ABSTRACT
Many chronic diseases are associated with decreased abundance of the gut commensal Faecalibacterium prausnitzii. This strict anaerobe can grow on dietary fibers, e.g., prebiotics, and produce high levels of butyrate, often associated to epithelial metabolism and health. However, little is known about other F. prausnitzii metabolites that may affect the colonic epithelium. Here, we analyzed prebiotic cross-feeding between F. prausnitzii and intestinal epithelial (Caco-2) cells in a “Human-oxygen Bacteria-anaerobic” coculture system. Inulin-grown F. prausnitzii enhanced Caco-2 viability and suppressed inflammation- and oxidative stress-marker expression. Inulin-grown F. prausnitzii produced excess butyrate and fructose, but only fructose efficiently promoted Caco-2 growth. Finally, fecal microbial taxonomy analysis (16S sequencing) from healthy volunteers (n = 255) showed the strongest positive correlation for F. prausnitzii abundance and stool fructose levels. We show that fructose, produced and accumulated in a fiber-rich colonic environment, supports colonic epithelium growth, while butyrate does not.
KEYWORDS: Gut bacteria, dysbiosis, fructose, intestinal epithelium, Faecalibacterium, inulin
Introduction
The human gut microbiota is characterized by a vast collection of bacterial species that colonize the gastrointestinal (GI) tract and plays a pivotal role in human health and disease.1 The gut microbiota composition is highly dynamic and complex, with high intra- and inter-individual diversity.2,3 A disturbed gut microbiota homeostasis is termed “dysbiosis” and, typically, characterized by a combination of increased potentially pathogenic bacteria and decreased beneficial bacterial abundances.3,4 Dysbiosis is associated with an increasing number of human diseases, including colon cancer, type-2 diabetes mellitus and inflammatory bowel disease (IBD).5 Especially in IBD, dysbiosis is illustrated by decreased bacterial diversity with significant shifts of microbial populations, e.g. decreased abundances of Bifidobacterium and Faecalibacterium and increased numbers of adherent-invasive Escherichia coli (AIEC) and mucin-degrading Ruminococcus gnavus.5–8 A consistent observation is the decreased abundance of the strictly anaerobic, butyrate-producing species Faecalibacterium prausnitzii, one of the most dominant commensal microbes in the human gut.9,10 Importantly, F. prausnitzii is one of the many commensals that has been selectively modulated in order to restore the microbiota composition and improve intestinal health.10 In this respect, F. prausnitzii is an attractive target to restore intestinal homeostasis, since it can metabolize a variety of dietary components, including dietary fibers.11
Most dietary carbohydrates are efficiently degraded in the small intestine to di- and monosaccharides, which are subsequently absorbed in the small intestine, with very little spill-over into the colon.12 In contrast, dietary fibers reach the colon without being processed in the proximal GI tract and serve as essential carbon and energy sources for gut bacteria.13 Non-digestible food components that have the ability to selectively promote growth and metabolism of commensal gut bacteria are defined as prebiotics.14 Relevant examples include inulin-type fructans (ITFs), pectins and resistant starch. Inulin consists of oligo- or polymers of D-fructose molecules (fructans) with one D-glucose molecule at the terminus of each polysaccharide chain. A number of commensal bacteria, including bifidobacteria and faecalibacteria, are able to degrade the glycosidic bonds between fructose monomers in inulin and are therefore sensitive to this class of prebiotics.15
As a consequence of luminal content metabolism, F. prausnitzii is able to produce large amounts of short-chain fatty acids (SCFAs), especially butyrate, which are secreted into the intestinal lumen.16 Butyrate is specifically known for its anti-inflammatory and anti-carcinogenic properties, serving also as a preferred energy source for the intestinal epithelium.17 Additionally, butyrate contributes to immune system activation and enhances gut barrier function.18–20 However, besides the production of SCFAs, little is known about the metabolites generated by F. prausnitzii through prebiotic degradation, which may be beneficial for the human intestinal epithelium.21,22
The aim of this study was to dissect host-microbe cross-feeding (e.g., syntrophy) of metabolites generated from prebiotics making use of the “Human-oxygen Bacteria-anaerobic” (HoxBan) in vitro coculture system in order to investigate the molecular actions and benefits of prebiotics to intestinal epithelial health.
Results
Metabolism of prebiotics by F. prausnitzii decreases inflammatory markers in Caco-2 cells
Earlier, we reported that glucose-grown F. prausnitzii exerts anti-inflammatory and anti-oxidant effects on Caco-2 intestinal epithelial cells when cocultured in the HoxBan system.23 As glucose is not a prominent carbon and energy source for bacteria in the colon, we here aimed to fully replace glucose with prebiotics, e.g., inulin, pectin or resistant starch, in the bacterial compartment and analyze the effect on cocultured intestinal epithelial cells. Thus, Caco-2 intestinal epithelial cells were cultured in the HoxBan system with or without F. prausnitzii (the latter condition termed “monoculture”). Figure 1a shows a schematic representation of the HoxBan system. Inulin-grown F. prausnitzii significantly reduced mRNA levels of inflammatory (NOS2) and oxidative stress (HMOX1) markers in Caco-2 cells when compared to Caco-2 monocultures (Figure 1b,c), as observed earlier for glucose-grown F. prausnitzii.23 Similar trends were observed for pectin- or resistant starch-grown F. prausnitzii, but not as pronounced as for inulin (Figure 1b,c).
Figure 1.
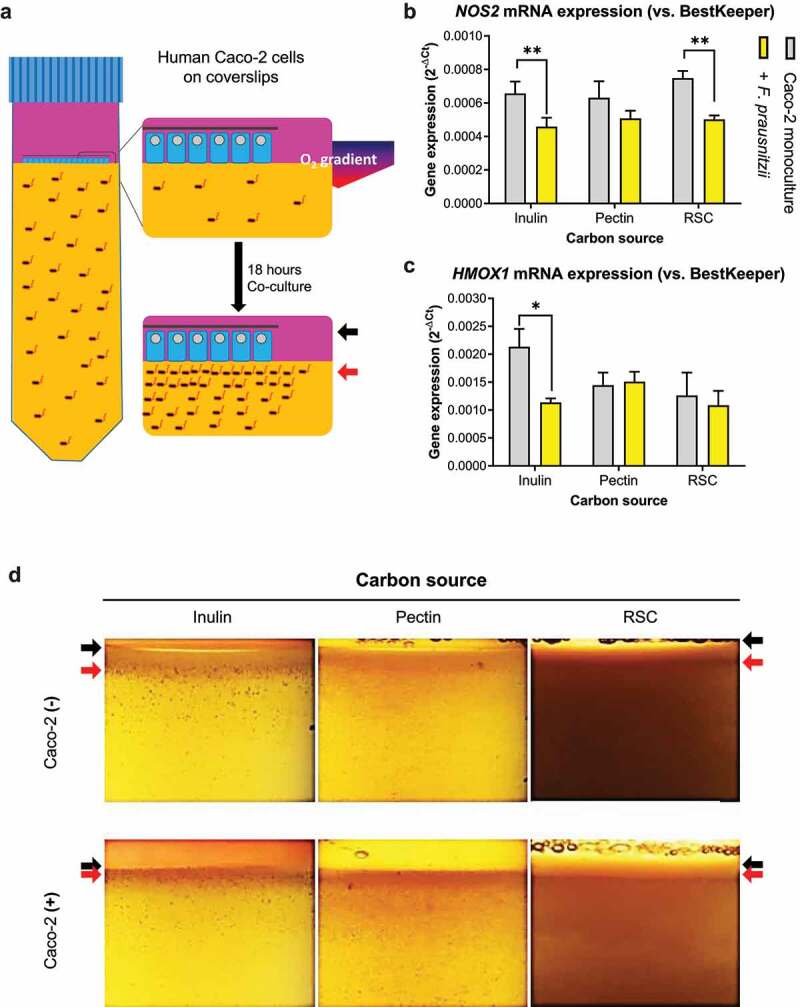
Metabolism of prebiotics by F. prausnitzii decreases inflammatory markers in Caco-2 cells. (a) Schematic representation of the HoxBan system, showing the oxic-anoxic interphase created between the Caco-2 monolayer (in human compartment, pink colored) and bacterial compartment (yellow color, representing the YCFA medium). The zoom schematically shows rim formation after 18-hour co-culture, where black arrows indicate the coverslip and red arrows indicate bacterial rim localization. (b-c) Effect of different prebiotic carbon sources on F. prausnitzii-regulated expression of NOS2 (inflammation marker, B) and HMOX1 (oxidative stress marker, C) in Caco-2 cells. Especially inulin-grown F. prausnitzii suppresses NOS2 and HMOX1 expression. (d) Prebiotic-grown F. prausnitzii in the absence and presence of Caco-2 cells. In the absence of Caco-2 cells (top panels), F. prausnitzii growth is enhanced (red arrow) in the upper part of the bacterial compartment forming a rim below the oxic-anoxic interphase (black arrow). In the presence of Caco-2 cells (bottom panels), F. prausnitzii growth is further enhanced (red arrow) and colonies appear closer to the oxic-anoxic interphase where the Caco-2 cells reside (black arrow). All experiments were performed with two biological replicates, each with an N = 3. *P < .05; **P < .01
Bacterial growth in the HoxBan tube was assessed by visual inspection of colony formation close to the coverslip with the Caco-2 cell monolayer (Figure 1d). Both in the absence and presence of Caco-2 cells, F. prausnitzii growth is enhanced (forming a rim of colonies, red arrows in Figure 1d) close to the interphase between the bottom anaerobic (bacterial) compartment and the upper oxygenated (human cell) compartment (black arrows in Figure 1d). This was observed for all fibers, as also observed earlier for glucose-grown F. prausnitzii in the HoxBan system.23 Importantly, the distance between the F. prausnitzii rim and the coverslip in the human cell compartment was smaller in all conditions in the presence of Caco-2 cells (red arrows in bottom panels Figure 1d) when compared to empty coverslips (red arrows in top panels Figure 1d). Thus, Caco-2 cells promote growth of fiber-fed F. prausnitzii closer to the epithelial oxygen-anaerobic interphase.
1. Digestion of inulin and pectin by F. prausnitzii promotes Caco-2 viability even in the complete absence of glucose
The culture medium for Caco-2 cells in the HoxBan system (e.g., DMEM) contains 4.52 g/L (25 mM) glucose, which may partly penetrate into the bacterial compartment and also support growth of F. prausnitzii. Indeed, omitting glucose completely from the bacterial compartment in the HoxBan system, while maintaining normal glucose levels in the human (upper) compartment, still led to bacterial growth and a non-significant decrease in NOS2 and HMOX1 mRNA levels in Caco-2 cells (Supplementary Figure S1A-D). Aiming to determine whether fiber-fed F. prausnitzii can provide sufficient essential metabolites for Caco-2 cells, glucose was excluded completely from both (human cell and bacterial) compartments of the HoxBan system, and only inulin or pectin was provided in the bottom (bacterial) compartment (Figure 2). Caco-2 cells show 85.9% cell viability after 16 h monoculture in the HoxBan system with glucose in the lower (bacterial) compartment, which drops to 71.1% and 71.7% when glucose is replaced by inulin or pectin, respectively. However, Caco-2 cell viability significantly increased (to 80.0%) when F. prausnitzii was cocultured on inulin (Figure 2a). Similar trends in promoting Caco-2 cell viability were observed for pectin- and glucose-grown F. prausnitzii (79.9% and 87.8%, respectively). When compared to glucose, basal mRNA levels of NOS2 and HMOX1 were strongly reduced in Caco-2 cells monocultured on inulin or pectin, which were not significantly changed in the presence of F. prausnitzii (Figure 2b-c). Again, bacterial growth was observed closer to the coverslip with Caco-2 cells when compared to empty coverslips (Figure 2d), similar as observed for conditions where glucose was present in the human (upper) compartment (Figure 1d).
Figure 2.
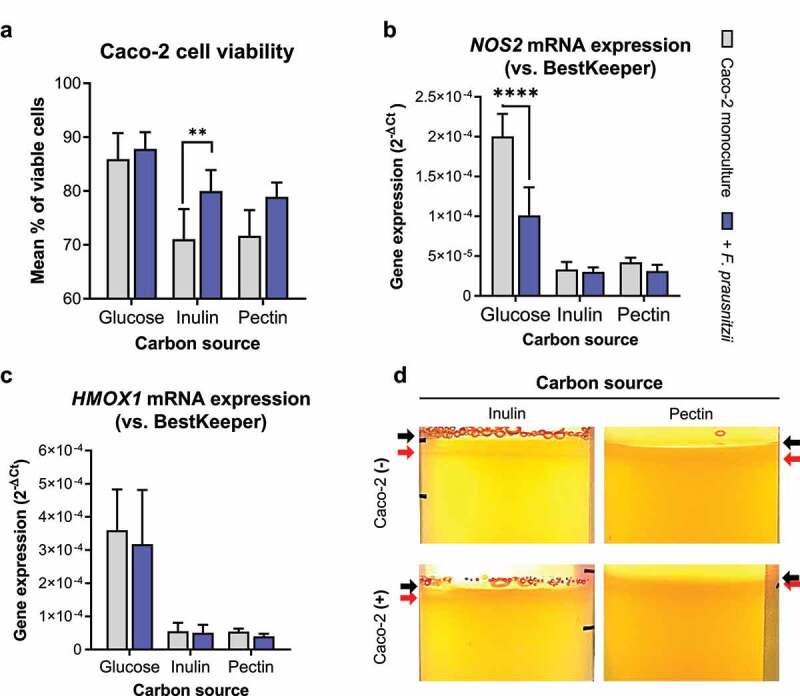
F. prausnitzii grown on inulin improves Caco-2 cell viability when cocultured in the complete absence of glucose. (a) Caco-2 cell viability after 18 h coculture without (gray bars) or with (blue bars) F. prausnitzii in the HoxBan system containing glucose, inulin or pectin as sole carbon and energy source. (b, c) Corresponding mRNA levels of NOS2 (b) and HMOX1 (c) in the F. prausnitzii-Caco-2 coculture shown in A. Basal levels of NOS2 and HMOX1 are reduced in Caco-2-F. prausnitzii cocultures on inulin and pectin when compared to glucose, with no significant additional effect of the presence of F. prausnitzii. (d) In the complete absence of glucose, inulin- or pectin-grown F. prausnitzii forms a growth rim in the upper part of the bacterial compartment (red arrows), which is closer to the coverslip (black arrows) containing Caco-2 cells (bottom panels) compared to empty coverslips (top panels). All experiments were performed with two biological replicates, each with an N = 3 (inulin) and 2 (pectin). **P < .01; ****P < .0001
Inulin-grown F. prausnitzii produces excess fructose, which is a carbon source for Caco-2 cells.
We next aimed to determine which metabolite(s) produced from inulin by F. prausnitzii could support Caco-2 cell growth and viability. First, F. prausnitzii was cultured in anaerobic batch cultures on inulin as sole carbon and energy source (Figure 3). Exponential growth was observed during the first 24 h, after which F. prausnitzii entered a stationary growth phase (Figure 3a). Bacterial growth was accompanied by a drop in pH of the medium from 6.5 to 5.5 (Figure 3b), indicating high metabolic activity of F. prausnitzii in both growth phases. Qualitative analysis of inulin and its breakdown products by High pH Anion Exchange Chromatography (HPAEC) revealed that medium-sized polymers (elute at 25–35 min) are first converted to smaller fructose polymers (elute around 25 min), which peak after 24 h F. prausnitzii growth (Figure 3c, right inset). The larger fructose polymers (elute at 35–40 min) are subsequently consumed when F. prausnitzii enters the stationary growth phase (between 24 and 48 h). Interestingly, the concentration of fructose monomers in the medium increases in a time-dependent manner, especially accumulating in the stationary growth phase of F. prausnitzii between 24 and 48 h (Figure 3c, fructose elutes at 5 min, see also red asterisks in left inset). Butyrate levels also increased in a time-dependent manner in the medium up to 10 mM after 24 h of F. prausnitzii culture. In contrast to fructose levels, butyrate levels did not significantly increase further during the stationary phase (Figure 3d). Propionate (~8 mM) and acetate (~30 mM) are standard components of the YFCA culture medium for F. prausnitzii and remained relatively stable throughout the exponential and stationary growth phases, although after 24 hours a drop was seen in the acetate concentration, due to consumption together with fructose (Figure 3e,f).
Figure 3.
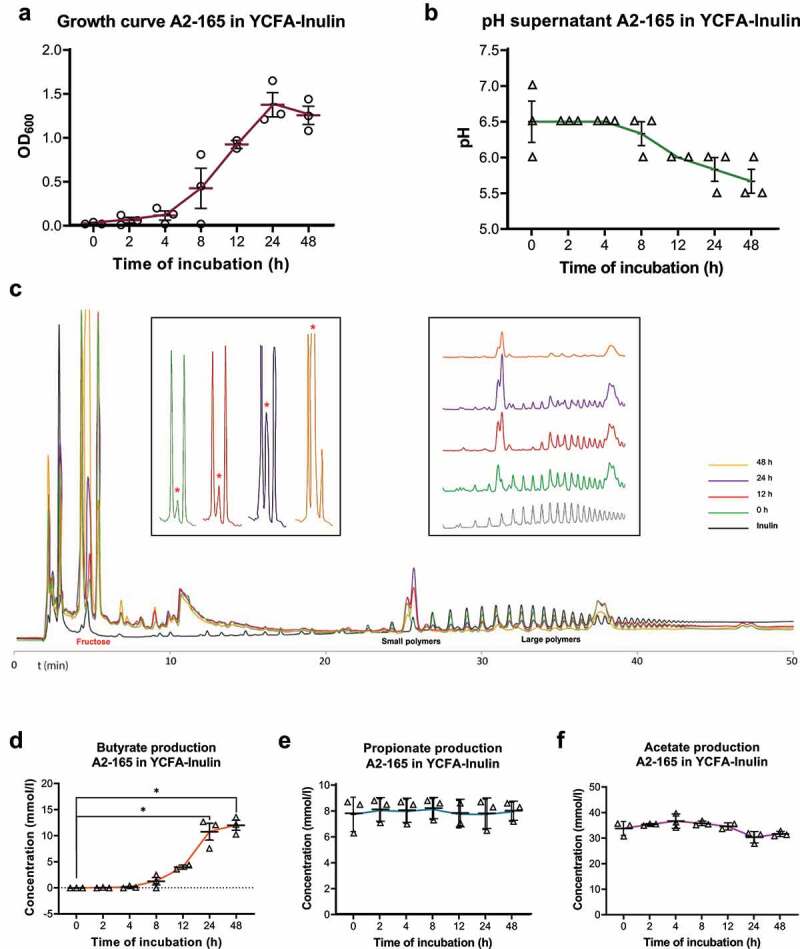
F. prausnitzii produces excess fructose and butyrate from inulin. F. prausnitzii was grown in bacterial broth containing inulin as sole sugar source and analyzed for (a) bacterial growth, (b) medium acidification, (c) inulin metabolism and fructose production, as well as the production of the short-chain fatty acids (SCFAs) butyrate (d), propionate (e) and acetate (f). Time-dependent growth of F. prausnitzii is associated with medium acidification, decrease in inulin (large polymers in C) and increase in fructose (asterisks in inset in C) and butyrate (D). Note that fructose levels increase particularly when F. prausnitzii is in the stationary growth phase (24–48 h), while butyrate levels increase most during the exponential growth phase (12–24 h). Propionate and acetate are endogenous components of the bacterial broth (at 8 and 30 mM, respectively) and their concentrations do not significantly change during F. prausnitzii growth on inulin. All experiments were performed with two biological replicates, each with an N = 3. *P < .05
Fructose, and not butyrate, supports proliferation of Caco-2 cells
Next, we analyzed whether the metabolites produced by inulin-fed F. prausnitzii, e.g., fructo-oligosaccharides (FOS), fructose and/or butyrate, as well as inulin itself can support growth of Caco-2 cells. Caco-2 cells were cultured for 5 days in DMEM medium without carbon source or supplemented with glucose (= standard medium as reference), fructose, FOS, inulin or butyrate (Figure 4a). Moreover, to tested the effect of butyrate on cell proliferation in the presence or absence of glucose (Figure 4b,c). Caco-2 cell proliferation was monitored in real-time using the xCELLigence system. Cell index (as a measure of cell proliferation) of Caco-2 cells grown on glucose increased exponentially after an initial lag phase and entered the stationary phase after approximately 110 h culture (Figure 4a, blue line). A similar growth profile was observed for Caco-2 cells grown on fructose (Figure 4a, purple line). In contrast, the cell index of Caco-2 cells grown in DMEM supplemented with FOS (Figure 4a, rose line) or inulin (Figure 4a, light green line) was similar to Caco-2 cells grown in a glucose-free medium (Figure 4a, red line). The maximum rate of cell proliferation (in cell index/hour; inset on Figure 4) was similar for Caco-2 cells grown on glucose and fructose, while FOS did not induce cell proliferation above cells in the glucose-free conditions.
Figure 4.
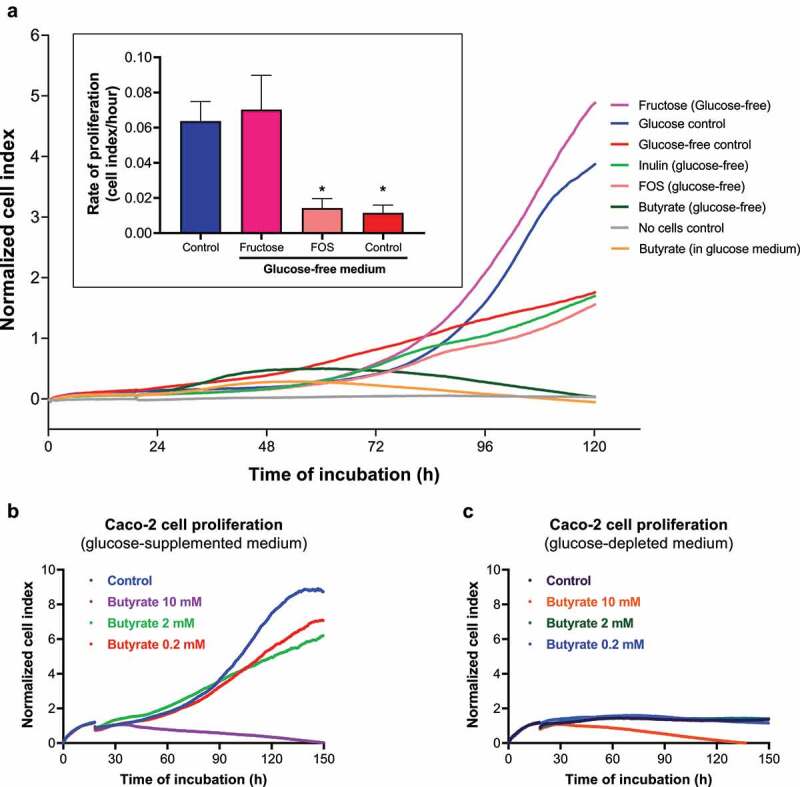
Caco-2 cells grow on fructose, but not on butyrate or inulin. (a) Caco-2 cells were cultured in a real-time cell analyzer (RTCA, xCELLigence) to monitor cell growth on different carbon/energy sources, e.g., glucose (as positive control, dark blue line), fructose (pink line), butyrate (dark green), inulin (light green) or fructose-oligosaccharide (FOS, rose line) and compared to cell cultured in the absence of a carbon and energy source (bright red line). The inset in A shows the maximum growth rate (in Δcell index/h) of Caco-2 cells on glucose (blue bar), fructose (purple bar) and FOS (rose bar) when compared to the no added carbon source (red bar). Caco-2 cells grow at a similar pace on glucose and fructose. In contrast, butyrate causes only an initial small increase in cell index (green line between 24–48 h), after which the cell index goes down, suggesting cell death. In fact, butyrate blocks growth of Caco-2 on glucose (Orange line). Butyrate decreases Caco-2 cell proliferation in a dose-dependent manner both in (b) glucose-supplemented and (c) glucose-depleted media. All experiments were performed with two biological replicates, each with an N = 3 (glucose and inulin) and 2 (pectin). *P < .05
Remarkably, low concentrations of butyrate (2 mM) in combination with glucose (Figure 4b), only slightly and transiently promoted Caco-2 cell proliferation in the early lag phase (up to 48 h), after which the cell index actually decreased in a dose-dependent manner, suggesting a cytotoxic effect on Caco-2 cells. Importantly, butyrate as a sole carbon source (Figure 4c) did not promote Caco-2 cell proliferation at any of the tested concentrations (0.2, 2 or 10 mM), with growth curves resembling the one of glucose-depleted control. Together, these data indicate that butyrate (in the presence or absence of glucose) is not a primary growth substrate for Caco-2 cells.
Inulin-grown F. prausnitzii enhances expression of the fructose transporter SCL2A5 in Caco-2 cells.
Fructose regulates its own cellular absorption by increasing transcription of, among others, SCL2A2 and SLC2A5, genes encoding the fructose transporters GLUT2 and GLUT5, respectively.24,25 SLC2A2 and SLC5A2 mRNA levels were indeed significantly enhanced in Caco-2-F. prausnitzii HoxBan cocultures when inulin was the only carbon source present, when compared to glucose as sole carbon source (Figure 5a). Such significant SLC2A5 upregulation was not observed when pectin was provided as sole carbon source, although upregulation of SLC2A2 was significant in these conditions. Furthermore, significant upregulation of the butyrate transporter MCT1 (encoded by the SLC16A1 gene) was observed in pectin-HoxBan cocultures, compared to the glucose condition (Figure 5b). Gene expression levels of the other butyrate transporter SMCT-1 (encoded by the gene SLC5A8) were undetectable. Taken together with previous results, this shows that F. prausnitzii produces fructose monomers from inulin and that Caco-2 cells use the fructose as carbon and energy source.
Figure 5.
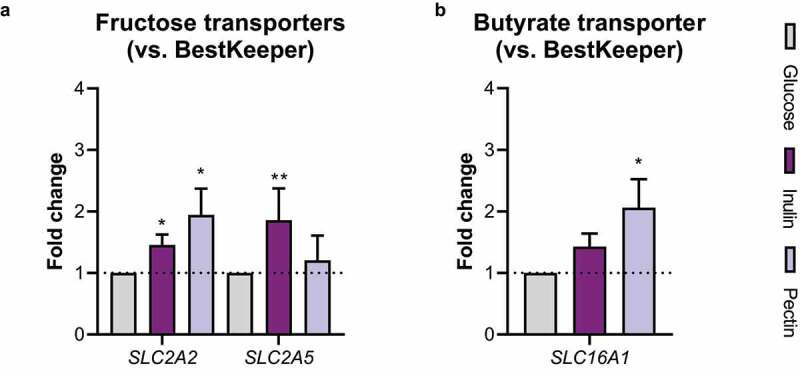
Inulin-grown F. prausnitzii increases expression of fructose transporters in Caco-2 cells. Caco-2 cells were cocultured with F. prausnitzii in the HoxBan system containing either glucose, inulin or pectin as sole carbon and energy source and analyzed for gene expression of A) the fructose transporters SLC2A2 and SLC2A5 (encoding GLUT2 and GLUT5 respectively) and B) the butyrate transporter SLC16A1 (encoding MCT1). Inulin-grown F. prausnitzii significantly induced gene expression of both fructose transporters. All experiments were performed with two biological replicates, each with an N = 3. *P < .05; **P < .01
Fecal concentrations of fructose and glucose associate with levels of F. prausnitzii in a population-based cohort
Concentrations of monosaccharides, e.g. glucose and fructose, are typically assumed to be low in the colon.12,26 Still, the above observation of bacterial fructose production from fibers prompted us to specifically analyze fructose and glucose levels in an unbiased metabolome analysis of 255 fecal samples from a population cohort, and analyze their putative correlation with the taxonomical composition of the microbiota (Figure 6). In order to account for potential confounders, we first investigated the relation between host- and dietary factors and fecal glucose and fructose levels. In total, we found 9 factors associated with the levels of the metabolites (Suppl. Table S3). These factors were then used as a covariate in the linear regression model to determine the residual-corrected association between the relative abundance o F. prausnitzii and glucose or fructose levels. Fructose and glucose were readily detected in stool samples of the study population. Remarkably, linear regression analysis (represented in a volcano plot; Figure 6a) of 112 bacterial species reveals that F. prausnitzii abundance showed the most significant positive correlation with stool fructose levels (R = 0.19, P(fdr)= 0.01 Figure 6b). Abundance of only one other species, Eubacterium ramulus, also correlated positively with fructose levels in stool, while 4 species correlated negatively to fructose levels in the stool (e.g., Methanobrevibacter sp., Methanobrevibacter smithii, Akkermansia muciniphila and Alistipes senegalensis). F. prausnitzii abundance also positively correlated with glucose levels in stool (R = 0.28 and P(fdr)= 4.3 × 10−6; Supplementary Figure S3), though the highest significance for a positive association with glucose stool levels was observed for Eubacterium rectale.
Figure 6.
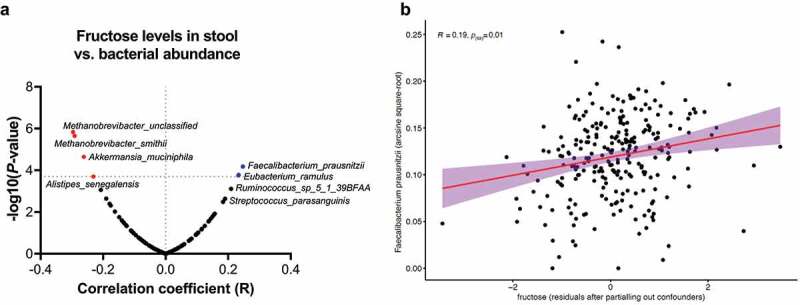
Fecal fructose levels positively correlate with F. prausnitzii abundance. (a) Volcano plot showing bacterial species whose relative abundance correlates significantly with fecal fructose levels in a population cohort (n = 255). Transformed F. prausnitzii levels most strongly correlate positively with fecal fructose levels (R = 0.19, p(fdr) = 0.01; B), while A. muciniphila is amongst species that show a significant negative correlation with fecal fructose
Discussion
In this study, we show that the dominant colonic commensal microbe F. prausnitzii produces excess butyrate and fructose when metabolizing the prebiotic inulin. Moreover, fructose was readily detected in 255 fecal samples of a large population cohort and positively correlated with F. prausnitzii abundance. Caco-2 intestinal epithelial cells use fructose as a carbon and energy source, whereas they do not grow on butyrate. When cocultured in vitro in the HoxBan system with inulin as sole carbon and energy source, the strict anaerobe F. prausnitzii enhances Caco-2 cell viability, induces expression of fructose and butyrate transporters and suppresses inflammatory and oxidative stress signaling in the human intestinal epithelial cells. Thus, bacterial breakdown of fibers/prebiotics to simple sugars/monosaccharides in the colon provides a constant fuel for epithelial cell proliferation while suppressing inflammation and oxidative stress. These data call for a reevaluation of the relative contribution of different bacterial metabolites generated from fibers/prebiotics, including butyrate and fructose, to gut health.
The prevailing concept of the action of prebiotics is that colonic bacteria feed on them thereby producing high amounts of short-chain fatty acids (SCFA), including butyrate that is a preferred energy source for colonocytes. Additionally, butyrate exerts various anti-inflammatory actions to promote gut health. Indeed, seminal work in the early 1980s showed that the rate of oxygen consumption is higher when primary human colonocytes are treated for 60 min with butyrate, when compared to glucose.27,28 Subsequent studies have supported these observations, for instance by using human intestinal epithelial cell lines (Caco-2 and T84). Treating these cells with butyrate in the presence of glucose increased oxygen consumption and saturation by increasing mitochondrial activity and activation of the oxygen-sensitive pathway hypoxia-inducible factor 1α (HIF1α).29,30 Together, these data indeed show that butyrate is an efficient energy source for colonocytes. However, exposure times are typically between minutes and several hours and do not establish butyrate as a carbon source for macromolecule biosynthesis to promote cell growth and proliferation. In human cells, butyrate is oxidized by mitochondria (β-oxidation) and yields acetyl-CoA as an end-product, which efficiently fuels ATP production.31 Human cells cannot convert acetyl-CoA to glucose, the universal carbon source for macromolecule biosynthesis in mammalian cells, consequently excluding biosynthetic functions of butyrate in intestinal epithelial cells.32,33 In fact, in vitro treatment of colonic crypts, crypt derived organoids and noncancerous cell lines (NCM460 and FHC) shows that butyrate reduces cell proliferation and induced cellular senescence.34–36 Fructose, on the other hand, is efficiently converted to glucose in the intestinal epithelium.37,38
Inulin is a fructose polymer with a glucose molecule at the terminus of each chain. Degradation of inulin and FOS by commensal gut bacteria has been extensively studied. Some butyrate-producing bacteria, like F. prausnitzii, can grow on inulin as primary carbon and energy source,39 others depend on cross-feeding of smaller metabolites generated by for instance bifidobacterial.40 Inulin-supplemented in vitro coculture of Lactobacillus acidophilus (IBB 801), Lactobacillus paracasei (8700:2) and Bifidobacterium longum (LMG 11047) resulted in release of free fructose to the media, together with lactate and acetate.41 Furthermore, in vitro fermentations of inulin by diluted stool samples showed a peak of fructose production after 8 h of incubation, which coincided with acidification of the media and decreased at 24 h of culture (Suppl. Figure S2A and B). These experiments demonstrated that the cross-feeding between different bacterial species still results in accumulation of fructose that is made available to intestinal epithelial cells. In our work, we found that fructose production was particularly increased when inulin-grown F. prausnitzii entered the stationary growth phase. In contrast, butyrate levels did not further increase then. A recent study made a similar observation39 with another strain of F. prausnitzii (DSM 17677®). These results imply that butyrate production from inulin requires (exponential) bacterial growth, while this is not the case for fructose production. F. prausnitzii likely secretes enzymes that breakdown the inulin polymers, enzymes that continue doing so when bacterial growth ceases. Upregulation of β-fructofuranosidase, as well as coupled ABC transporter protein, have been described in Roseburia intestinalis (a Firmicutes gut commensal and butyrate producer, as F. prausnitzii) during growth on inulin.42,43 Nevertheless, this has not yet described for F. prausnitzii grown on inulin-type fructans. The fecal fructose levels are likely a result of such bacterial fiber fermentation, since dietary monosaccharides are effectively absorbed in the small intestine. Interestingly, of all bacteria detected, F. prausnitzii abundance showed the strongest positive correlation to fructose levels in fecal samples, which may suggest that F. prausnitzii is a contributing factor in producing fructose, or monosaccharides in general, in the colon from dietary fibers and thereby cross-feeding to the colonic epithelium.
The presence of fructose in feces also implies that it is always available as a carbon and energy source for the human colonic epithelium and not completely used by other bacteria. Cross-feeding of metabolites produced by inulin-grown F. prausnitzii to Caco-2 intestinal epithelial cells was reproduced in vitro using the HoxBan system, which allows coculturing of strict anaerobes like F. prausnitzii and oxygen-requiring human intestinal epithelial cells.23 Caco-2 cell viability was improved when cocultured with inulin-grown F. prausnitzii, when compared to conditions with bacteria. We were not able to detect significant amounts of fructose in the medium after coculture in the HoxBan system, but this is likely a result of efficient absorption of the produced fructose by the Caco-2 cells. Gene expression of SLC2A2 and SLC2A5 (encoding the fructose transporters GLUT2 and GLUT5) is induced by fructose,12 which was also observed in Caco-2 cells cocultured with inulin-grown F. prausnitzii. GLUT5 is the main fructose transporter situated at the apical membrane of intestinal epithelial cells, and its deletion completely abrogates transepithelial fructose transport, highlighting its physiological importance.44,45 In contrast, GLUT2, situated at the basolateral membrane, is not specifically upregulated by high luminal fructose contents, as it serves multiple substrates (also glucose and galactose) and its deletion only modestly impairs fructose absorption.46 In line with this, the longstanding notion that GLUT2 is the main apical fructose transporter has been excessively scrutinized.24 Our data also indicated that GLUT5 is most sensitive to inulin with an approximately 2-fold increase in expression in inulin-grown Caco-2/F. prausnitzii cocultures. Interestingly, inulin-grown F. prausnitzii also suppressed expression of NOS2 and HMOX1, as sensitive markers of inflammation and oxidative stress. Lastly, we observed that fructose and butyrate transporters are also upregulated in Caco-2 cells in coculture with pectin-grown F. prausnitzii. However, further research is needed to understand the cross-feeding from pectin metabolites to intestinal epithelial cells.
Various clinical trials have been conducted to increase the abundance of beneficial bacteria like F. prausnitzii and A. muciniphila by dietary factors (reviewed by Verhoog et al. (2019)). Most of these studies reported that diets enriched for fructo-oligosaccharides (FOS), inulin or raffinose enhance F. prausnitzii levels that, based on our data, would in turn provide multiple carbon and energy sources to the colonic epithelium and improve gut health.
In conclusion, this study shows that fermentation of inulin by F. prausnitzii produces excess fructose, a carbon and energy source that is readily available for the human colonic epithelium. As F. prausnitzii levels are decreased in various metabolic and digestive diseases, including IBD,47–52 which are also characterized by a leaky gut, promoting F. prausnitzii abundance and/or metabolism by administration of prebiotics like inulin may provide the highly required carbon and energy sources to support effective epithelial regeneration.
Materials & methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Klaas Nico Faber k.n.faber@umcg.nl.
Materials availability
TaqMan primers and probes for RT-qPCR generated in this study are described in Table 1. F. prausnitzii (strain A2-165) was kindly provided by S. Duncan and H. Flint (Rowett, UK).
Data and Code Availability
The raw metagenomics sequencing data used for this study are available from the European Genome-phenome Archive data repository: 1000IBD cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001004194) and LifeLines DEEP cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001001991). Due to participant confidentiality, the datasets are available upon reasonable request to the University Medical Center of Groningen (UMCG) and LifeLines, respectively. The metabolomics data supporting the current study have not yet been deposited in a public repository because of participant confidentiality, but are available from the corresponding author on request.
Experimental model and subjects details
Cell lines and bacterial strains
Human epithelial colon adenocarcinoma cells (Caco-2, male, ATCC®, HTB-37TM) were used as representative of intestinal epithelial cells. Cells were incubated at 37°C and 5.0% CO2. As previously described in Sadaghian Sadabad et al (2015), Caco-2 cells were cultured in Glutamax™ Dulbecco’s Modified Eagle Medium (DMEM, ThermoFisher Scientific Inc) with 25 mM glucose and supplemented with 10% fetal calf serum (FCS, Invitrogen), 1% non-essential amino acids (NEAA, Gibco) and 1% PSF antibiotic cocktail (penicillin (10 U/ml), streptomycin sulfate (100 μg/ml) and fungi zone; Lonza). For use in the HoxBan system, cells were seeded on coverslips until 70–80% confluence in a 12-well plate (4 cm2/well), for 24 hours. About 1 hour before transferring the coverslip-attached Caco-2 cells to the coculture system, the medium in each well was discarded and replaced by PSF-free DMEM medium.
F. prausnitzii (A2-165) was cultured anaerobically at 37°C starting from −80°C glycerol stocks (20% glycerol in YCFAG medium) in YCFAG medium, which contains 2.5 g/l yeast extract, 10.0 g/l casitone, fatty acids (9 mM propionate, 1 mM isobutyrate, 1 mM isovalerate and 1 mM valerate), 33 mM sodium acetate and 4.52 g/l (20 mM) glucose (detailed medium composition can be found in Sadaghian Sadabad et al (2015)). In specific experiments, glucose was omitted from YCFAG medium or replaced by 4.52 g/l of specified fibers, e.g., inulin (I; Raftiline® HP, CAS number 9005–80-5, ORAFTI S.A.), apple pectin (P; catalog number 93854 Sigma-Aldrich) or resistant starch (RSC; catalog number S4126, Sigma-Aldrich). Media is further described on Suppl. Table S1. F. prausnitzii was inoculated from frozen stocks at a dilution of 1:1,000 in 5 ml YCFAG, -I, -P or -RSC for 14–16 hours until an optical density (at 600 nm) of 0.8.
Human studies
Population-based fecal metagenomics and fecal metabolomics data were derived from the LifeLines-DEEP cohort available at the time of conducting this research, a Dutch general population-based cohort study consisting of individuals from the Northern part of the Netherlands.53 Metagenomics and untargeted metabolomics data were available from a subset of this cohort (n = 255, 114 males (44.7%, average age 46.8 (S.D. 12.8), average BMI 25.17 (S.D. 3.8)) for which microbiome data was also available and after excluding subjects with inflammatory bowel diseases. Participants were requested to collect and freeze fecal samples at home. Samples were then picked up and transported on dry ice and stored at −80°C until further analysis. All participants provided written informed consent prior to sample collection (IRB ref. M12.113965).
Methods details
HoxBan coculture system
As described by Sadaghian Sadabad et al. (2015), anaerobically-grown F. prausnitzii was inoculated in YCFAG liquid broth, from which 1 ml was used to inoculate 1 liter of autoclaved agar-based (1% agar) YCFAG, -I, -P or -RSC (pH = 6.5) and cooled-down to approximately 40°C. For each 50 ml Falcon test tube, 40 ml of this mixture was added in an anaerobic cabinet and transferred to a laminar flow cabinet and opened at ambient air. 10 ml of 37°C PSF-free DMEM was added to each tube. Next, the Caco-2 cells grown on coverslips were laid upside-down on the top of the bacteria-containing agar medium. The screw caps of the Falcon tubes were kept loosely tightened, to allow oxygen entry into the system for the Caco-2 cells. Coculture took place for 18 hours at 37°C and 5% CO2, after which coverslips were collected and processed for downstream analyses.
RNA isolation and gene expression quantification
RNA was isolated from Caco-2 cells using TRIzol (Sigma-Aldrich) according to the manufacturer’s protocol (Thermo-Scientific), followed by quantification of RNA using a NanoDrop 2000 c spectrophotometer. To synthesize complementary DNA (cDNA), 2.5 ng of RNA was added to a final volume of 50 μl, 10% reaction RT Buffer (final concentration 50 mM Tris-HCl, 50 mM KCl, 3 mM MgCl2, 5 mM DTT), 10% dNTP mix (final concentration 1 mM dATP, dGTP, dTTP, dCTP, Sigma-Aldrich), 2% random primers (0.01 μg/μl; Sigma-Aldrich), 2% M-MLV RT (100 U; Invitrogen) and 1.5% RNAse OUT (30 U; Invitrogen). cDNA synthesis was performed on a thermal cycler (Bio-Rad T100) for 10 minutes at 25°C followed by 60 minutes at 37°C and 5 minutes at 95°C. Prior to quantitative real-time PCR, cDNA solution for each sample was diluted 20-fold in RNAse-free water.
Gene expression of inducible nitric oxide synthase (NOS2), heme oxygenase 1 (HMOX1), interleukin-1 beta (IL1B), glucose transporter 2 (GLUT2) (SLC2A2) and glucose transporter 5 (GLUT5) (SLC2A5) were measured by TaqMan-based quantitative Real-Time PCR (RT-PCR). BestKeeper Index analysis of the housekeeping genes 18S, GAPDH, RPII and ACTB was used for normalization of gene expression levels of genes of interest (software available at https://www.gene-quantification.de/bestkeeper.html). The sequence and description of the probes and primers used in this study are presented in Supplementary Table S2. Each sample was prepared in duplicates, in a reaction mix (final volume 20 μl) containing 0.2 μM fluorescent probes, 0.936 μM forward and reverse primers, 10 μl qPCR reaction buffer (Eurogenic), 4.48 μl RNAase-free water and 4 μl cDNA. Amplification was performed after 10 minutes of heating at 95°C followed by a 40-times repeated cycle, consisting of 15 seconds at 95°C and 1 min at 60°C, using a StepOnePlus (AB, Applied Biosystems) PCR system.
Cell viability assay
Viability of Caco-2 cells was assessed by Trypan blue staining. Trypan blue solution (0.2%, Sigma-Aldrich) was added to the coverslip-attached Caco-2 cells (500 μl/well) for 1 minute, after which Caco-2 cells were fixed for 10 minutes at RT using 4% paraformaldehyde (PFA; Sigma-Aldrich) in phosphate-buffered saline solution (PBS; catalog 10010023, Gibco®, ThermoFisher Scientific). Adherent cells were rinsed 4 times with PBS and fixed on top of glass plates using mounting medium (Thermo-Fisher Scientific). Quantification of cell viability was performed by manually counting Trypan blue-positive and – negative cells using light microscopy (at 200x magnification). Cell viability was expressed as the mean percentage of viable cells per field.
Proliferation assay
The xCELLigence® RTCA-DP system was used to assess Caco-2 cell proliferation, by monitoring electrical resistance buildup on well surface. 2,000 cells were seeded in an xCELLigence® 16-well and attachment was allowed overnight on standard DMEM medium, in a standard CO2 cell culture incubator. After the attachment phase, medium was replaced by either new standard or glucose-depleted DMEM medium with different carbon-sources in duplicate. Glucose, fructose, fructo-oligosaccharides (FOS) and inulin were added to a final concentration of 4.52 g/l, which is equal to the standard sugar concentration in the standard YCFAG bacterial agar in the HoxBan coculture system. Butyrate (Sigma-Aldrich) was added to concentrations ranging from 0.2 to 10 mM. The 16-well plate was placed in the xCELLigence system and cell index was measured every 15 min for 5 days, medium and conditions being refreshed every 2 days. Rate of cell proliferation (expressed as cell index/hour) was assessed by calculating the slope of growth curve at exponential phase for each treatment.
Inulin breakdown by F. prausnitzii
From an overnight culture of F. prausnitzii in YCFAG (described above), 50 μl was inoculated in 5 ml YCFAI medium and statically incubated for 18 hours under anaerobic conditions. The resultant bacterial suspension was again diluted (1:100) in 50 ml YCFAI, and kept in anaerobic conditions at 37°C. Aliquots of 1 ml (in duplicates) were taken from this inoculum at indicated time points (0, 2, 4, 8, 12, 24 and 48 hours) and pH measured, followed by centrifugation (13,000 × g for 5 min) to remove bacteria. The supernatant was collected and stored at −20°C prior to fructose or SCFAs analysis by high-performance liquid chromatography (HPLC).
Carbohydrate semi-quantitative analysis
1.0 ml supernatant aliquots (as described above) were transferred to a 1.5 ml tube and centrifuged at 13,000 × g at RT, the supernatants were diluted 1:5 in MilliQ water for analysis. Samples were analyzed by High pH Anion Exchange Chromatography (HPAEC) on an ICS300 system at the University of Groningen, Dept. Microbial Physiology, eluted with a 55 min linear gradient of NaOAc (30–600 mM) in 100 mM NaOH. Carbohydrates were detected with a Pulsed Amperometric Detector, using a quadruple potential waveform.54 A qualitative standard of equimolar glucose (G), fructose (F), sucrose (GF), kestose (GF2) and nystose (GF3) (0.1 mM) was used to identify FOS peaks. Confirmation of fructose and fructo-oligosaccharide peaks in samples was achieved by spiking 500 μl supernatant sample with 100 μl with 0.28 mM fructose or 100 μl 1 mg/mL small polymer inulin (Sensus, Roosendaal, The Netherlands).
Short-chain fatty acid (SCFA) analysis
SCFA in bacterial supernatants were quantified as described by Moreau et al (2003) with minor modifications. Supernatant samples were thawed on ice and 85 μl was diluted in 415 μl MilliQ water. A 7-point calibration curve in Milli-Q was stored in aliquots at −80 °C with final concentrations between 0.0 and 8.0 mM for acetate and between 0.0 to 4.0 mM for propionate and butyrate. 100 μl internal standard solution (1 mg/ml 2-ethylbutyrate in Milli-Q), 20 μl 20% (w/v) sulphosalicylic acid solution and two drops 37% HCl (approximately 100 μl) were added to the 500 µl diluted supernatant samples and calibration samples. The samples were centrifuged at 16,100 × g for 20 min at 4°C, and the supernatant was transferred to a glass tube containing a spatula tip of sodium chloride (approximately 30 mg). 2 ml of diethyl ether was added and the sample was vortexed for 10 min at RT and centrifuged at 3,000 × g for 10 min at 4°C. From the clear upper layer, a 500 μl aliquot was taken and transferred to a glass GC-vial. Subsequently, 50 μl of MBTSTFA + 1% TBDMCS (N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide + 1% tertbutyldimetheylchlorosilane) was added and left to derivatize overnight at room temperature. 3 μl derivatized sample was injected into the GC-MS (7890A GC System and 5975 C inert XI EI/CI MSD with an EI inert 350 source, Agilent Technologies, Santa Clara, USA). Analysis was carried out in a split mode with an inlet split ratio of 20:1. Samples were analyzed in SIM acquisition mode; acetate at m/z 117, propionate at m/z 131, butyrate at m/z 145 and 2-ethylbutyrate at m/z 175. Injector, source and quadrupole temperatures were 280°C, 230°C and 150°C, respectively. A Zebron capillary GC column of 30 mm x 0.25 mm, 0.25 μm film thickness was used (ZB-1, Phenomenex, Torrance, USA). The GC oven was programmed as follows: 40°C held for 0 min, increased to 70°C at 5°C/min, held at 70°C for 3.5 min, increase to 160°C at 20°C/min, increased to 280°C at 35°C/min and finally held at 280°C for 3 min with a total run time of 20.43 min. The flow was set a 1.0 ml/min with helium as carrier gas. Data processing was carried out with MassHunter Workstation Software (MassHunter, Agilent Technologies).
Fecal human microbiota analysis
Metagenomics of fecal samples
Taxonomy of the fecal microbiome was characterized in a high-resolution fashion using whole-genome metagenomic shotgun sequencing (MGS). Microbial DNA extraction from frozen fecal samples and MGS sequencing using the Illumina HiSeq platform was performed as described previously.55 Genomic library preparation was performed using the Nextera XT Library preparation kit. Trimmomatic (v.0.32) was used to remove adapters and trim the ends of metagenomic reads.49,56 Cleaned metagenomic reads were processed through a previously published bioinformatics pipeline.49 Taxonomic compositions were profiled using the software tool MetaPhlAn2 and were expressed as relative abundances in the microbiome samples.57
Metabolomics of fecal samples
Human data was obtained from the LifeLines-DEEP study cohort. In short, this cohort comprises 1,500 participants from the northern parts of the Netherlands for which multiple data layers (genetics, microbiome, phenotypes, etc.) are available. In this cohort, 255 participants’ fecal metabolites were measured. Levels of specific metabolites of interest (fructose, glucose and butyrate) in fecal samples were extracted from an untargeted Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) analysis performed by Metabolon®, Inc., Morrisville, NC, USA. Full methodological details on the complete procedure can be found elsewhere.58 In brief, fecal samples were lyophilized and extracted at a constant per-mass basis. Recovery standards were added prior to the first step in the extraction process for quality-control (QC) purposes. Proteins and macromolecule derivatives were removed from the samples using methanol precipitation under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000). Next, samples were split into five different fractions: two fractions for analysis by two separate reverse phases (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one fraction for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one fraction for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one fraction reserved as back-up. Metabolites profiles were derived from untargeted metabolomic experiments and each metabolite was expressed as a relative proportion represented as areas under the curve (AUC) of mass-spectrometry experiments, therefore, we could not estimate the total concentrations of fructose or glucose present in the fecal samples.
Quantification and statistical analysis
Metabolomics of fecal samples
Three types of controls were used and analyzed together with all experimental samples: 1) a pooled matrix sample, generated by extracting a small volume of each experimental sample, and serving as technical replicate throughout the dataset, 2) extracted water samples serving as process blanks, and 3) a cocktail of QC standards. Metabolites were identified by comparison to a reference library containing chemical standards. Area under the curve (AUC) analysis was performed for peak quantification and normalized to day median values. Control and curation procedures were performed to ensure high quality of the data and true chemical assignment, while removing artifacts and background noise. A total of 1,192 metabolites were measured in 255 fecal samples.
Metabolome values were log-10 transformed and centered around the mean (mean = zero and standard deviation = 1). Metabolomic values below the lower limit of detection (LLoD) were transformed as half of the lowest value observed for each individual metabolite. Regarding the metagenomic data, gut bacteria were analyzed at species-level. Bacterial species present in less than 20% of fecal samples were removed. Relative abundances of bacterial species were arcsine square root-transformed before entry into analysis.
Statistical analyses
Data analysis and data visualization were performed using GraphPad Prism® (GraphPad, version 8.0), with experimental data expressed as means (μ) ± standard deviations (SD). Different conditions were compared using the two-way ANOVA test with Bonferroni post-hoc comparisons for the difference between two separate conditions. P-values ≤ 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
RKW is supported by a research grant from the Seerave Foundation and by the collaborative TIMID project (LSHM18057-SGF) financed by the PPP allowance made available by Top Sector Life Sciences & Health to Samenwerkende Gezondheidsfondsen (SGF) to stimulate public–private partnerships and co-financing by health foundations that are part of the SGF. Furthermore, we thank Hermi Kingma and Rebecca Heiner for the SCFA analysis.
Funding Statement
The research position of RRF was supported by the Graduate School of Medical Sciences (GSMS) of the University of Groningen, the Netherlands. The research position of ARB was supported by a JSM MD-PhD trajectory grant (grant no. 17-57) from the Junior Scientific Masterclass (JSM) of the University of Groningen, the Netherlands. AS was supported by ZonMW-MKMD (grant no. 114021513). The funders had no role in the design of the study, collection, analysis, or interpretation of data or in writing the manuscript.
Disclosure statement
RKW acted as consultant for Takeda and received unrestricted research grants from Takeda. RKW received unrestricted research grants from Johnson & Johnson Pharmaceuticals, Tramedico and Ferring, and received speaker fees from AbbVie, MSD, Olympus and AstraZeneca. GD received research grant from Royal DSM and speaker’s fees from Janssen Pharmaceuticals, Pfizer and Abbvie. All other authors have no conflicts of interest to declare.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. Internet] 2015. [cited 2016 Nov 3]; 12:205–18. Available from(4)http://www.nature.com/doifinder/10.1038/nrgastro.2015.34 [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB. Diversity of the human intestinal microbial flora. Science. Internet] 2005. [cited 2018 Apr 23]; 308(5728):1635–1638. Available from http://www.sciencemag.org/cgi/doi/10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. Internet] Available from. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schippa S, Conte M. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6(12):5786–5805. Internet] Available from. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146(6):1489–1499. Internet] Available from. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115(1):182–205. Internet] Available from. doi: 10.1016/s0016-5085(98)70381-6. [DOI] [PubMed] [Google Scholar]
- 7.Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci. 2007;104(34):13780–13785. Internet] Available from. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut Internet] 2011; 60:631–637. Available from(5) http://www.ncbi.nlm.nih.gov/pubmed/21209126 [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Shen J, Ran ZH. Association between faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract. [Internet] 2014 [cited 2017 Jun 12]; 2014:872725. Available from. http://www.ncbi.nlm.nih.gov/pubmed/24799893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A Internet] 2008. [cited 2020 May 13]; 105:16731–16736. Available from(43) http://www.ncbi.nlm.nih.gov/pubmed/18936492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moens F, Rivière A, Selak M, De Vuyst L. Inulin-type fructan degradation capacity of interesting butyrate-producing colon bacteria and cross-feeding interactions of Faecalibacterium prausnitzii DSM 17677T with bifidobacteria. Arch Public Heal Internet] 2014; 72. Available from. doi: 10.1186/2049-3258-72-s1-o6. [DOI] [Google Scholar]
- 12.Ferraris RP, Yasharpour S, Lloyd KCK, Mirzayan R, Diamond JM. Luminal glucose concentrations in the gut under normal conditions. Am J Physiol Internet] 1990. [cited 2021 Feb 15]; 259:G822–37. Available from. http://www.ncbi.nlm.nih.gov/pubmed/2240224. [DOI] [PubMed] [Google Scholar]
- 13.Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70(6):443–459. Internet] Available from. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 14.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr Internet] 1995; 125:1401–1412. Available from(6): http://www.ncbi.nlm.nih.gov/pubmed/7782892 [DOI] [PubMed] [Google Scholar]
- 15.Kolida S, Gibson GR. Prebiotic capacity of inulin-type fructans. J Nutr. 2007;137(11):2503S–2506S. Internet] Available from. doi: 10.1093/jn/137.11.2503s. [DOI] [PubMed] [Google Scholar]
- 16.Sh D, Barcenilla A, Cs S, Se P, Flint HJ. Acetate utilization and butyryl coenzyme a (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68(10):5186–5190. Internet] Available from. doi: 10.1128/aem.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. Internet] Available from. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 18.Venegas DP, De La Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases [Internet]. Frontiers Media S.A. 2019; cited 2021 Mar 17 Available from https://pubmed.ncbi.nlm.nih.gov/30915065/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Poul E, Loison C, Struyf S, Springael J-Y, Lannoy V, Decobecq M-E, Brezillon S, Dupriez V, Vassart G, Van Damme J, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;27828:25481–25489Internet] Available from 10.1074/jbc.m301403200 [DOI] [PubMed] [Google Scholar]
- 20.Wang H-B, Wang P-Y, Wang X, Wan Y-L, Liu Y-C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein claudin-1 transcription. Dig Dis Sci. 2012;57(12):3126–3135. Internet] Available from. doi: 10.1007/s10620-012-2259-4. [DOI] [PubMed] [Google Scholar]
- 21.Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol. 2016;428(16):3230–3252. Internet] Available from. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, Wolvers D, Watzl B, Szajewska H, Stahl B, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr Internet] 2010; 104 Suppl: S1–63. Available from. S1–S63.http://www.ncbi.nlm.nih.gov/pubmed/20920376 [DOI] [PubMed] [Google Scholar]
- 23.Sadaghian Sadabad M, von Martels JZH, Khan MT, Blokzijl T, Paglia G, Dijkstra G, Harmsen HJM, Faber KN. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci Rep Internet] 2016. [cited 2017 May 23]; 5:17906. Available from. doi: 10.1038/srep17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferraris RP, Choe JY, Patel CR. Intestinal absorption of fructose [Internet]. Annu. Rev. Nutr. 2018. [cited 2021 Feb 9];38(1):41–67. Available from https://pubmed.ncbi.nlm.nih.gov/29751733/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones HF, Butler RN, Brooks DA. Intestinal fructose transport and malabsorption in humans [Internet]. Am. J. Physiol. - Gastrointest. Liver Physiol.2011 [cited 2021 Feb 9]; 300. Available from. https://pubmed.ncbi.nlm.nih.gov/21148401/ [DOI] [PubMed] [Google Scholar]
- 26.Di Rienzi SC, Britton RA. Adaptation of the gut microbiota to modern dietary sugars and sweeteners [Internet]. Adv. Nutr. 2020. [cited 2021 Feb 15];11(3):616–629. Available from https:///pmc/articles/PMC7231582/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roediger WEWW. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83(2):424-9. [PubMed] [Google Scholar]
- 28.Roediger WEW. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. Internet] 1980. [cited 2020 Mar 27]; 21(9):793–798. Available from http://gut.bmj.com/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe Internet] 2015. [cited 2017 May 23]; 17:662–671. Available from (5)http://linkinghub.elsevier.com/retrieve/pii/S1931312815001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fagundes RR, Taylor CT. Determinants of hypoxia inducible factor (HIF) activity in the intestinal mucosa. J Appl Physiol [Internet] 2017. [cited 2017 May 21].jap.00203.2017. Available from http://www.ncbi.nlm.nih.gov/pubmed/28408694 [DOI] [PubMed] [Google Scholar]
- 31.Blouin J-M-M, Penot G, Collinet M, Nacfer M, Forest C, Laurent-Puig P, Coumoul X, Barouki R, Benelli C, Bortoli S. Butyrate elicits a metabolic switch in human colon cancer cells by targeting the pyruvate dehydrogenase complex. Int J Cancer. Internet] (11): 2011. [cited 2021 Apr 13]; 128:2591–2601. Available fromhttp://doi.wiley.com/10.1002/ijc.25599 [DOI] [PubMed] [Google Scholar]
- 32.De Figueiredo LF, Schuster S, Kaleta C, Fell DA. Can sugars be produced from fatty acids? A test case for pathway analysis tools. Bioinformatics. Internet] 2008. [cited 2020 Mar 31]; 24:2615–2621. Available from(22) http://www.ncbi.nlm.nih.gov/pubmed/18806269 [DOI] [PubMed] [Google Scholar]
- 33.Kaleta C, de Figueiredo LF, Werner S, Guthke R, Ristow M, Schuster S. In silico evidence for gluconeogenesis from fatty acids in humans. PLoS Comput Biol. 2011;7(7):e1002116. doi: 10.1371/journal.pcbi.1002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Kim R, Gunasekara DB, Reed MI, DiSalvo M, Nguyen DL, Bultman SJ, Sims CE, Magness ST, Allbritton NL. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. CMGH. 2018;5:113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng H, Safratowich BD, Wang TTY, Hamlin SK, Johnson LK. Butyrate inhibits deoxycholic‐acid‐resistant colonic cell proliferation via cell cycle arrest and apoptosis: a potential pathway linking dietary fiber to cancer prevention. Mol Nutr Food Res. Internet] 2020. [cited 2020 Mar 30]. 1901014. Available from 64(8) http://www.ncbi.nlm.nih.gov/pubmed/32003143 [DOI] [PubMed] [Google Scholar]
- 36.Ocadiz-Ruiz R, Al P, Mm H, Ding L, Er F, JL M. ZBP-89 function in colonic stem cells and during butyrateinduced senescence. Oncotarget. 2017;8(55):94330–94344. doi: 10.18632/oncotarget.21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino B, Fernández-Díaz CM, Cózar-Castellano I, Perdomo G. Intestinal fructose and glucose metabolism in health and disease. Nutrients. 2020;12(1):94. doi: 10.3390/nu12010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang C, Hui S, Lu W, Cowan AJ, Morscher RJ, Lee G, Liu W, Tesz GJ, Birnbaum MJ, Rabinowitz JD. The small intestine converts dietary fructose into glucose and organic acids. Cell Metab. 2018;27(351–361.e3):351–361.e3. doi: 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moens F, De Vuyst L. Inulin-type fructan degradation capacity of Clostridium cluster IV and XIVa butyrate-producing colon bacteria and their associated metabolic outcomes. Benef Microbes. 2017;8(3):473–490. doi: 10.3920/BM2016.0142. [DOI] [PubMed] [Google Scholar]
- 40.Moens F, Weckx S, De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int J Food Microbiol. 2016;231:76–85. Internet] Available from. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Moens F, Verce M, De Vuyst L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int J Food Microbiol. F M, M V, L DV 2017; Internet] [cited 2021 Aug 6]; 241:225–236. Available from https://pubmed.ncbi.nlm.nih.gov/27810444/ [DOI] [PubMed] [Google Scholar]
- 42.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer C-D, Young P, Rucklidge G, Ramsay AG, et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Sciences (New York) [Internet] 2011. [cited 2021 Mar 17]; 108:4672–4679. Available from]. www.pnas.org/cgi/doi/10.1073/pnas.1000091107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut [Internet]. 2016. [cited 2018 Mar 30]. Available from. http://www.ncbi.nlm.nih.gov/pubmed/27446020 [DOI] [PMC free article] [PubMed]
- 44.Patel C, Douard V, Yu S, Gao N, Ferraris RP. Transport, metabolism, and endosomal traffickingdependent regulation of intestinal fructose absorption. FASEB J. Internet] 2015. [cited 2021 Mar 22]; 29(9):4046–4058. Available from https://pubmed.ncbi.nlm.nih.gov/26071406/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barone S, Fussell SL, Singh AK, Lucas F, Xu J, Kim C, Wu X, Yu Y, Amial H, Seidler U, et al. Slc2a5 (Glut5) is essential for the absorption of fructose in the intestine and generation of fructose-induced hypertension. J Biol Chem Internet] 2009. [cited 2021 Mar 22]; 284(8):5056–5066. Available from https://pubmed.ncbi.nlm.nih.gov/19091748/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gouyon F, Caillaud L, Carrière V, Klein C, Dalet V, Citadelle D, Kellett GL, Thorens B, Leturque A, Brot-Laroche E. Simple-sugar meals target GLUT2 at enterocyte apical membranes to improve sugar absorption: a study in GLUT2-null mice. J Physiol. Internet] 2003. [cited 2021 Mar 22]; 552(3):823–832. Available from https://pubmed.ncbi.nlm.nih.gov/12937289/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease [Internet]. Clin. J. Gastroenterol. 2018. [cited 2020 Apr 10];11(1):1–10. Available from http://www.ncbi.nlm.nih.gov/pubmed/29285689 [DOI] [PubMed] [Google Scholar]
- 48.Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. 2019;9(1):1–10. doi: 10.1038/s41598-019-49452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Siles M, Martinez-Medina M, Surís-Valls R, Aldeguer X, Sabat-Mir M, Duncan SH, Flint HJ, Garcia-Gil LJ. Changes in the abundance of faecalibacterium prausnitzii phylogroups i and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis. 2016;22(1):28–41. doi: 10.1097/MIB.0000000000000590. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Siles M, Martinez-Medina M, Abellà C, Busquets D, Sabat-Mir M, Duncan SH, Aldeguer X, Flint HJ, Garcia-Gil LJ, Elkins CA. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015;81(21):7582–7592. doi: 10.1128/AEM.02006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63(8):1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 53.Tigchelaar EF, Zhernakova A, Dekens JAM, Hermes G, Baranska A, Mujagic Z, Swertz MA, Muñoz AM, Deelen P, Cénit MC, et al. Cohort profile: lifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open Internet] 2015;(8): [cited 2020 Apr 10]; 5:e006772. Available fromhttp://www.ncbi.nlm.nih.gov/pubmed/26319774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rocklin RD, Clarke AP, Improved Long-Term WM. Reproducibility for pulsed amperometric detection of carbohydrates via a new quadruple-potential waveform. Anal Chem. 1998;70(8):1496–1501. doi: 10.1021/ac970906w. [DOI] [Google Scholar]
- 55.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science (80-) [Internet] 2016. [cited 2021 Apr 6]; 352(6285) :565–569. Available fromhttps://pubmed.ncbi.nlm.nih.gov/27126040/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Trimmomatic: UB. A flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12(10):902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 58.Zierer J, Ma J, Kastenmüller G, Mangino M, Long T, Telenti A, Rp M, Ks S, JT B, CJ S, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50(6):790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw metagenomics sequencing data used for this study are available from the European Genome-phenome Archive data repository: 1000IBD cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001004194) and LifeLines DEEP cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001001991). Due to participant confidentiality, the datasets are available upon reasonable request to the University Medical Center of Groningen (UMCG) and LifeLines, respectively. The metabolomics data supporting the current study have not yet been deposited in a public repository because of participant confidentiality, but are available from the corresponding author on request.


