Machine learning–enabled perception-based detection of protein biomarkers was achieved in gynecologic cancer patient biofluids.
Abstract
Conventional molecular recognition elements, such as antibodies, present issues for developing biomolecular assays for use in certain technologies, such as implantable devices. Additionally, antibody development and use, especially for highly multiplexed applications, can be slow and costly. We developed a perception-based platform based on an optical nanosensor array that leverages machine learning algorithms to detect multiple protein biomarkers in biofluids. We demonstrated this platform in gynecologic cancers, often diagnosed at advanced stages, leading to low survival rates. We investigated the detection of protein biomarkers in uterine lavage samples, which are enriched with certain cancer markers compared to blood. We found that the method enables the simultaneous detection of multiple biomarkers in patient samples, with F1-scores of ~0.95 in uterine lavage samples from patients with cancer. This work demonstrates the potential of perception-based systems for the development of multiplexed sensors of disease biomarkers without the need for specific molecular recognition elements.
INTRODUCTION
Current biomolecular identification methodologies rely heavily on one-to-one recognition via specific proteins and nucleic acids such as antibodies, peptides, and aptamers to bind analytes (1–5). However, the development of highly sensitive and specific binding moieties in quantities sufficient to detect target molecules with one-to-one recognition has multiple challenges that delay the development of a robust, versatile, and cost-effective platform for multiple analyte detection. The challenges of using antibodies include long-term stability/robustness, transient/real-time applications, and production difficulties, especially when many different antibodies must be developed (6–8). Hence, technologies that replace antibodies could enhance the development of certain types of point-of-care assays, medical devices such as wearable sensors, and diagnostics in underresourced settings (9, 10).
Perception-based machine learning (ML) platforms, modeled after the complex olfactory system, can isolate individual signals through an array of relatively nonspecific receptors (11). Each receptor captures certain features, and the overall ensemble response is analyzed by the neural network in our brain, resulting in perception. Biofluids such as blood, urine, saliva, and sweat are indicative of physiological conditions and enable biomarker detection in their native state (12, 13). Recent advances in ML methodologies have made complex algorithms more accessible, facilitating the integration of perception systems into materials science (14, 15). We believe that perception-based sensors can be developed to enable the successful, multiplexed detection of analytes without the need for antibodies.
Previous attempts to develop perception-based sensing platforms have had limited success. Prior works include “electronic nose” technologies (16–19) for gas sensing, based on conducting polymers, DNA-decorated field-effect transistors (20), and other technologies based on protein recognition using simple data analytic techniques (21). “Optical” noses have been developed as well (22, 23). However, these developments are limited in their ability to detect molecules such as proteins and in physiological conditions and complex biofluids. To overcome limitations associated with one-to-one recognition elements, we are investigating the development of a perception-based methodology that uses ML processes coupled with a sensor array, where each element exhibits moderate selectivity for a wide range of molecules.
The prognosis and quality of life of patients with cancer are strongly affected by the ability to accurately diagnose diseases at an early stage. One such example is ovarian cancer, the fifth leading cause of cancer-related deaths among women in the United States and first among gynecologic malignancies (24), with 22,000 new cases and 14,000 deaths per year (24). The 5-year relative survival rate for patients diagnosed with ovarian cancer is 44% (25), while detection at stage I can increase the 5-year survival rate to more than 90% (26). However, there are no methods to date that achieve early, accurate diagnoses or are there strategies to rapidly determine patient response to treatment to inform the choice of therapy.
To detect gynecologic cancers, such as high-grade serous ovarian carcinoma (27–29) and endometrial cancers (30, 31), U.S. Food and Drug Administration–approved serum biomarkers such as cancer antigen 125 (CA-125) and human epididymis protein 4 (HE4) have been used as well as ultrasonography. However, these methods lack the sensitivity to detect early-stage cancer and have had little impact on survival (32, 33). A recent study of uterine lavage (or uterine washings, fluids removed from the uterus after perfusion with saline) found substantially higher levels of biomarkers, such as HE4, CA-125, chitinase-3-like protein 1 (YKL-40), and mesothelin, than those found in serum (34). Therefore, the use of uterine lavage has the potential to improve early detection.
Single-wall carbon nanotubes (SWCNTs) have unique optical properties and sensitivity that make them valuable as sensor materials (35). SWCNTs emit near-infrared (NIR) photoluminescence with distinct narrow emission bands that are exquisitely sensitive to the local environment (36). In addition, the emission is photostable, enabling quantitative and long-term monitoring of small molecules, proteins, nucleic acids, and enzymatic activities both in vitro and in vivo (37–41). Individual SWCNT species (or chiralities) have distinct bandgaps, which contribute to their varying sensitivities to redox phenomena (42, 43). Their emission bands also respond to the local dielectric and electrostatic charge environment, resulting in solvatochromic shifting (44, 45). Coatings such as DNA can confer not only colloidal stability in an aqueous solution but also selectivity by modulating the surface coverage and bandgap (46). The use of DNA-wrapped SWCNTs (DNA-SWCNTs) has been used for the detection of a wide range of analytes in biological media, including in live cells and animals (41, 47).
In this study, we investigate a perception-based sensing system to detect multiple biomarkers in human biofluids (Fig. 1). We developed a DNA-SWCNT–based photoluminescent sensor array wherein the optical responses were used to train ML models to detect gynecologic cancer biomarkers HE4, CA-125, and YKL-40 in laboratory-generated samples and patient fluids. Distinct changes in fluorescent peak position and intensity values from each DNA-SWCNT combination were observed in response to the protein analytes. ML algorithms support vector machine (SVM), random forest (RF), and artificial neural network (ANN) enabled the prediction of both the presence (classification) and concentration (regression) of each biomarker. In uterine lavage samples, the classification results were highly accurate, producing F1-scores of ~0.95 in laboratory-generated samples and classification successes of 100% for HE4 and CA-125 and 91% for YKL-40 in cancer patient samples. This work suggests that a nanosensor/perception-based sensing system can accurately detect multiple disease biomarkers in patient biofluids.
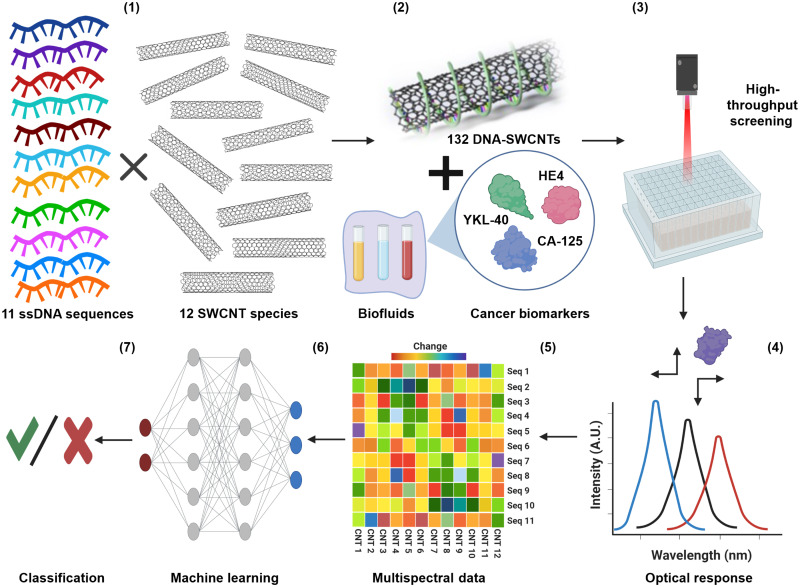
Fig. 1. Perception-based nanosensor platform for protein biomarkers.
(1) Eleven single-stranded DNA oligonucleotides wrap SWCNT chiralities to form DNA-SWCNT sensor complexes. (2) The array of sensors is incubated in the sample of interest. (3) The optical response of the sensors is interrogated by high-throughput NIR spectroscopy. (4) The spectroscopic data are fitted to determine the wavelength and intensity of each sensor emission band. (5) The sensor responses are processed into a feature vector (FV) training set. A.U., arbitrary units. (6) ML algorithms are trained and validated for each target protein and their combinations. Seq, sequence; CNT, Carbon nanotubes. (7) Prediction results are evaluated.
RESULTS
DNA-SWCNT array
We characterized multiple DNA-SWCNT complexes to form the basis of a sensor array. Eleven DNA sequences [(AT)11, (AT)15, (AT)20, (GT)12, (ATT)4, (TCT)5, T3C3T3C3T3, C3T9C3, C3T3C9, CT2C3T2C, and (AC)15] were chosen because many of them are recognition sequences of specific SWCNT chiralities, which suggest ordered wrapping on their surface, while others confer some degree of specificity to proteins or other analytes (48–51). Twelve semiconducting SWCNT species present in the HiPCO preparation [(6,5), (8,4), (10,3), (7,5), (7,6), (8,3), (9,5), (9,4), (8,6), (8,7), (10,2), and (9,7)] were evaluated because of their high concentrations in the sample and bright photoluminescence in the serum/water optical window of 900 to 1400 nm (fig. S1, A and B). The combinatorial possibilities of 12 SWCNT species and 11 DNA sequences resulted in the formation of 132 distinct DNA-SWCNT complexes that were investigated within the context of a sensor array. The DNA-SWCNT complexes exhibited high colloidal stability and strong photoluminescence, as previously reported (52–54). We characterized the complexes using ultraviolet-visible NIR (UV-Vis-NIR) absorbance, NIR fluorescence spectroscopy, atomic force microscopy (AFM), and zeta potential measurements (fig. S1). The measurements confirmed the emissive properties of at least 12 SWCNT chiralities (fig. S1B), a highly negative zeta potential of DNA-SWCNT complexes formed with all 11 DNA sequences (fig. S1C), and a DNA banding pattern along the SWCNT surface for all sequences (fig. S1, D to H).
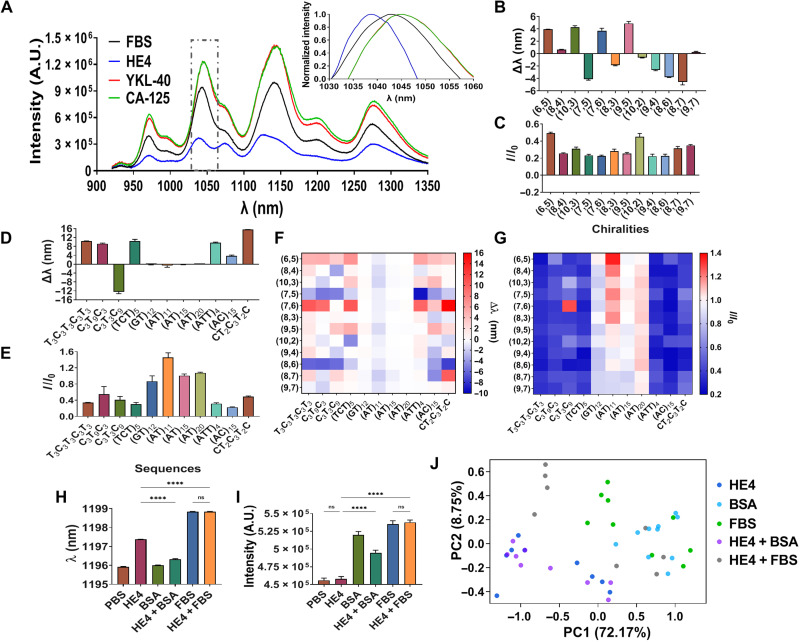
The optical responses of the DNA-SWCNT complexes to known gynecologic cancer biomarkers were assessed via spectroscopy. High-throughput NIR spectroscopy (in the range of 900 to 1400 nm) was conducted on all DNA-SWCNT complexes introduced to laboratory-generated samples of the protein biomarkers HE4, CA-125, and YKL-40 in 10% fetal bovine serum (FBS) solutions (to provide a relevant background of interferent molecules). The spectroscopic bands of all SWCNT chiralities were fitted to extract peak wavelength shift (Δλ) and intensity ratio (I/I0) with respect to a control sample in 10% FBS. As a representative example, the (7,5) chirality emission peak blue-shifted (Δλ < 0), and its intensity was attenuated (I/I0 < 1) in response to HE4 (Fig. 2A), while brightening and red shifting were observed upon exposure to CA-125 and YKL-40 (Fig. 2A and inset). Similar analyses found diverse optical responses to single biomarkers across SWCNT chiralities (Fig. 2, B and C) and DNA wrappings (Fig. 2, D and E). There were no obvious correlations between the response and conditions in which they were challenged (Fig. 2, F and G, and fig. S2).
Fig. 2. DNA-SWCNT optical responses to gynecologic cancer biomarkers.
(A) Representative spectra of DNA-SWCNT complexes in response to cancer protein biomarkers. Inset: Normalized spectrum of the (7,5) chirality. (B) Wavelength modulation of (AC)15-SWCNT complexes upon incubation with 100 nM HE4; n = 3. (C) Intensity modulation of (AC)15-SWCNT complexes upon incubation with HE4; n = 3. (D) Wavelength modulation of DNA-(7,6) complexes upon incubation with HE4; n = 3. (E) Intensity modulation of DNA-(7,6) complexes upon incubation with HE4; n = 3. (F) Heatmap of total wavelength modulations of DNA-SWCNT complexes upon incubation with HE4; n = 3. (G) Heatmap of total intensity modulations of DNA-SWCNT complexes upon incubation with HE4; n = 3. (H) Wavelength of (AT)11-(8,6) complex upon incubation with phosphate-buffered saline (PBS), HE4, bovine serum albumin (BSA), and FBS in PBS; n = 3, means ± SEM; ****P < 0.0001, unpaired t test. (I) Intensity of (AT)11-(8,6) complex upon incubation with PBS, HE4, BSA, and FBS in PBS; n = 3, means ± SEM; ****P < 0.0001, unpaired t test. “ns” denotes not significant. (J) Principal components analysis (PCA) plot of the DNA-SWCNT response to HE4 versus interferents.
To study the physical properties of the DNA-SWCNT complexes that could contribute to the distinct responses, we analyzed the SWCNT surface charge and DNA wrapping patterns on the SWCNT surface. Zeta potential measurements of the DNA-SWCNTs showed that surface charge varied between approximately −44 and −55 mV, depending on the DNA sequence (fig. S1C), likely a result of differences in DNA packing densities. To further investigate, we conducted AFM, which revealed substantially differences in the density of observable height maxima/peaks on the SWCNTs of approximately 40% (fig. S1, D to H), ascribable to the DNA. These findings suggest that the unique responses of each DNA-SWCNT to the proteins are likely due, in part, to the distinct DNA wrapping patterns on each SWCNT chirality, in addition to structural differences between the biomarkers such as size, charge, hydrophobicity, and the level of glycosylation (table S1) (55, 56).
Next, we investigated the specificity of the DNA-SWCNTs by examining the response to HE4 in the presence of interferents [i.e., bovine serum albumin (BSA) and FBS]. We found that some DNA-SWCNTs responded differently to the analyte and interferents, but the specificity of any one complex appeared marginal (Fig. 2, H and I). To assess the distinctness of DNA-SWCNT responses to a protein biomarker versus interferents, we applied principal components analysis (PCA). The analysis of DNA-SWCNT responses to HE4 and interferent proteins failed to separate distinct optical responses of HE4 (Fig. 2J). We thus concluded that more sophisticated data analyses were needed to determine whether the DNA-SWCNT array could correctly identify analytes within a complex environment.
ML feature vector construction
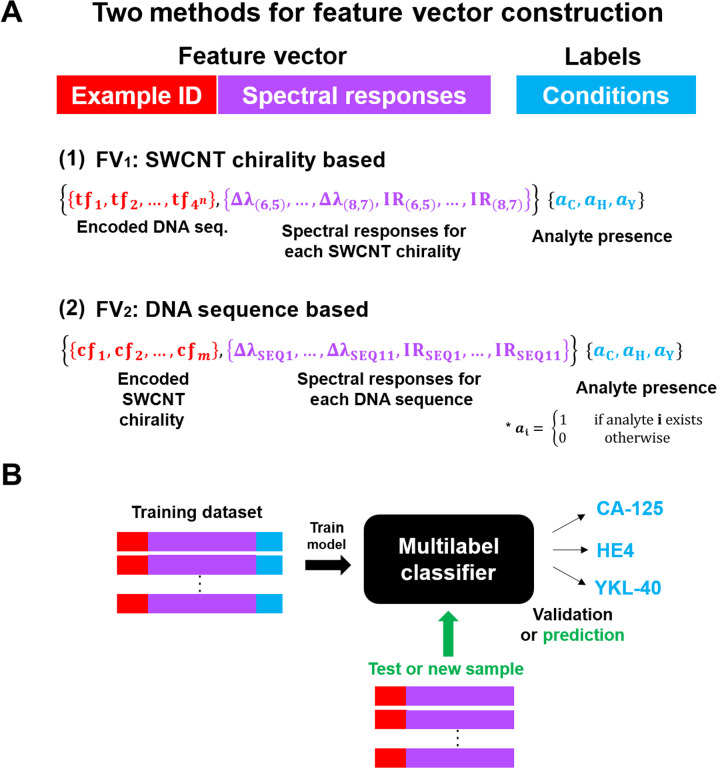
To differentiate the biomarkers via the DNA-SWCNT optical responses, we investigated several ML strategies. We tested two different feature vector (FV) methods to represent experimentally measured data matrices composed of DNA sequences and SWCNT chiralities (Fig. 3A). Each vector was constructed with two components: “Example ID,” SWCNT chirality or DNA sequence, and “Features,” the DNA-SWCNT complex emission intensity and wavelength response. In addition, the vector corresponds to a specific label that indicates the presence of each biomarker in the sample. The first FV (FV1) is focused on chirality [Fig. 3A, (1)] and uses DNA sequences as the example IDs and chirality-dependent optical responses as features. Underlying this choice of feature is the hypothesis that the spectroscopic response of multiple SWCNTs in combination with a single DNA sequence is sufficient to determine the presence or concentration of biomarkers. DNA sequences were encoded into IDs as either bigram or trigram term frequency vectors (48). Therefore, the total number of features is 40 using a bigram representation (16 + 2 × 12) and 88 using a trigram frequency vector (64 + 2 × 12).
Fig. 3. FV construction.
(A) The FV contains two parts, example encoding (red) and optical response–based features (purple), with each vector corresponding to a label that indicates the biomarker conditions (blue). The total features of FV1 are described by 4n + 2M, where tf denotes an n-gram term frequency vector (i.e., n = 2 in bigram and n = 3 in trigram), and M denotes the number of chiralities. The total features of FV2 are described by M + 2N, where cf denotes SWCNT chirality features, N denotes the number of sequences and M denotes the number of chiralities. a is an indicator function for the analyte presence (either 0 or 1). The subscripts C, H, and Y represent CA-125, HE4, and YKL-40, respectively. (B) Each FV is processed by a multilabel classifier (black box) to classify (detect) each biomarker. IR is the intensity ratio and defined as IR = I/I0.
The second FV (FV2) uses chiralities as the example IDs [Fig. 3A, (2)] combined with sequence-dependent optical responses as features. Underlying the FV2 is the hypothesis that a single SWCNT in combination with a number of DNA sequences is sufficient to determine the presence or concentration of biomarkers. SWCNTs were represented using the one-hot encoding (“1” for specific chirality and “0” for the other chiralities) (57); hence, the total number of features used for FV2 is 34 (12 + 2 × 11).
Input data formatted according to FV1 and FV2 were used to train several classification algorithms for the detection of individual biomarkers or combinations thereof (Fig. 3B). Three ML algorithms—SVM, RF, and ANN—were trained using an initial dataset and were evaluated by 10-fold cross-validation. Bayesian optimization was used for hyperparameter tuning. The resulting F1-scores were used to assess model performance (fig. S3).
Classification model training and validation
We investigated the potential for the platform to detect the presence/absence of a single biomarker, HE4, using binary classification algorithms. We introduced the DNA-SWCNT complexes to solutions of HE4 and background/interferents FBS, BSA, and mixtures, all in phosphate-buffered saline (PBS). We classified the data using several approaches such as bi-class (±HE4), multiclass (HE4, HE4 ± FBS, FBS, HE4 ± BSA, BSA), and multilabel (±HE4 and ±FBS and ±BSA). Criteria for excluding certain FVs were wavelength shifts higher than 20 nm or poor peak fitting, both most likely caused by low signal intensities. We found that RF resulted in better F1-scores than ANN and SVM (>0.93) (fig. S4A). The performance of biclass classifiers was slightly better than multiclass and multilabel classifiers. Overall, the algorithms provided high F1-scores (>0.92). While using FV2, all algorithms provided high F1-scores (1.0 for biclass and 0.9 to 1.0 for multiclass/multilabel classification). The high values of F1-scores raised concerns with overfitting, which could occur with small sample sets. Another concern is the high initial concentration of analytes in the training sets.
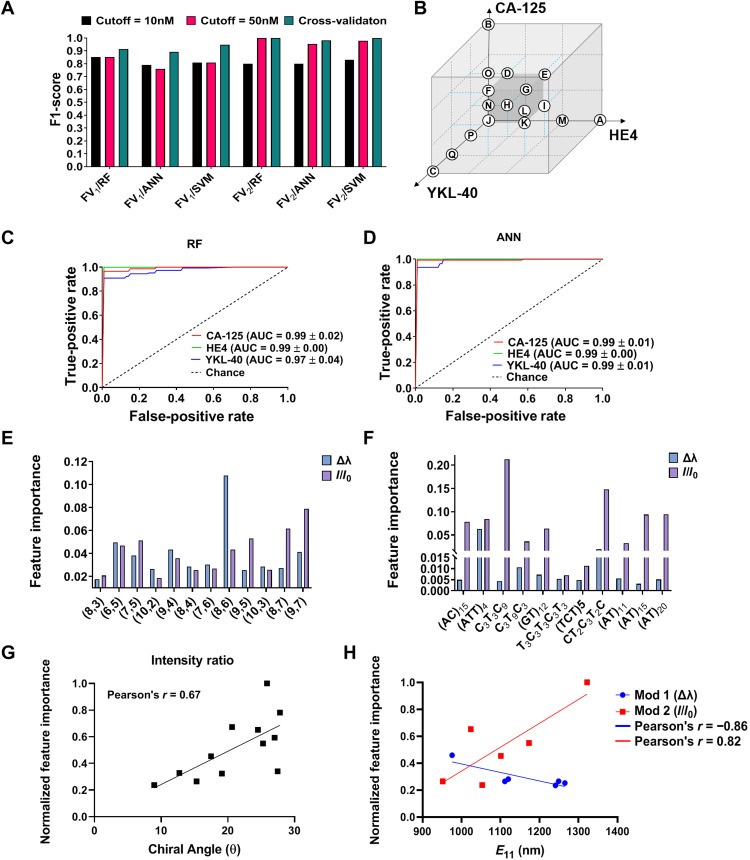
To alleviate those concerns and determine the detection limit of the platform for HE4 classification using the model trained with high concentrations, we tested against several lower HE4 concentrations. Figure 4A shows F1-scores for the three algorithms using both FVs for 10 and 50 nM HE4 thresholds (as these concentrations are relevant to cancer diagnosis). Both FVs generated high values for F1-scores on cross-validation at 50 nM HE4 concentration (F1-score > 0.89 in FV1 and F1-score > 0.98 in FV2) and in the range of 0.79 to 0.85 at 10 nM HE4. While both FVs continued to predict with high F1-scores, the performance of FV2 was better than FV1 and provided good results for all three algorithms.
Fig. 4. ML results and analysis.
(A) F1-scores of three algorithms for each FV corresponding to different thresholds. (B) Schematic of training/testing method for different concentrations of biomarker combinations. (C) Receiver operating characteristic (ROC) of each biomarker via RF model. Area under the curve (AUC) values for each biomarker. (D) ROC of each biomarker via ANN model. AUC values for each biomarker. (E) Feature importance of SWCNT chiralities generated by FV1. (F) Feature importance of DNA sequences generated by FV2. (G) Intensity change feature importance versus SWCNT chiral angle. (H) Normalized feature importance values of wavelength shift (Δλ) and intensity change versus SWCNT emission wavelength.
To investigate the potential for the platform to detect other/multiple cancer biomarkers, we optimized multilabel classification methodologies. We trained the ML algorithms using the optical response of the DNA-SWCNT complexes to a single and multiple combinations of HE4, CA-125, and YKL-40 with various concentrations, ranging from 0 to 100 nM (Fig. 4B and table S2). To generate as comprehensive a dataset as possible, we screened over 17 different biomarker combinations, which resulted in more than 200 examples for each FV. We incubated the DNA-SWCNT complexes in FBS and PBS to assess the biomarkers in complex environments. We constructed three types of multilabel ML models: adaptive algorithm, binary relevance, and label powerset (58, 59). Cross-validation results (fig. S4B) show that the F1-scores using FV2 were substantially higher (>0.96) compared to FV1 (>0.68) across all the models, with RF and ANN outperforming SVM (with F1-scores of 0.97). To validate the F1-scores of the top-performing algorithms, we generated receiver operating characteristic (ROC) curves for the three biomarkers (Fig. 4, C and D). The areas under the curve (AUCs) were all greater than 0.97. Individual analyses of each biomarker showed high F1-scores for HE4 (1 and 0.99 in RF and ANN, respectively), CA-125 (1 and 0.91 in RF and ANN, respectively), and YKL40 (0.96 and 0.84 in RF and ANN, respectively). These results demonstrate the ability of the model to detect single and multiple biomarkers in mixtures with high precision (fig. S4C). In addition, the accuracy of detection was mostly high, depending on the biomarker (fig. S4D).
On the basis of these results, we decided to proceed with FV2 as the FV used for the classification and RF and ANN as the ML algorithms. The better performances of FV2 suggest that the collection of spectroscopic features from a single SWCNT, in combination with a number of DNA sequences, is better than a FV that comprises data from a single DNA sequence on a number of SWCNTs.
To evaluate the concentration of each biomarker in each sample, we also conducted a regression analysis. Regression results of RF and ANN using FV2 achieved R2 values of 0.93 and 0.92, respectively (shown in fig. S4E).
ML feature importance
To understand the relationship between the nanosensor array composition and the ML predictions, we used feature importance analysis to investigate the DNA-SWCNT properties that influence the prediction. We extracted the feature importance values from the algorithms using both FV1 and FV2 (Fig. 4, E and F). We found that the relative importance of nanotube chiralities on the marker prediction appeared to correlate with chiral angle of the nanotube species, as defined by Pearson’s correlation coefficient (Fig. 4G). There also appeared to be some dependence on nanotube mod (fig. S5, A and B), wherein nanotube chirality vectors (n,m) calculated via mod(n-m, 3) gives a value of 1 or 2 for semiconducting carbon nanotubes (60, 61). We also found some correlation between the importances of wavelength shifting responses of mod 1 chiralities with the nanotube optical bandgap (E11) (r = −0.86) and intensity responses of mod 2 chiralities with optical bandgap (r = 0.82) (Fig. 4H and fig. S5, C to F). These correlations suggest that nanotube structure contributed to the differences in the optical responses of the nanosensors that enabled enough response diversity to result in positive predictive value.
Among DNA wrapping sequences, C3T3C9 and CT2C3T2C presented the highest and second highest feature importance values, respectively (Fig. 4F). The intensity ratio feature exhibited higher importance values than the wavelength shifting responses across all sequences. Using this feature importance analysis, we narrowed down the array to the five most important DNA sequences [(AC)15, (AT)11, (AT)15, CT2C3T2C, and T3C3T3C3T3] to reduce the number of features and, therefore, the number of experimental conditions. The optimized model generated F1-scores of 0.98 for classification and R2 of 0.78 for regression. The combined results suggest that the sensitivity of this platform for the biomarkers is dependent on both the nanotube structure and the unique morphology of the DNA adhesion on the nanotubes due to sequence-dependent π-π stacking of the base pairs on the graphitic sidewall of the SWCNTs.
Uterine lavage patient samples
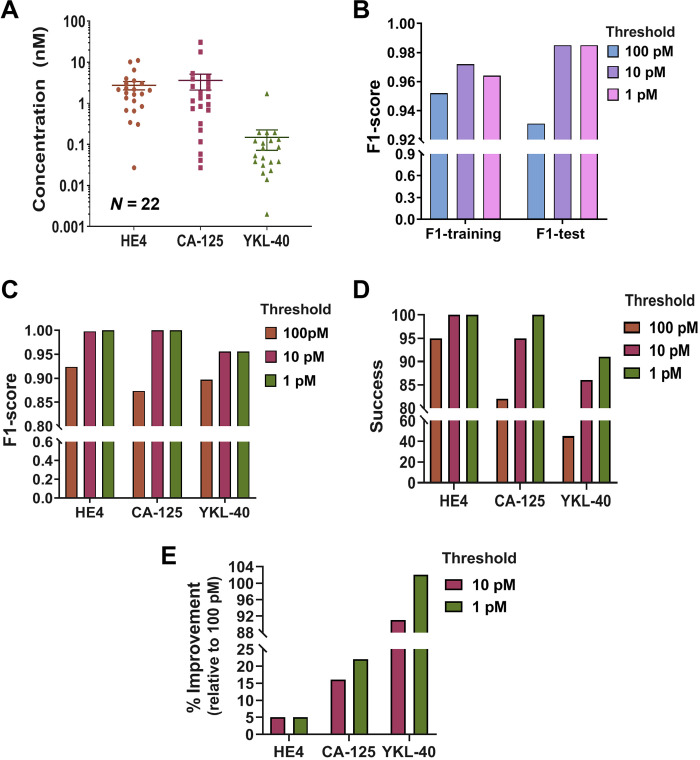
Uterine washing samples were collected from consenting cancer patients with diagnoses of several gynecologic conditions, including ovarian and endometrial cancers (fig. S6) (30, 62–64). To investigate the ability of the platform to detect multiple biomarkers in a patient biofluid sample, we tested the optimized platform in uterine washings. We incubated the DNA-SWCNT complexes in uterine lavage samples (N = 22). The conventional clinical laboratory measurements showed a high biomarker distribution (Fig. 5A), with mean concentration values of HE4, CA-125, and YKL-40 equaling 2.75 ± 0.63 nM, 3.62 ± 1.52 nM, and 0.15 ± 0.08 nM, respectively. Because of the subnanomolar range of the biomarkers in the patient samples, we retrained the algorithms with lower concentrations of all biomarkers (1 pM to 100 nM) and used the sensor array responses from the uterine lavage patient samples as a test set (Fig. 5B). The F1-score of the training set was improved as the threshold was decreased to less than 100 pM (0.95 increased to 0.97). In addition, the F1-score of the test set was strongly improved (0.93 increased to 0.99). This result indicates that several sample concentrations of less than 100 pM were inaccurately classified with the higher threshold. Note that there was a negligible difference between the F1-scores when using a 10 or 1 pM threshold. This may be due to the fact that there was only one measurement of less than 10 pM. We further evaluated the F1-score of individual biomarker predictions (Fig. 5C). When the threshold was decreased to 10 pM (>0.95), there were substantial improvements in the sensitivities to HE4, CA-125, and YKL-40 (0.92 increased to 1, 0.87 increased to 1, and 0.89 increased to 0.95 in HE4, CA-125, and YKL-40, respectively).
Fig. 5. Biomarker detection in uterine lavage samples.
(A) Concentrations of HE4, CA-125, and YKL-40 were measured by enzyme-linked immunosorbent assay in uterine lavage samples. (B) Classification F1-scores for detection of the three biomarkers in lavage samples from training and test datasets, applying different protein concentration thresholds. (C) Classification F1-scores of nanosensor detection of each biomarker, applying different protein concentration thresholds. (D) The success of the detection of each biomarker via classification, applying different concentration thresholds. (E) Improvement in classification success, relative to the threshold of 100 pM.
To evaluate the prediction strength, we examined the success of classification results for each threshold by comparing them to the actual levels of each biomarker measured by the clinical laboratory (Fig. 5D). We defined success as the percentage of correct classification (either true positive or true negative) for each biomarker. In all the biomarkers, the classification prediction was strongly improved when the threshold was decreased to less than 100 pM (Fig. 5E). HE4 presented the most successful classifications and showed improvement from 95 to 100% success with both 10 and 1 pM thresholds. CA-125 was initially predicted with 82% success but substantially improved to 100% success when the threshold was changed to 1 pM. The most substantial change was observed with YKL-40, from 50% success with a 100 pM threshold to 91% success with a 1 pM threshold. The platform was able to accurately classify the patient biofluid samples, indicated by the high F1-scores and classification success values.
DISCUSSION
Current diagnostic methodologies for protein biomarkers normally use one-to-one recognition assays, mainly using antibodies. Here, we described a new approach for the detection of multiple biomarkers in biofluids for disease diagnosis using an artificial molecular perception system. We developed an array of relatively nonspecific DNA-SWCNT sensors, containing individual hybrids of 132 DNA-SWCNTs. The use of multiple SWCNT chiralities enabled us to generate a large set of sensors that could be interrogated rapidly via high-throughput NIR spectroscopy to form a wide diversity of responses when they were exposed to different target proteins. On the basis of several studies (34, 65–67), we initially targeted gynecologic cancer biomarkers HE4, CA-125, and YKL-40.
The advantages of the method include the high optical sensitivity of SWCNTs to diverse analytes and the ability to modify their environmental sensitivities/specificities. We introduced a diverse set of SWCNT environmental responsivities via surface coatings of different DNA sequences that modulated the optical bandgaps and surface morphologies. Commercially available and well-characterized mixtures of SWCNT chiralities enable fast and robust preparation of multiple DNA-SWCNT complexes. These mixtures provide a broad group of features/examples for ML that can be fine-tuned later, based on feature importance analysis, eliminating the need to develop specific DNA-SWCNT complexes of isolated chiralities. ML algorithms enabled training from DNA-SWCNT spectral response data to detect biomarkers in both laboratory-generated samples and cancer patient uterine lavage samples. Feature importance analysis showed that the intensity ratios contributed most to platform predictions, as compared to wavelength shifts. We believe that this phenomenon may result from the fact that SWCNT intensity exhibits greater sensitivity to more physicochemical phenomena than emission wavelength (39, 42, 68).
Notably, the classification success rate in patient samples was high even in subnanomolar ranges, with a rate of 100% for HE4 and CA-125 and 91% in YKL-40. These results support the conclusion that the perception mode of sensing can successfully generate accurate predictions.
Ideally, classification and regression algorithms would be trained with sensor data using biomarker concentrations at a similar order of magnitude as known concentrations from published clinical data. However, in some cases, there is no way to know the biomarker concentration in advance; several iterations may be needed to optimize the sensitivity of the platform. Combining detection and quantification will allow this technology to better screen and categorize patients based on the levels of markers for early detection.
This platform could be continuously improved by increasing the sizes of datasets and analyzing feature importance. For example, interpreting the feature importance values can aid with DNA sequence design and expanding the library of DNA-SWCNT complexes. In addition, expanding the spectroscopic range may increase the number of SWCNT chiralities and, thus, sensors that can be measured. While increasing the number of features (nanosensors) may contribute to the sensitivity of the platform, the number of examples (conditions) should be increased as well (to prevent overfitting). We also recognize the need to increase the number of patient samples to continually validate and increase the robustness of the model.
We believe that this platform can be translated to the clinic for use in laboratory medicine or point-of-care settings. While the development of the platform requires high-throughput NIR screening for the training of ML algorithms, we showed that comprehensive analysis of both the FV construction and feature importance values can reduce the required DNA-SWCNT array to detect biomarkers in serum samples. By studying the feature importance values obtained from the algorithms trained with FV2 (sequence-based), we found that the intensity ratios contributed more to the overall prediction than the wavelength shifts. These insights have important outcomes for potentially translating this platform to the clinic. For example, using single SWCNT chiralities and multiple DNA sequences or analyzing only intensity ratio for several SWCNT chiralities, we can reduce the optical configuration to few excitation wavelengths, requiring filters and single-channel detectors, rather than an advanced spectrometer. This enables the use of simpler, more portable optical instrumentation. However, note that the reduction of chiralities also reduces the number of examples and can increase the risk of overfitting. To avoid this problem, the number of examples should always be maximized and compared to the number of features.
Last, because of the flexibility and the nonspecific nature of the individual sensor elements, the proposed platform is not restricted to ovarian cancer biomarkers and can potentially be trained to detect other disease biomarkers without the need to engineer different arrays of nanosensors. This platform enables antibody-free detection that would be useful when an especially robust or long-term measurement is needed, such as in wearable/implantable devices, point-of-care diagnostics, and for underresourced situations where cold chain storage may not be available.
MATERIALS AND METHODS
Materials
SWCNTs produced by the high-pressure carbon monoxide (HiPco) process were purchased from Unidym (Sunnyvale, CA, USA). Single-stranded DNA oligonucleotides [T3C3T3C3T3, C3T9C3, C3T3C9, (TCT)5, (GT)12, (AT)11, (AT)15, (AT)20, (ATT)4, (AC)15, and CT2C3T2C] were purchased from Integrated DNA Technologies (Coralville, IA, USA). HE4 was purchased from RayBiotech. Human CA-125, also known as MUC16, and human YKL-40 were purchased from R&D Systems. BSA was purchased from Sigma-Aldrich. FBS was purchased from Thermo Fisher Scientific. Uterine washings from the Institutional Review Board–consented patients with cancer were provided by the Department of Laboratory Medicine at Memorial Sloan Kettering.
Preparation of DNA-SWCNT complexes
SWCNTs were mixed with a specific DNA oligonucleotide at a 1:2 mass ratio, respectively, in 1 ml of IDTE buffer. The sample was ultrasonicated continuously for 45 min at 40% of maximum amplitude, using a 3-mm titanium tip (Sonics Vibra-Cell). The mixture was ultracentrifuged (Sorvall Discovery 90SE) for 30 min at 250,000g. The top 80% of the supernatant was collected, and the concentration of suspended SWCNTs was determined by UV-Vis-NIR spectrophotometry (JASCO V-670) using the extinction coefficient A910 = 0.02554 liter mg−1 cm−1; where the path length l is 1 cm. To remove excess free DNA, 300 μl of the sample was filtered twice using a 100-kDa Amicon centrifuge filter (Millipore) at 5000g for 10 min. Following filtration, the DNA-SWCNT complexes were tested at a concentration of 5 mg/liter of SWCNT in 100 μl of solution in 96-well plates. The zeta potential of the DNA-SWCNT complexes was measured using a Zetasizer ZSP (Malvern). The samples were diluted to a concentration of 0.5 mg/liter using double-distilled water.
NIR fluorescence spectroscopy of DNA-SWCNTs
NIR fluorescence spectroscopy was used to measure the photoluminescence emission from the DNA-SWCNT complexes, as described previously (69). For solution measurements, spectra were acquired using an apparatus built in-house consisting of a continuous-wave 730-nm diode laser with an output power of 2 W or a SuperK EXTREME supercontinuum white-light laser source connected to a Varia variable bandpass filter accessory capable of tuning the output from 490 to 825 nm with a bandwidth of 20 nm (NKT Photonics). The laser was injected into a multimode fiber that was fed into the back of an Olympus IX-71 inverted microscope where it passed through a 20× LCPlan N, 20×/0.45 objective (Olympus, USA) and a dichroic mirror (875-nm cutoff; Semrock). The light was f-number matched to the spectrometer using several lenses and injected into an IsoPlane spectrograph (Princeton Instruments) with a slit width of 410 μm, which dispersed the emission using a 86 g mm−1 gating with a 950-nm blaze wavelength coupled to a NIRvana 2D InGaAs NIR detector (Princeton Instruments) or a Shamrock 303 spectrometer with the Andor iDus 1D InGaAs Array Camera (Oxford Instruments). An HL-3-CAL EXT halogen calibration light source (Ocean Optics) was used to correct for wavelength-dependent features in the emission intensity arising from the excitation power, spectrometer, detector, and other optics. A Hg/Ne pencil-like calibration lamp (Newport) was used to calibrate spectrometer wavelengths. Data were obtained from each well of a 96-well plate using the custom LabVIEW (National Instruments) code. Another custom program, written in MATLAB (MathWorks) software was used to subtract background, correct for abnormalities in excitation profiles, and fit the data with Lorentzian functions. Smoothing, where applicable, was done by applying a Savitzky-Golay filter.
Atomic force microscopy
DNA-SWCNT complexes were plated on a freshly cleaved mica substrate (Structure Probe, Inc) for 4 min before washing with 10 ml of distilled water and blowing dry with argon gas. An Asylum Research MFP-3D-Bio instrument equipped with an Olympus AC240TS AFM probe in alternating current mode was used. Data were acquired at 2.93 nm pixel −1 x-y resolution and 15.63 pm of z resolution. The images were analyzed using Gwyddion software. To measure height or length distributions, at least 20 ROIs were analyzed.
ML method development
The dataset comprises the photoluminescence spectra of each combination of DNA-SWCNT complex exposed to different combinations of a small number of analytes (HE4, CA-125, YKL-40, BSA, and FBS). That is, we had total N·M·L combinations where N is the number of DNA sequences, M is the number of SWCNT chiralities, and L is the number of analyte combinations. The spectra were analyzed to yield two parameters for each SWCNT type: the wavelength peak shift (Δλ) and intensity ratio (IR)
| (1) |
and
| (2) |
where λ0 and I0 are the wavelength and intensity of a control sample (DNA-SWCNT without analyte); λi and Ii are the wavelength and intensity of DNA-SWCNT with analyte combination, i.
Input and output (target) variables were identified for the ML algorithms. The input variables include DNA sequence, SWCNT chirality, and the two spectroscopically measured parameters (Δλi, IR). The output variable either represents the presence (for classification) or concentration (for regression) of each analyte. Three classification approaches were examined: biclass (±biomarker), multiclass (±biomarker combination), and multilabel (±each biomarker).
To train the models, categorical data (such as SWCNT chirality and analyte type) were transformed to numeric values, using the one-hot encoding technique (57). DNA sequences were encoded as term-frequency vectors, using subsets of two or three bases as a term and calculating the frequency of that term in the sequence (48). Figure S3 depicts the overall scheme for the input feature construction. Two FVs were constructed to emphasize the sensitivity of each component in the DNA/SWCNT complex and find a balance between the number of features and examples. For each FV, one component of the DNA/SWCNT complex was encoded as an ID of the example, while the other components’ responses were defined as features. In FV1, the DNA sequences were encoded as the IDs, and the chirality-dependent optical responses were encoded as features. In FV2, the SWCNT chiralities were encoded as the IDs, and the sequence-dependent optical responses were encoded as features.
Three algorithms—SVM, RF, and ANN—were trained and tested with FV1 and FV2 for both classification and regression. Each model was evaluated by 10-fold cross-validation. All ML algorithms were implemented using the Scikit-learn ML library (58). To find hyperparameters that maximize performance, Bayesian hyperparameter optimization was implemented using HyperOpt (70).
Each model was evaluated by the produced F1-score and accuracy values for classification and R2 value for regression. Accuracy (Eq. 3) calculates the percentage of correctly classified examples. F1-score, which is a composite value of precision (Eq. 4) and recall (Eq. 5), gives a measure of accuracy but takes the false positives and negatives into account as well (Eq. 6)
| (3) |
| (4) |
| (5) |
| (6) |
CA-125 concentration unit conversion
The concentration unit of CA-125 used in the clinic, units per milliliter, was converted to nanomolar by titrating CA-125 and measuring using an ARC i2000 instrument. A linear concentration curve with R2 = 0.9997 was generated. The resulting unit conversion is as follows: [U/ml] = 0.18*[pM].
Statistical analysis
In vitro experiments were analyzed by two-sided t tests. Reported P values were assigned ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05, and exact P values are reported in the captions.
Acknowledgments
We would like to thank the Molecular Cytology Core Facility at Memorial Sloan Kettering Cancer Center and B. Wang for acquiring the DNA-SWCNT AFM images. We would like to thank Biorender.com for serving as a platform for the scientific illustration.
Funding: This work was supported in part to D.A.H. by the NIH New Innovator Award (DP2-HD075698), NCI (R01-CA215719), the Cancer Center Support Grant (P30 CA008748), the National Science Foundation CAREER Award (1752506), the Honorable Tina Brozman Foundation for Ovarian Cancer Research, the American Cancer Society Research Scholar Grant (GC230452), the Pershing Square Sohn Cancer Research Alliance, the Expect Miracles Foundation–Financial Services Against Cancer, the Cycle for Survival’s Equinox Innovation Award in Rare Cancers, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center, the Kelli Auletta Fund, and the Center for Molecular Imaging and Nanotechnology of Memorial Sloan Kettering Cancer Center, Functional Genomics Initiative, the Alan and Sandra Gerry Metastasis, and Tumor Ecosystems Center. Z.Y. was supported by the Ann Schreiber Mentored Investigator Award (Ovarian Cancer Research Fund) and Young Investigator 2019 (Kaleidoscope of Hope). Y.Y. was supported by a Dean’s Fellowship at Lehigh University. A.J.’s contributions are part of the NHI initiative at Lehigh University. M.Z. acknowledges NIST internal funding for support. D.A.L. is supported by U.S. Department of Defense Award: W81XWH-15-1-0429, and Arnold Chavkin and Laura Chang.
Author contributions: Z.Y., Y.Y., A.J., M.Z., and D.A.H. conceived the research and the experimental design. Z.Y., Y.Y., E.A., A.H.S., Q.C., and W.C. performed the experiments and data analysis. D.A.L., M.F., and L.R. obtained and handled the patient samples. A.J., M.Z., and D.A.H. supervised the research. Z.Y., E.A., and D.A.H. wrote the original manuscript. All authors provided input and feedback for manuscript preparation.
Competing interests: D.A.H. is co-founder and officer with an equity interest in Goldilocks Therapeutics Inc., LipidSense Inc., and Nirova Biosense Inc. and a member of the scientific advisory boards of Concarlo Holdings LLC, Nanorobotics Inc., and Mediphage Bioceuticals Inc. D.A.L. has a consulting/advisory role for Tesaro/GSK and Merck and receives research funding to the institution from Merck, Tesaro, Clovis Oncology, Regeneron, Agenus, Takeda, Immunogen, VBL Therapeutics, Genentech, Celsion, Ambry, and Splash Pharmaceuticals. He also is a co-founder with an equity interest in Nirova BioSense Inc. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sample datasets and scripts are available at the following repository (https://doi.org/10.5281/zenodo.5495768). They are also available at https://bitbucket.org/jagotagrouplehigh/machine-perception-nanosensor-platform.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 and S2
REFERENCES AND NOTES
- 1.Holford T. R., Davis F., Higson S. P., Recent trends in antibody based sensors. Biosens. Bioelectron. 34, 12–24 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Lipshutz R. J., Fodor S. P., Gingeras T. R., Lockhart D. J., High density synthetic oligonucleotide arrays. Nat. Genet. 21, 20–24 (1999). [DOI] [PubMed] [Google Scholar]
- 3.C. Chandola, M. Neerathilingam, Aptamers for targeted delivery: Current challenges and future opportunities, in Role of Novel Drug Delivery Vehicles in Nanobiomedicine (IntechOpen, 2020). [Google Scholar]
- 4.Ni S., Zhuo Z., Pan Y., Yu Y., Li F., Liu J., Wang L., Wu X., Li D., Wan Y., Zhang L., Yang Z., Zhang B.-T., Lu A., Zhang G., Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mater. Interfaces 13, 9500–9519 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Kim J., Campbell A. S., de Avila B. E., Wang J., Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 37, 389–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M., Reproducibility crisis: Blame it on the antibodies. Nature 521, 274–276 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Bradbury A., Pluckthun A., Reproducibility: Standardize antibodies used in research. Nature 518, 27–29 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Yoo H., Jo H., Oh S. S., Detection and beyond: Challenges and advances in aptamer-based biosensors. Mater. Adv. 1, 2663–2687 (2020). [Google Scholar]
- 9.Shrivastava S., Trung T. Q., Lee N. E., Recent progress, challenges, and prospects of fully integrated mobile and wearable point-of-care testing systems for self-testing. Chem. Soc. Rev. 49, 1812–1866 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Fan R., Andrew T. L., Perspective—Challenges in developing wearable electrochemical sensors for longitudinal health monitoring. J. Electrochem. Soc. 167, 037542 (2020). [Google Scholar]
- 11.Malnic B., Hirono J., Sato T., Buck L. B., Combinatorial receptor codes for odors. Cell 96, 713–723 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Veenstra T. D., Conrads T. P., Hood B. L., Avellino A. M., Ellenbogen R. G., Morrison R. S., Biomarkers: Mining the biofluid proteome. Mol. Cell. Proteomics 4, 409–418 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Stern E., Vacic A., Rajan N. K., Criscione J. M., Park J., Ilic B. R., Mooney D. J., Reed M. A., Fahmy T. M., Label-free biomarker detection from whole blood. Nat. Nanotechnol. 5, 138–142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang L., Tsao D. Y., The code for facial identity in the primate brain. Cell 169, 1013–1028.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta S., Stevens C. F., Navlakha S., A neural algorithm for a fundamental computing problem. Science 358, 793–796 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Persaud K., Dodd G., Analysis of discrimination mechanisms in the mammalian olfactory system using a model nose. Nature 299, 352–355 (1982). [DOI] [PubMed] [Google Scholar]
- 17.Freund M. S., Lewis N. S., A chemically diverse conducting polymer-based "electronic nose". Proc. Natl. Acad. Sci. 92, 2652–2656 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson A. D., Baietto M., Applications and advances in electronic-nose technologies. Sensors 9, 5099–5148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurs P. C., Bakken G., McClelland H., Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem. Rev. 100, 2649–2678 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Staii C., Johnson A. T., Chen M., Gelperin A., DNA-decorated carbon nanotubes for chemical sensing. Nano Lett. 5, 1774–1778 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Zhou H., Baldini L., Hong J., Wilson A. J., Hamilton A. D., Pattern recognition of proteins based on an array of functionalized porphyrins. J. Am. Chem. Soc. 128, 2421–2425 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Hayasaka T., Lin A., Copa V. C., Lopez L. P., Loberternos R. A., Ballesteros L. I. M., Kubota Y., Liu Y., Salvador A. A., Lin L., An electronic nose using a single graphene FET and machine learning for water, methanol, and ethanol. Microsyst. Nanoeng. 6, 50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shumeiko V., Paltiel Y., Bisker G., Hayouka Z., Shoseyov O., A nanoscale paper-based near-infrared optical nose (NIRON). Biosens. Bioelectron. 172, 112763 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Vaughan S., Coward J. I., Bast R. C. Jr., Berchuck A., Berek J. S., Brenton J. D., Coukos G., Crum C. C., Drapkin R., Etemadmoghadam D., Friedlander M., Gabra H., Kaye S. B., Lord C. J., Lengyel E., Levine D. A., McNeish I. A., Menon U., Mills G. B., Nephew K. P., Oza A. M., Sood A. K., Stronach E. A., Walczak H., Bowtell D. D., Balkwill F. R., Rethinking ovarian cancer: Recommendations for improving outcomes. Nat. Rev. Cancer 11, 719–725 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maringe C., Walters S., Butler J., Coleman M. P., Hacker N., Hanna L., Mosgaard B. J., Nordin A., Rosen B., Engholm G., Gjerstorff M. L., Hatcher J., Johannesen T. B., McGahan C. E., Meechan D., Middleton R., Tracey E., Turner D., Richards M. A., Rachet B., ICBP Module 1 Working Group, Stage at diagnosis and ovarian cancer survival: Evidence from the international cancer benchmarking partnership. Gynecol. Oncol. 127, 75–82 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Raamanathan A., Simmons G. W., Christodoulides N., Floriano P. N., Furmaga W. B., Redding S. W., Lu K. H., Bast R. C., McDevitt J. T., Programmable bio-nano-chip systems for serum CA125 quantification: Toward ovarian cancer diagnostics at the point-of-care. Cancer Prev. Res. 5, 706–716 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han C., Bellone S., Siegel E. R., Altwerger G., Menderes G., Bonazzoli E., Egawa-Takata T., Pettinella F., Bianchi A., Riccio F., Zammataro L., Yadav G., Marto J. A., Penet M. F., Levine D. A., Drapkin R., Patel A., Litkouhi B., Ratner E., Silasi D. A., Huang G. S., Azodi M., Schwartz P. E., Santin A. D., A novel multiple biomarker panel for the early detection of high-grade serous ovarian carcinoma. Gynecol Oncol. 149, 585–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaroli D., Coukos G., Scholler N., Beyond CA125: The coming of age of ovarian cancer biomarkers. Are we there yet? Biomark. Med 3, 275–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coll-De La Rubia E., Martinez-Garcia E., Dittmar G., Gil-Moreno A., Cabrera S., Colas E., Prognostic biomarkers in endometrial cancer: A systematic review and meta-analysis. J. Clin. Med. 9, 1900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutt S., Tailor A., Ellis P., Michael A., Butler-Manuel S., Chatterjee J., The role of biomarkers in endometrial cancer and hyperplasia: A literature review. Acta Oncol. 58, 342–352 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Brown P. O., Palmer C., The preclinical natural history of serous ovarian cancer: Defining the target for early detection. PLoS Med. 6, e1000114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buys S. S., Partridge E., Black A., Johnson C. C., Lamerato L., Isaacs C., Reding D. J., Greenlee R. T., Yokochi L. A., Kessel B., Crawford E. D., Church T. R., Andriole G. L., Weissfeld J. L., Fouad M. N., Chia D., O’Brien B., Ragard L. R., Clapp J. D., Rathmell J. M., Riley T. L., Hartge P., Pinsky P. F., Zhu C. S., Izmirlian G., Kramer B. S., Miller A. B., Xu J.-L., Prorok P. C., Gohagan J. K., Berg C. D.; PLCO Project Team , Effect of screening on ovarian cancer mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening randomized controlled trial. JAMA 305, 2295–2303 (2011). [DOI] [PubMed] [Google Scholar]
- 34.D. La. evine, Detection of ovarian cancer. U.S. Patent 20,130,078,319 (2013).
- 35.Zaporotskova I. V., Boroznina N. P., Parkhomenko Y. N., Kozhitov L. V., Carbon nanotubes: Sensor properties. A review. Modern Electronic Mater. 2, 95–105 (2016). [Google Scholar]
- 36.Heller D. A., Pratt G. W., Zhang J., Nair N., Hansborough A. J., Boghossian A. A., Reuel N. F., Barone P. W., Strano M. S., Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proc. Natl. Acad. Sci. U.S.A. 108, 8544–8549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Boghossian A. A., Barone P. W., Rwei A., Kim J.-H., Lin D., Heller D. A., Hilmer A. J., Nair N., Reuel N. F., Strano M. S., Single molecule detection of nitric oxide enabled by d(AT)15DNA adsorbed to near infrared fluorescent single-walled carbon nanotubes. J. Am. Chem. Soc. 133, 567–581 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Harvey J. D., Jena P. V., Baker H. A., Zerze G. H., Williams R. M., Galassi T. V., Roxbury D., Mittal J., Heller D. A., A carbon nanotube reporter of microRNA hybridization events in vivo. Nat. Biomed. Eng. 1, 0041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaari Z., Cheung J. M., Baker H. A., Frederiksen R. S., Jena P. V., Horoszko C. P., Jiao F., Scheuring S., Luo M., Heller D. A., Nanoreporter of an enzymatic suicide inactivation pathway. Nano Lett. 20, 7819–7827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galassi T. V., Antman-Passig M., Yaari Z., Jessurun J., Schwartz R. E., Heller D. A., Long-term in vivo biocompatibility of single-walled carbon nanotubes. PLOS ONE 15, e0226791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams R. M., Lee C., Galassi T. V., Harvey J. D., Leicher R., Sirenko M., Dorso M. A., Shah J., Olvera N., Dao F., Levine D. A., Heller D. A., Long-term in vivo biocompatibility of single-walled carbon nanotubes. Sci. Adv. 4, eaaq1090 (2018).29675469 [Google Scholar]
- 42.Horoszko C. P., Jena P. V., Roxbury D., Rotkin S. V., Heller D. A., Optical voltammetry of polymer-encapsulated single-walled carbon nanotubes. J. Phys. Chem. C 123, 24200–24208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka Y., Hirana Y., Niidome Y., Kato K., Saito S., Nakashima N., Experimentally determined redox potentials of individual (n,m) single-walled carbon nanotubes. Angew. Chem. Int. Ed. Engl. 48, 7655–7659 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Heller Daniel A., Jeng Esther S., Yeung T.-K., Martinez Brittany M., Moll Anthonie E., Gastala Joseph B., Strano Michael S., Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science 311, 508–511 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Roxbury D., Jena P. V., Shamay Y., Horoszko C. P., Heller D. A., Cell membrane proteins modulate the carbon nanotube optical bandgap via surface charge accumulation. ACS Nano 10, 499–506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng M., Jagota A., Strano M. S., Santos A. P., Barone P., Chou S. G., Diner B. A., Dresselhaus M. S., McLean R. S., Onoa G. B., Samsonidze G. G., Semke E. D., Usrey M., Walls F. J., Structure-based carbon nanotube sorting by sequence-dependent DNA assembly. Science 302, 1545–1548 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Jena P. V., Roxbury D., Galassi T. V., Akkari L., Horoszko C. P., Iaea D. B., Budhathoki-Uprety J., Pipalia N., Haka A. S., Harvey J. D., Mittal J., Maxfield F. R., Joyce J. A., Heller D. A., A carbon nanotube optical reporter maps endolysosomal lipid flux. ACS Nano 11, 10689–10703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Zheng M., Jagota A., Learning to predict single-wall carbon nanotube-recognition DNA sequences. Npj Comput. Mater. 5, 3 (2019). [Google Scholar]
- 49.Landry M. P., Ando H., Chen A. Y., Cao J., Kottadiel V. I., Chio L., Yang D., Dong J., Lu T. K., Strano M. S., Single-molecule detection of protein efflux from microorganisms using fluorescent single-walled carbon nanotube sensor arrays. Nat. Nanotechnol. 12, 368–377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ao G., Streit J. K., Fagan J. A., Zheng M., Differentiating left- and right-handed carbon nanotubes by DNA. J. Am. Chem. Soc. 138, 16677–16685 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Yang Y., Shankar A., Aryaksama T., Zheng M., Jagota A., Quantification of DNA/SWCNT solvation differences by aqueous two-phase separation. Langmuir 34, 1834–1843 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Zheng M., Jagota A., Semke E. D., Diner B. A., McLean R. S., Lustig S. R., Richardson R. E., Tassi N. G., DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater. 2, 338–342 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Roxbury D., Tu X., Zheng M., Jagota A., Recognition ability of DNA for carbon nanotubes correlates with their binding affinity. Langmuir 27, 8282–8293 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Manohar S., Tang T., Jagota A., Structure of homopolymer DNA−CNT hybrids. J. Phys. Chem. C 111, 17835–17845 (2007). [Google Scholar]
- 55.Expacy (https://web.expasy.org/protparam/).
- 56.UniProt (www.uniprot.org/uniprot).
- 57.D. M. Harris, S. Harris, Introductory digital design & computer architecture curriculum, in 2013 IEEE International Conference on Microelectronic Systems Education (MSE), Austin, TX, USA, 2 to 3 June 2013. [Google Scholar]
- 58.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay E., Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 59.Zhang M., Zhou Z., A review on multi-label learning algorithms. IEEE Trans. Knowl. Data Eng. 26, 1819–1837 (2014). [Google Scholar]
- 60.Hamada N., Sawada S.-I., Oshiyama A., New one-dimensional conductors: Graphitic microtubules. Phys. Rev. Lett. 68, 1579–1581 (1992). [DOI] [PubMed] [Google Scholar]
- 61.Sfeir M. Y., Optical spectroscopy of individual single-walled carbon nanotubes of defined chiral structure. Science 312, 554–556 (2006). [DOI] [PubMed] [Google Scholar]
- 62.D. Daley-Brown, G. Oprea-Ilies, A. Quarshie, R. R. Gonzalez-Perez, Emerging biomarkers and clinical implications in endometrial carcinoma, in Role of Biomarkers in Medicine (InTech, 2016). [Google Scholar]
- 63.Li J., Dowdy S., Tipton T., Podratz K., Lu W.-G., Xie X., Jiang S.-W., HE4 as a biomarker for ovarian and endometrial cancer management. Expert. Rev. Mol. Diagn. 9, 555–566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mutz-Dehbalaie I., Egle D., Fessler S., Hubalek M., Fiegl H., Marth C., Widschwendter A., HE4 is an independent prognostic marker in endometrial cancer patients. Gynecol. Oncol. 126, 186–191 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Barnabas G. D., Bahar-Shany K., Sapoznik S., Helpman L., Kadan Y., Beiner M., Weitzner O., Arbib N., Korach J., Perri T., Katz G., Blecher A., Brandt B., Friedman E., Stockheim D., Jakobson-Setton A., Eitan R., Armon S., Brand H., Zadok O., Aviel-Ronen S., Harel M., Geiger T., Levanon K., Microvesicle proteomic profiling of uterine liquid biopsy for ovarian cancer early detection. Mol. Cell. Proteomics 18, 865–875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maritschnegg E., Heitz F., Pecha N., Bouda J., Trillsch F., Grimm C., Vanderstichele A., Agreiter C., Harter P., Obermayr E., Vergote I., Zeillinger R., Speiser P., Uterine and tubal lavage for earlier cancer detection using an innovative catheter: A feasibility and safety study. Int. J. Gynecol. Cancer 28, 1692–1698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maritschnegg E., Wang Y. X., Pecha N., Horvat R., Van Nieuwenhuysen E., Vergote I., Heitz F., Sehouli J., Kinde I., Diaz L. A., Papadopoulos N., Kinzler K. W., Vogelstein B., Speiser P., Zeillinger R., Lavage of the uterine cavity for molecular detection of müllerian duct carcinomas: A proof-of-concept study. J. Clin. Oncol. 33, 4293–4300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heller D. A., Jin H., Martinez B. M., Patel D., Miller B. M., Yeung T. K., Jena P. V., Hobartner C., Ha T., Silverman S. K., Strano M. S., Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nat. Nanotechnol. 4, 114–120 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Roxbury D., Jena P. V., Williams R. M., Enyedi B., Niethammer P., Marcet S., Verhaegen M., Blais-Ouellette S., Heller D. A., Hyperspectral microscopy of near-infrared fluorescence enables 17-chirality carbon nanotube imaging. Sci. Rep. 5, 14167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Bergstra, D. Yamins, D. D. Cox, Making a science of model search: Hyperparameter optimization in hundreds of dimensions for vision architectures, in Proceedings of the 30th International Conference on International Conference on Machine Learning - Volume 28 (JMLR.org: Atlanta, GA, USA, 2013), pp I–115–I–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 and S2