity (Spearman’s correlation coefficient; r = -0.603, p = 0.008), then the effects of D-tagatose on oral streptococci were analyzed in vitro. In chemically defined medium (CDM) containing D-tagatose as the sole carbohydrate source, S. mutans and S. gordonii each showed negligible biofilm formation, whereas significant biofilms were formed in cultures of S. oralis. Furthermore, even in the presence of glucose, S. mutans and S. gordonii showed growth suppression and decreases in the final viable cell count in a D-tagatose concentration-dependent manner. In contrast, no inhibitory effects of D-tagatose on the growth of S. oralis were observed. To investigate species-specific inhibition by D-tagatose, the metabolomic profiles of D-tagatose-treated S. mutans, S. gordonii, and S. oralis cells were examined. The intracellular amounts of pyruvate-derived amino acids in S. mutans and S. gordonii, but not in S. oralis, such as branched-chain amino acids and alanine, tended to decrease in the presence of D-tagatose. This phenomenon indicates that D-tagatose inhibits growth of those bacteria by affecting glycolysis and its downstream metabolism. In conclusion, the present study provides evidence that D-tagatose is abundant in saliva of individuals with good oral health. Additionally, experimental results demonstrated that D-tagatose selectively inhibits growth of the oral pathogens S. mutans and S. gordonii. In contrast, the oral commensal S. oralis seemed to be negligibly affected, thus highlighting the potential of administration of D-tagatose as an oral prebiotic for its ability to manipulate the metabolism of those targeted oral streptococci.
Keywords: d-tagatose, biofilm, transcriptomics, metabolomics (OMICS), Streptococcus mutans (S. mutans), Streptococcus gordonii, Streptococcus oralis
Introduction
Dental caries and periodontal disease are prevalent polymicrobial infections found in the human oral cavity, which often lead to tooth loss and have effects on general health (Lamont and Hajishengallis, 2015; Johansson et al., 2016; Bernabe et al., 2020). Such infections develop when dental biofilm shifts from a symbiotic to dysbiotic state (Lamont et al., 2018) and disruption of biofilm showing symbiosis has been found to be associated with its maturation over time (Mira et al., 2017). During periodontal pathogenic biofilm formation, early colonizers such as streptococci adhere to salivary pellicles on tooth surfaces, followed by accumulation of intermediate colonizers, including Fusobacterium nucleatum and periodontopathic bacteria such as Porphyromonas gingivalis, in the biofilm (Kuboniwa and Lamont, 2010). In particular, Streptococcus gordonii supports progression of periodontal disease due to its symbiotic relationship with F. nucleatum and P. gingivalis via metabolites (Periasamy and Kolenbrander, 2009; Kuboniwa et al., 2009b; Sakanaka et al., 2015). On the other hand, during formation of cariogenic biofilm, cariogenic bacteria represented by the major strain Streptococcus mutans become dominant due to multiple factors related to lifestyle and a diet rich in sugars (Selwitz et al., 2007; Gross et al., 2012). In both cases, oral streptococci, which are early colonizers, play a critical role.
Recent studies have shown phenotypic and metabolic heterogeneity in related key species, including Streptococcus oralis, a typical oral commensal bacterium, S. mutans, a cariogenic bacterium, and S. gordonii, which functions as an accessory pathogen in periodontopathic biofilm (Periasamy and Kolenbrander, 2010; Hajishengallis, 2015; Thurnheer and Belibasakis, 2018). Furthermore, other reports have shown competitive relationships between these oral streptococci, as S. mutans was found to inhibit the growth of S. gordonii by producing the bacteriocin mutacin and S. gordonii inhibited mutacin production by producing the bacteriocin challisin (Kreth et al., 2011). Additionally, because of the high acid resistance of S. mutans, it has shown better survival in low pH biofilm as compared to acid-sensitive streptococci (Kreth et al., 2011). On the other hand, S. mutans is highly sensitive to hydrogen peroxide produced by S. gordonii and S. oralis (Kreth et al., 2008; Redanz et al., 2018).
Presently, understanding of how to control these species remains limited, though that is considered essential for maintaining oral health. In oral streptococci, glycolysis is a crucial metabolic pathway for energy acquisition and production of organic acids, thus xylitol, a sugar alcohol, is widely used as a sugar substitute to reduce caries risk, because it cannot be metabolized by oral streptococci (L, 1995; Makinen, 2010; Milgrom et al., 2012). However, the preventive effect of xylitol is inhibited by other monosaccharides (Gauthier et al., 1984), while it has also been shown to have side effects in the gastrointestinal tract (Oku and Nakamura, 2007).
D-tagatose is a ketohexose, 4-epimer of fructose that has lower calories and a reduced glycemic effect as compared to sucrose (Donner et al., 1999; Oh, 2007; Guerrero-Wyss et al., 2018; Roy et al., 2018). It has been reported that D-tagatose showed effects on the metabolome of pig intestinal microflora, especially flora in the cecum and colon, and caused increases in butyric acid and valeric acid (Lærke et al., 2000). Other reports have also shown species-specific growth inhibitory effects on Phytophthora spp. by D-tagatose (Chahed et al., 2020; Corneo et al., 2021). Based on these findings, this artificial sweetener is expected to be applied in various contexts, such as diabetes management, pesticides, and prebiotics for the intestines. Additionally, D-tagatose has been reported to inhibit the growth of S. mutans (Sawada et al., 2015; Hasibul et al., 2018), though there are few studies regarding its effects on other types of oral bacteria. Furthermore, no known reports of comprehensive analyses of tagatose-induced dynamic changes in oral bacteria using metabolomics and transcriptomics have been presented.
In the present study, metabolomic profiling of human saliva samples showed that D-tagatose was associated with good oral hygiene. Furthermore, results of in vitro examinations of the effects of D-tagatose on physiology, as well as transcriptomic and metabolomic profiles of the oral streptococci S. mutans, S. gordonii, and S. oralis highlight its potential use as an oral prebiotic for altering the metabolism of those oral streptococci.
Materials and Methods
Saliva Metabolome
Data for the present study were obtained by reanalysis of raw data from our previous study (Kuboniwa et al., 2016) using MS-DIAL version 3.90 (Tsugawa et al., 2015). Briefly, that previous study was conducted from 2013 to 2014 with approval from the Osaka University Research Ethics Committee, and in accordance with the principles of the Helsinki Declaration and STROBE guidelines for human observational studies. Nineteen participants provided written informed consent prior to enrolment. Each subject was asked to refrain from eating, drinking, or toothbrushing for at least one hour prior to undergoing the procedures. Five calibrated and licensed dentists evaluated plaque accumulation at four sites per tooth for all present, and the average per tooth was used for the total score, which was defined as modified plaque index (mPlI) (Sakanaka et al., 2017). mPlI was not obtained in that manner for one of the subjects, and all participants except for this subject were asked to expectorate unstimulated whole saliva over a 10-minute period into a 50-mL tube (Corning, Corning, NY, USA) that was kept on ice. Subsequently, the 18 samples were frozen with liquid nitrogen and stored at −80°C until analysis. Metabolomics raw data were obtained using gas chromatography coupled with mass spectrometry (GCMS-QP2010 Ultra, Shimazu).
Bacterial Strains and Growth Conditions
The strains used in this study are listed in Table 1. All were grown statically at 37°C under aerobic conditions in brain-heart infusion broth (BHI; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) or, when necessary, in chemically defined medium (CDM), developed according to a previously reported method (Loo et al., 2000). Briefly, CDM consisting of 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, 0.2% casamino acids, and 100 μM MnCl2 4H2O (pH 7.4) was supplemented with filter-sterilized vitamins (0.04 mM nicotinic acid, 0.1 mM pyridoxine HCl, 0.01 mM pantothenic acid, 1 μM riboflavin, 0.3 μM thiamin HCl, 0.05 μM D-biotin), amino acids (4 mM L-glutamic acid, 1 mM L-arginine HCl, 1.3 mM L-cysteine HCl, 0.1 mM L-tryptophan), and MgSO4 7H2O (2 mM).
Table 1.
Bacterial strains used in this study.
| Strains | Description | Source or Ref. |
|---|---|---|
| Streptococcus gordonii DL1 Challis | Wild type | Pakura and Walczak |
| Streptococcus mutans OMZ175 | Wild type | B. Guggenheim |
| Streptococcus mutans UA159 | Wild type | University of Alabama |
| Streptococcus oralis ATCC9811 | Wild type | American Type Culture Collection |
Saliva Preparation
Saliva was collected in a sterile centrifuge tube on ice from six healthy donors and pooled, as described previously (Kuboniwa et al., 2009a). Dithiothreitol (Sigma-Aldrich, St. Louis, MO) was added to a 2.5 mM final concentration, then the saliva was gently stirred on ice for 10 minutes and centrifuged at 3,000 × g for 20 minutes at 4°C. The clarified saliva supernatant was decanted, 3 volumes of distilled water was added, and the 25% saliva was filtered through a 0.20 μm pore size filter and frozen in 40 ml aliquots. Immediately prior to use, the sterile saliva was thawed at 37°C; the slight precipitate was pelleted at 1,430 × g for 5 min, and the clear 25% saliva supernatant was used in experiments.
Biofilm Assay
S. oralis ATCC9811, S. mutans OMZ175, UA159, and S. gordonii Challis DL1 cells that reached the late logarithmic growth phase were collected, then subjected to centrifugation (7,670 × g, 4°C, 7 minutes). Cells (108 CFU) were then stained with hexidium iodide (HI; Molecular Probes, Carlsbad, CA, USA) and cultured in CDM containing 0.8% D-glucose, sucrose, D-tagatose, or no sugar at 37°C under aerobic conditions for 24 hours using the eight-well LAB-TEK Chamber Slide System (Nalge Nunc International, Naperville, IL, USA) after coating with 25% sterile human saliva. Biofilm formed on the bottom surface of each chamber was observed using a confocal laser scanning microscope [(CLSM), TCS SP8; Leica Microsystems GmbH, Wetzlar, Germany]. Images of 10 random fields of view in each chamber were obtained, then analyzed using the Imaris 7.0.1 software package (BitplaneAG, Zurich, Switzerland).
Growth Study
S. oralis ATCC9811, S. mutans OMZ175, UA159, and S. gordonii Challis DL1 cells (108 CFU) were cultured under aerobic conditions at 37°C in BHI without or with D-tagatose at a concentration of 0.1%, 0.5%, 1%, 5%, or 10%. Similarly, the bacteria were cultured in CDM containing 0.2% D-glucose without or with D-tagatose at a concentration of 1% or 5%. Optical density at 600 nm (OD600) was determined using an ultraviolet-visible spectrophotometer (UV-1600; Shimadzu Corporation, Kyoto, Japan). To investigate whether D-tagatose is utilized by S. oralis, S. mutans, and/or S. gordonii, aerobic cultures were performed at 37°C in CDM without or with 0.8% D-tagatose or D-glucose.
Viability Assay
Bacterial cells were cultured at 37°C for 48 hours under aerobic conditions in CDM containing 0.2% D-glucose without or with D-tagatose at a concentration of 1% or 5%, then stained using a LIVE/DEAD® BacLight™ Bacterial Viability Kit (Molecular Probes, Eugene, OR), according to the manufacturer’s instructions. Stained cells were observed using a multi-label counter (Wallac 1420 ARVO MX; Perkin Elmer, Waltham, Massachusetts) (excitation wavelength 485 nm, emission wavelength 535 nm). Viable cell counts were calculated based on a calibration curve.
Extracellular Polysaccharide (EPS) Production Assay
S. mutans OMZ175 was cultured under aerobic conditions at 37°C for 48 hours in CDM containing 0.2% D-glucose without or with 1% or 5% D-tagatose. The cells were then collected by centrifugation (7,670 × g, 4°C, 7 minutes) and washed with 10 mM phosphate-buffered saline (PBS, pH 7.4). EPS was stained with concanavalin A (Con A) and wheat germ agglutinin (WGA), and bacterial cells with HI, which were then washed and suspended in PBS. Bacterial suspensions (OD600 = 4.0) in 10 random fields were observed with CLSM. Obtained images were analyzed using the Imaris 7.0.1 software package.
Intracellular Metabolite Measurement
S. oralis ATCC9811, S. mutans UA159, and S. gordonii Challis DL1 were initially adjusted to an OD600 of 0.8, then grown aerobically at 37°C in CDM containing 0.2% D-glucose without or with 1% or 5% D-tagatose. After three hours of incubation, bacterial cells were harvested by centrifugation (7,670 × g, 4°C, 7 minutes), washed with 1 ml of ultrapure water, and immediately frozen by liquid nitrogen. Ribitol was added as an internal standard for normalization, then 1.3 ml of acetonitrile was added to remove protein. After thoroughly mixing the samples, centrifugation was performed at 20,630 × g for 3 minutes at 4°C. The upper 1.6 ml was evaporated in a centrifugal concentrator for 120 minutes at room temperature and freeze-dried overnight. One hundred μl of 20 mg/mL methoxyamine hydrochloride in pyridine and 50 μl N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA, Sigma-Aldrich) were added to the samples as derivatizing agents, then they were thoroughly mixed and centrifuged at 15,490 × g for 3 minutes at 25°C, after which the upper 50 μl of each sample was transferred to a vial. Measurement of extracted metabolites was performed using a GC-MS/MS-TQ8040 device (Shimadzu). Obtained GC/MS data were analyzed with an ABF converter (https://www.reifycs.com/AbfConverter/index.html) and the MS-DIAL software package, ver. 4.60 (http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/index.html).
RNA Extraction and Sequencing
Triplicate samples used for extracting total RNA were cultured for 30 minutes under the same conditions as for those subjected to metabolomics analysis. Bacteria harvested by centrifugation were immediately frozen and homogenized with zirconia beads at 1,500 rpm for 5 minutes at 4°C with use of a ShakeMaster NEO (BioMedical Science, Tokyo, Japan). Total RNA was isolated using TRIzol reagent (Life Technologies, Inc.) and an RNeasy kit (Qiagen). RNA quality was evaluated based on test concentration (Nanodrop 1000), agarose gel electrophoresis (AGE), and BioAnalizer results. Quality-controlled RNA samples were reversed into cDNA and then used to construct a sequencing library, sequenced using an Illumina NovaSeq 6000 by Rhelixa Co., Ltd. (Tokyo, Japan).
Statistical Analysis
All pairwise comparisons of groups were performed using a two-tailed t-test. Comparisons of multiple groups, except for growth curve analyses, were performed using one-way analysis of variance with post hoc paired comparisons conducted with Dunnett’s test. All statistical analyses were done with SPSS version 26 (IBM Japan, Tokyo, Japan), with a p value of 0.05 considered to be statistically significant. Metabolite set enrichment analysis (MSEA) was performed as previously described (Sakanaka et al., 2017). Differentially expressed genes (DEGs) were identified using the Bioconductor package edgeR. Gene Ontology (GO) enrichment analysis of DEGs was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) of the NIH.
Results
Saliva Metabolome
Data analyzed from our previous study (Kuboniwa et al., 2016) using the new software package MS-DIAL showed 114 annotated metabolites, with D-tagatose among them. The correlation between D-tagatose and mPlI, considered to be an indicator of oral hygiene status, was evaluated in 18 subjects for whom mPlI results were available, which showed that D-tagatose was significantly negatively correlated with mPlI (Spearman’s correlation coefficient; r = -0.603, p = 0.008; Figure 1). This suggested that tagatose may play a role in maintaining a low level of plaque and thus promotes good hygiene in the oral cavity.
Figure 1.
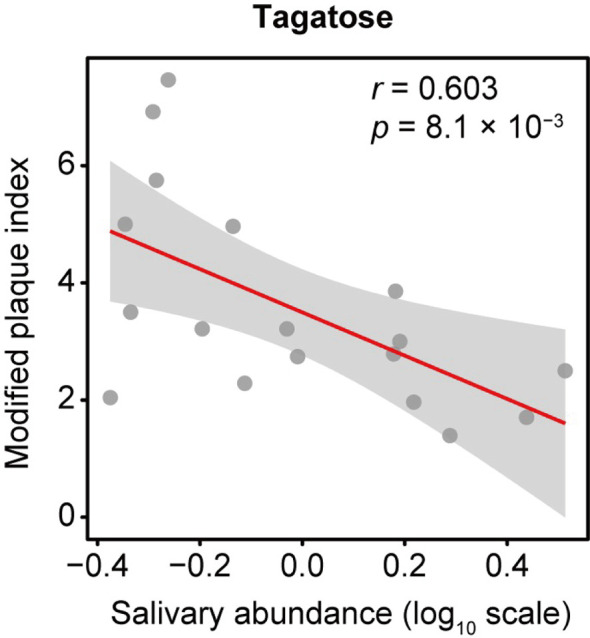
Association of salivary tagatose with modified plaque index (mPlI). The correlation between D-tagatose and mPlI, considered to be an indicator of oral hygiene status, was evaluated in 18 subjects for whom mPlI data were available. D-tagatose was found to be significantly negatively correlated with mPlI (Spearman’s correlation coefficient; r = -0.603, p = 0.008; ). It is suggested that tagatose may play a role in maintaining a low level of plaque and thus good hygiene in the oral cavity.
Effects of D-Tagatose on Planktonic Growth
To focus on the potential of D-tagatose as a prebiotic, its effects on the physiology of multiple oral streptococci were investigated. First, growth of S. gordonii, S. mutans, and S. oralis in BHI containing D-tagatose was examined, which showed that the lag phase of growth of S. gordonii and S. mutans was prolonged in the presence of D-tagatose at a concentration of 0.5% or higher (Figures 2A, 3A). On the other hand, S. oralis growth was unaffected by D-tagatose at a concentration of 1% or lower. However, growth was inhibited 5% and 10% D-tagatose, though inhibition of S. oralis growth was limited as compared to that seen with S. gordonii and S. mutans (Figure 4A).
Figure 2.
Bacterial growth of S. mutans. All data are shown as the mean ± S.D. of triplicate experiments. (A) Growth of S. mutans OMZ175 in BHI containing various concentrations of D-tagatose. (B, D) Growth of S. mutans OMZ175 and UA159 in CDM containing no sugar, 0.8% D-glucose, or 0.8% D-Tagatose. (C, E) Growth of S. mutans OMZ175 and UA159 in CDM containing 0.2% D-glucose with 0%, 1%, or 5% D-tagatose.
Figure 3.

Bacterial growth of S. gordonii. All data are shown as the mean ± S.D. of triplicate experiments. (A) Growth of S. gordonii in BHI containing various concentrations of D-tagatose. (B) Growth of S. gordonii in CDM containing no sugar, 0.8% D-glucose, or 0.8% D-Tagatose. (C) Growth of S. gordonii in CDM containing 0.2% D-glucose with 0%, 1%, or 5% D-tagatose.
Figure 4.

Bacterial growth of S. oralis. All data are shown as the mean ± S.D. of triplicate experiments. (A) Growth of S. oralis in BHI containing various concentrations of D-tagatose. (B) Growth of S. oralis in CDM containing no sugar, 0.8% D-glucose, or 0.8% D-Tagatose. (C) Growth of S. oralis in CDM containing 0.2% D-glucose with 0%, 1%, or 5% D-tagatose.
Growth of these species in the presence of D-tagatose as the sole energy source was examined. S. gordonii, S. mutans, and S. oralis growth was not supported in CDM containing 0.8% D-tagatose instead of 0.8% D-glucose (Figures 2B, D, 3B, 4B), same as in CDM with no energy source, indicating that D-tagatose is not used by these species as an energy source. When cultured in CDM containing D-glucose, D-tagatose inhibited the growth of S. gordonii and S. mutans cells in the exponential phase, though the final OD600 of S. mutans was elevated (Figures 2C, E, 3C), while no such inhibitory effect on growth of S. oralis was observed (Figure 4C). These results suggested that D-tagatose selectively inhibits planktonic growth of S. gordonii and S. mutans.
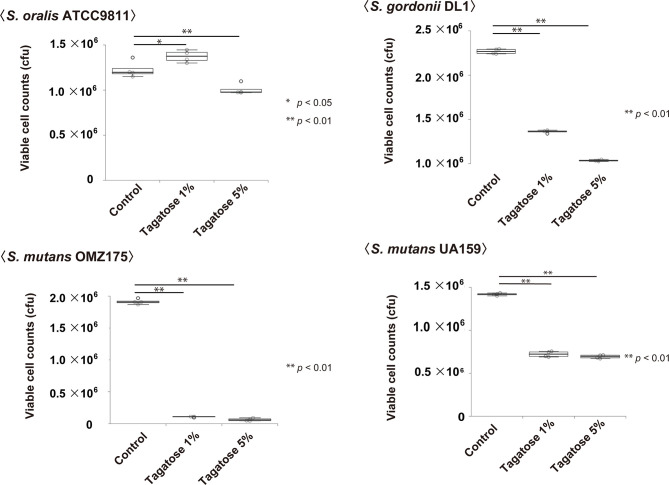
Effects of D-Tagatose on Bacterial Viability
Assays of bacterial viability in the presence of D-tagatose showed significantly decreased numbers of viable S. gordonii and S. mutans cells, which occurred in a dose-dependent manner (Figure 5). In contrast, the number of viable cells of S. oralis was increased when incubated with 1% D-tagatose in CDM, while there was only a negligible decrease in the presence of 5% D-tagatose (Figure 5). Accordingly, D-tagatose was shown to selectively suppress the viability of S. gordonii and S. mutans, and had very limited effects on S. oralis.
Figure 5.
Effects on viable cells of S. mutans, S. gordonii, and S. oralis. Cells were incubated in CDM containing 0.2% glucose with 0%, 1%, or 5% tagatose for 48 hours. Statistical differences were calculated using Dunnett’s test. *p < 0.05, **p < 0.01.
Effects of D-Tagatose on Production of EPS in S. mutans
To examine the increase in final OD600 of S. mutans in the presence of both glucose and tagatose, EPS production assays were performed. Since not only viable cell count but also extracellular products could have effects on OD600 in UV-visible spectrophotometer readings (Takahashi et al., 2004), EPS production by S. mutans under a D-tagatose stress condition was confirmed. Confocal imaging results showed that the ratio of EPS/cell was altered by treatment with D-tagatose (Figure 6A), as the treated groups exhibited a significant increase in relative quantity of EPS as compared with the control, in a dose-dependent manner (Figure 6B). These findings indicated that tagatose impairs the viability of S. mutans, though accelerates its EPS production in the presence of glucose, corresponding to the change in final OD600 seen with D-tagatose addition.
Figure 6.
Effects of D-tagatose on S. mutans EPS production. (A) Representative CLSM images showing typical architecture of planktonic cells (red) and EPS (green) following reconstruction with Imaris software. (B) Relative amounts of EPS to cells. Ten fields per sample were randomly recorded with CLSM and the relative amount of EPS was calculated using IMARIS software. Control, without tagatose. **P < 0.01.
Effects of D-Tagatose on Biofilm Formation
The effects of D-tagatose on biofilm formation by the tested strains were examined using biofilm assay results as well as examinations of confocal imaging data of biofilms cultured in CDM containing glucose, sucrose, or tagatose as the sole carbon source. When cultured in CDM containing D-tagatose, S. gordonii and S. mutans each showed negligible biofilm formation, whereas significantly increased biofilm formation was noted when grown in CDM containing D-glucose (Figures 7, 8). Treatment with sucrose resulted in robust biofilm formation by S. mutans, whereas biofilms formed by S. gordonii were relatively fragile with some detached cells observed. Interestingly, S. oralis formed significantly increased biofilm not only when the cells were treated with D-glucose but also when exposed to D-tagatose (Figure 9), suggesting that this strain may have a capacity for utilization of D-tagatose, in contrast to S. gordonii and S. mutans.
Figure 7.
Effects of D-Tagatose on S. mutans biofilm formation. (A, C) Representative CLSM images showing typical architecture of biofilms following reconstruction with Imaris software. S. mutans cells were stained with hexidium iodide (red), and incubated with saccharides or nothing was added. (B, D) Biovolume analysis of S. mutans. Ten fields per sample were randomly recorded with CLSM and S. mutans biovolume was quantified using IMARIS software. Control, containing no sugar. **P < 0.01.
Figure 8.
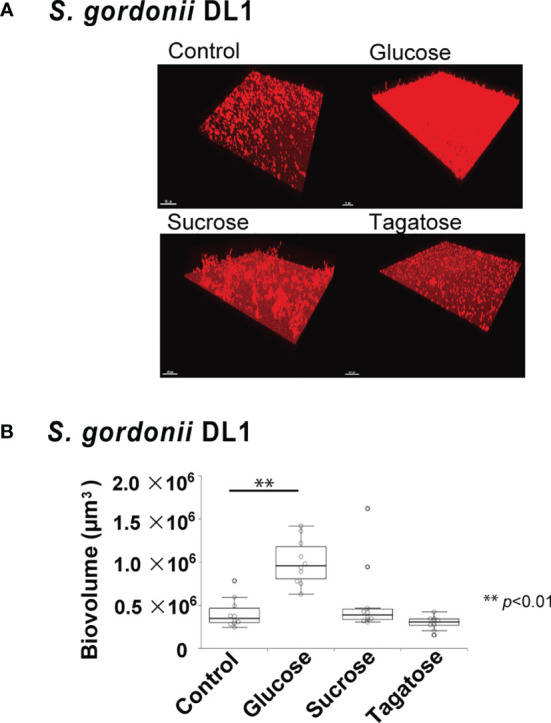
Effects of D-Tagatose on S. gordonii biofilm formation. (A) Representative CLSM images showing typical architecture of biofilms following reconstruction with Imaris software. S. gordonii cells were stained with hexidium iodide (red), and incubated with saccharides or nothing was added. (B) Biovolume analysis of S. gordonii. Ten fields per sample were randomly recorded with CLSM and S. gordonii biovolume was quantified using IMARIS software. Control, containing no sugar. **P < 0.01.
Figure 9.

Effects of D-Tagatose on S. oralis biofilm formation. (A) Representative CLSM images showing typical architecture of biofilms following reconstruction with Imaris software. S. oralis cells were stained with hexidium iodide (red), and incubated with saccharides or nothing was added. (B) Biovolume analysis of S. oralis. Ten fields per sample were randomly recorded with CLSM and S. oralis biovolume was quantified using IMARIS software. Control, containing no sugar. **P < 0.01.
RNA-Seq Analysis
To gain insight into the species-specific effects of D-tagatose, RNA-seq analysis of the examined species in the presence of tagatose was performed. Bacterial cells were incubated in CDM containing D-tagatose for 30 minutes, then total RNA was extracted from each sample. The results showed detection of 199 S. mutans genes (116 up-regulated, 83 down-regulated), 38 S. gordonii genes (35 up-regulated, three down-regulated), and 12 S. oralis genes (four up-regulated, eight down-regulated) as DEGs (FDR <0.05, |log2(fold change)| >1) when the cells were incubated with 5% D-tagatose. Volcano plots of DEGs also showed that D-tagatose altered the transcriptome profiles of each of these three streptococci (Figure S1).
DEGs of S. mutans and S. gordonii when incubated with 5% D-tagatose are listed in Table S1. GO analysis of S. mutans showed that genes related to the histidine biosynthetic and lactose catabolic processes via D-tagatose-6-phosphate were up-regulated, whereas those related to the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS) were down-regulated (p <0.05) (Table S2). Also, GO analysis of S. gordonii showed that genes related to lactose metabolism, galactose catabolism, and PTS were up-regulated (p <0.05) (Table S2 and Figure S1), while KEGG pathway analysis revealed that fructose-PTS genes (SMU_114,115, SGO_1111-1113) were up-regulated in both S. mutans and S. gordonii (Figure S1). These changes in gene expression indicate that S. mutans and S. gordonii take up D-tagatose via fructose-PTS (Figure 10), and showed that variations in gene expression for these two species are similar.
Figure 10.
Expressions of transporter genes. Gene expression levels of fructose-specific PTS and amino acid ABC transporters are shown. The three squares show Log2-transformed fold change values for tagatose-incubated cells calculated as compared to the control S. mutans (left square), S. gordonii (center square), and S. oralis (right square), according to a reported color scale.
S. oralis DEGs are also presented in Table S1. There was no information in DAVID regarding S. oralis ATCC9811, thus GO analysis was not performed. The expression of fructose-specific PTS genes in S. oralis was not altered by 5% D-tagatose (Figure S1). On the other hand, that changed gene expressions related to the ATP binding cassette (ABC) transporter. Additionally, a polar amino acid ABC transporter permease gene (SOR_1115) was up-regulated (Figure 10), while sugar ABC transporter genes (SOR_0121, 1899) were down-regulated.
Alteration of Metabolomic Profile
Metabolomic analysis was also performed to observe the effects of changes in gene expression profiles on the tested strains. Principal component analysis illustrated differences in intracellular metabolite profiles between the strains with or without D-tagatose (Figure 11). MSEA was done to explore metabolic pathways altered by D-tagatose, and a comparison of those results between the control and D-tagatose groups showed alterations in glycolysis and alanine metabolism (Table S3). Glycolysis is a series of metabolism reactions in which glucose is oxidized to pyruvate, then converted to organic acids, alanine, branched-chain amino acids (BCAAs), and others. A variety of factors are involved in reprogramming the glycolysis pathway, for example, shift of lactate fermentation to BCAA production for pH homeostasis (Matsui and Cvitkovitch, 2010). Five metabolites were detected as fermentation products in S. oralis, S. mutans, and S. gordonii (lactate, alanine, leucine, isoleucine, valine) as the result of glycolysis (Figure 12). D-tagatose significantly increased lactate in S. mutans, while that showed an increasing tendency in S. gordonii (p = 0.067). In contrast, D-tagatose did not increase lactate in S. oralis (p = 0.421). Alanine was significantly decreased in S. mutans and S. gordonii, while it was increased in S. oralis. Furthermore, D-tagatose significantly decreased leucine, isoleucine, and valine in S. mutans and S. gordonii, but not in S. oralis. Together, these results suggested that D-tagatose has effects on energy production via the PTS system, as well as BCAAs and alanine production via the pyruvate oxidation pathway, leading to disruption of metabolic homeostasis.
Figure 11.
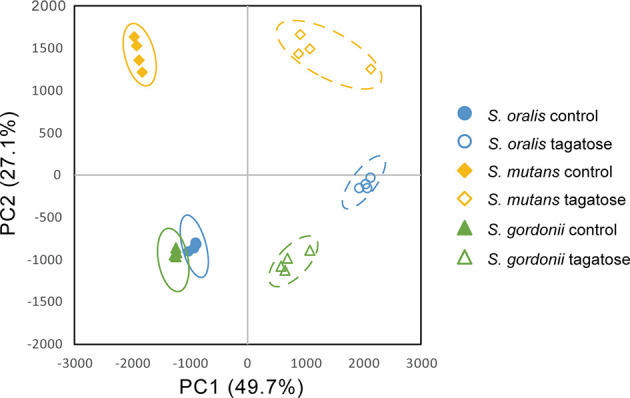
Principal component analysis of intracellular metabolome of S. gordonii, S. mutans, and S. oralis. Score plots for PC1 and PC2 are shown.
Figure 12.
Contents of tagatose and downstream metabolites from pyruvate. Metabolites were extracted from S. mutans, S. gordonii, and S. oralis cells after three hours of incubation in CDM with or without tagatose, followed by determination of those levels using GC-MS. To minimize the effects of different growth stages, the OD600 value was set to 0.8 before starting the culture. Bars represent the mean ± S.D. (n = 4). P-values were determined with Student’s t-test.
Discussion
Results of this study indicate that D-tagatose causes species-specific transcriptomic and metabolomic changes in S. mutans, S. gordonii, and S. oralis, with different inhibitory properties demonstrated with each strain. D-tagatose is a non-cariogenic sugar known to suppress growth of S. mutans. The present findings provide further evidence in that regard, as transcriptome and metabolome changes in oral streptococci including S. mutans caused by D-tagatose have not been previously reported. The tested strains did not grow in the presence of D-tagatose, though its uptake by each strain was demonstrated. The diverse transcriptome variations due to D-tagatose noted among the test strains suggests that the mechanisms of its uptake are different.
Fructose-specific PTS, whose genes are up-regulated by D-tagatose in S. mutans and S. gordonii, consumes ATP and converts fructose to fructose-1-phosphate, and S. mutans has two operons that encode components of fructose-specific PTS (FruI, FruCD) (Wen et al., 2001). D-tagatose was found to increase the expression levels of fluI and fruCD in S. mutans by about 1.45 and 33-34 fold, respectively, while fruA, a fructose-specific PTS gene encoded by S. gordonii and S. oralis, was up-regulated by approximately 11-fold in S. gordonii, but negligibly in S. oralis. Transcription of fruI and fruA is allosterically controlled by FruR, a DeoR family transcriptional regulator, and that transcription is repressed in the absence of the inducer fructose-1-phosphate (Yoon et al., 2021). No direct uptake activity of D-tagatose via PTS was detected in the present study, though our other results suggested that D-tagatose phosphorylated by FruI, FruA, or FruCD may accumulate in S. mutans and S. gordonii, and then function as an inducer of fructose-specific PTS gene expression. Accumulation of phosphorylated saccharides is known to cause sugar-phosphate stress, which results in harmful effects towards gram-negative organisms (Vanderpool and Gottesman, 2004; Maleki et al., 2017).
Accumulation of fructose-1-phosphate in S. mutans increases cell lysis and envelope DNA release (Zeng and Burne, 2019). Fructose-specific PTS is also involved in xylitol uptake (Wen et al., 2001). Xylitol is non-acidogenic sugar alcohol known for its inhibitory effect on S. mutans, which takes it up via fructose-specific PTS, causing accumulation of xylitol-5-phosphate that cannot be metabolized, resulting in triggering of sugar-phosphate stress (L, 1995; Kakuta et al., 2003). These facts support the speculation that S. mutans and S. gordonii uptake D-tagatose via fructose-specific PTS, and accumulate phosphorylate D-tagatose.
The present GO analysis results showed that the D-tagatose-6-phosphate pathway, a branch of galactose metabolism, was activated in S. mutans and S. gordonii. Transcription of the lactose operon (lacABCD), a D-tagatose-6-phosphate pathway gene cluster, is allosterically regulated by LacR, a DeoR family transcriptional regulator, which becomes inactivated by the presence of D-tagatose-6-phosphate (Rooijen et al., 1993). Thus, these results suggest that S. mutans and S. gordonii convert D-tagatose to D-tagatose-6-phosphate. However, no growth of S. mutans or S. gordonii was seen in the present of D-tagatose, even though both showed a metabolic pathway to obtain energy from D-tagatose-6-phosphate. Additional research is needed to assess whether S. mutans and S. gordonii acquire ATP from D-tagatose.
Analyses of metabolomics data indicate that changes in the metabolomic profile of glycolytic end-metabolites due to D-tagatose treatment varied among the examined bacterial species. Amino acid pools such as alanine and BCAAs were decreased in S. mutans and S. gordonii, but did not in S. oralis. In support of those findings, previous reports have noted that D-tagatose caused species-specific growth inhibition of Phytophthora spp. and decreased amino acid content in Phytophthora infestans (Chahed et al., 2020; Chahed et al., 2021). In S. oralis, the polar amino acid transporter gene (SOR_1115) was up-regulated, whereas SMU_1179c and SGO_1037, orthologous genes of SOR_1115, were down-regulated. These gene expression changes may trigger changes in BCAAs and alanine pools, since glutamate and its substrate glutamine are polar amino acids, and streptococci utilize glutamate for synthesis of alanine and BCAAs. Alanine and glutamate are utilized for synthesis of peptidoglycan constituting cell walls (Chapot-Chartier and Kulakauskas, 2014; Huang et al., 2020), while BCAAs are used for synthesis of membrane branched-chain fatty acids (Kaneda, 1963; Kaneda, 1977; Santiago et al., 2012). In addition, BCAAs have been shown to activate the global regulator CodY for transcriptional reprogramming (Sonenshein, 2007; Santiago et al., 2012). Therefore, BCAAs and alanine are essential for cell replication and adaptation to environmental changes. On the other hand, a previous report indicated that accumulation of organic acids lowers intracellular pH and suppresses glycolysis (Dashper and Reynolds, 2000). Among the present test strains, only S. oralis has the L-lactate oxidase gene (lctO) and can oxidize lactate to pyruvate under aerobic conditions. As for Streptococcus pneumoniae, it has been reported that LctO and pyruvate oxidase (SpxB) may work collaboratively to efficiently obtain ATP (Taniai et al., 2008). Thus, it is possible that such an orchestrated action also contributes to avoiding accumulation of lactate in S. oralis. In addition, in consideration of the findings showing alanine increase by D-tagatose in S. oralis, it is possible that lactate is reused for alanine synthesis via LctO. Therefore, results indicating different metabolome profiles among the tested bacterial species reveal a link to the species-specific inhibitory effect of D-tagatose noted in this study.
Our analyses of clinical saliva samples showed D-tagatose as a substance that characterizes individuals with good oral hygiene. This result appears to be well supported by previous findings indicating that chewing gum containing D-tagatose reduced the number of aerobic and anaerobic bacteria in saliva (Nagamine et al., 2020). D-tagatose is an extremely rare monosaccharide in nature, though can be produced not only chemically, but also biologically using enzymes such as L-arabinose isomerase and D-tagatose 4-epimerase (Kim, 2004; Oh, 2007; Shin et al., 2020). Therefore, the origin of D-tagatose detected in saliva is unknown, though it cannot be denied that it may be derived from a metabolite of oral bacteria. In other words, it is likely that the diversity of the oral microbiome is involved in the presence of D-tagatose in saliva.
In summary, the present study provides evidence that D-tagatose is abundant in the saliva of individuals with good oral health, and the experimental results also demonstrated selectively inhibited growth of the oral pathogens S. mutans and S. gordonii by D-tagatose. In contrast, the oral commensal S. oralis seemed to be negligibly affected, highlighting the potential of use of D-tagatose as an oral prebiotic for manipulating the metabolism of these oral streptococci.
Data Availability Statement
The data presented in the study are deposited in the DNA Data Bank of Japan Sequence Read Archive repository, accession number DRA012703, https://ddbj.nig.ac.jp/public/ddbj_database/dra/fastq/DRA012/DRA012703/.
Ethics Statement
The data presented in the study are deposited in the DNA Data Bank of Japan Sequence Read Archive repository, accession number DRA012703.
Author Contributions
SM contributed to design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. MK contributed to conceptions, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. AS contributed to design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. EH contributed to design, data acquisition, analysis, and interpretation, critically revised the manuscript. AI and YI contributed to data acquisition, critically revised the manuscript. AA contributed to design, data analysis and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Funding
This research is funded by a Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Challenging and Pioneering Research (grant 20K20393) and JSPS KAKENHI grants (grants 18H04068, 19H03862, 18K17281, 20K18827).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Miho Kakiuchi for her assistance with data acquisition. The authors have no potential conflicts of interest to declare with respect to authorship and/or publication of this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.767944/full#supplementary-material
References
- Bernabe E., Marcenes W., Hernandez C. R., Bailey J., Abreu L. G., Alipour V., et al. (2020). Global, Regional, and National Levels and Trends in Burden of Oral Conditions From 1990 to 2017: A Systematic Analysis for the Global Burden of Disease 2017 Study. J. Dent. Res. 99 (4), 362–373. doi: 10.1177/0022034520908533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahed A., Lazazzara V., Moretto M., Nesler A., Corneo P. E., Barka E. A., et al. (2021). The Differential Growth Inhibition of Phytophthora spp. Caused by the Rare Sugar Tagatose Is Associated With Species-Specific Metabolic and Transcriptional Changes. Front. Microbiol. 12, 711545. doi: 10.3389/fmicb.2021.711545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahed A., Nesler A., Navazio L., Baldan B., Busato I., Ait Barka E., et al. (2020). The Rare Sugar Tagatose Differentially Inhibits the Growth of Phytophthora infestans and Phytophthora Cinnamomi by Interfering With Mitochondrial Processes. Front. Microbiol. 11, 128. doi: 10.3389/fmicb.2020.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapot-Chartier M.-P., Kulakauskas S. (2014). Cell Wall Structure and Function in Lactic Acid Bacteria. Microbial Cell Factories 13(Suppl 1), S9. doi: 10.1186/1475-2859-13-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo P. E., Nesler A., Lotti C., Chahed A., Vrhovsek U., Pertot I., et al. (2021). Interactions of Tagatose With the Sugar Metabolism are Responsible for Phytophthora infestans Growth Inhibition. Microbiol. Res. 247:126724. doi: 10.1016/j.micres.2021.126724 [DOI] [PubMed] [Google Scholar]
- Dashper S. G., Reynolds E. C. (2000). Effects of Organic Acid Anions on Growth, Glycolysis, and Intracellular pH of Oral Streptococci. J. Dental Res. 79 (1), 7. doi: 10.1177/00220345000790011601 [DOI] [PubMed] [Google Scholar]
- Donner T. W., Wilber J. F., Ostrowski D. (1999). D-Tagatose, a Novel Hexose: Acute Effects on Carbohydrate Tolerance in Subjects With and Without Type 2 Diabetes. Diabetes Obes. Metab. 1, 7. doi: 10.1046/j.1463-1326.1999.00039.x [DOI] [PubMed] [Google Scholar]
- Gauthier L., Vadeboncceur C., Mayrand D. (1984). Loss of Sensitivity to Xylitol by Streptococcus mutans LG-1. Caries Res. 18, 7. doi: 10.1159/000260779 [DOI] [PubMed] [Google Scholar]
- Gross E. L., Beall C. J., Kutsch S. R., Firestone N. D., Leys E. J., Griffen A. L. (2012). Beyond Streptococcus mutans: Dental Caries Onset Linked to Multiple Species by 16S rRNA Community Analysis. PloS One 7 (10), e47722. doi: 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Wyss M., Duran Aguero S., Angarita Davila L. (2018). D-Tagatose Is a Promising Sweetener to Control Glycaemia: A New Functional Food. BioMed. Res. Int. 2018:8718053. doi: 10.1155/2018/8718053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. (2015). Periodontitis: From Microbial Immune Subversion to Systemic Inflammation. Nat. Rev. Immunol. 15 (1), 30–44. doi: 10.1038/nri3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasibul K., Nakayama-Imaohji H., Hashimoto M., Yamasaki H., Ogawa T., Waki J., et al. (2018). D-Tagatose Inhibits the Growth and Biofilm Formation of Streptococcus mutans. Mol. Med. Rep. 17 (1), 843–851. doi: 10.3892/mmr.2017.8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Hernandez-Valdes J. A., Kuipers O. P., Kok J. (2020). Lysis of a Lactococcus lactis Dipeptidase Mutant and Rescue by Mutation in the Pleiotropic Regulator CodY. Appl. Environ. Microbiol. 86 (8), e02937–19. doi: 10.1128/AEM.02937-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I., Witkowska E., Kaveh B., Lif Holgerson P., Tanner A. C. (2016). The Microbiome in Populations With a Low and High Prevalence of Caries. J. Dent. Res. 95 (1), 80–86. doi: 10.1177/0022034515609554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta H., Iwami Y., Mayanagi H., Takahashi N. (2003). Xylitol Inhibition of Acid Production and Growth of Mutans Streptococci in the Presence of Various Dietary Sugars Under Strictly Anaerobic Conditions. Caries Res. 37 (6), 404–409. doi: 10.1159/000073391 [DOI] [PubMed] [Google Scholar]
- Kaneda T. (1963). Valine as a Source of the Branched Short Chain Precursor in the Biosynthesis If Iso-C14, Iso-C15, Iso-C16 and Iso-C17 Fatty Acids by Bacillus subtitlis. Biochem. Biophys. Res. Commun. 10 (3), 5. doi: 10.1016/0006-291x(63)90431-5 [DOI] [PubMed] [Google Scholar]
- Kaneda T. (1977). Fatty Acids of the Genus Bacillus: An Example of Branched-Chain Preference. Bacteriol Rev. 41 (2), 28. doi: 10.1128/br.41.2.391-418.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. (2004). Current Studies on Biological Tagatose Production Using L-Arabinose Isomerase: A Review and Future Perspective. Appl. Microbiol. Biotechnol. 65 (3), 243–249. doi: 10.1007/s00253-004-1665-8 [DOI] [PubMed] [Google Scholar]
- Kreth J., Merritt J., Qi F. (2011). Antagonistic, Synergistic, and Counteroffensive Strategies for Streptococcal Interspecies Interactions. Oral. Microbial Communities 22, 13. doi: 10.1128/9781555817107.ch22 [DOI] [Google Scholar]
- Kreth J., Zhang Y., Herzberg M. C. (2008). Streptococcal Antagonism in Oral Biofilms: Streptococcus sanguinis and Streptococcus gordonii Interference With Streptococcus mutans. J. Bacteriol 190 (13), 4632–4640. doi: 10.1128/JB.00276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Amano A., Hashino E., Yamamoto Y., Inaba H., Hamada N., et al. (2009. a). Distinct Roles of Long/Short Fimbriae and Gingipains in Homotypic Biofilm Development by Porphyromonas gingivalis. BMC Microbiol. 9:105. doi: 10.1186/1471-2180-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Hendrickson E. L., Xia Q., Wang T., Xie H., Hackett M., et al. (2009. b). Proteomics of Porphyromonas gingivalis Within a Model Oral Microbial Community. BMC Microbiol. 9:98. doi: 10.1186/1471-2180-9-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Lamont R. J. (2010). Subgingival Biofilm Formation. Periodontol 2000 52 (1), 38–52. doi: 10.1111/j.1600-0757.2009.00311.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Sakanaka A., Hashino E., Bamba T., Fukusaki E., Amano A. (2016). Prediction of Periodontal Inflammation via Metabolic Profiling of Saliva. J. Dent. Res. 95 (12), 1381–1386. doi: 10.1177/0022034516661142 [DOI] [PubMed] [Google Scholar]
- L T. (1995). Xylitol: A Review of its Action on Mutans Streptococci and Dental Plaque−its Clinical Significance. Int. Dental J. 45, 16. [PubMed] [Google Scholar]
- Lærke H. N., Jensen B. B., Højsgaard S. (2000). In Vitro Fermentation Pattern of D-Tagatose Is Affected by Adaptation of the Microbiota From the Gastrointestinal Tract of Pigs. Nutrient Metab. 130, 8. doi: 10.1093/jn/130.7.1772 [DOI] [PubMed] [Google Scholar]
- Lamont R. J., Hajishengallis G. (2015). Polymicrobial Synergy and Dysbiosis in Inflammatory Disease. Trends Mol. Med. 21 (3), 172–183. doi: 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont R. J., Koo H., Hajishengallis G. (2018). The Oral Microbiota: Dynamic Communities and Host Interactions. Nat. Rev. Microbiol. 16 (12), 745–759. doi: 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo C. Y., Corliss D. A., Ganeshkumar N. (2000). Streptococcus gordonii Biofilm Formation: Identification of Genes That Code for Biofilm Phenotypes. J. Bacteriol 182 (5), 9. doi: 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinen K. K. (2010). Sugar Alcohols, Caries Incidence, and Remineralization of Caries Lesions: A Literature Review. Int. J. Dent. 2010:981072. doi: 10.1155/2010/981072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki S., Hrudikova R., Zotchev S. B., Ertesvag H. (2017). Identification of a New Phosphatase Enzyme Potentially Involved in the Sugar Phosphate Stress Response in Pseudomonas fluorescens. Appl. Environ. Microbiol. 83 (2), e02361–16. doi: 10.1128/AEM.02361-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui R., Cvitkovitch D. (2010). Acid Tolerance Mechanisms Utilized by Streptococcus mutans. Future Microbiol. 5 (3), 15. doi: 10.2217/fmb.09.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgrom P., Soderling E. M., Nelson S., Chi D. L., Nakai Y. (2012). Clinical Evidence for Polyol Efficacy. Adv. Dent. Res. 24 (2), 112–116. doi: 10.1177/0022034512449467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A., Simon-Soro A., Curtis M. A. (2017). Role of Microbial Communities in the Pathogenesis of Periodontal Diseases and Caries. J. Clin. Periodontol 44 (Suppl 18), S23–S38. doi: 10.1111/jcpe.12671 [DOI] [PubMed] [Google Scholar]
- Nagamine Y., Hasibul K., Ogawa T., Tada A., Kamitori K., Hossain A., et al. (2020). D-Tagatose Effectively Reduces the Number of Streptococcus mutans and Oral Bacteria in Healthy Adult Subjects: A Chewing Gum Pilot Study and Randomized Clinical Trial. Acta Med. Okayama 74 (4), 11. doi: 10.18926/AMO/60369 [DOI] [PubMed] [Google Scholar]
- Oh D. K. (2007). Tagatose: Properties, Applications, and Biotechnological Processes. Appl. Microbiol. Biotechnol. 76 (1), 1–8. doi: 10.1007/s00253-007-0981-1 [DOI] [PubMed] [Google Scholar]
- Oku T., Nakamura S. (2007). Threshold Transitory Diarrhea Induced by Ingestion of Xylitol and Lactitol in Young Male and Female Adults. J. Nutr. Sci. Vitaminol 53, 8. doi: 10.3177/jnsv.53.13 [DOI] [PubMed] [Google Scholar]
- Periasamy S., Kolenbrander P. E. (2009). Mutualistic Biofilm Communities Develop With Porphyromonas gingivalis and Initial, Early, and Late Colonizers of Enamel. J. Bacteriol 191 (22), 6804–6811. doi: 10.1128/JB.01006-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy S., Kolenbrander P. E. (2010). Central Role of the Early Colonizer Veillonella Sp. In Establishing Multispecies Biofilm Communities With Initial, Middle, and Late Colonizers of Enamel. J. Bacteriol 192 (12), 2965–2972. doi: 10.1128/JB.01631-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redanz S., Cheng X., Giacaman R. A., Pfeifer C. S., Merritt J., Kreth J. (2018). Live and Let Die: Hydrogen Peroxide Production by the Commensal Flora and its Role in Maintaining a Symbiotic Microbiome. Mol. Oral. Microbiol. 33 (5), 337–352. doi: 10.1111/omi.12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijen R., Dechering K. J., Niek C., Wilmink J., Vos W. (1993). Lysines 72, 80 and 213 and Aspartic Acid 210 of the Lactococcus lactis LacR Repressor are Involved in the Response to the Inducer Tagatose-6-Phosphate Leading to Induction of lac Operon Expression. Protein Engineer Design Selection 6 (2), 6. doi: 10.1093/protein/6.2.201 [DOI] [PubMed] [Google Scholar]
- Roy S., Chikkerur J., Roy S. C., Dhali A., Kolte A. P., Sridhar M., et al. (2018). Tagatose as a Potential Nutraceutical: Production, Properties, Biological Roles, and Applications. J. Food Sci. 83 (11), 2699–2709. doi: 10.1111/1750-3841.14358 [DOI] [PubMed] [Google Scholar]
- Sakanaka A., Kuboniwa M., Hashino E., Bamba T., Fukusaki E., Amano A. (2017). Distinct Signatures of Dental Plaque Metabolic Byproducts Dictated by Periodontal Inflammatory Status. Sci. Rep. 7:42818. doi: 10.1038/srep42818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka A., Kuboniwa M., Takeuchi H., Hashino E., Amano A. (2015). Arginine-Ornithine Antiporter ArcD Controls Arginine Metabolism and Interspecies Biofilm Development of Streptococcus gordonii. J. Biol. Chem. 290 (35), 21185–21198. doi: 10.1074/jbc.M115.644401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago B., MacGilvray M., Faustoferri R. C., Quivey R. G., Jr. (2012). The Branched-Chain Amino Acid Aminotransferase Encoded by ilvE is Involved in Acid Tolerance in Streptococcus mutans. J. Bacteriol 194 (8), 2010–2019. doi: 10.1128/JB.06737-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada D., Ogawa T., Miyake M., Hasui Y., Yamaguchi F., Izumori K., et al. (2015). Potent Inhibitory Effects of D-Tagatose on the Acid Production and Water-Insoluble Glucan Synthesis of Streptococcus mutans GS5 in the Presence of Sucrose. Acta Med. Okayama 69 (2), 7. doi: 10.18926/AMO/53339 [DOI] [PubMed] [Google Scholar]
- Selwitz R. H., Ismail A. I., Pitts N. B. (2007). Dental Caries. Lancet 369 (9555), 51–59. doi: 10.1016/s0140-6736(07)60031-2 [DOI] [PubMed] [Google Scholar]
- Shin K.-C., Lee T.-E., Seo M.-J., Kim D. W., Kang L.-W., Oh D.-K. (2020). Development of Tagaturonate 3-Epimerase Into Tagatose 4-Epimerase With a Biocatalytic Route From Fructose to Tagatose. ACS Catalysis 10 (20), 12212–12222. doi: 10.1021/acscatal.0c02922 [DOI] [Google Scholar]
- Sonenshein A. L. (2007). Control of Key Metabolic Intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5 (12), 917–927. doi: 10.1038/nrmicro1772 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Imazato S., Russell R. R. B., Noiri Y., Ebisu S. (2004). Influence of Resin Monomers on Growth of Oral Streptococci. J. Dent. Res. 83 (4), 5. doi: 10.1177/154405910408300406 [DOI] [PubMed] [Google Scholar]
- Taniai H., Iida K., Seki M., Saito M., Shiota S., Nakayama H., et al. (2008). Concerted Action of Lactate Oxidase and Pyruvate Oxidase in Aerobic Growth of Streptococcus pneumoniae: Role of Lactate as an Energy Source. J. Bacteriol 190 (10), 3572–3579. doi: 10.1128/JB.01882-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurnheer T., Belibasakis G. N. (2018). Streptococcus oralis Maintains Homeostasis in Oral Biofilms by Antagonizing the Cariogenic Pathogen Streptococcus mutans. Mol. Oral. Microbiol. 33 (3), 234–239. doi: 10.1111/omi.12216 [DOI] [PubMed] [Google Scholar]
- Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., et al. (2015). MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 12 (6), 523–526. doi: 10.1038/nmeth.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool C. K., Gottesman S. (2004). Involvement of a Novel Transcriptional Activator and Small RNA in Post-Transcriptional Regulation of the Glucose Phosphoenolpyruvate Phosphotransferase System. Mol. Microbiol. 54 (4), 1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x [DOI] [PubMed] [Google Scholar]
- Wen Z. T., Browngardt C., Burne R. A. (2001). Characterization of Two Operons That Encode Components of Fructose-Specifc Enzyme II of the Sugar:Phosphotransferase System of Streptococcus mutans. FEMS Microbiol. Lett. 205 (2), 337–342. doi: 10.1111/j.1574-6968.2001.tb10969.x [DOI] [PubMed] [Google Scholar]
- Yoon C. K., Kang D., Kim M. K., Seok Y. J. (2021). Vibrio cholerae FruR Facilitates Binding of RNA Polymerase to the fru Promoter in the Presence of Fructose 1-Phosphate. Nucleic Acids Res. 49 (3), 1397–1410. doi: 10.1093/nar/gkab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Burne R. A. (2019). Essential Roles of the sppRA Fructose-Phosphate Phosphohydrolase Operon in Carbohydrate Metabolism and Virulence Expression by Streptococcus mutans. J. Bacteriol 201 (2), e00586–18. doi: 10.1128/JB.00586-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the DNA Data Bank of Japan Sequence Read Archive repository, accession number DRA012703, https://ddbj.nig.ac.jp/public/ddbj_database/dra/fastq/DRA012/DRA012703/.