Abstract
Cse4p is a structural component of the core centromere of Saccharomyces cerevisiae and is a member of the conserved CENP-A family of specialized histone H3 variants. The histone H4 allele hhf1-20 confers defects in core centromere chromatin structure and mitotic chromosome transmission. We have proposed that Cse4p and histone H4 interact through their respective histone fold domains to assemble a nucleosome-like structure at centromeric DNA. To test this model, we targeted random mutations to the Cse4p histone fold domain and isolated three temperature-sensitive cse4 alleles in an unbiased genetic screen. Two of the cse4 alleles contain mutations at the Cse4p-H4 interface. One of these requires two widely separated mutations demonstrating long-range cooperative interactions in the structure. The third cse4 allele is mutated at its helix 2-helix 3 interface, a region required for homotypic H3 fold dimerization. Overexpression of wild-type Cse4p and histone H4 confer reciprocal allele-specific suppression of cse4 and hhf1 mutations, providing strong evidence for Cse4p-H4 protein interaction. Overexpression of histone H3 is dosage lethal in cse4 mutants, suggesting that histone H3 competes with Cse4p for histone H4 binding. However, the relative resistance of the Cse4p-H4 pathway to H3 interference argues that centromere chromatin assembly must be highly regulated.
The centromere is a specialized chromatin structure that has evolved to ensure the accurate segregation of replicated chromosomes during mitosis and meiosis. It is composed of centromeric DNA (CEN DNA) plus a complex assembly of chromosomal and kinetochore proteins (12, 13, 22). The centromere performs at least three essential functions during the cell division cycle. First, it provides the site of attachment for spindle microtubules, thus enabling the accurate segregation of replicated sister chromatids. Second, it serves to prevent premature sister chromatid separation during segregation (6). Third, the kinetochore complex monitors the integrity of microtubule attachment and activates the spindle checkpoint pathway in the presence of spindle damage (44). Thus, identifying the proteins of the centromere and defining their functions are critical for understanding the essential process of chromosome segregation.
The budding yeast Saccharomyces cerevisiae provides a simple model system for both genetic and biochemical studies of centromere structure and function. The CEN DNA of S. cerevisiae is entirely contained within a sequence of about 125 bp of DNA present on all 16 chromosomes. Each CEN DNA is made up of three conserved sequence elements, CDEI, CDEII, and CDEIII, which are bound by sequence-specific DNA binding proteins and additional kinetochore proteins (reviewed in references 21, 22, and 33). This complex centromere assembly produces a distinctive chromatin structure in vivo. At each centromere, the CEN DNA region exists as a nuclease-resistant chromatin domain spanning a total of 180 to 220 bp. This core centromere is delimited on each side by strong nuclease-hypersensitive sites and is flanked by arrays of positioned nucleosomes (7, 17). These mapping experiments led to the hypothesis that the core centromere contained a nucleosome-like structure (reviewed in reference 41).
Recent evidence suggests that the nuclease resistant core centromere contains a specialized variant nucleosome particle. Key to this hypothesis has been the identification of a conserved family of centromere-specific histone H3 protein variants found in a wide range of organisms including mammals (35, 48), yeast (45, 47), worms (11), and flies (18). The founding member of this family is the mammalian centromere-specific protein CENP-A (35, 48). CENP-A is a 16-kDa protein with roughly 65% similarity to histone H3 over its histone fold domain. The results of site-directed mutational studies of CENP-A are consistent with the properties expected for a histone fold and have shown that this domain is necessary for localization of the protein to the kinetochore (35, 42). CENP-A has recently been localized to the inner plate of the kinetochore, and there is a strong correlation between the presence of a functional mammalian centromere and CENP-A protein, although a direct test of the role of CENP-A in centromere function has not yet been completed (50, 52).
The homologue of CENP-A in S. cerevisiae is encoded by the gene CSE4. It was first identified in a screen for mutants defective for chromosome segregation (47) and subsequently through two independent genetic screens based on the fidelity of mitotic chromosome transmission (4, 45). CSE4 encodes a 27-kDa protein composed of a N-terminal domain with a unique sequence and a C-terminal domain consisting of a histone fold motif with >65% similarity to histone H3. Both the N-terminal and histone fold domains of Cse4p are essential for function (23, 30). The histone folds of both Cse4p and CENP-A share distinct sequence features, including a short peptide insertion and the unusual presence of a tryptophan residue within the first loop of the fold. Cse4p also has properties expected of an H3-like histone, since it elutes biochemically with the major core histones during column chromatography (47) and it interacts genetically with histone H4 (45).
Several observations indicate that Cse4p is a specific structural component of the S. cerevisiae centromere (30). First, core centromere chromatin is disrupted in cse4 mutants when cells are shifted to restrictive temperatures. Second, Cse4p is physically localized to the CEN DNA as determined by in vivo cross-linking and chromatin immunoprecipitation. It fails to cross-link to the DNA located in the nucleosomes surrounding the core domain or to several other noncentromeric DNA locations examined. Finally, as visualized by immunofluorescence microscopy, Cse4p is localized in the nucleus in discrete foci whose number and movements during the cell division cycle are consistent with those expected for CEN DNA.
Histone H4 is also likely to be a component of the S. cerevisiae core centromere. In a previous genetic screen for histone H4 mutants specifically defective in mitotic chromosome transmission, we identified a novel temperature-sensitive H4 mutant, hhf1-20, which at semipermissive temperatures has a 60-fold increase in the frequency of chromosome loss. At restrictive temperatures hhf1-20 cells arrest at medial nuclear division. Increased CSE4 gene dosage suppresses the essential defect caused by the hhf1-20 allele (45). Furthermore, cells expressing hhf1-20 also show a disruption of core centromere chromatin at restrictive temperatures, and this defect is suppressed by overexpression of wild-type Cse4p in the hhf1-20 mutants (30).
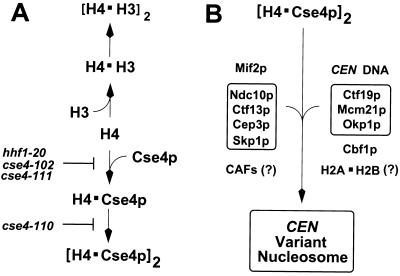
These genetic and biochemical results led us to propose a model in which the core centromere of S. cerevisiae is assembled around a specialized variant nucleosome containing at least Cse4p and histone H4 (30). This model makes strong predictions regarding genetic interactions between Cse4p, histone H4, and histone H3. To investigate these predictions we have constructed a set of cse4 point mutants by both site-directed and random mutagenesis of the DNA sequence encoding the Cse4p histone fold. Here we report the characterization of these new cse4 alleles, their interactions with histone H4, and their effects on centromere function. The results of these experiments show that Cse4p-histone H4 interactions are essential for centromere function in S. cerevisiae and suggest a highly regulated pathway for the assembly of the core centromere chromatin.
MATERIALS AND METHODS
Strains and plasmids.
The relevant plasmid constructs used in this study are listed in Table 1, and the genotypes of yeast strains are listed in Table 2. All DNA and bacterial manipulations were by standard protocols (39). Yeast growth media, as well as transformation, sporulations, and DNA isolation protocols were carried out as described previously (1).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype | Source or reference |
|---|---|---|
| pPY3-4 | 2μm URA3 CSE4 | This study |
| pPY11 | 2μm URA3 HHT1 | This study |
| pPY12 | CEN6 ARS4 TRP1 GAL1:CSE4 | This study |
| pPY13 | CEN6 ARS4 TRP1 CSE4 | This study |
| pPY18 | CEN6 ARS4 TRP1 cse4-101 | This study |
| pPY19 | CEN6 ARS4 TRP1 cse4-102 | This study |
| pPY20 | CEN6 ARS4 TRP1 cse4-103 | This study |
| pPY21 | CEN6 ARS4 TRP1 cse4-104 | This study |
| pLG2 | CEN6 ARS4 TRP1 cse4-106 | This study |
| pLG3 | CEN6 ARS4 TRP1 cse4-107 | This study |
| pLG4 | CEN6 ARS4 TRP1 cse4-108 | This study |
| pLG8 | 2μm LEU2 CSE4 | This study |
| pLG11 | CEN6 ARS4 TRP1 cse4-109 | This study |
| pLG12 | CEN6 ARS4 TRP1 cse4-110 | This study |
| pLG26 | CEN6 ARS4 TRP1 cse4-111 | This study |
| pLG31 | CEN6 ARS4 TRP1 cse4-112 | This study |
| pLG39 | 2μm URA3 GAL1:HHF1 | This study |
| pLG41 | 2μm URA3 GAL1:HHT1 | This study |
| pLG48 | 2μm LEU2 cse4-110 | This study |
| pLG49 | 2μm LEU2 cse4-111 | This study |
| pLG50 | 2μm LEU2 cse4-102 | This study |
| pKF71 | HIS3 GAL1:CEN6 | 15 |
| pPM178 | CEN6 ARS4 TRP1 CSE4-HA | 30 |
| pTK56 | CEN6 ARS4 TRP1 cse4-102-HA | This study |
| pTK57 | CEN6 ARS4 TRP1 cse4-110-HA | This study |
| pTK58 | CEN6 ARS4 TRP1 cse4-111-HA | This study |
| pMS337 | CEN4 ARS1 LEU2 HHT1 HHF1 | 31 |
| pMS352 | CEN4 ARS1 LEU2 HHT1 hhf1-20 | 45 |
| pRS425 | 2μm LEU2 | 43 |
| pRS426 | 2μm URA3 | 43 |
TABLE 2.
Yeast strains used in this study
| Strain | Relevant genotype |
|---|---|
| MSY535 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1 hhf1-10 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| MSY554 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1 hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| MSY559 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1HHF1 Δ(HHT2-HHF2) |
| MSY713 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1 hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) rad9::URA3 |
| MSY715 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1 hhf1-20 Δ(HHT1-HHF1) Δ(HHT2-HHF2) bub2::URA3 |
| MSY717 | MATa ura3-52 leu2-3,112 trp1Δ1 his3Δ200 cse4::LEU2 pPY9 |
| MSY718 | MATα ura3-52 leu2-3,112 trp1Δ1 his3Δ200 cse4::LEU2 pPY9 |
| MSY781 | MATa ura3-52 leu2-3,112 lys2Δ200 HHT1 hhf1-36::LEU2 Δ(HHT1-HHF1) Δ(HHT2-HHF2) |
| MSY819a | MATa ura3-52 leu2-3,112 trp1Δ1 his3Δ200 cse4::LEU2 mad1-1 pPY12 |
| MSY824 | MATa ura3-52 leu2-3,112 trp1Δ1 his3Δ200 cse4::LEU2 rad9::URA3 pPY12 |
| s30b | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 ctf13-30 |
| MSY1167 | MATa ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 ctf13-30 HIS3::pKF71 |
| MSY1358 | MATa ade2-101 can1-200 his3-11,15 leu2-3,112 trp1-1 ura3-1 cse4::LEU2 pPY9 |
| MSY1398 | MATa ade2-101 can1-200 his3-11,15 leu2-3,112 trp1-1 ura3-1 cse4::LEU2 HIS3::pKF71 pPY9 |
| MSY1542 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(HHT1-HHF1) Δ(HHT2-HHF2) HIS3::pKF71 pMS352 |
| MSY1543 | MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(HHT1-HHF1) Δ(HHT2-HHF2) HIS3::pKF71 pMS337 |
Provided by Daniel Burke.
Provided by Forrest Spencer (15).
Site directed mutagenesis.
All site-directed mutants used in this study were created using the pAlter system (Promega). The oligonucleotide sequence used to delete the KDQ insertion in Cse4p was 5′ TTGACTGCCAACGTAAATCAGTTGTAAACTCGTCTGTAA 3′. The underlined letters mark the boundary of the deleted nucleotides. The oligonucleotide used to change the tryptophan to phenylalanine was 5′ CGCCATTGACTGAAAACGTAAATC 3′. Oligonucleotides 180 (5′ CAATAATCCTACCTGATACGCTTCG 3′) and 179 (5′ CTTGCAAATGGAACTTTGGAGATTA 3′) were used to create cse4-111 and cse4-112. Boldface type indicates change from the wild-type sequence.
Mutagenesis and library construction.
The substrate for mutagenesis of CSE4 was a 1.8-kb EcoRI-PstI fragment of pPY12 that contains a GAL1-CSE4 fusion gene. Oligonucleotides that annealed to the GAL1 promoter region and approximately 200 bp downstream of the 3′ end of CSE4 were used as primers for the PCR. Mutagenesis was performed essentially as described (32) to generate a library of PCR fragments. This library was used to repair gaps in pPY13 and introduce changes in the histone fold region of Cse4p. Analysis of the phenotype of the mutants was performed by plasmid shuffle (8). Five cse4(Ts) alleles were recovered into DH5α bacteria as described (38). Plasmid DNA was purified from the bacteria and used to transform MSY718. All five plasmids conferred temperature-sensitive growth. Sequence analysis revealed that two of the plasmids were identical and thus define four independent mutants.
Western blots.
Wild-type, cse4-102, cse4-110, and cse4-111 constructs containing an in-frame hemagglutinin (HA) tag in the N terminus of the protein were constructed using a KpnI-BstBI fragment from pPM178 (30). These were introduced as the sole source of Cse4p by plasmid shuffle into MSY1358 and serve to complement the null mutant as assayed by growth. Cultures were grown at a permissive temperature, and then half the cultures were shifted to 37°C for 4 h. Cells were harvested and lysates were prepared by glass bead disruption and 0.3 M ammonium sulfate extraction buffer. A non-HA-tagged wild-type Cse4p strain served as a negative control. Proteins were quantitated using the Bio-Rad microassay, and a total of 100 μg of protein from each lysate was run on a sodium dodecyl sulfate–12.5% polyacrylamide gel. The proteins were electroblotted onto nitrocellulose and blocked with 10% dry milk in Tris-buffered saline overnight. Enhanced chemiluminescence was performed essentially as described by the manufacturer (Amersham Life Science) using the HA.11 antibody (1:1,000; BAbCO) as the primary antibody against the HA tag and goat anti-mouse coupled to horseradish peroxidase (Amersham) as the secondary antibody.
Flow cytometry and immunofluorescence.
Flow cytometry and cell cycle analysis were carried out as described previously (28). Overnight cultures were diluted to 5 × 106 cells/ml and incubated at either 28 and 33°C or 28 and 37°C. These cultures were maintained for 3 or 5 h at those temperatures. One-milliliter samples for flow cytometry were taken every hour and analyzed on a FACScan fluorescence-activated cell sorter (Becton Dickinson). At the termination of the time course, 5 to 10 ml of each culture was prepared for immunofluorescence as described (37). Indirect immunofluorescence was performed using YOL34 (rat α-tubulin) as the primary antibody and goat α-rat conjugated to Texas Red as the secondary antibody. Nuclear DNA was visualized by the addition of DAPI (4′,6′-diamidino-2-phenylindole) (0.4 μg/ml; Boehringer Manheim).
Checkpoint analysis.
Strains containing either HHF1, hhf1-20, CSE4, GAL1::CSE4, or the cse4 point mutants alone or in combination with a rad9Δ, bub2Δ, mad1-1, or mad1Δ mutation were grown to mid-log phase (5 × 106 to 1 × 107) at 28°C. The cultures were shifted to 37°C, and aliquots were plated on yeast-peptone-dextrose plates and grown at 28°C. Plating efficiency was determined from the number of colonies formed at 28°C after 3 days of growth divided by the total number of cells plated. Cell cycle arrest was monitored by microscopy and flow cytometry.
Chromatin Immunoprecipitations.
In vivo cross-linking, chromatin immunoprecipitation, and DNA fragment mapping were performed as described (29, 30). Cultures were grown to a concentration of 2 × 107 cells/ml at 28°C and then shifted to 37°C for 4 h before cross-linking. Strains MSY1517 through MSY1520 were grown in yeast-peptone-dextrose and the histone overexpression strains MSY1475 through MSY1482 were grown in SC-URA medium with galactose as the carbon source (1). Cross-linked chromatin was precipitated with purified anti-HA immunoglobulin G monoclonal antibody (12CA5) at a final concentration of 4 μg/ml per reaction mixture.
Transcriptional read-through assays.
pKF71 was obtained from the Spencer laboratory and was described previously (15). The plasmid was digested with BamHI and used to transform MSY1358 or MX1-4C. Strains were maintained in galactose-containing medium to ensure the test centromere remained inactive. The level of transcription off the GAL1 promoter in wild-type, cse4-110, and cse4-111 was determined using pJK101 (10). o-Nitrophenyl-β-d-galactopyranoside assays were performed essentially as described (1). The protein concentration of the lysates was determined by the Bio-Rad protein microassay using bovine serum albumin as the standard. The specific activity was calculated as follows: optical density at 420 nm × 0.850/ (0.0045 × protein concentration × extract volume × time).
Molecular modeling.
The structures of the S. cerevisiae Cse4p and histone H4 proteins were modeled on the X-ray crystal structures of the nucleosome using Sybyl (Tripos). Identical interpretations were obtained using either the histone octamer data set (Protein Data Bank entry 1HIO [2]) or the nucleosome data set (Protein Data Bank entry 1AOI [26]). The amino acid differences present in the yeast proteins were introduced into the crystal structures and optimized by local energy minimization around the set of yeast-specific residues.
RESULTS
Site-directed cse4 mutations.
The first loop in the histone fold domain of Cse4p has two obvious features that distinguish it from histone H3: (i) a three-amino-acid insertion and (ii) the substitution of tryptophan (W178) for the corresponding phenylalanine found in H3. Both of these alterations are characteristic of the CENP-A family of H3 variants, occurring in Cse4p in S. cerevisiae, several variants in Caenorhabditis elegans, and mammalian CENP-A. The position of the short amino acid loop insertion is conserved in all family members, but its exact length and sequence vary. In Cse4p the insertion comprises the three amino acids lysine, aspartate, and glutamine (KDQ) at positions 171 to 173. Tryptophan is an unusual amino acid in histone proteins and is not present in the major core histones. Thus, given its location in loop 1, it could potentially play a role in the centromere-specific functions of the family. The roles of these features in directing the localization of CENP-A to human centromeres have been tested, and neither deleting the extra loop residues nor replacing the CENP-A tryptophan with phenylalanine affected CENP-A localization to the centromere (42). We were able to test the full biological function of these same features in Cse4p by site-directed mutagenesis and gene replacement in S. cerevisiae.
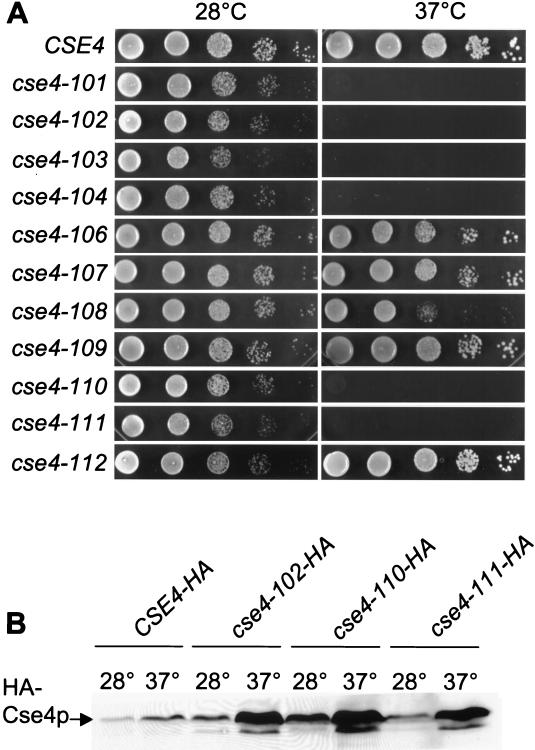
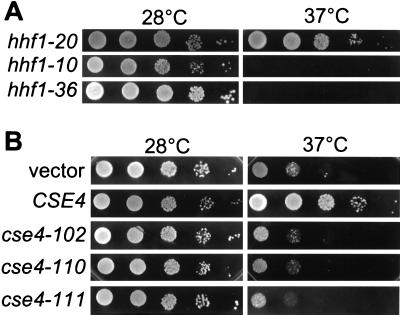
To determine whether the KDQ insertion was required for protein function, the codons for the insertion were deleted from the gene to create the allele cse4-106. To determine if the tryptophan was required for function, it was changed to its H3 equivalent phenylalanine (W178F) to create the allele cse4-109. These alleles were subcloned into pRS314 and introduced into MSY1358 by plasmid shuffle as the only source of Cse4p protein. Cells expressing cse4-106 or cse4-109 were wild type in a variety of assays, including growth, temperature sensitivity (Fig. 1A), hhf1-20 suppression, and centromere transcriptional read-through (see below and Fig. 4A). These results are entirely consistent with those reported for CENP-A and show that neither the KDQ insertion nor the conservative W178 substitution is essential for Cse4p function in mitotic growth.
FIG. 1.
Conditional growth and protein levels of Cse4p point mutants. (A) The growth phenotypes of the wild-type plasmid shuffle strain and the histone fold Cse4p mutants at the permissive (28°C) and restrictive (37°C) temperature are shown. Tenfold serial dilutions of each strain were spotted onto synthetic complete medium minus tryptophan and grown for 2 days at the indicated temperatures. The location of the point changes in the mutants can be found in Table 1. (B) Mutant Cse4 proteins are stable at the restrictive temperature. Crude lysates were made from the indicated strains grown at 28°C and then shifted to 37°C for 4 h. Equal amounts of protein were run on a sodium dodecyl sulfate–12.5% polyacrylamide gel and electroblotted to nitrocellulose. Western blot analyses were performed using and anti-HA antibody (HA.11).
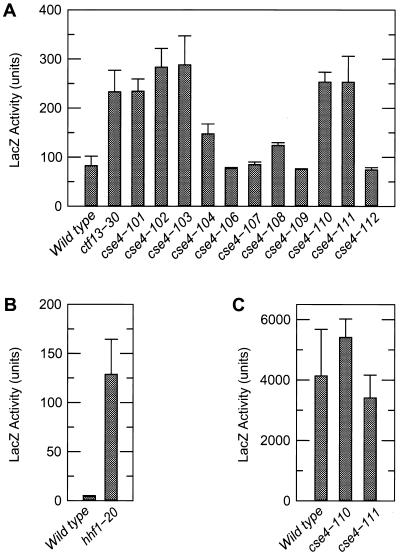
FIG. 4.
Transcriptional read-through of CEN DNA in cse4 and hhf1-20 mutants. For each panel the indicated strains were grown in synthetic complete galactose medium lacking histidine, and cell extracts were prepared after 18 h of growth at the semipermissive temperature. The units for levels of β-galactosidase activity (LacZ Activity) are nanomoles of ONPG cleaved per milligram of lysate protein per minute. (A) LacZ expression was determined in wild-type, ctf13-30, and isogenic cse4 mutant strains grown at 33°C. In these strains GAL1::LacZ expression requires transcription through a CEN DNA insert located in an intron of the reporter gene. The CEN DNA insert partially blocks transcription in wild-type cells, giving low levels of expression. In ctf13-30 mutants this block is compromised, giving increased expression (14). The values represent the average of two to three independent cultures. (B) Transcription through CEN DNA was determined for isogenic HHF1 and hhf1-20 strains as described above. Cells were grown at a semipermissive temperature of 34°C, and the values represent the average of three to four independent cultures. (C) LacZ expression was determined for wild-type and two cse4 mutant strains as described above. However, in these strains the GAL1::LacZ reporter gene is carried on a 2μ plasmid and lacks the CEN DNA insert. In all panels, error bars show standard deviations.
Isolation of cse4 histone fold mutations.
To identify essential functional residues in Cse4p, we carried out an unbiased genetic screen for temperature-sensitive cse4 mutants. A library of cse4 mutants was constructed by targeting random mutations to the DNA sequence encoding the histone fold domain using PCR misincorporation. The fold domain mutations were then recovered by gap repair into the full-length CSE4 sequence to generate a mutant library in yeast (see Materials and Methods). The final constructs also carried a triple-HA epitope tag in the unique N-terminal domain of Cse4p to facilitate assays of protein expression.
Four independent temperature-sensitive mutants were recovered from a screen of 3,200 library colonies and designated cse4-101, -102, -103, and -104 (Fig. 1A). The library plasmid was recovered from each isolate and sequenced to determine its amino acid sequence (Table 3). Each of these initial alleles carried mutations producing two amino acid substitutions. Since both cse4-101 and cse4-104 shared the amino acid substitution L196S, we constructed the single L196S substitution allele cse4-110. As shown in Fig. 1A, the L196S substitution is sufficient to confer the temperature-sensitive phenotype. Next, the two substitutions in the cse4-102 allele were separated into the single substitution alleles cse4-107 and cse4-108. As shown in Fig. 1A, neither of these single point mutations produced a tight temperature-sensitive lethal phenotype. A slight growth defect is evident in cse4-108, but this allele has little effect on centromere integrity (see Fig. 4A below). Thus, the essential defect in cse4-102 requires both substitutions. Finally, the two substitutions in cse4-103, I156V and L193Q, were separated into the single point mutant alleles cse4-111 and cse4-112. As shown in Fig. 1A, the single L193Q substitution in cse4-111 is sufficient to confer the temperature-sensitive phenotype, whereas the I156V substitution in cse4-112 gives wild-type growth.
TABLE 3.
Cse4p amino acid substitutions in cse4 alleles
| Allele | Substitution(s) |
|---|---|
| cse4-101 | V194A, L196S |
| cse4-102 | L175S, M217T |
| cse4-103 | I156V, L193Q |
| cse4-104 | L196S, I229V |
| cse4-106 | Δ172-174 |
| cse4-107 | L175F |
| cse4-108 | M217T |
| cse4-109 | W178F |
| cse4-110 | L196S |
| cse4-111 | L193Q |
| cse4-112 | I156V |
Mutant Cse4p proteins are stable.
Since Cse4p is an essential gene product, rapid degradation of the mutant proteins at restrictive temperatures would provide a trivial explanation for the temperature sensitivity of the mutants. Therefore, we determined the relative levels of Cse4p present in the wild-type and mutant strains at permissive and restrictive temperatures. Lysates were prepared from cultures grown at 28°C or shifted to 37°C for 4 h, and the expression of Cse4p was assayed by Western blotting using the anti-HA antibody, HA.11. As shown in Fig. 1B, Cse4p from the cse4 mutants was not degraded at restrictive temperatures. On the contrary, the mutants contain more protein than wild-type cells at both permissive and restrictive temperatures. At present we do not know the reasons for this increased accumulation, but in any case the temperature-sensitive phenotypes of cse4-102, -110, and -111 cannot be due to the simple absence of protein.
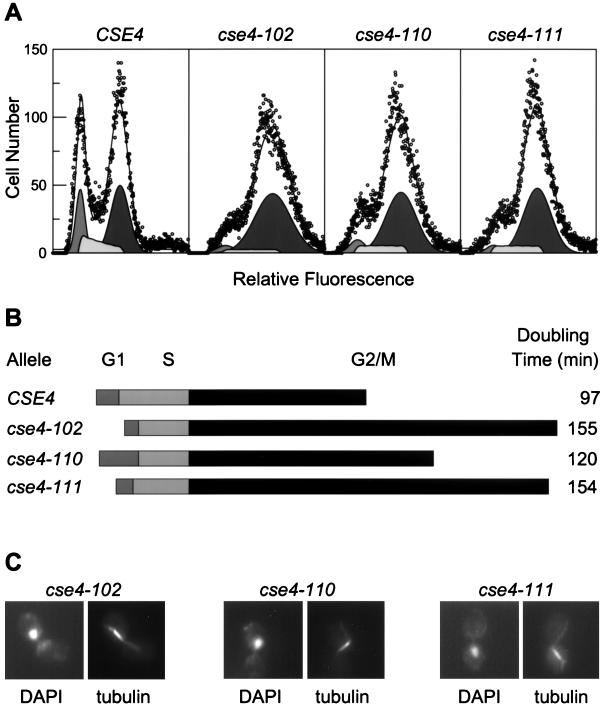
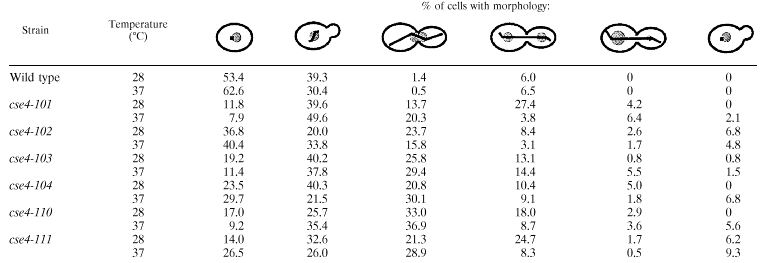
The cse4 histone fold mutants are impaired for nuclear division.
Several lines of evidence indicate that these new cse4 histone fold mutants are defective for centromere function. First, they have a cell division cycle defect at mitosis. As shown in the DNA histograms of Fig. 2A, cse4 mutants show a marked accumulation of cells with a 2C DNA content even at permissive temperatures, reflecting a delay in cell cycle progressions through G2/M (Fig. 2A and B). This arrest is exacerbated at the restrictive temperature (Table 4). The most abundant class of aberrant cells found in the mutant strains is a large budded cell with a single nucleus and a bipolar spindle (Fig. 2C). This type of cell is rarely found in the wild-type strain but can range up to 36% in the mutants (Table 4). Often the mitotic spindle in mutant cells is not properly aligned with respect to the bud. These results are similar to those reported previously for cse4-1 and hhf1-20 (45, 47).
FIG. 2.
G2/M arrest in cse4 mutants. (A) Flow cytometry of cse4 strains in exponential cultures grown at 28°C. Wild-type and cse4 mutant strains were stained for DNA content with propidium idodide and analyzed by flow cytometry. Representative histograms of cell number versus relative fluorescence are shown. In each panel the circles show the channel data points and the solid line shows the overall fit of the data to a model of G1, S, and G2/M phase cell populations. Below each plot the individual G1, S, and G2/M model fits are shown at half scale. (B) Cell division cycle profiles. The relative lengths of the cell cycle phases were approximated using culture doubling times and the percentage of cells in each phase as estimated from the flow cytometry histograms. The estimates are the mean of six independent measurements for each mutant. The profiles are aligned at the S/G2/M boundary for comparison of the relative lengths of the G2/M cell cycle phase. (C) Morphology of cse4 cells accumulated at restrictive temperature. The nuclear and spindle morphology are shown for the class of cse4-102, cse4-110, and cse4-111 mutant cells that accumulate following arrest at the restrictive temperature (Table 2). Each pair of micrographs illustrates the nuclear staining (DAPI) and the spindle staining (tubulin) visualized with an antitubulin antibody.
TABLE 4.
Morphology of cse4 mutants
 |
cse4 and hhf1-20 mutants engage the spindle checkpoint pathway.
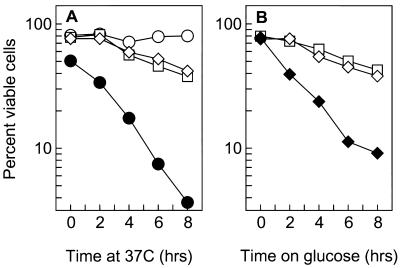
The second line of evidence suggesting that the cse4 and hhf1-20 mutants are impaired for centromere function is that they specifically activate the spindle-kinetochore checkpoint pathway. The integrity of spindle attachment to the kinetochore is monitored by a checkpoint pathway which includes the product of the BUB and MAD genes (20, 24, 36, 51). In the presence of spindle or kinetochore damage, the checkpoint pathway serves to negatively regulate cell cycle progression at G2/M until the damage is repaired. The consequence of disrupting the checkpoint pathway in the presence of an incomplete kinetochore or spindle is that cells fail to arrest division and lose viability as a result of the damage. As shown in Fig. 3 both hhf1-20 and a conditional GAL1::CSE4 null allele are hypersensitive to disruption of the kinetochore-spindle checkpoint pathway, but not to disruption of the DNA damage checkpoint pathway. Flow cytometry and analysis of cell morphology confirmed that the decreased viability in the double mutants was accompanied by a loss of cell cycle checkpoint arrest (data not shown). The cse4 point mutants were difficult to examine in temperature shift experiments because of their constitutive G2/M defect even at permissive temperatures. However, cse4 mad1Δ double mutants showed reduced viabilities consistent with an activation of the spindle checkpoint in the cse4 single mutant cells.
FIG. 3.
hhf1-20 and cse4 mutants activate the spindle-kinetochore checkpoint pathway. (A) Checkpoint control in hhf1-20 mutants. Cultures were grown to mid-log phase at 28°C and then shifted to 37°C, the restrictive temperature for hhf1-20 cells. The kinetics of cell viability are shown following the shift to the restrictive temperature. Open circle, MSY559 (HHF1); open square, MSY554 (hhf1-20); open diamond, MSY556 (hhf1-20 rad9::URA3); filled circle, MSY713 (hhf1-20 bub2::URA3). (B) Checkpoint control in GAL1::CSE4 conditional null mutants. Cultures were grown to mid-log phase in galactose medium and then shifted to raffinose medium to turn off CSE4 expression. The kinetics of cell viability are shown following the shift to raffinose medium. Open square, MSY753 (GAL1::CSE4); open diamond, MSY824 (GAL1::CSE4 rad9::URA3); filled diamond, MSY819 (GAL1::CSE4 mad1).
cse4 and hhf1-20 mutants disrupt core centromere structure in vivo.
The third line of evidence for a kinetochore defect in the cse4 histone fold mutants comes from an in vivo assay of kinetochore function (15). The rationale for this assay is that placement of a CEN DNA sequence within the intron of an artificial GAL1::lacZ reporter reduces the expression of β-galactosidase by assembling a kinetochore complex and blocking the transcribing polymerase. Mutations in CEN DNA, or in known kinetochore proteins, disrupt this complex giving increased transcriptional read-through and expression of lacZ (15). Therefore, to assay kinetochore function in vivo, we integrated this reporter construct into the set of cse4 and hhf1-20 mutants and measured β-galactosidase activities in cells grown at semipermissive temperatures. The results of these assays are shown in Fig. 4. The known kinetochore mutant ctf13-30 serves as a positive control in these assays (15) and routinely expressed levels of β-galactosidase activity two- to threefold higher than wild-type control cultures. All of the cse4(Ts) mutants produced increased expression of lacZ, consistent with a disruption of the kinetochore complex. Furthermore, cse4 mutants that were not temperature-sensitive did not give increased transcriptional read-through (Fig. 4A). We also found that the histone H4 mutant hhf1-20 gave increased lacZ expression in cells grown at semipermissive temperatures (Fig. 4B).
Since cse4 and hhf1-20 mutations affect proteins of the histone family, the increased expression of lacZ in the read-through assay could have been the result of alterations in GAL1 promoter activity and not the centromere complex. Therefore, we assayed the expression of a GAL1::lacZ construct lacking CEN DNA. As shown in Fig. 4C, there was no change in the level of expression of β-galactosidase among mutant and wild-type strains. Therefore, we conclude that cse4 and hhf1-20 affect the integrity of the CEN DNA kinetochore complex directly and not the basal transcription of the reporter. Based on their nuclear and spindle morphologies, their activation of the spindle-kinetochore checkpoint arrest, and their disruption of a CEN DNA block to transcription, we conclude that the lethal defect in these cse4(Ts) mutants is in centromere structure and function.
The locations of cse4 mutations are at sites of histone-histone interaction.
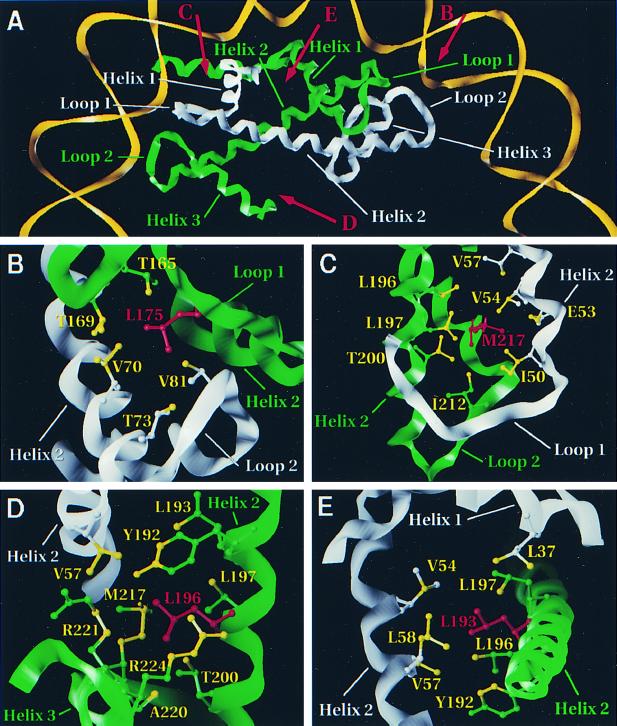
To begin to understand how the cse4 mutations disrupt centromere function, we mapped the locations of the amino acid substitutions in the mutants onto a molecular model of the Cse4p-H4 dimer based on the crystal structure of the nucleosome (Fig. 5). The histone fold motif (2) consists of two short alpha-helical segments (helix 1 and helix 3) connected to a long central alpha-helix (helix 2) by two loop and beta-strand segments (loop 1 and loop 2). Two histone fold domains pair head-to-tail across their long central helical regions, forming paired-element motifs where loop 1 from one chain runs antiparallel to loop 2 from the other chain (Fig. 5A).
FIG. 5.
Model of the locations of cse4 amino acid substitutions. The protein structures of the Cse4p-H4 dimer were modeled based on the X-ray crystal structures of histones H3 and H4 in the nucleosome (Materials and Methods). In each panel structures and labels referring to Cse4p are shown in green and those referring to histone H4 are shown in white. The wild-type residue that is mutated in each cse4 allele is shown in red. Atoms that are within 5 Å of the side chain of the mutated residue are colored yellow. (A) A partial nucleosome structure depicting a Cse4p-H4 dimer and one turn of DNA is shown. The view is down the superhelical axis of the DNA, perpendicular to the pseudodyad axis of the nucleosome which is within the plane of the illustration extending from left to right. Approximate vectors for the points of view shown in panels B, C, D, and E are indicated by the red arrows. (B) The region surrounding L175, one of the two amino acids mutated in cse4-102, is shown. The view is looking down between loop 1 of Cse4p and loop 2 of histone H4. (C) The region around M217, the second amino acid mutated in cse4-102, is shown. The view is looking down just behind loop 1 of histone H4 onto helix 3 of Cse4p. Foreground residues in helix 1 of histone H4 have been clipped to expose the interior region. (D) The region around L196, the residue mutated in cse4-110, is shown. The view is looking up towards the dyad axis along helix 3 of Cse4p. (E) The region around L193, the residue mutated in cse4-111, is shown. The view looks down through the dimer where helix 2 of Cse4p and helix 2 of histone H4 cross.
The three amino acid substitutions in cse4-102 and cse4-111 all involve residues in Cse4p that are predicted to be in close association with the histone fold domain of H4. Residue L175 in loop 1 of Cse4p resides in a hydrophobic environment created by residues V70, T73, and V81 from histone H4 (Fig. 5B). Residue M217 in helix 3 of Cse4p is in close proximity to amino acids I50, E53, V54, and V57 from helix 2 of histone H4 (Fig. 5C). Thus, both the L175F and M217T mutations in cse4-102 are predicted to affect its interaction with histone H4 and perhaps the binding of the Cse4p-H4 dimer to DNA (3, 26). Residue L193 of Cse4p is located near the middle of the long helix 2 domain where it meets with helix 2 of histone H4. L193 projects into a hydrophobic region formed by neighboring Cse4p residues plus amino acids L37, V54, V57, and L58 from histone H4 (Fig. 5E). Thus, the L193Q substitution in cse4-111 would also be predicted to disrupt the Cse4p-H4 interface.
The mutation in cse4-110 is different. Residue L196 is located on helix 2 of Cse4p and participates in interactions stabilizing the packing of helix 2 and helix 3 of Cse4p. It is close to neighboring residues on helix 2 but also to amino acids M217, A220, R221, and R224 from helix 3 of Cse4p. Only V57 from histone H4 is nearby this region (Fig. 5D). The helix 2-helix 3 structure is critical for the homotypic dimerization of H3 fold domains through the assembly of a stable four-helix bundle (2, 26). Thus, the major defect caused by the L196S mutation in cse4-110 is predicted to be primarily a disruption of Cse4p dimerization and not its interaction with histone H4.
Mutant Cse4p protein remains bound to CEN DNA at restrictive temperatures.
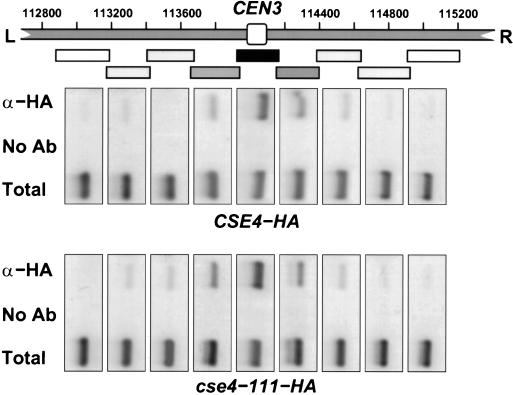
The locations of the amino acid substitutions in Cse4p suggest that at restrictive temperatures H4-Cse4p and Cse4p-Cse4p interactions are defective. Thus, the most-aggressive hypothesis for the temperature-sensitive lethal phenotype of the cse4 mutants was that the centromere nucleosome becomes unstable and physically dissociates from the CEN DNA at elevated temperatures. To test this hypothesis, we analyzed the association of Cse4p with CEN DNA by in vivo cross-linking and chromatin immunoprecipitation (ChIP). Cells expressing either wild-type or mutant HA-tagged Cse4p were grown at 28°C, shifted to 37°C for 4 h, and then analyzed by ChIP. Cse4p remained specifically bound to CEN DNA at restrictive temperatures whether mutant or wild type (data not shown). Furthermore, the distribution of cross-linked Cse4p within the CEN3 region was unaltered. As shown in Fig. 6, both wild-type Cse4-HAp and mutant Cse4-111-HAp continue to localize to the core CEN3 DNA fragment following growth at restrictive temperatures. Similar results were obtained with cse4-102 and cse4-110 (data not shown). These results show that preexisting centromeres do not suffer a major loss of Cse4p from the core region when mutants are shifted to restrictive temperatures.
FIG. 6.
Chromatin immunoprecipitation of CEN3 DNA with wild-type and mutant Cse4 protein. The region of chromosome III from approximately bp 112,800 to 115,200 is illustrated in the diagram. The location of CEN3, containing the CDEI, CDEII, and CDEIII sequences, is indicated by the open box in the center of the region separating the left (L) and right (R) arms of chromosome III. Cells expressing epitope-tagged Cse4 protein from either the wild-type gene (CSE4-HA) or the mutant cse4-111 allele (cse4-111-HA) were grown at the restrictive temperature for 4 h. Formaldehyde cross-linked chromatin was prepared and immunoprecipitated with anti-HA antibody specific for Cse4-HAp (α-HA) or mock treated (No Ab). The coimmunoprecipitated DNAs and total input DNA (Total) were analyzed by PCR with primers corresponding to short, overlapping intervals flanking CEN3. A map of the amplified intervals is provided by location of the boxes shown below the chromosome III diagram. The sets of PCR products from these reactions are shown in the images located below the corresponding sequence interval. Mutant Cse4-111p remains specifically associated with CEN3 DNA at the restrictive temperature. Similar results were obtained for cse4-102 and cse4-110.
CSE4 suppression of hhf1-20.
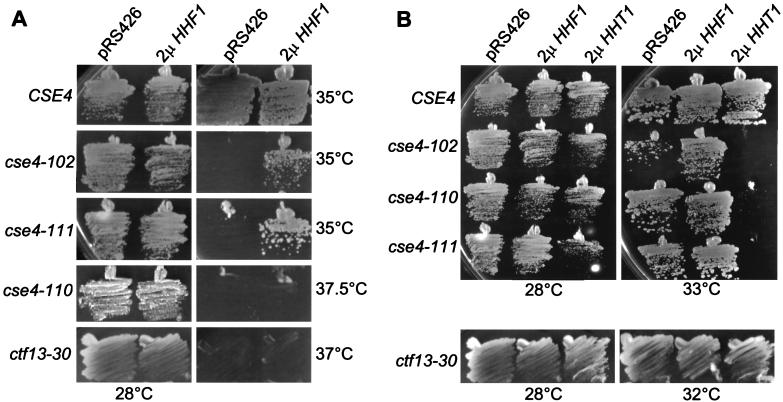
A second way in which defects in H4-Cse4p and Cse4p-Cse4p protein-protein interactions might be lethal is during the assembly of new centromeres when these interactions are first established. In that case, we reasoned that overexpression of wild-type Cse4p should suppress the mutation in hhf1-20 by driving the assembly of H4-Cse4p dimers. To test this prediction, high-copy CSE4 was introduced into hhf1-20 cells and two unrelated temperature-sensitive histone H4 mutants, hhf1-36 and hhf1-10, as controls. The hhf1-36 allele encodes a Y72G substitution within the histone fold domain of H4, altering its interaction with H2A-H2B dimers. Mutants expressing hhf1-36 are temperature sensitive and arrest at restrictive temperatures in G1 due to a failure to transcribe the G1 cyclin genes (40). The hhf1-10 allele encodes a histone H4 protein with four substitutions within the N-terminal domain, K5Q, K8Q, K12Q, and K16Q. Cells expressing hhf1-10 are temperature sensitive, and they delay in the cell cycle at G2/M due to activation of the DNA damage checkpoint pathway (28). As shown in Fig. 7A, high-copy CSE4 is able to suppress hhf1-20 but is unable to suppress the temperature-sensitive lethality of either hhf1-10 or hhf1-36.
FIG. 7.
Cse4p suppression of hhf1 mutations. (A) Cse4p is a specific suppressor of hhf1-20. The individual hhf1 mutant yeast strains indicated on the left of each row were transformed with a high-copy plasmid expressing CSE4. Tenfold serial dilutions of each strain were spotted on synthetic complete medium lacking uracil and grown at permissive (28°C) and restrictive (37°C) temperatures. (B) High-copy expression of mutant cse4 alleles does not suppress hhf1-20. hhf1-20 cells were transformed with five different plasmids containing the CSE4 alleles indicated to the left of each row: vector (pRS425), CSE4 (pLG8), cse4-102 (pLG50), cse4-110 (pLG48), or cse4-111 (pLG49). Tenfold serial dilutions were spotted onto synthetic complete medium lacking leucine and grown at permissive (28°C) and restrictive (37°C) temperatures.
The structures of the cse4(Ts) alleles described above suggest defects in either Cse4p-H4 interactions or Cse4p-Cse4p dimerization. Either defect would be predicted to compromise the ability of the mutant cse4 allele to suppress hhf1-20, provided that suppression is relevant to centromere function. As shown in Fig. 7B this is the case; none of the cse4 alleles were able to suppress hhf1-20, nor did they show any synthetic dosage lethality.
Histone H4 suppression of cse4.
The simplest interpretation of the suppression of hhf1-20 by high-copy CSE4 is that an increased concentration of Cse4p drives its interaction with the mutant histone H4 protein. This model predicts that the reciprocal should also be true: cse4 alleles should be suppressed by increased expression of histone H4. To test this hypothesis we placed expression of HHF1 under the control of the GAL1 promoter and introduced this construct into isogenic wild-type and cse4 mutant strains on a multicopy plasmid. When these strains were grown on glucose, where the GAL1::HHF1 gene is repressed, there was no change in the temperature sensitivity of the cse4 mutants (not shown). However, on galactose, where histone H4 is overexpressed, we observed an allele-specific pattern of suppression. As illustrated in Fig. 8A, overexpression of histone H4 was able to suppress the temperature-sensitive lethality of cse4-102 and cse4-111, but not cse4-110. This pattern of suppression matches precisely that predicted from the structures of the individual cse4 alleles. The mutations in both cse4-102 and cse4-111 are within the Cse4p-histone H4 interface, while the mutation in cse4-110 is not in a position to greatly impair the interaction of Cse4p with histone H4. Thus, this allele-specific suppression of cse4 by histone H4 provides compelling genetic evidence for the interaction of these two histone proteins as an essential part of centromere function in vivo.
FIG. 8.
Genetic interactions of histones H3 and H4 with cse4 alleles. (A) Overexpression of histone H4 suppresses cse4-102 and cse4-111 but not cse4-110. The histone H4 gene HHF1 was placed under control of the GAL1 promoter in plasmid pLG39. The CSE4 wild-type, cse4 mutant, and ctf13-30 strains indicated on the left of each row were transformed with either vector alone (pRS426) or pLG39 carrying the GAL1::HHF1 construct (2μ HHF1). Strains were grown on synthetic complete galactose medium at either permissive (28°C) or minimal restrictive (35, 37, or 37.5°C) temperatures. (B) Overexpression of histone H3 is dosage lethal in cse4 mutants. The histone H3 gene HHT1 was placed under control of the GAL1 promoter in plasmid pLG41. The CSE4 wild-type, cse4 mutant, and ctf13-30 strains were transformed with vector (pRS426) or pLG41 (2μ HHT1) and grown at either permissive (28°C) or semipermissive (33 or 32°C) temperatures.
Histone H3 is dosage lethal in cse4 mutants.
An important implication of the variant nucleosome model of the centromere is that Cse4p should be in competition with histone H3 for binding to histone H4. If Cse4p and histone H4 interact through their respective histone fold domains, then the association of histone H3 with histone H4 must block the binding of Cse4p. We reasoned that overexpression of histone H3 might titrate available histone H4 molecules and enhance its competition with Cse4p. To test this hypothesis, we placed the histone H3 gene HHT1 under control of the GAL1 promoter and introduced the construct into isogenic wild-type and cse4 mutant cells. As shown in Fig. 8B, induction of GAL1::HHT1 has no effect on the growth of wild-type strains at a variety of temperatures. In contrast, overexpression of histone H3 has a strong dominant-negative effect on all three of the cse4 mutants. Impaired growth is evident even at 28°C, and the mutant cells overexpressing histone H3 die at 30 to 33°C instead of 37°C. Overexpression of HHT1 was not generally dosage lethal with centromere mutations, as it did not affect the growth profile of ctf13-30 mutants (Fig. 8B).
Finally, we used ChIP to ask whether overexpression of H3 or H4 disrupts the binding of Cse4p to CEN3 DNA. If overexpression of histone H3 partially blocks the assembly of new centromeres, we reasoned that the CEN3 signal would be altered by less than twofold, a difference that is below the current resolution of our assays. However, if overexpression disrupted preexisting centromeres, then the change in CEN3 immunoprecipitation might be significantly larger. As shown in Fig. 9, neither the dosage lethality of HHT1 overexpression nor the dosage suppression of HHF1 over-expression caused any major change in the association of Cse4p protein with CEN3 DNA in cse4 mutants. Thus, the most likely target of H3 and H4 overexpression is the centromere nucleosome assembly pathway.
FIG. 9.
ChIP of CEN3 DNA with Cse4 protein following overexpression of histones H3 or H4. Strains expressing epitope-tagged Cse4 protein from wild-type, cse4-102, and cse4-110 alleles were grown on galactose medium to induce overexpression of either histone H3 (top panel) or histone H4 (bottom panel) from the GAL1 promoter carried on 2μ multicopy plasmids (+). Control cells contained the 2μ vector only (−). Cultures were grown for 4 h at the temperatures indicated, and the in vivo cross-linking of Cse4 protein with the core CEN3 DNA fragment was determined by ChIP as described in the legend to Fig. 6. The cse4-102 and cse4-110 mutants are able to form colonies at 33°C on galactose medium with vector alone but are inviable at that temperature when histone H3 is overexpressed. Conversely, cse4-102 cells are inviable at 35°C on galactose medium but can form colonies at that temperature when histone H4 is overexpressed. The cse4-110 mutant is poorly suppressed by histone H4 overexpression and fails to give colonies at 37°C with either vector or histone H4 overexpression. The association of Cse4 protein with CEN3 DNA was unaffected by overexpression of either histone H3 or histone H4 in the mutant strains.
DISCUSSION
Several lines of evidence argue that histones H4 and Cse4p play important roles in the assembly and function of the core centromere of S. cerevisiae. First, both cse4 and hhf1-20 mutants are defective for mitotic chromosome transmission and arrest with a characteristic G2/M phenotype (4, 45, 47), a phenotype shared by the new cse4(Ts) mutants described above. Also, as shown here, this arrest phenotype is imposed by activation of the spindle-kinetochore checkpoint pathway in the mutants, implying that histone H4 and Cse4p are required for normal kinetochore function. Furthermore, both cse4 and hhf1-20 mutants disrupt the ability of the centromere complex to block transcriptional elongation, suggesting a direct role in centromere chromatin structure in vivo. Finally, in vivo crosslinking experiments showed that Cse4p is physically located at CEN DNA within the core centromere (30).
Taken together, these results lead to the simple hypothesis that Cse4p and histone H4 interact through their respective histone fold domains to form a nucleosome-like structure at the centromere. We undertook an unbiased genetic screen for Cse4p histone fold mutations to challenge this hypothesis directly, and the results of the experiments reported here provide strong support for the model. First, dosage suppression by CSE4 is allele specific and has no effect on temperature-sensitive alleles of histone H4 that do not affect the centromere pathway. Second, point mutations within the histone fold domain of Cse4p are defective for centromere function in vivo. Third, overexpression of wild-type histone H4 was able to suppress the lethality of cse4 mutations and this suppression was also allele specific, affecting only those cse4 alleles in which the amino acid substitutions were at the Cse4p-H4 interface. This reciprocal allele-specific suppression argues that Cse4p and histone H4 interact through their histone fold domains to accomplish one or more essential steps in centromere structure and function in vivo.
Site-directed mutations targeted to the CENP-A gene first highlighted the importance of cooperative interactions across the histone fold domain (42). The cse4-102 allele provides a striking example of such a long-range cooperative interaction. This allele encodes two amino acid substitutions located at opposite ends of the Cse4p histone fold. Either mutation alone has little or no effect on growth or centromere integrity, but together they result in a temperature-sensitive lethal phenotype and a loss of the block to transcriptional read-through of CEN DNA. A second important conclusion from the CENP-A analysis was that the helix 2-helix 3 region of the fold is important in the assembly of CENP-A at the centromere (42). The cse4-110 allele confirms this conclusion for Cse4p. In the nucleosome, the helix 2-helix 3 structure of the H3 fold is critical for the homotypic dimerization of H3 in the (H3-H4)2 tetramer (2, 26). The L196S mutation in cse4-110 is located at the center of the interactions stabilizing the helix 2-helix 3 structures. This substitution presumably disturbs the integrity of Cse4p dimerization and formation of the (Cse4p-H4)2 tetramer in the centromere nucleosome-like complex.
Surprisingly, none of the mutations in the cse4(Ts) alleles altered codons for Cse4p-specific residues. The mutations at L175, L193, and L196 all affect invariant leucines conserved throughout the histone H3 family of proteins. Residue M217 in Cse4p differs from the corresponding I124 in S. cerevisiae histone H3; however, methionine occurs at this position in the major histone H3 proteins of other organisms and it is not characteristic of the CENP-A variant proteins. Interestingly, Keith et al. found that no individual Cse4p-specific residue was essential for centromere function using chimeric gene constructs that substituted portions of the histone H3 gene for the corresponding region of CSE4 (23). Thus, as the Cse4p/CENP-A family evolved to assume a specialized role in centromere chromatin, it accumulated multiple cooperative substitutions spread across the protein while continuing to maintain highly conserved properties of the H3 histone fold.
The dominant-negative phenotype of histone H3 overexpression in cse4 mutants has important implications for the assembly pathway of the centromere. The simplest interpretation of this interference is that histone H3 competes with Cse4p for binding to histone H4 (Fig. 10A). In this model, cse4 mutations that impair formation of the (Cse4p-H4)2 tetramer would shift the equilibrium to the histone H3 pathway, leading to a defect in centromere function. Overexpression of histone H3 would further antagonize Cse4p-H4 assembly by competing for histone H4, exacerbating the centromere defect of the cse4 mutants. Although this simple model provides a useful framework, it fails to explain the remarkable buffering capacity of the Cse4p branch of the pathway. Histone H3 is an abundant nuclear protein and must be highly expressed to provide two molecules for each of the roughly 100,000 nucleosomes that package the yeast genome. Currently, the exact stoichiometry of Cse4p in the cell is not known, but it is not abundant (T. Kalashnikov and M. M. Smith, unpublished data), perhaps on the order of two molecules at the centromere of each of the 16 yeast chromosomes. Despite this low abundance, high-copy overexpression of histone H3 from the GAL1 promoter is not deleterious in CSE4 wild-type cells (Fig. 8B) (see also reference 27), and in cse4 cells its expression only reduces the maximal permissive temperature of the mutants. How, then, does the Cse4p assembly pathway withstand the competition of excess histone H3?
FIG. 10.
A model of the Cse4p-H4 assembly pathway. (A) A simple model for the interactions of histones H3 and H4 with Cse4p is shown. In this model, histones Cse4p and H3 compete for the binding of histone H4. The structural and genetic results reported here suggest that the formation of the H4-Cse4p dimer is impaired in hhf1-20, cse4-102, and cse4-111 mutants, while formation of the (H4-Cse4p)2 tetramer is impaired in the cse4-110 mutant. In those mutants, overexpression of histone H3 further antagonizes the formation of the centromere variant nucleosome, leading to dosage lethality. (B) Cooperative downstream interactions would favor the assembly of a CEN variant nucleosome in the face of histone H3 competition for histone H4. Additional known centromere components are illustrated. It is not known whether histone H2A-H2B dimers participate in the structure, or whether specialized chromatin assembly factors (CAFs) may be required.
One attractive mechanism for buffering Cse4p would be to separate H3-H4 assembly from Cse4p-H4 assembly in either time or space (for a recent summary, see reference 18). Both the transcription and the translation of histone H3 and H4 in S. cerevisiae are cell cycle regulated, and their expression decreases rapidly outside of early S phase (14, 19, 25; reviewed in reference 34). Currently nothing is known about Cse4p translation; however, its mRNA is present constitutively at low abundance throughout the cell division cycle (P. Yang and M. M. Smith, unpublished data). Thus, once out of S phase, the concentrations of free histone H3 and H4 may fall to levels comparable to the concentration of free Cse4p. Consistent with this idea, if CENP-A is expressed in mammalian cells under the cell cycle regulation of the major histone H4 genes, it is incorporated generally into the chromatin (42).
Highly cooperative downstream binding interactions would provide another potential mechanism for driving the Cse4p-H4 assembly pathway (Fig. 10B). Centromere assembly is known to be dependent on CEN DNA and multiple protein complexes, consistent with strong cooperative binding among these components and CEN DNA (16, 29, 33, 46). Thus, cooperative binding of the (Cse4p-H4)2 tetramer with CEN DNA and other centromere proteins would serve to drive the H4-Cse4p branch of the pathway against a concentration of histone H3. Finally, there may be novel chromatin assembly factors or structural arrangements in the cell that have evolved to specifically catalyze the assembly of the centromere nucleosome (5, 9, 18, 49). We anticipate that further genetic and biochemical studies designed to test the predictions of these models will provide important insights into the mechanisms by which centromeres are specified and assembled.
ACKNOWLEDGMENTS
We thank Forrest Spencer and Daniel Burke for plasmids and strains; Pamela Meluh, David Auble, Michael Christman, David Allis, Daniel Burke, and Van Moudrianakis for helpful discussions; and William Ross for expert technical assistance with the flow cytometry.
This work was supported by NIH grant GM28920 to M.M.S.
REFERENCES
- 1.Adams A, Gottschling D E, Kaiser C, Stearns T. Methods in yeast genetics: a laboratory course manual. Cold Spring HarborNew York, N.Y.: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 2.Arents G, Burlingame R W, Wang B C, Love W E, Moudrianakis E N. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arents G, Moudrianakis E N. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker R E, Harris K, Zhang K. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basrai M A, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggins S, Murray A W. Sister chromatid cohesion in mitosis. Curr Opin Cell Biol. 1998;10:769–775. doi: 10.1016/s0955-0674(98)80120-8. [DOI] [PubMed] [Google Scholar]
- 7.Bloom K S, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 8.Boeke J D, Trueheart J, Natsoulis B, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 9.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 10.Brent R, Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984;312:612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- 11.Buchwitz B J, Ahmad K, Moore L L, Roth M B, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 12.Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- 13.Craig J M, Earnshaw W C, Vagnarelli P. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp Cell Res. 1999;246:249–262. doi: 10.1006/excr.1998.4278. [DOI] [PubMed] [Google Scholar]
- 14.Cross S L, Smith M M. Comparison of the structure and cell cycle expression of mRNAs encoded by two histone H3-H4 loci in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:945–954. doi: 10.1128/mcb.8.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doheny K F, Sorger P K, Hyman A A, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espelin C W, Kaplan K B, Sorger P K. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funk M, Hegemann J H, Philippsen P. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:153–160. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S, Ahmad K, Platero J S, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hereford L M, Osley M A, Ludwig I J R, McLaughlin C S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- 20.Hoyt M A, Totis L, Roberts B T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 21.Hyland K M, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyman A A, Sorger P K. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 23.Keith K C, Baker R E, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig J R, II, McLaughlin C S. Proceedings of the Berkeley Workshop on Recent Advances in Yeast Molecular Biology: Recombinant DNA. Berkeley: University of California; 1982. Periodic synthesis of histone proteins through the cell cycle of Saccharomyces cerevisiae as determined by centrifugal elutriation; pp. 113–121. [Google Scholar]
- 26.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 27.Meeks-Wagner D, Wood J S, Garvik B, Hartwell L H. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986;44:53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- 28.Megee P C, Morgan B A, Smith M M. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 29.Meluh P B, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo crosslinking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 31.Morgan B A, Mittman B A, Smith M M. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 33.Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osley M A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 35.Palmer D K, O'Day K, Wener M H, Andrews B S, Margolis R L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 38.Robzyk K, Kassir Y. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Santisteban M S, Arents G, Moudrianakis E N, Smith M M. Histone octamer function in vivo: mutations in the dimer-tetramer interfaces disrupt both gene activation and repression. EMBO J. 1997;16:2493–2506. doi: 10.1093/emboj/16.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulman I, Bloom K S. Centromeres: an integrated protein/DNA complex required for chromosome movement. Annu Rev Cell Biol. 1991;7:311–336. doi: 10.1146/annurev.cb.07.110191.001523. [DOI] [PubMed] [Google Scholar]
- 42.Shelby R D, Vafa O, Sullivan K F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skibbens R V, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 45.Smith M M, Yang P, Santisteban M S, Boone P W, Goldstein A T, Megee P C. A novel histone H4 mutant defective for nuclear division and mitotic chromosome transmission. Mol Cell Biol. 1996;16:1017–1026. doi: 10.1128/mcb.16.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorger P K, Severin F F, Hyman A A. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan K F, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vafa O, Sullivan K F. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Burke D J. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warburton P E, Cooke C A, Bourassa S, Vafa O, Sullivan B A, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan K F, Poirier G G, Earnshaw W C. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]