R-loops lead to chronic inflammation upon DNA damage.
Abstract
How DNA damage leads to chronic inflammation and tissue degeneration with aging remains to be fully resolved. Here, we show that DNA damage leads to cellular senescence, fibrosis, loss-of-tissue architecture, and chronic pancreatitis in mice with an inborn defect in the excision repair cross complementation group 1 (Ercc1) gene. We find that DNA damage-driven R-loops causally contribute to the active release and buildup of single-stranded DNAs (ssDNAs) in the cytoplasm of cells triggering a viral-like immune response in progeroid and naturally aged pancreata. To reduce the proinflammatory load, we developed an extracellular vesicle (EV)-based strategy to deliver recombinant S1 or ribonuclease H nucleases in inflamed Ercc1−/− pancreatic cells. Treatment of Ercc1−/− animals with the EV-delivered nuclease cargo eliminates DNA damage-induced R-loops and cytoplasmic ssDNAs alleviating chronic inflammation. Thus, DNA damage-driven ssDNAs causally contribute to tissue degeneration, Ercc1−/− paving the way for novel rationalized intervention strategies against age-related chronic inflammation.
INTRODUCTION
The great majority of premature aging-like (progeroid) syndromes are associated with inborn defects in genome maintenance, supporting the notion that DNA damage drives aging and causally contributes to age-related diseases (1–3). To counteract DNA damage, mammalian cells rely on a series of partially overlapping DNA repair systems to remove DNA lesions that may block transcription or DNA replication. For bulky helix-distorting damage, such as the main ultraviolet (UV)–induced lesions, the principal repair mechanism is the evolutionarily conserved nucleotide excision repair (NER) pathway (4, 5). NER operates via a “cut and patch” type of mechanism involving ~30 proteins that recognize and remove helical distortions throughout the genome, i.e., global genome NER, or selectively from the transcribed strand of active genes, i.e., transcription-coupled NER (6, 7). In humans, defects in NER are causally linked to mutagenesis and cancer initiation as in the cancer-prone syndrome xeroderma pigmentosum (XP; complementation groups XP-A to XP-G) or to developmental and neuronal abnormalities as seen in a heterogeneous group of premature aging-like (progeroid) syndromes, including the Cockayne syndrome (affected genes: Csa and Csb), trichothiodystrophy (affected genes: Xpb and Xpd), or the XPF-ERCC1 syndrome (XFE; affected genes: Ercc1 and Xpf) (2). We and others have recently shown that DNA damage–driven inflammation contributes substantially in NER progeria and age-related degenerative diseases, but the mechanisms remain unresolved (8–11). Xeroderma pigmentosum F - excision repair cross complementation group (XPF-ERCC1) is a heterodimeric, structure-specific endonuclease complex required for lesion excision in NER (4, 5) that plays an analogous role in the repair of highly cytotoxic DNA interstrand cross-links (ICLs) (12).
Using ERCC1-defective animal models of the human progeroid syndrome XFE (13), we show that persistent DNA damage leads to the premature onset of chronic pancreatitis in the DNA repair–deficient animals. The gradual accumulation of irreparable DNA lesions triggers the formation of R-loops that causally contribute to the release and buildup of single-stranded DNA (ssDNA) fragments in the cytoplasm of cells, stimulating a viral-like immune response in progeroid Ercc1−/− and naturally aged pancreata. To lessen the proinflammatory load, we developed an extracellular vesicle (EV)–based strategy to deliver recombinant ribonuclease (RNase) H or S1 nuclease in inflamed Ercc1−/− pancreatic cells in vitro and in vivo. We find that treatment with the EV-delivered nuclease cargo rapidly removes R-loops and the ssDNA moieties in the cytoplasm of pancreatic cells, alleviating the proinflammatory response seen in progeroid Ercc1−/− animals. Thus, the development of rationalized intervention regimens against DNA damage–driven cytoplasmic ssDNAs may be a promising therapeutic strategy against chronic inflammation and tissue degeneration in NER progeria and likely during aging.
RESULTS
Persistent DNA damage associates with chronic pancreatitis in Ercc1−/− mice
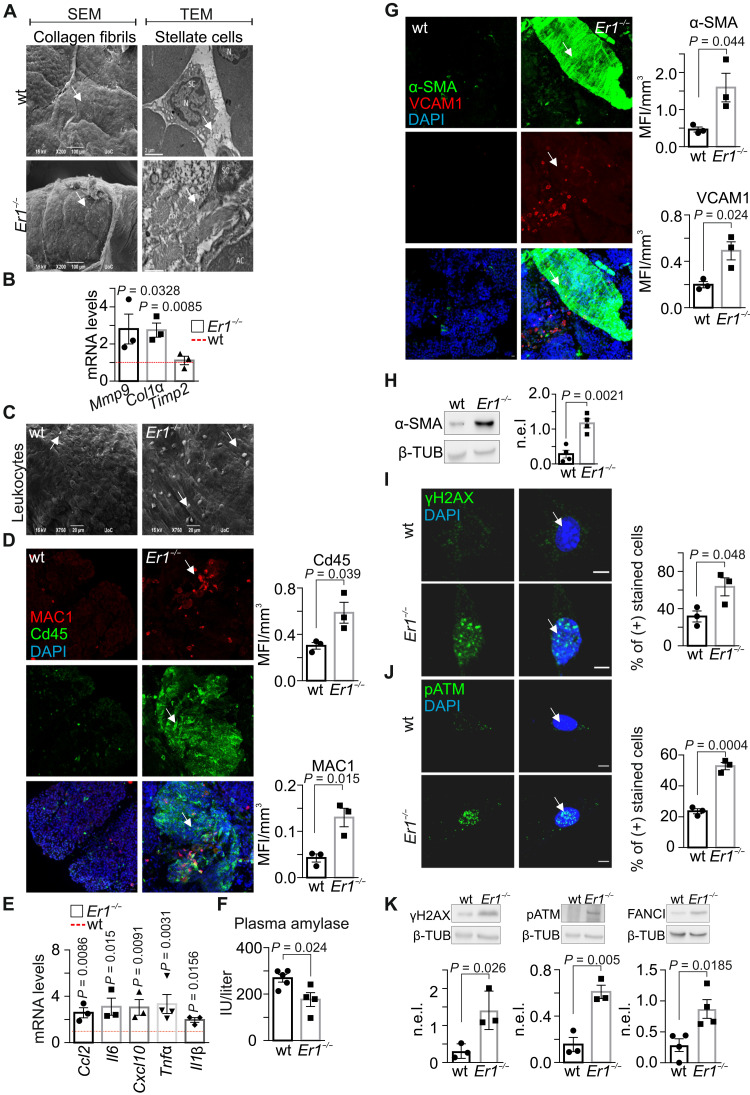
DNA damage causally contributes to aging, as exemplified by the growing list of progeroid DNA repair–deficient syndromes with age-related metabolic and endocrine defects (1, 10, 14–16). To gain insight into the causal contribution of DNA damage in tissue-degenerative changes in the pancreas, we studied the pancreata of 15-day-old DNA repair–deficient Ercc1−/− and age-matched wild-type (wt) littermate control mice. Scanning and transmission electron micrographs (SEM and TEM) reveal loss-of-tissue architecture and fibrosis with frequent and significantly denser collagen fibrils, observed in the vicinity of stellate cells in Ercc1−/− pancreata (Fig. 1A) but not in the pancreata of the NER-defective Csbm/m, Xpa−/−, or Xpc−/−, known to manifest a milder phenotype, or the wt corresponding control animals (fig. S1A). Fibrosis in Ercc1−/− pancreata is accompanied by an increase in the mRNA levels of collagen 1a (Col1a) and matrix metalloproteinase 9 (Mmp-9) genes and no changes in the mRNA levels of the tissue inhibitor of metalloproteinases 2 (Timp2) gene (Fig. 1B). Likewise, we find a significant increase in the mRNA levels of Col1a and Mmp-9 genes in the pancreata of severely progeroid P15 Csbm/m;Xpc−/− double-mutant (DKO) mice. Consistently, our analysis revealed normative mRNA levels for the Timp2 gene in the DKO or single-mutant pancreata (fig. S1B). Further SEM studies, as well as immunostaining against CD45 (for leukocytes) or MAC1 (for macrophages), and quantitative polymerase chain reaction (qPCR) analysis reveal the presence of lymphocytic infiltrates (Fig. 1, C and D) and a significant increase in Ccl2, Il6, Cxcl10, Tnf-α, and Il1β mRNA levels (Fig. 1E) in P15 Ercc1−/− pancreata compared to wt controls. Amylase, a pancreatic enzyme that catalyzes the hydrolysis of starch into smaller carbohydrate molecules, is typically released into the bloodstream in case of inflammation. We find that the pancreatic tissue amylase levels are comparable between Ercc1−/− and wt pancreata (fig. S1, C and D). However, the serum amylase levels are significantly lower in the DNA repair–deficient animals compared to wt controls (Fig. 1F), indicating the premature onset of chronic pancreatitis and permanent tissue damage (17). Immunostaining and Western blotting for α–smooth muscle actin (α-SMA), a marker for the activation of pancreatic stellate cells, reveal the marked increase in α-SMA protein levels in Ercc1−/− pancreata (Fig. 1, G and H) and in primary pancreatic cells (PPCs) (fig. S1E) compared to corresponding wt controls. Likewise, we find that the vascular cell adhesion molecule 1 (VCAM-1), a cytokine-inducible endothelial CAM, is overexpressed in Ercc1−/− pancreata (Fig. 1G). Phosphorylated histone H2AX (γH2AX)–containing foci accumulate at sites of DNA breaks (18). Consistently, the number of γH2AX-positive nuclei was significantly higher in the DNA repair–defective Ercc1−/− PPCs (Fig. 1I) compared to wt control cells. We also find marked differences in the number of positively stained nuclei for pATM (phosphorylated ataxia talangiectasia-mutated), a central mediator of the DNA damage response (DDR; Fig. 1J), and an increase in the protein levels of γH2AX, pATM, as well as Fanconi anemia complementation group I (FANCI), known to be involved in the repair of DNA ICLs (Fig. 1K; as indicated) (19). Thus, an inborn ERCC1 defect leads to persistent DNA damage accumulation associated with aberrant tissue morphological changes and chronic inflammation in Ercc1−/− pancreata.
Fig. 1. Pancreatic fibrosis and inflammation in Ercc1−/− mice.
(A) SEM (left) and TEM (right) of collagen (arrows) in wt and Ercc1−/− (Er1−/−) pancreas. N, nucleus; col, collagen; AC, acinar cell; SC, stellate cell. (B) Mmp9, Col1a, and Timp2 mRNA levels in Er1−/− and wt pancreata. (C) Infiltrated leukocytes (arrows) in wt and Er1−/− pancreata (SEM). (D) Immunostaining of CD45- and MAC1-positive cells infiltrating wt and Er1−/− pancreata (n = 3). (E) Ccl2, Il6, Cxcl10, Τnfα, and Il1β mRNA levels in Er1−/− compared to wt pancreata (n = 3). (F) Plasma amylase levels [international units per liter (IU/liter)] in wt and Er1−/− mice. (G) Immunostaining of α-SMA and VCAM1 in wt and Er1−/− pancreata (n = 3). (H) α-SMA protein levels in wt and Er1−/− whole-cell pancreas extracts (n = 4). (I) Immunostaining of γH2AX and (J) pATM in wt and Er1−/− PPCs (n = 3). (K) γH2AX, pATM, and FANCI protein levels in of wt and Er1−/− whole-cell pancreas extracts (n ≥ 3). In (B) and (E), the red dashed lines depict the wt levels. The graphs in (D) and (G) represent the mean fluorescence intensity (MFI) per cubic micrometer of tissue; in (I) and (J), the percentage of positive-stained cells; and in (H) and (K), the β-tubulin (β-TUB)–normalized protein expression levels (n.e.l.) in Er1−/− pancreata compared to wt controls. All tissues and cells are derived from P15 mice. Scale bars, 5 μm. Error bars indicate SEM among n ≥ 3 replicates.
Ercc1−/− pancreatic cells become senescent but show no signs of apoptosis
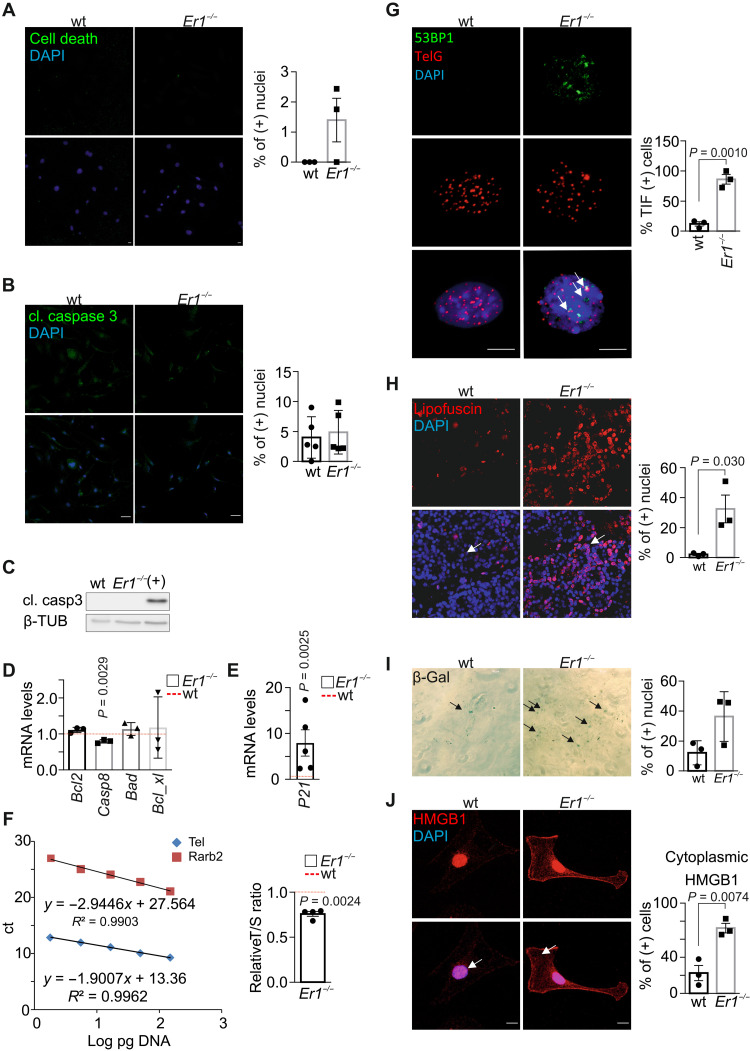
In response to DNA damage, mammalian cells undergo cellular senescence or apoptosis to suppress tumorigenesis (20). In situ detection of fragmented DNA assay [terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL)] revealed no significant differences in the percentage of TUNEL-positive Ercc1−/− and wt PPCs (Fig. 2A and fig. S2A). Likewise, staining and Western blotting for cleaved caspase 3 detected few, if any, apoptotic cells in Ercc1−/− PPCs (Fig. 2, B and C). In support, the mRNA levels of Bcl2, Caspase 8, Bad, and Bcl_xl proapoptotic genes are comparable in Ercc1−/− and wt pancreata (Fig. 2D). However, we find a higher number of annexin V (+)/propidium iodide (PI) (+) cells in Ercc1−/− pancreata, indicating the presence of ruptured cell membranes that typically associate with necrotic cell death (fig. S2B). Further analysis in Ercc1−/− pancreata revealed an increase in the mRNA levels of the senescence-associated cyclin-dependent kinase inhibitor P21 (Fig. 2E), a ~30% shorter telomere length (Fig. 2F), and an increase in the number of telomere dysfunction–induced foci (TIFs) (21), as evidenced by the higher number of 53BP1 foci that colocalized with telomeres in Ercc1−/− PPCs (Fig. 2G), while Tert mRNA levels are comparable to wt controls (fig. S2C). Consistent with previous findings, further analysis of Ercc1−/− metaphase chromosome spreads revealed that P15 PPCs do not accumulate telomere fusions, indicating that the progeroid Ercc1−/− cells have not yet reached the critical telomere length (fig. S2D) (22). In P15 Ercc1−/− pancreata, we find a ~16-fold increase in lipofuscin pigment known to accumulate with aging (Fig. 2H) and a higher number of senescence-associated β-galactosidase–positive (SA-β-gal+) Ercc1−/− cells compared to corresponding wt controls (Fig. 2I). High-mobility group box 1 protein (HMGB1) is a ubiquitous damage-associated molecular pattern protein in the nucleus that promotes inflammation when released extracellularly (23, 24). We find a significant accumulation of HMGB1 in the cytoplasm and in the cellular membrane of Ercc1−/− PPCs compared to wt control cells (Fig. 2J). Consistent with the release of HMGB1 in the extracellular milieu of senescent cells (25), Western blotting reveals a decrease in the intracellular HMGB1 protein levels along with a marked increase of HMGB1 protein levels in the culture media of Ercc1−/− PPCs (Fig. 3A). Together, our findings indicate that the DNA repair defect triggers cellular senescence and necrotic cell death but no apoptosis in Ercc1−/− pancreata and PPCs.
Fig. 2. Er1−/− pancreatic cells present senescent, but not apoptotic, characteristics.
(A) TUNEL in wt and Ercc1−/− (Er1−/−) PPCs (n = 3). (B) Immunostaining of cleaved (cl.) caspase 3 in wt and Er1−/− PPCs (n = 5). (C) Cleaved caspase 3 (casp3) in wt and Er1−/− pancreas whole-cell extracts. UV-irradiated (20 J/m2) wt BMDM protein extracts were used as positive control (+) (n = 3). (D) Bcl2, Casp8, Bad, and Bcl_xl mRNA levels in Er1−/− compared to wt pancreata (n = 3). (E) P21 mRNA levels in Er1−/− compared to wt pancreata (n = 5). (F) Telomere length in wt and Er1−/− pancreata compared to a single-copy gene (Rarb2). The graphs depict the standard curves for telomere (Tel) and Rarb (left) and the relative telomere to Rarb [telomere/single copy gene (T/S)] ratio in wt and Er1−/− pancreata (right, n = 4). ct, cycle threshold. (G) Immunofluorescence in situ hybridization of 53BP1 with telomeric DNA (TelG) in wt and Er1−/− PPCs. Arrows denote TIFs on telomeres. The graph depicts the mean percentage of cells containing ≥2 TIFs ± SD, n = 3. (H) Lipofuscin staining, as depicted by GL13 binding (SenTraGor), in wt and Er1−/− pancreata (n = 3). (I) SA-β-gal assay in wt and Er1−/− PPCs (n = 3 technical replicates). (J) Immunostaining of cytoplasmic HMGB1 in wt and Er1−/− PPCs (white arrows; n = 3). In (D) and (E), the red dashed lines represent the wt levels. The graphs in (H) and (I) depict the percentage of positive-stained cells and, in (J), cytoplasm-positive–stained cells. Tissues and cells are derived from P15 mice. Scale bars, 5 μm. Error bars indicate SEM among n ≥ 3 replicates.
Fig. 3. Ercc1 deletion drives a viral-like response and cytoplasmic ssDNA accumulation.
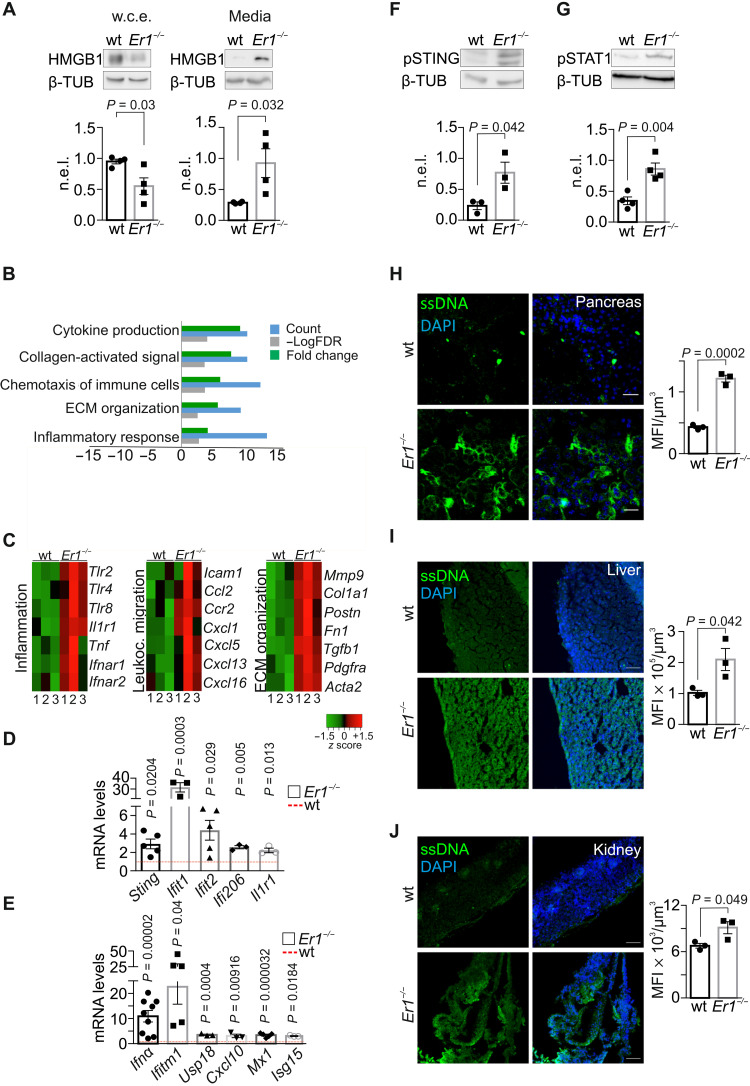
(A) HMGB1 in whole-cell extracts (w.c.e.) and in the culture media of wt and Er1−/− PPCs (n = 4). (B) Overrepresented biological processes (Gene Ontology) of the differentially expressed genes in Er1−/− compared to wt pancreata [meta–false discovery rate (FDR) ≤ 0.01, fold change ≥ ±1.5, n = 3]. Processes with up-regulated genes (green) and down-regulated genes (red). (C) Heatmaps of representative genes associated with inflammation, leukocyte migration, and extracellular matrix (ECM) organization up-regulated in Er1−/− (1 to 3) compared to wt (1 to 3) pancreata. (D) mRNA levels of selected overrepresented genes in wt and Er1−/− pancreata. (E) mRNA levels of overrepresented Ifnα signature genes in wt and Er1−/− pancreata. (F) Phosphorylated STING (pSTING) and (G) phosphorylated STAT1 (pSTAT1) in whole-cell extracts of wt and Er1−/− pancreata (n = 3 to 4). (H) Immunostaining of ssDNA in wt and Er1−/− pancreas, (I) liver, and (J) kidney (n = 3). The graphs in (A), (F), and (G) represent the β-TUB n.e.l. of proteins in wt and Er1−/− pancreata and, in (H) to (J), the MFI per cubic micrometer of tissue. In (D) and (E), the red dashed lines represent the wt levels. Tissues and cells are derived from P15 mice. Scale bars, 40 μm. Error bars indicate SEM among n ≥ 3 replicates.
The ERCC1-XPF defect triggers a viral-like immune response associated with the accumulation of cytoplasmic ssDNA moieties
To assess the senescence-associated gene expression changes triggered by persistent DNA damage in the pancreas signaling (26, 27), we performed RNA sequencing (RNA-seq) profiling in 15-day-old Ercc1−/− and age-matched wt pancreata. The analysis revealed 4403 differentially expressed genes [meta–false discovery rate (FDR) ≤ 0.01, fold change ≥ ±1.5; 2405 up-regulated genes; 1998 down-regulated genes; data S1]. Within this set of genes, gene ontology classification was able to discern a set of seven biological processes containing a significantly disproportionate number of genes relative to those expected in the murine genome (FDR ≤ 0.05). These processes were ranked by their relative enrichment score (see Materials and Methods) and associated with cytokine production, antigen presentation, immune cell chemotaxis, and proinflammatory responses (Fig. 3B). Further analysis within the overrepresented biological processes revealed the up-regulation of proinflammatory genes and genes associated with leukocyte migration and extracellular matrix organization (Fig. 3C). qPCR confirmed the increase in the mRNA levels of several proinflammatory and interferon (IFN) response genes, i.e., Sting, Ifit1, Ifit2, Ifi206, Il1r1, Ifna, Ifitm1, Usp18, Cxcl10, Mx1, and Isg15 in Ercc1−/− pancreata (Fig. 3, D and E), as well as the increase in the mRNA levels of Sting, Ifitm1, Mx1, and Ifit1 genes in the pancreata of Csbm/m;Xpc−/− DKO mice (fig. S1B). Western blot analysis revealed an increase in the protein levels of pSTING (phosphorylated form of stimulator of IFN genes; Fig. 3F) and STING (fig. S3A) known to act as a sensor of cyclic deoxyguanosine monophosphate and as an adaptor protein mediating the IFN response, once activated by DNA sensors such as cGAS (8). Further analysis revealed an increase in pSTAT1 (phosphorylated signal transducer and transcription activator) protein levels, known to mediate the cellular response to IFNs (Fig. 3G) (28).
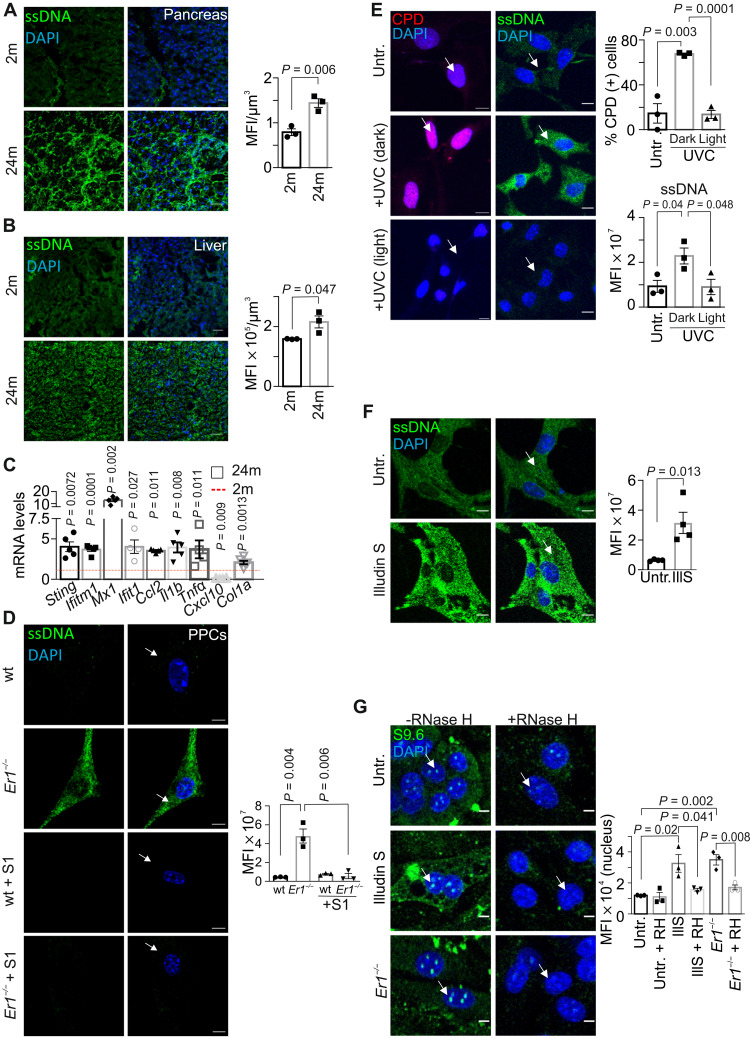
The observed IFN response and the increase in the mRNA levels of several genes associated with nucleic acid sensing, i.e., Sting, Irf1, Ddx58, Ddx60, and Tlr3 (fig. S3B) (8), prompted us to test for the presence of DNA fragments in the cytoplasm of Ercc1−/− PPCs. Immunofluorescence (IF) studies with antibodies that are preferentially reactive against ssDNA or double-stranded DNA (dsDNA) revealed the accumulation of cytoplasmic ssDNA moieties in the pancreas (Fig. 3H), liver (Fig. 3I), and kidney (Fig. 3J) of P15 Ercc1−/− animals as well as in Ercc1−/− PPCs (fig. S3C) and in bone marrow–derived macrophages (BMDM) (fig. S3D). We find no evidence of cytoplasmic dsDNAs in Ercc1−/− PPCs (fig. S3E) or in the kidney (fig. S3F) and liver (fig. S3G) of Ercc1−/− mice. Further IF studies revealed that ssDNAs also accrue in the cytosol of the NER-defective Csbm/m and Xpc−/− pancreatic cells, albeit to a lesser extent (fig. S3H). Notably, we find that cytoplasmic ssDNAs also accumulate in the pancreata and livers of the 24-month-old naturally aged mice (Fig. 4, A and B) followed by an increase in the mRNA levels of proinflammatory and IFN response genes, i.e., Sting, Ifitm1, Mx1, Ifit1, Ccl2, Il1b, Tnfα, Cxcl10, and Col1a (Fig. 4C); the latter is consistent with the immune response against cytosolic DNA fragments (8). To further confirm the presence of cytoplasmic ssDNAs in Ercc1−/− PPCs, cells were treated with mung bean S1 endonuclease, which specifically degrades ssDNA, leaving the duplex DNA intact (29). Treatment with the S1 nuclease led to the complete removal of ssDNAs from the cytoplasm of Ercc1−/− PPCs (Fig. 4D).
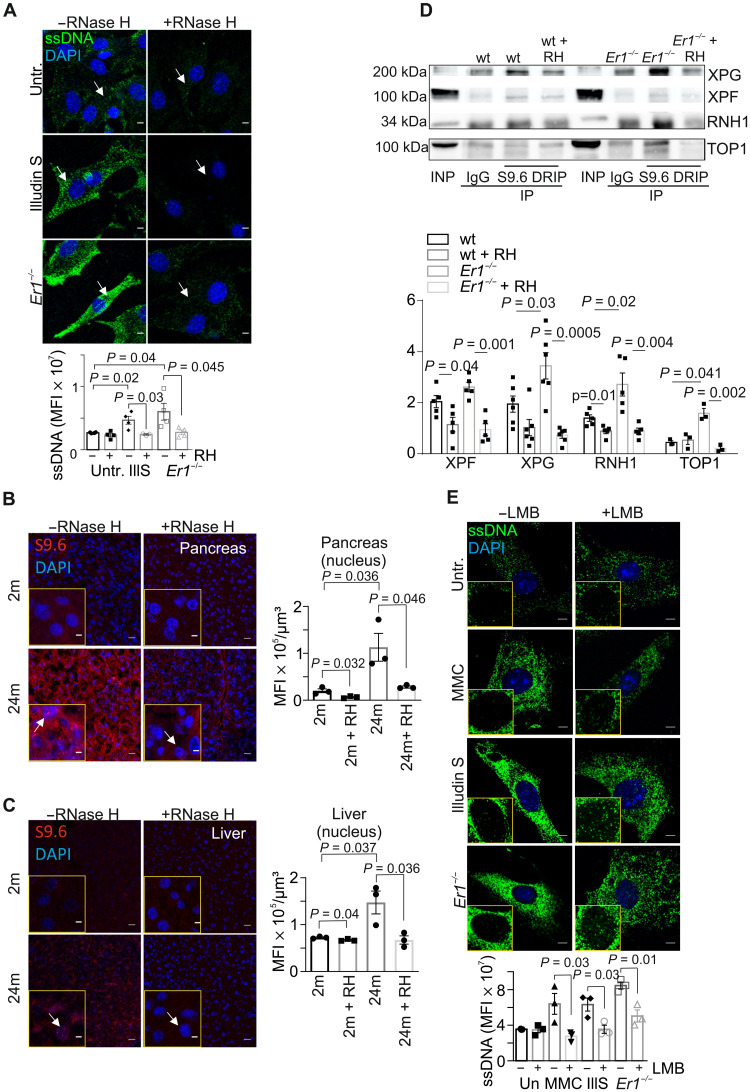
Fig. 4. DNA damage–induced cytoplasmic ssDNA accumulation is instigated by R-loops.
(A) Immunostaining of ssDNA in 2-month (2m)– and 24-month (24m)–old wt pancreata (n = 3). (B) IF detection of ssDNA in 2 month (2m)– and 24 month (24m)–old wt livers (n = 3). (C) mRNA levels of selected genes in 24-month-old wt pancreata. The red dashed line represents the levels in 2-month-old wt pancreata (n = 4 to 5). (D) Immunostaining of ssDNA (arrows) in wt and Ercc1−/− (Er1−/−) PPCs with or without S1 nuclease treatment (n = 3). (E) Immunostaining of cyclobutane-pyrimidine dimers (CPD) (left; red) or ssDNA (right; green) in untreated (untr.) CPD+ transgenic PPCs, UV-irradiated (15 J/m2) and kept in the dark or under white light (1.5 hours of incubation; n = 3). The graphs depict the percentage of CPD-positive–stained cells and the ssDNA MFI per cell, respectively. (F) Immunostaining of ssDNA in untreated and Illudin S–treated (IllS) (30 ng/ml; 3 hours) wt PPCs (n = 4). (G) Immunostaining of R-loops (S9.6 antibody) in Er1−/− and untreated or Illudin S–treated (30 ng/ml; 3 hours) wt PPCs in the absence or presence of transfected recombinant RNase H (n = 3). The graphs in (A) and (B) depict the MFI per cubic micrometer of tissue; in (D) and (F), the MFI per cell; and in (G), the MFI per nucleus. Unless otherwise stated, tissues and cells are derived from P15 mice. Scale bars, 5 μm; in (A) and (B), 40 μm. Error bars indicate SEM among n ≥ 3 replicates.
DNA damage–induced R-loops trigger the accumulation of cytoplasmic ssDNAs
Owing to the DNA repair defect in Ercc1−/− pancreata, we reasoned that the presence of irreparable DNA lesions causally contribute to the generation of ssDNAs in the cytoplasm of Ercc1−/− PPCs. In support, treatment of wt PPCs with the potent genotoxin mitomycin (MMC), which generates DNA ICLs, or with UV-C irradiation (15 J/m2) led to the rapid accumulation of ssDNAs in the cytoplasm (fig. S4, A and B). Inactivation of ATM kinase with KU-55933 inhibitor or of ataxia-telangiectasia and Rad3-related (ATR) kinase with NU6027 had no detectable impact on cytoplasmic ssDNAs in MMC- or UV-treated cells (fig. S4, A and B; as shown), indicating that the accumulation of cytoplasmic ssDNAs is driven by DNA damage itself and it does not require canonical DDR signaling. To further establish that nuclear DNA damage causally contributes to the release of ssDNAs in the cytoplasm, we isolated PPCs from cyclobutane pyrimidine dimer (CPD)–photolyase transgenic mice that ubiquitously express the marsupial CPD-specific photolyase transgene (30). The CPD photolyase can directly revert UV-induced CPDs into undamaged bases using visible light energy in a reaction known as photoreactivation. Exposure of CPD-photolyase transgenic PPCs to UV irradiation (15 J/m2) and subsequent incubation in the dark (CPD photolyase is inactive and UV-induced CPDs persist) led to the accumulation of ssDNA moieties in the cytoplasm of these cells (Fig. 4E; as indicated). ssDNAs were absent when UV-treated, CPD-photolyase transgenic PPCs were also exposed to the photoreactivating light (photolyase is active and UV-induced CPDs are repaired) (Fig. 4E; as indicated). Further studies revealed that treatment of wt PPCs with Illudin S, a fungal sesquiterpene known to induce transcription-blocking DNA adducts, recapitulates the accumulation of cytoplasmic ssDNAs seen in Ercc1−/− PPCs or in MMC- and UV-treated PPCs (Fig. 4F), indicating that irreparable DNA lesions interfering with transcription may underlie the observed release of ssDNAs in the cytoplasm of PPCs. Persistent DNA lesions often hamper the progression of RNA polymerase II on actively transcribed genes, leading to the gradual buildup of R-loops in cells. In line, confocal microscopy studies revealed the accumulation of R-loops in Ercc1−/− and Illudin S–treated wt PPCs (Fig. 4G). Detection of RNA by the S9.6 antibody was recently shown to generate artifacts when imaging RNA:DNA hybrids (31). To exclude that the S9.6 IF signal observed arises from dsRNA rather than RNA:DNA hybrids, we transfected PPCs with the RNA:DNA hybrid–specific nuclease, RNase H; the latter removed RNA-DNA hybrids, leading to the substantial decrease of R-loops in these cells (Fig. 4G; as indicated). Next, we sought to test whether DNA damage–driven R-loops causally contribute to the release of ssDNA moieties in the cytoplasm of Illudin S–treated and Ercc1−/− PPCs. Consistently, we find that ssDNAs are removed from the cytoplasm of Illudin S–treated or Ercc1−/− PPCs when these cells are transfected with RNase H to eliminate R-loops (Fig. 5A). Likewise, R-loops accumulate significantly in MMC-treated PPCs (fig. S4C), and RNase H–mediated removal of R-loops leads to the substantial elimination of cytoplasmic ssDNAs in MMC-treated PPCs (fig. S4D). Consistent with the accumulation of cytoplasmic ssDNAs in the 24-month-old pancreata and livers, we find that R-loops also accumulate in the cell nuclei of pancreata, livers, and kidneys of the 24-month-old naturally aged mice compared to 2-month-old young adult animals (Fig. 5, B and C, and fig. S4E). In addition to the nuclear accumulation of R-loop structures, we find that RNA:DNA hybrids also accumulate in the cytoplasm pancreatic cells in 24-month-old kidneys and livers. However, this fraction is mostly colocalized with mitochondria (32). The S9.6 IF signal was efficiently removed by RNase H treatment but remained unaffected upon treatment with ribonucleases RNase A or T1, indicating that the signal is not derived from dsRNA or ssRNA moieties, respectively (fig. S4F). Cytosolic ssDNAs also accumulate in serum-starved or hydroxyurea-treated Ercc1−/− PPCs [1% fetal bovine serum (FBS); fig. S5, A and B], suggesting that DNA damage–associated ssDNAs can accumulate in the cytoplasm of these cells independently of DNA replication.
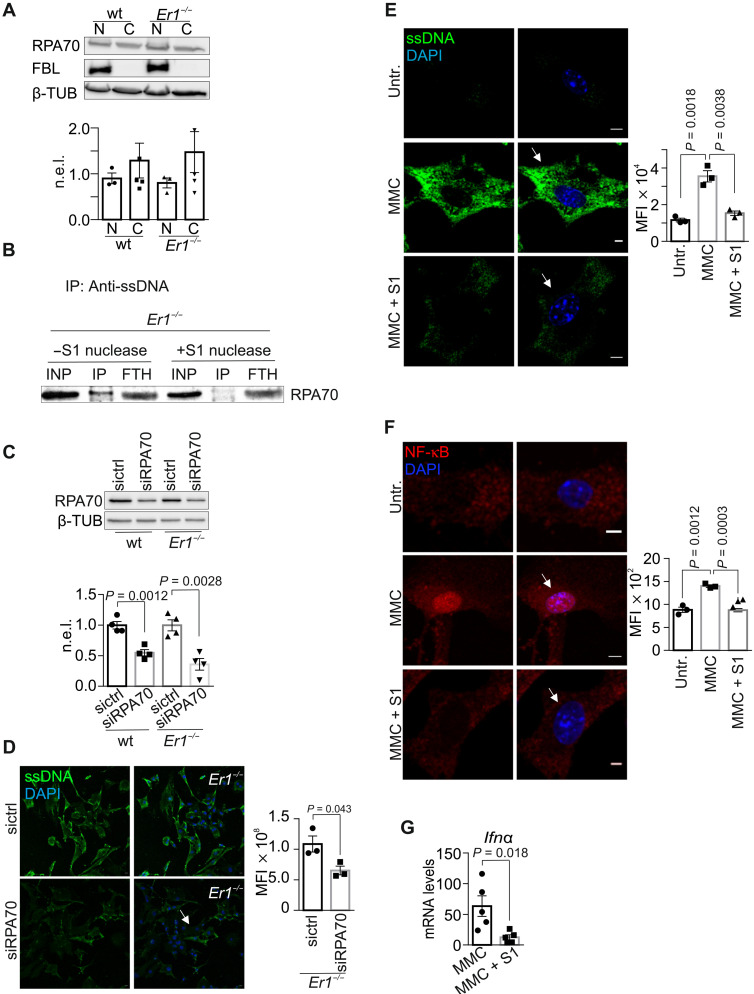
Fig. 5. RPA mediates the nucleocytoplasmic transport of ssDNA moieties.
(A) Immunostaining of ssDNA in Ercc1−/− (Er1−/−) and untreated or Illudin S–treated wt PPCs in the absence or presence of transfected recombinant E. coli RNase H (n = 3 to 4). (B) Immunostaining of R-loops, by means of S9.6 antibody staining, in 2- and 24-month-old naturally aged pancreata in the absence or presence of transfected recombinant RNase H (n = 3). (C) S9.6 immunostaining of R-loops in 2- and 24-month-old livers in the absence or presence of transfected recombinant RNase H (n = 3). (D) S9.6 immunoprecipitation (DRIP) followed by Western blotting for XPG, XPF, RNase H1 (RNH1), and DNA topoisomerase 1 (TOP1) in wt and Er1−/− pancreatic nuclear extracts with and without RNase H (RH) treatment (n = 6). (E) Immunostaining of ssDNA (green) in Er1−/−, untreated, MMC- (10 μg/ml; 4 hours), and Illudin S–treated (30 ng/ml; 3 hours) wt PPCs, cultured with or without the nuclear export inhibitor leptomycin B (LMB; 40 nM). Image inlays show the nuclear ssDNA signal (green) (n = 3). The graphs depict the MFI per cell in (A), per nucleus in (B) and (C), and per cytoplasm in (E). Unless otherwise stated, all tissues and cells are derived from P15 mice. Scale bars, 5 μm. Error bars indicate SEM among n ≥ 3 replicates.
To further explore the origin of cytoplasmic ssDNAs, we treated wt pancreatic cells with trans-retinoic acid (tRA) and Illudin S, to induce transcription of tRA-responsive genes, in the presence of transcription-blocking DNA lesions. We then performed qPCR on ssDNA fragments isolated from untreated and tRA/Illudin S–treated PPCs, where the second strand was synthesized with the Klenow fragment of DNA polymerase I (fig. S5C). Our analysis revealed the presence of ssDNAs originating from the tRA-inducible genes Rarb and Stra6 (33) in tRA/Illudin S–treated PPCs compared to untreated controls, indicating that cytoplasmic ssDNAs are generated by cotranscriptional R-loops. The β-actin gene was used as a control because it is a tRA-nonresponsive gene that was previously shown to accumulate R-loops (34, 35). Next, we explored the possibility of long interspersed nuclear element 1 (LINE1) derepression by assessing LINE1 transcripts in wt and Ercc1−/− pancreata, as well as in untreated and tRA/Illudin S–treated PPCs (36). qPCR analysis revealed no evidence to support the presence of LINE1 open reading frame 1 (ORF1), ORF2, 5′ untranslated region (5′UTR), and 3′UTR transcripts (fig. S5D).
To further investigate how R-loops lead to the generation of ssDNAs, we used a DNA-RNA immunoprecipitation (DRIP) approach coupled to Western blotting, to isolate RNA:DNA hybrids from wt and Ercc1−/− pancreata, in the presence or absence of RNase H. In agreement with previous findings, this approach revealed that XPG (xeroderma pigmentosum G) and XPF nucleases are bound together with DNA topoisomerase 1 (TOP1) to R-loops in wt and Ercc1−/− pancreatic cells (Fig. 5D and fig. S5E; as indicated) (33, 37, 38). Our finding that, in Ercc1−/− cells, the XPF DRIP signals are lower than those of XPG supports previous observations indicating that XPF is unstable in the absence of ERCC1 (39, 40). The latter makes XPF the limiting factor during the endonuclease-mediated resolution of R-loops in Ercc1−/− cells. Consistent with the increased amount of RNA:DNA hybrids in Ercc1-deficient pancreata (Fig. 4G), we also find higher levels of RNase H1 bound to R-loops in Ercc1−/− cells compared to wt controls (Fig. 5D and fig. S5E; as indicated).
ssDNAs are released into the cytoplasm via an active nucleocytoplasmic transport mechanism
Micronuclei containing whole or fragmented chromosomes may passively diffuse into the cytoplasm when the nuclear envelope breaks down in mitotically dividing cells (41). The lack of 4′,6-diamidino-2-phenylindole (DAPI)–stained chromatin fragments in the cytoplasm of Ercc1−/− PPCs along with the presence of cytoplasmic ssDNAs in hydroxyurea-treated Ercc1−/− PPCs or in Ercc1−/− livers and kidneys, where the great majority of cells typically do not divide, prompted us to test whether ssDNAs are actively released in the cytoplasm of Ercc1−/− cells. Analysis of nucleocytoplasmic protein shuttling is greatly facilitated by the use of leptomycin B (LMB), an inhibitor of CRM1-dependent nuclear export that does not interfere with protein transport into the nucleus. We find that treatment of Ercc1−/− PPCs and MMC- or Illudin S–treated wt PPCs with LMB leads to the marked decrease of ssDNAs in the cytoplasm that concurrently accumulate in the nuclei of these cells (Fig. 5E and fig. S5F). This finding suggests that cytoplasmic ssDNAs are of nuclear origin and that they are released into the cytoplasm of DNA repair–deficient cells via an active nucleocytoplasmic transport mechanism. Replication protein A (RPA) binds preferentially to ssDNA during DNA replication, repair, or recombination in eukaryotic cells (42). Western blotting of nuclear and cytoplasmic protein extracts in wt and Ercc1−/− pancreata revealed that RPA70, the DNA binding subunit of RPA (from now on referred to as RPA), is located both in the nucleus and the cytoplasm of Ercc1-deficient PPCs (Fig. 6A). We find that RPA binds cytoplasmic ssDNAs in Ercc1−/− PPCs and that the RPA-ssDNA interaction is abolished when Ercc1−/− PPC cytoplasmic protein extracts are treated with the mung bean S1 nuclease (Fig. 6B). Consistently, we find higher levels of RPA bound to ssDNA in the cytoplasm of MMC-treated compared to untreated PPCs (fig. S5G), as indicated by anti-ssDNA immunoprecipitation experiments in cytoplasmic extracts of equal quantity. Next, we performed a small interfering RNA (siRNA)–mediated knockdown of RPA to test for the functional relevance of RPA to the accumulation of cytoplasmic ssDNAs in Ercc1−/− PPCs (Fig. 6C). We find that RPA knockdown limits substantially the accumulation of ssDNAs in the cytoplasm of Ercc1−/− PPCs (Fig. 6D). Thus, DNA damage–driven ssDNAs are bound by RPA and are actively transported to the cytoplasm. Inhibition of nuclear export or abrogation of RPA via RNA interference–mediated knockdown substantially impedes their release and accumulation in the cytoplasm.
Fig. 6. In vitro and in vivo effect of ssDNA abrogation in Er1−/− pancreata.
(A) RPA70 in nuclear and cytoplasmic extracts from wt and Er1−/− pancreata (n = 4). (B) Immunoprecipitation (IP) using anti-ssDNA in untreated or S1 nuclease–treated cytoplasmic extracts from Er1−/− pancreata for RPA70. The input (INP) and flow through (FTH) are 1/20 of the extract used (n = 3). (C) RPA70 in whole-cell extracts of wt and Er1−/− PPCs, untreated, or incubated with scrambled (sictrl) or siRNA against Rpa70 (siRPA70) (n = 4). (D) Immunostaining of ssDNA in wt and Er1−/− PPCs, untreated, or incubated with scrambled (sictrl) or siRNA against Rpa70 (siRPA70) (n = 3). (E) Immunostaining of ssDNA upon recombinant S1 nuclease transfection in untreated and MMC-treated (10 μg/ml; 4 hours) wt PPCs (n = 3). (F) Immunostaining of NF-κB upon recombinant S1 nuclease transfection in untreated and MMC-treated (10 μg/ml; 4 hours) wt PPCs (n = 3). (G) Ifnα mRNA levels in MMC-treated wt PPCs, untreated, or transfected with S1 nuclease (n = 5). The graphs in (A) and (C) represent the β-TUB– or fibrillarin (FBL)–normalized expression of RPA70 levels (n.e.l.) in Er1−/− compared to corresponding wt controls, as indicated. The graphs in (D) to (F) depict the MFI per cell. All tissues and cells are derived from P15 mice. Scale bars, 5 μm. Error bars indicate SEM among n ≥ 3 replicates.
To further investigate the kinetics of R-loop–induced cytoplasmic ssDNA accumulation and genome instability, we treated wt PPCs with tRA and Illudin S to induce transcription in the presence of transcription-blocking DNA lesions. Next, we performed a series of IF studies for genome-bound and cytoplasmic RPA70, cytoplasmic ssDNA, RNA:DNA hybrids, and γH2AX+;53BP1+ foci (fig. S6, A to F). Using a preextraction, fixation, and immunostaining approach, we find that, upon tRA/Illudin S treatment, the genome-bound RPA70 mean fluorescence intensity (MFI) levels increase over time until the 16-hour time point and remain stable thereafter (fig. S6A). Similar kinetics were observed for cytoplasmic RPA70, whose MFI levels were measured in the presence of cyclohexamide to exclude any signal derived from newly translated protein (fig. S6B). Consistently, we find that cytoplasmic ssDNAs reach their maximum MFI levels at 16 hours of treatment (fig. S6C). These findings are in agreement with a previous work on the global exhaustion of RPA (43) and indicate that, as RPA-bound ssDNA is translocated into the cytoplasm, less RPA70 becomes available to bind the continuously increasing amount of R-loops (fig. S6D), further contributing to genome instability (fig. S6E). The latter is expected to lead to increasing amounts of RPA70-free ssDNA moieties, which will not be transferred into the cytoplasm (fig. S6, B and C). Similar findings were observed in tRA/Illudin S–treated cells cultured in the presence of ATR inhibitor (ATRi). In this condition, RNA:DNA hybrids continue to accumulate, generating more γH2AX+;53BP1+ foci, further contributing to DNA damage (fig. S6, A to F).
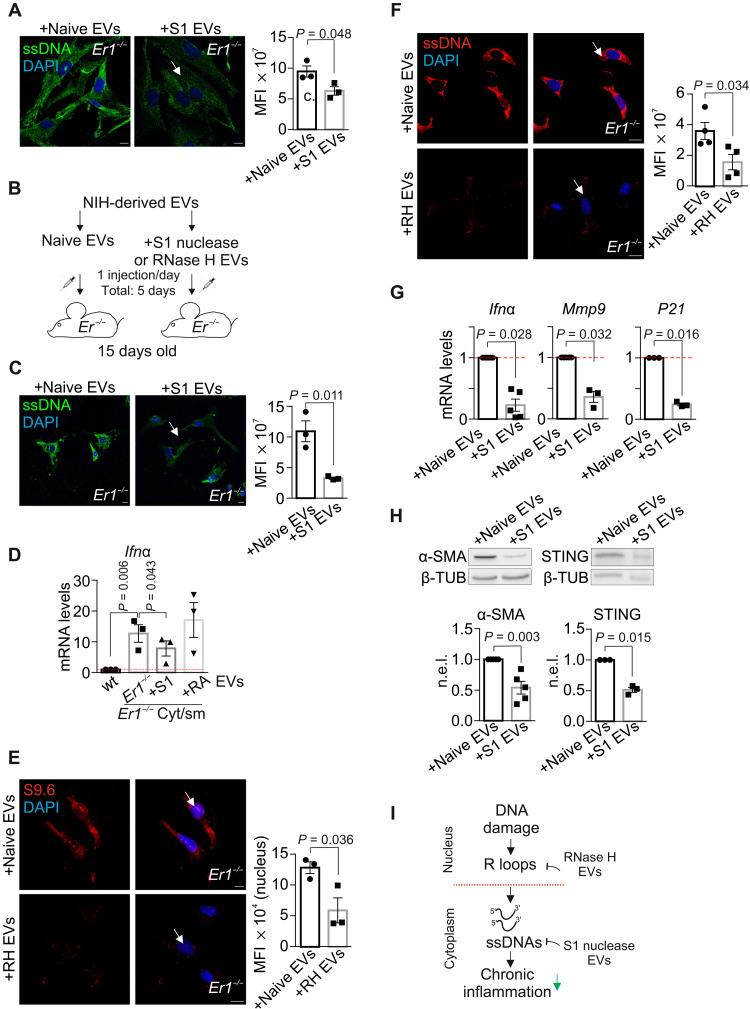
An EV-based nuclease cargo eradicates cytoplasmic ssDNAs and the proinflammatory response in Ercc1−/− pancreata
Cells respond to cytosolic DNA introduced into the cytoplasm by triggering an innate immune response. We, therefore, reasoned that the removal of ssDNAs from the cytoplasm of Ercc1−/− PPCs would be beneficial for reducing the proinflammatory load in inflamed cells, thereby improving the outcome of chronic pancreatitis in Ercc1−/− mice. To test this, we first transiently transfect recombinant mung bean S1 nuclease into MMC-treated wt PPCs and corresponding untreated control cells. We find that mung bean S1–mediated degradation of MMC-induced ssDNAs (Fig. 6E) prevents the nuclear translocation of nuclear factor κB (NF-κB), a potent regulator of the innate immune response against foreign pathogens (Fig. 6F) (44). Moreover, the nuclease-mediated removal of cytosolic ssDNAs lowers the Ιfnα mRNA levels compared to those observed in untreated wt cells (Fig. 6G). Thus, removal of cytosolic ssDNAs substantially dampens the proinflammatory response seen in Ercc1−/− PPCs.
EVs are lipid bilayer–delimited particles that are produced in the endosomal compartment of most eukaryotic cells and are important drivers of intercellular communication. EVs are released into the bloodstream and are known to discharge their content into far distant recipient cells and tissues to exert multiple physiological stimuli (45). First, we used an EV-based strategy to deliver recombinant S1 nuclease to remove the cytoplasmic ssDNAs in Ercc1−/− PPCs. We find that treatment with naive (i.e., not loaded) or mung bean S1 nuclease-loaded EVs substantially diminishes the accumulated ssDNAs in the cytoplasm of Ercc1−/− PPCs (Fig. 7A). Next, we embarked on a physiological application by injecting intraperitoneally P15 Ercc1−/− mice with naive or mung bean S1 nuclease-loaded EVs for five consecutive days (Fig. 7B). We find that treatment of Ercc1−/− mice with EV-based S1 nuclease cargo leads to the substantial removal of cytoplasmic ssDNAs (Fig. 7C). To test that cytoplasmic ssDNAs in Ercc1−/− pancreata causally contribute to the observed IFNα response (Figs. 3E and 7C), we next prepared National Institutes of Health (NIH) 3T3–derived EVs carrying the cytoplasmic nucleic acid fraction from wt (EVswt cyto) or Ercc1−/− (EVsErcc1 cyto) pancreata. Consistently, we find a substantial increase in the Ifna mRNA levels of PPCs treated for 24 hours with EVsErcc1 cyto but not with EVswt cyto. Ifna mRNA levels substantially decrease when PPCs are treated with EVsErcc1 cyto carrying cytoplasmic DNA already pretreated with the mung bean S1 nuclease but not with the RNase A (Fig. 7D; as indicated). Our finding that DNA damage–driven R-loops causally contribute to the release of cytoplasmic ssDNAs in Ercc1−/− pancreata prompted us to test whether an EV-mediated delivery of recombinant RNase H could remove R-loops and the accumulated cytosolic ssDNAs in Ercc1−/− pancreata. Intraperitoneal injections of P15 Ercc1−/− mice with naive or recombinant Escherichia coli RNase H–loaded EVs for two consecutive days revealed the substantial removal of R-loops (Fig. 7E), leading to a decrease in cytoplasmic ssDNAs (Fig. 7F) in Ercc1−/− pancreata. We found that treatment of Ercc1−/− mice with EVs carrying RNase H together with S1 nuclease (RH + S1) has an additive effect on the removal of RNA:DNA hybrids and cytoplasmic ssDNA fragments, compared to animals treated with naive EVs or EVs loaded with scramble protein (fig. S6, G and H). Similar to PPCs, treatment of Ercc1−/− mice with S1 nuclease– or RH + S1–loaded EVs leads to the marked decrease in the mRNA levels of Ifna, Mmp9, and P21 genes (Fig. 7G and fig. S6I; as indicated) and to the substantial decrease in the protein levels of α-SMA and STING in Ercc1−/− pancreata (Fig. 7H). Thus, DNA damage–driven R-loops trigger the accumulation of cytoplasmic ssDNAs leading to the activation of proinflammatory responses associated with chronic inflammation in Ercc1−/− pancreata. An EV-based delivery of S1 nuclease and/or RNase H cargo in mice can eliminate cytoplasmic ssDNAs, reducing the proinflammatory load in Ercc1−/− PPCs and pancreata. The latter has important ramifications for the development of efficient rationalized intervention strategies against chronic inflammation in NER progeria and likely also with aging (Fig. 7I).
Fig. 7. Delivery of an EV-mediated nuclease cargo alleviates the proinflammatory response in Er1−/− pancreata.
(A) Immunostaining of ssDNA in Er1−/− PPCs exposed to naive or EVs loaded with S1 nuclease (S1 EVs) (n = 3). (B) Experimental design of injections in wt or Er1−/− mice, with NIH cell line–derived naive or S1 EVs. (C) Immunostaining of ssDNAs in PPCs from Er1−/− mice injected with naive or S1 EVs (n = 3). (D) Ifnα mRNA levels in wt PPCs treated with EVs carrying the cytoplasmic nucleic acid fraction from wt (wt cyto) or Er1−/− (Er1−/− cyto) pancreata, treated with S1 nuclease (+S1) or RNase A (+RA) (n = 3). Cyt/sm, cytoplasm. (E) S9.6 immunostaining in PPCs from Er1−/− mice injected with naive or EVs loaded with RNase H (RH EVs) (n = 3). (F) Immunostaining of ssDNA in PPCs from Er1−/− mice injected with naive or RH EVs (n = 3). (G) Ifnα, mmp9, and p21 mRNA levels from pancreata of wt and Er1−/− mice injected with naive or S1 EVs (n ≥ 3). (H) α-SMA (n = 5) and STING (n = 3) protein levels in pancreas whole-cell extracts of Er1−/− mice injected with naive or S1 EVs (β-TUB–normalized). (I) A schematic graph on the functional contribution of DNA damage–driven R-loops in chronic inflammation. The graphs depict MFI per cell in (A), (B), and (F) and MFI per nucleus in (E). The red dashed lines in (D) and (G) represent the wt levels. All tissues and cells are derived from P15 mice. Scale bars, 5 μm. Error bars indicate SEM among n ≥ 3 replicates.
DISCUSSION
Whereas the links between genome instability and enhanced cancer predisposition are well established, less is known about how DNA damage leads to the premature onset of age-related pathological features in man and accompanied animal models with mutations in DNA repair genes. Here, we provide evidence that persistent DNA damage triggers the formation of R-loops, leading to the accumulation of cytoplasmic ssDNA moieties in Ercc1−/− pancreata or cells exposed to genotoxins. In turn, the presence of ssDNA fragments in the cytoplasm stimulates a proinflammatory response, leading to the premature onset of chronic pancreatitis and fibrosis in progeroid Ercc1−/− mice. In this work, cytosolic ssDNAs also accrue in inflamed naturally aged pancreata where fibrosis and chronic pancreatitis are known to be frequently observed with aging (46). Treatment with EVs carrying RNase H or S1 nucleases removes DNA damage–induced R-loops and the associated cytoplasmic ssDNAs substantially diminishing the proinflammatory response seen in progeroid Ercc1−/− pancreata.
DDR signaling is not required for the generation of cytoplasmic ssDNAs upon DNA damage as inhibition of ATM or ATR has no impact on the accumulation of ssDNA moieties in MMC- or UV-treated cells, respectively. However, the light-dependent removal of UV-induced CPDs from the genome of CPD-photolyase transgenic cells rapidly eliminates the great majority of cytosolic ssDNAs, indicating that cytoplasmic ssDNAs are of nuclear origin. We and others have recently shown that R-loops are increasingly formed in cells that are defective in DNA repair or RNA processing (47–49)—leading to DNA breaks (50), transcription stalling, and DDR activation—and that XPG and XPF are recruited on RNA:DNA hybrids under conditions that promote R-loop formation (33, 50). A central aspect of these findings is that the great majority of ssDNAs in the cytoplasm originate from DNA damage–driven R-loops. In support, mutations in RNase H2 that impair the efficient processing of RNA-DNA hybrids or ribonucleotides are known to trigger the activation of a robust proinflammatory response (51). Moreover, a defect in the R-loop processing enzyme SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1) was recently shown to trigger DNA replication fork–induced ssDNAs, genome instability, and inflammation (52).
Once generated, we find that ssDNAs are actively transported into the cytosol of pancreatic cells, bound by RPA. The latter could reflect a physiological response of the nucleus to remove by-products of DNA damage repair or irreversibly damaged genome fragments. Alternatively, ssDNAs may also be passively diffused into the cytosol of mitotically dividing cells when the nuclear envelope breaks down (41). Besides the nucleus, we find that RNA-DNA hybrids also accumulate in the cytoplasm of naturally aged liver, kidney, and pancreas cells. Endogenous RNA:DNA hybrids are known to be regulated by RNA polymerase III in human cells (53) and could be attributed to R-loops, to reverse-transcribed retroelements arising upon DNA damage (51, 54), or else, from the mitochondrial genome (55, 56). Previous work on the enzymatic removal of cytoplasmic RNA:DNA hybrids did not rescue the proinflammatory response, indicating that cytoplasmic RNA:DNA hybrids do not contribute in innate immune activation (51). We find no evidence of apoptosis in Ercc1−/− pancreatic cells. Instead, the DNA repair–deficient cells show marked signs of senescence. Upon DNA damage, senescent cells are known to secrete high levels of inflammatory cytokines and immune modulators associated with the senescence-associated secretory phenotype (SASP) response (26). SASP and the gradual buildup of immunogenic ssDNAs in the cytoplasm of pancreatic cells could further perpetuate the vicious cycle of DNA damage–driven immune activation, leading to the infiltration of immune cells, chronic inflammation, and fibrosis in Ercc1−/− pancreata.
So far, the lack of knowledge on the causal mechanisms underlying DNA damage–driven inflammation has hindered the development of rationalized intervention strategies against degenerative diseases, especially at older ages. EVs are nonimmunogenic carriers, allowing their therapeutic cargo to circulate for extended periods of time within the body (45). Here, we developed an EV-based therapeutic strategy aiming at diminishing chronic inflammation due to the improper accumulation of ssDNAs in the cytoplasm of damaged cells and prevent disease. In Ercc1−/− pancreata, we find that treating mice with EVs carrying the ssDNA-specific S1 nuclease is equally effective in reducing the proinflammatory signal with EVs carrying the RNA-DNA hybrid–specific RNase H. Thus, as DNA damage–driven R-loops and associated cytoplasmic ssDNA moieties accumulate over time, the proposed therapeutic scheme may be a promising strategy to combat chronic inflammation, thereby improving considerably the outcome of causally linked inflammatory diseases with aging (8, 57).
MATERIALS AND METHODS
Animal models
The generation and characterization of NER-deficient mice have been previously described (13, 58). Animals were kept on a regular diet and housed at the Institute of Molecular Biology and Biotechnology (IMBB) animal house, which operates in compliance with the “Animal Welfare Act” of the Greek government, using the “Guide for the Care and Use of Laboratory Animals” as its standard. As required by the Greek law, formal permission to generate and use genetically modified animals was obtained from the responsible local and national authorities. All animal studies were approved by an independent Animal Ethical Committee at the Foundation of Research and Technology - Hellas (FORTH).
Primary cells and treatments
PPCs were isolated from P15 animals. Briefly, pancreata were excised, minced, and incubated in collagenase type IV (2.5 mg/ml) at 37°C for 15 min. After centrifugation, cells were resuspended and cultured in standard medium containing Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, streptomycin (50 μg/ml), penicillin (50 U/ml) (Sigma-Aldrich), and 2 mM l-glutamine (Gibco) for 2 days. In BMDM cell cultures, the medium was supplemented with 30% L929-conditioned culture media for 5 days. For subsequent experiments, cells were rinsed with phosphate-buffered saline (PBS); exposed to UV-C irradiation (10 to 20 J/m2), MMC (10 μg/ml, 4 hours; AppliChem), Illudin S (30 ng/ml, 3 hours; Santa Cruz Biotechnology), and hydroxyurea (650 μM, 16 hours); and cultured at 37°C before subsequent experiments. Preincubation with ATM inhibitor (10 μM; Millipore), ATRi (10 μM; Millipore), and LMB (40 nM; Merck) started 1 hour before genotoxic treatments and lasted throughout the experiment.
Protein transfection and EV injections
For the protein transfection experiments (Pierce Protein Transfection Reagent, Thermo Fisher Scientific), 10 U of recombinant mung bean S1 nuclease (100 U/μl; Thermo Fisher Scientific) or RNase H (5 U/μl; New England Biolabs) was used according to the manufacturer’s instructions. For the RPA70 knockdown, 20 pmol of a stealth RNA oligo was used (MSS 229016, Thermo Fisher Scientific). PPCs were transfected with Lipofectamine 2000 (Invitrogen) and subsequently seeded in 6-well or 24-well plates, according to the manufacturer’s protocol. As a nontargeting control, the AllStars negative (Qiagen) was used. For protein cell extracts, tissues were digested with collagenase (2.5 mg/ml) at 37°C for 15 min. Collagenase was neutralized with 10% FBS, and samples were further washed with 1× PBS/1% bovine serum albumin (BSA). For the delivery of EVs, 13-day-old Er1−/− mice were injected intraperitoneally every 24 hours for 5 days with EVs isolated from media of 10 × 106 NIH-3T3 cells. EVs were purified using the differential ultracentrifugation protocol (33, 59). Briefly, culture medium was centrifuged sequentially at 300g (10 min), 2000g (10 min), and 10,000g (30 min) to remove dead cells and cell debris. EVs were purified with the final step of ultracentrifugation at 100,000g for 2 hours. For functional experiments, the EVs used were purified from cells at a quantity of five times the number of recipient cells. S1 nuclease and/or RNase H or RNase A (1 mg/ml; Santa Cruz Biotechnology) was loaded using 0.2% saponin. The EVs were then centrifuged for 1.5 hours at 100,000g to ensure the incorporation of the nuclease, passed from a 0.2-μm filter, and were injected intraperitoneally to mice. For the isolation of cytoplasmic DNA, wt and Ercc1−/− pancreata were lysed with NP-40 lysis buffer [10 mM tris-HCl (pH 7.9), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and protease inhibitors]. Cytoplasmic lysates were incubated with 0.2 M NaCl and proteinase K (0.1 mg/ml) per 50 μg of protein for 2 hours at 65°C. For the purification of DNA, the cytoplasmic extracts were loaded on phaselock tubes, with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) and incubated for 5 min at room temperature (RT) on a rotating wheel. The samples were centrifuged for 5 min at RT. The aqueous phase was isolated and incubated for 3 hours at −80°C with 0.2 M NaCl and 2.5 volumes of cold 100% ethanol. The samples were centrifuged for 25 min at 4°C. Eighty percent of ethanol was added to the pellet and centrifuged for 5 min at 4°C. The DNA pellets were air-dried. DNA (2.5 μg) was treated with 1 μl of S1 nuclease (100 U/μl) or 1 μl of RNase A (100 μg/μl) and incubated for 2 hours at 37°C. The DNA was loaded to the EVs using 0.2% saponin and incubated with the PPCs for 24 hours.
Immunofluorescence
For IF experiments of mouse tissues, minced tissues or cells were fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 1% BSA. After overnight incubation with primary antibodies, secondary fluorescent antibodies were added, and DAPI was used for nuclear counterstaining. Samples were visualized with an SP8 TCS laser scanning confocal microscope (Leica), and scanned sections were analyzed using the Fiji (ImageJ) software. Mouse livers, kidneys, and pancreata were embedded in optimal cutting temperature (OCT) compound, before frozen sectioning on a Cryotome CM 1850, permeabilization, blocking, and immunostaining as described above. For the SA-β-gal activity, the β-gal assay kit was used (Abcam Inc., ab65351) according to the manufacturer’s instructions. Briefly, Er1−/− and wt PPCs were fixed, washed with PBS, and stained in β-gal fixative solution at 37°C until β-gal staining became visible in either experimental or control plates (3 hours). Cells were washed in PBS, and β-gal–positive cells were counted in random fields in triplicate wells. For SenTraGor staining, cells were treated according to the manufacturer’s instructions (Arriani Pharmaceuticals, AR8850020). Briefly, fixed differentiated PPCs were washed with 50 and 70% EtOH incubated with SenTraGor reagent at RT, washed in EtOH, and incubated with primary anti-biotin antibody and fluorochrome-labeled secondary antibody. Cells were washed in PBS, and the lipofuscin-positive cells were counted in random fields in each of the six individual repeats using Fiji software.
SDS–polyacrylamide gel electrophoresis
For SDS–polyacrylamide gel electrophoresis (PAGE) analysis, cells were pelleted and tissues from Er1−/− and wt animals were homogenized in sucrose buffer [0.32 M sucrose, 15 mM Hepes-KOH, 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% BSA, 0.1% NP-40, and protease inhibitors (pH 7.9)]. Cell pellets were washed three times with 1× PBS. Cell pellets were then resuspended in NP-40 lysis buffer [10 mM tris-HCl (pH 7.9), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and protease inhibitors] and incubated for 10 min at 4°C. After centrifugation, the supernatant was kept as the cytoplasmic fraction, and pellets were resuspended in high-salt extraction buffer [10 mM Hepes-KOH (pH 7.9), 380 mM KCl, 3 mM MgCl2, 0.2 mM EDTA, 20% glycerol, and protease inhibitors] and incubated for 60 min at 4°C. The supernatant after centrifugation was kept as the nuclear fraction. For whole-cell extract preparations, cell pellets were resuspended in 150 mM NaCl, 50 mM tris (pH 7.5), 5% glycerol, 1% NP-40, and 1 mM MgCl and incubated on ice for 20 min.
Antibodies
Antibodies against FANCI (H102; IF: 1/50), amylase (G-10; WB: 1/500, IF: 1/100), goat anti-rat immunoglobulin G (IgG)–CFL 647 (sc-362293; IF: 1/1000), translocase of outer membrane, subunit 20 (TOM20) (sc-17764; IF: 1/100), XPF (sc-398032; WB: 1/500), and donkey anti-goat IgG–horse radish peroxidase (HRP) (sc-2020; WB: 1/5000) were from Santa Cruz Biotechnology. Antibodies against ssDNA (MAB3299; IF: 1/70, IP: 5 μg/1.5 mg of protein), dsDNA (MAB1293; IF: 1/100), S9.6 (MABE1095; IF: 1/100, DRIP: 8 μg/5 μg of DNA), γH2AX (05-636; IF: 1/12,000), goat anti-rabbit IgG-HRP (AP132P; WB: 1/10,000), and goat anti-mouse IgG-HRP (AP124P; WB: 1/10,000) were from Millipore. α-SMA (ab5694; WB: 1/500, IF: 1/50), HMGB1 (ab18256; WB: 1/1000, IF: 1/150), γH2AX (ab22551; WB: 1/500), GAPDH (glyceraldehyde phosphate dehydrogenase) (ab8245; WB: 1/4000), fibrilarin (FBL) (ab5821; WB: 1/2500), and β-tubulin (ab6046; WB: 1/1000) were from Abcam; cleaved caspase 3 (#9661; IF: 1/50, WB: 1/500), NF-κB p65 (#8242; WB: 1/500, IF: 1/100), pATM (#S1981; IF: 1/100), STING (#13647S; WB: 1/400), pSTING (#72971S; WB: 1/400), and pStat1 (#13647S; WB: 1/500) were from Cell Signaling Technology. VCAM (P8B1; IF: 1/200), CD45 (H5A5; IF: 1/200), and MAC1 (M1/70.15.11.5.2; IF: 1/200) were from the Developmental Studies Hybridoma Bank. 53BP1 (NB100-304; IF: 1300), RPA70 (NB100-2204; WB: 1/1000), TOP1 (NBP1-30482; WB: 1/1000), and goat anti-mouse IgM 550 (NB120-9167R; IF: 1/200) were from Novus Biologicals. Goat anti-mouse IgG Alexa Fluor 488 (A-11001; IF: 1/2000), goat anti-mouse IgG Alexa Fluor 555 (A-21422; IF: 1/2000), donkey anti-rabbit IgG Alexa Fluor 488 (A-21206; IF: 1/2000), donkey anti-rabbit IgG Alexa Fluor 555 (A-31572; IF: 1/2000), goat anti-rat IgG Alexa Fluor 555 (A-21434; IF: 1/2000), XPG (PA5-102437; WB: 1/1000), and DAPI (62247; IF: 1/20,000) were from Thermo Fisher Scientific, and CPD (TDM-2; IF: 1/50) was from Cosmo Bio. Antibody against RNase H1 (15606-1-AP; WB: 1/500) was from ProteinTech. Antibody against TRF1 (PA5-28423; IF: 1/100) was from Invitrogen.
SEM and TEM
For SEM, fresh pancreata were cut into small blocks. The tissue was fixed for 2 hours with 2% paraformaldehyde (PFA)–2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.42). Samples were postfixed overnight in 1% osmium tetroxide (OTO method) and dehydrated in a graded series of ethanol. Specimens were coated in gold, mounted on aluminum stubs, and examined with a JEOL JSM6390 LV SEM (Peabody, MA) using an accelerating voltage of 15 kV. For TEM, pancreata were washed in PBS and fixed in 2% PFA–2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.42) with 0.1% magnesium chloride and 0.05% calcium chloride. Afterward, the tissue was washed with sodium cacodylate buffer, the samples were fixed in 1% osmium tetroxide in sodium cacodylate buffer, and samples were dehydrated in ethanol gradient. Samples were then treated with propylene oxide and embedded in an epon/araldite resin mix. Ultrathin sections (50 to 100 nm) were taken on a Leica LKB2088 ultramicrotome and were examined under JEM 100C/JEOL/Japan TEM. Microphotographs were obtained with an ES500W Enlangshen camera and processed with the Digital Micrograph software (Gatan, Germany).
Annexin V–PI FACS analysis
Pancreata from 15-day-old Er1−/− and wt (n = 3) were minced and digested in 1× PBS/1% BSA/0.1% NaN3/collagenase (2.5 mg/ml). Homogenized tissue was further washed twice with ice-cold PBS and passed through a 100 μΜ wire mesh. The cells (106/ml) were resuspended in 1× binding buffer. Cells (105) were used for the staining, and 5 μl of annexin V–fluorescein isothiocyanate and 5 μl of PI were added per sample. The cells were vortexed gently and incubated for 15 min at RT in the dark. Four hundred microliters of 1× binding buffer was added to each sample. A FACSCalibur (BD Biosciences) was used, and data were analyzed using the FlowJo software (Tree Star).
TUNEL assay
The pancreatic cells or tissue were fixed for 30 min in 4% formaldehyde, washed for 30 min, and permeabilized in 0.1% Triton X-100 or 0.1% Na citrate for 2 or 8 min, respectively. The samples were washed for 30 min and mixed with 45 μl of TUNEL mix and 5 μl of enzyme per reaction for 1 hour at 37°C in the dark. Then, they were washed with PBS and counterstained for 10 min with DAPI. For positive control, PPCs were treated for 10 min with 2 U of micrococcal nuclease (MNase)/0.01 U of deoxyribonuclease (DNase) I. As a negative control, the cells were stained only with the labeling solution.
Telomere length measurement
The measurements of telomere length in pancreata were conducted as described previously (60). The pancreata were lysed, and nuclei were processed for DNA extraction. After phenol purification, the DNA was precipitated with 2.5 volumes of ice-cold 100% EtOH and 200 mM NaCl at −20°C overnight. DNA quantity was measured with nanodrop and diluted at a final concentration of 35 ng/μl. The PCR program for the telomeric amplification is as follows: 95°C for 10 min, (95°C for 15 s, 54°C for 2 min, and 72°C for 7 min) × 22 cycles, and 72°C for 7 min; and for the single-copy gene (Rarb): 95°C for 10 min, (95°C for 15 s, 58°C for 1 min, and 72°C for 30 s) × 30 cycles, 72°C for 5 min, and 10°C for forever. For the master mix, 15 mM tris-Cl (pH 8), 50 mM KCl, 2 mM MgCl2, 0.2 mM dNTPs (deoxyribonucleotide triphosphate), 5 mM dithiothreitol, 1% DMSO, 5× Sybr Green, and 1.25 U of Platinum Taq. The standard curve was prepared with the use of serial dilutions of a DNA sample of known quantity. The primer sequences used in qPCR are, for TelC-telG, GGTTTTTGAGGGTGAGGGTGAG (forward) and GGTGAGGGTGAGGGTTCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA (reverse) and, for Rarb, CCTGAGTCAGCAAGGACACT (forward) and CCTGAGTCAGCAAGGACACT (reverse).
Immunofluorescence in situ hybridization
PPCs were seeded on coverslips and fixed in 4% PFA/1× PBS for 10 min at 4°C. Immunostaining for 53BP1 was performed as described above; cells were then washed with 1× PBS and fixed again in 4% FA/1× PBS for 10 min at RT. After dehydration of the cells in increasing concentration of ethanol, 250 nM peanut agglutinin telomeric probe (TelC-Cy3; F1002) was added to the coverslip, in hybridization buffer [20 mM tris (pH 7.4), 70% formamide, salmon sperm DNA (0.1 μg/ml), and 2× standard saline citrate (SSC)]. The cells were denatured at 85°C for 10 min and then left at RT for 2 hours. Then, cells were washed in prewarmed 2× SSC/0.1% Tween 20, and nuclei were counterstained with DAPI.
Metaphase chromosome spreads
For the metaphase chromosome studies in wt and Er1−/− pancreatic cells, cells were arrested in colcemid (0.1 μg/ml) for 20 hours, harvested by trypsinization, incubated for 15 min at 37°C in 75 mM KCl, and fixed in freshly made methanol/acetic acid (3:1). Cells were dropped onto wet slides and air-dried overnight in a chemical hood. Fluorescence in situ hybridization was performed as described above.
RNA-seq and qPCR
Total RNA was isolated from wt and Er1−/− pancreata with the total RNA isolation kit (Qiagen) as described by the manufacturer. For RNA-seq, libraries were prepared using an Illumina TruSeq mRNA stranded sample preparation kit. Library preparation started with 1 μg of total RNA. After poly-A selection (using poly-T oligo-attached magnetic beads), mRNA was purified and fragmented using divalent cations under elevated temperature. The RNA fragments underwent reverse transcription using random primers. This is followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. After end repair and A-tailing, indexing adapters were ligated. The products were then purified and amplified (14 PCR cycles) to create the final cDNA libraries. After library validation and quantification (Agilent 2100 Bioanalyzer), equimolar amounts of library were pooled. The pool was quantified by using the Peqlab KAPA Library Quantification Kit and the Applied Biosystems 7900HT Sequence Detection System. The pool was sequenced by using an S2 flow cell on the Illumina NovaSeq6000 sequencer and the 2x100nt protocol. qPCR was performed with a Bio-Rad 1000-series thermal cycler according to the instructions of the manufacturer (Bio-Rad) as previously described (61). In case of the qPCR for the intronless Ifna gene, the RNA samples were treated with DNase (Promega). The generation of specific PCR products was confirmed by melting curve analysis and gel electrophoresis. Each primer pair was tested with a logarithmic dilution of a cDNA mix to generate a linear standard curve [crossing point (CP) plotted versus log of template concentration], which was used to calculate the primer pair efficiency (E = 10(−1/slope)). Hypoxanthine guanine phosphoribosyltransferase 1 (Hprt-1) mRNA was used as an external standard. For data analysis, the second derivative maximum method was applied: (E1gene of interest ΔCP (cDNA of wt mice − cDNA of Ercc1−/−) gene of interest)/(EGapdh ΔCP (cDNA wt mice − cDNA) gapdh). For cytoplasmic ssDNA qPCR, the Klenow fragment of DNA polymerase I was used to generate the second strand at 25°C for 1.5 hours in the presence of random hexamers and dNTPs.
The genes tested by qPCR are as follows: Il6 (forward: AGTTGCCTTCTTGGGACTGA; reverse: CAGAATTGCCATTGCACAAC), Ccl2 (forward: TGATCCCAATGAGTAGGCTGGAG; reverse: ATGTCTGGACCCATTCCTTCTTG), Cxcl10 (forward: ATGACGGGCCAGTGAGAATG; reverse: CATCGTGGCAATGATCTCAACA), Col1α (forward: CCTCAGGGTATTGCTGGACA; reverse: TCACGTCCAGATTCACCAGG), Mmp9 (forward: GTCATTCGCGTGGATAAGGAG; reverse: CACTGCAGGAGGTCGTAGG), Timp2 (forward: CGAGTTTATCTACACGGCCC; reverse: CAGGCTCTTCTTCTGGGTGA), Bcl2 (foward: TCGCAGAGATGTCCAGTCAG; reverse: ATGCCGGTTCAGGTACTCAG), Casp8 (forward: GGCCTCCATCTATGACCTGA; reverse: TGTGGTTCTGTTGCTCGAAG), Bad (forward: CGTGAGCTCCGAAGGATGAG; reverse: TCGCATCTGTGTTGCAGTG), Bcl_xl (forward: AGGGGCTTAGCTGCTGAAAG; reverse: GTGGACAAGGATCTTGGGGG), P21 (forward: TTGCACTCTGGTGTCTGAGC; reverse: TCTGCGCTTGGAGTGATAGA), Sting (forward: AAATAACTGCCGCCTCATTG; reverse: TGGGAGAGGCTGATCCATAC), Ifit1 (forward: CCAAGTGTTCCAATGCTCCT; reverse: GGATGGAATTGCCTGCTAGA), Ifit2 (foward: AGTACAACGAGTAAGGAGTCACT; reverse: AGGCCAGTATGTTGCACATGG), Ifi206 (forward: ACTGTGAGCCTGGGGATAAG; reverse: AGGGATCCATGAGAGTGAGC), II1r1 (forward: ACTGTTGCCTGAGGTCTTGG; reverse: CAGCTGAAGCCTCCCATATC), Ifnα (forward: CTGCTGGCTGTGAGGACATA; reverse: GGCTCTCCAGACTTCTGCTC), Ifitm1 (forward: GACAGCCACCACAATCAACAT; reverse: CCCAGGCAGCAGAAGTTCAT), Usp18 (forward: GAACTCTTTGCCGTGATTGC; reverse: CCAAGAGATAGGCCGTTTCC), Mx1 (forward: GACCATAGGGGTCTTGACCAA; reverse: AGACTTGCTCTTTCTGAAAAGCC), Isg15 (forward: GGTGTCCGTGACTAACTCCAT; reverse: TGGAAAGGGTAAGACCGTCCT), Irf8 (forward: CCAAACTCATTCTGGTGCAG; reverse: AAAGGGTCTCTGGTGTGAGG), Irf1 (forward: GGAAGGGAAGATAGCCGAAG; reverse: GGGCTGTCAATCTCTGGTTC), Tnfα (forward: CACGTCGTAGCAAACCACC; reverse: GATAGCAAATCGGCTGACGG), Gapdh (forward: ACTGAGCAAGAGAGGCCCTA; reverse: TATGGGGGTCTGGGATGGAA), Il1β (forward: GCCCATCCTCTGTGACTCAT; reverse: TTGTCGTTGCTTGGTTCTCC), Tert (forward: TCAAGCTGGCTGCTCATTCT; reverse: GGATGGTCATTGTCGCCTCT), Rarb (forward: GGATTAGAGTTTTTGTGTGTTG; reverse: TACCCCACCAATACCCAAACA), Stra6 (forward: ATATGGTGGCGCTCGTAGTG; reverse: TCTGCAATGGTCCTGACACC), b-actin_pr (forward: GAGGGGAGAGGGGGTAAA; reverse: GAAGCTGTGCTCGCGG), Β-actin (forward: TAGGGTCGCGGTGTG; reverse: GGTTCCCAATACTGTGTACTC), L1 ORF1 (forward: ATCTGTCTCCCAGGTCTGCT; reverse: AGTGCTGCGTTCTGATGATG), L1 ORF2 (forward: GCTTCGGTGAAGTAGCTGGA; reverse: TTCGTTAGAGTCACGCCGAG), L1 5′UTR (forward: CTGCCTTGCAAGAAGAGAGC; reverse: AGTGCTGCGTTCTGATGATG), and L1 3′UTR (forward: AGCCAAATGGATGGACCTGG; reverse: AAGGAGGGGCATAGTGTCCA).
Data analysis
For RNA-seq, the data were downloaded as FASTQ files, and their quality was checked with FASTQC, a quality control tool for high-throughput sequence data: www.bioinformatics.babraham.ac.uk/projects/fastqc. A warning for overrepresenting sequences related to hydrolytic enzymes was ignored because of the physiology of the pancreas. The data were aligned to mm10 genome assembly available from the University of California Santa Cruz via Bowtie2. The differentially expressed genes were identified with metaseqR. Count normalization was performed on the basis of edgeR algorithm. Statistical analysis was performed with DESeq2, edgeR, and NOISeq algorithms. Differentially expressed genes found by all three algorithms were used for downstream analysis. Significant overrepresentation of pathways and gene networks was determined by Gene Ontology (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes pathways (www.genome.jp/kegg/pathway.html). Data analysis was performed with the PANTHER Classification System using the overrepresentation test. The P values were determined by the Fisher’s exact test, and fold enrichment refers to the observed over the expected number within the reference list (Mus musculus) that map to the annotation data category. For all analyses, unless otherwise indicated, P values were calculated by two-tailed Student’s t test, and the significance is set at P value ≤ 0.05.
DRIP assay
DRIP analysis was based on the protocol by Goulielmaki et al. (33). Briefly, DRIP was performed without a cross-linking step. Nuclei were isolated using 0.5% NP-40 buffer and resuspended in TE buffer supplemented with 0.5% SDS and 100 mg of proteinase K. Genomic DNA was isolated after the addition of potassium acetate (1 M) and isopropanol precipitation. DNA was sonicated on ice for 3 min using the Covaris S220 Focused Ultrasonicator. Samples were then treated with RNase H (10 U/5 μg of DNA) at 37°C overnight. Samples were immunoprecipitated with S9.6 antibody (8 μg of antibody/5 μg of DNA) overnight at 4°C, followed by incubation for 3 hours with protein G–Sepharose beads (Millipore) and washed sequentially. The complexes were eluted and resolved on 8 to 12% SDS-PAGE.
ssDNA immunoprecipitation assay
For coimmunoprecipitation assays, untreated or MMC-treated PPCs from wt mice or pancreata from wt and Er1−/− mice were fixed in 1% formaldehyde at RT for 2.5 min, and cells were lysed using the NP-40 lysis buffer [10 mM tris-HCl (pH 7.9), 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and protease inhibitors]. Cytoplasmic lysates (1.5 to 3 mg) were diluted threefold by adding ice-cold HENG buffer [10 mM Hepes-KOH (pH 7.9), 1.5 mM MgCl2, 0.25 mM EDTA, and 20% glycerol] and precipitated with 5 to 10 μg of anti-ssDNA antibody (MAB3299) o/n at 4°C followed by incubation for 3 hours with protein G Sepharose beads (Millipore). Immunoprecipitates were washed five times [10 mM Hepes-KOH (pH 7.9), 300 mM KCl, 0.3% NP-40, 1.5 mM MgCl2, 0.25 mM EDTA, 20% glycerol, and protease inhibitors], eluted, and resolved on 8 to 12% SDS-PAGE.
Acknowledgments
Funding: The Horizon 2020 ERC Consolidator grant “DeFiNER” (GA 64663); the ERC PoC “Inflacare” (GA 874456); the Horizon 2020 Marie Curie ITN “aDDRess” (GA 812829), “HealthAge” (GA 812830), and “Chromatin3D (GA 622934); the Santé Foundation; ELIDEK grant 631, and EDBM MIS: 5048456 supported this work.
Author contributions: Conceptualization: G.A.G. and K.S. Methodology: O.C., K.S., E.G., G.N., A.A.-C., K.G., A.Z., P.T., and J.A. Investigation: O.C., K.S., E.G., G.N., A.A.-C., K.G., A.Z., P.T., and J.A. Visualization: O.C., K.S., E.G., G.N., and K.G. Supervision: G.A.G. Writing—Original draft: G.A.G. and K.S. Writing—Review and editing: G.A.G. and K.S.
Competing interests: O.C., K.S., E.G., G.N., K.G., and G.A.G. are inventors on a patent application related to this work filed by the Foundation of Research and Technology–Hellas, application number EP21386046.3, submission number: 10064400, filed 10 August 2021. The authors declare that they have no other competing interests.
Data and materials availability: The RNA-seq data (E-MTAB-10393) are deposited in ArrayExpress (www.ebi.ac.uk/arrayexpress/). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Chatzidoukaki O., Goulielmaki E., Schumacher B., Garinis G. A., DNA damage response and metabolic reprogramming in health and disease. Trends Genet. 36, 777–791 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Rieckher M., Garinis G. A., Schumacher B., Molecular pathology of rare progeroid diseases. Trends Mol. Med. 27, 907–922 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Schumacher B., Pothof J., Vijg J., Hoeijmakers J. H. J., The central role of DNA damage in the ageing process. Nature 592, 695–703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apostolou Z., Chatzinikolaou G., Stratigi K., Garinis G. A., Nucleotide excision repair and transcription-associated genome instability. Bioessays 41, e1800201 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kamileri I., Karakasilioti I., Garinis G. A., Nucleotide excision repair: New tricks with old bricks. Trends Genet. 28, 566–573 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Hanawalt P. C., Subpathways of nucleotide excision repair and their regulation. Oncogene 21, 8949–8956 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Marteijn J. A., Lans H., Vermeulen W., Hoeijmakers J. H., Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Chatzinikolaou G., Karakasilioti I., Garinis G. A., DNA damage and innate immunity: Links and trade-offs. Trends Immunol. 35, 429–435 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Stratigi K., Chatzidoukaki O., Garinis G. A., DNA damage-induced inflammation and nuclear architecture. Mech. Ageing Dev. 165, 17–26 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Karakasilioti I., Kamileri I., Chatzinikolaou G., Kosteas T., Vergadi E., Robinson A. R., Tsamardinos I., Rozgaja T. A., Siakouli S., Tsatsanis C., Niedernhofer L. J., Garinis G. A., DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 18, 403–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilstra J. S., Robinson A. R., Wang J., Gregg S. Q., Clauson C. L., Reay D. P., Nasto L. A., Croix C. M. St., Usas A., Vo N., Huard J., Clemens P. R., Stolz D. B., Guttridge D. C., Watkins S. C., Garinis G. A., Wang Y., Niedernhofer L. J., Robbins P. D., NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J. Clin. Invest. 122, 2601–2612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niedernhofer L. J., Odijk H., Budzowska M., van Drunen E., Maas A., Theil A. F., de Wit J., Jaspers N. G., Beverloo H. B., Hoeijmakers J. H., Kanaar R., The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24, 5776–5787 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedernhofer L. J., Garinis G. A., Raams A., Lalai A. S., Robinson A. R., Appeldoorn E., Odijk H., Oostendorp R., Ahmad A., van Leeuwen W., Theil A. F., Vermeulen W., van der Horst G. T., Meinecke P., Kleijer W. J., Vijg J., Jaspers N. G., Hoeijmakers J. H., A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Garinis G. A., van der Horst G. T., Vijg J., Hoeijmakers J. H., DNA damage and ageing: New-age ideas for an age-old problem. Nat. Cell Biol. 10, 1241–1247 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulielmaki E., Ioannidou A., Tsekrekou M., Stratigi K., Poutakidou I. K., Gkirtzimanaki K., Aivaliotis M., Evangelou K., Topalis P., Altmuller J., Gorgoulis V. G., Chatzinikolaou G., Garinis G. A., Tissue-infiltrating macrophages mediate an exosome-based metabolic reprogramming upon DNA damage. Nat. Commun. 11, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huerta Guevara A. P., McGowan S. J., Kazantzis M., Stallons T. R., Sano T., Mulder N. L., Jurdzinski A., van Dijk T. H., Eggen B. J. L., Jonker J. W., Niedernhofer L. J., Kruit J. K., Increased insulin sensitivity and diminished pancreatic beta-cell function in DNA repair deficient Ercc1 mice. Metabolism 117, 154711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh H. C., Kwon C. I., El Haji I. I., Easler J. J., Watkins J., Fogel E. L., McHenry L., Sherman S., Zimmerman M. K., Lehman G. A., Low serum pancreatic amylase and lipase values are simple and useful predictors to diagnose chronic pancreatitis. Gut Liver 11, 878–883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A., H2AX: The histone guardian of the genome. DNA Repair 3, 959–967 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Sato K., Ishiai M., Toda K., Furukoshi S., Osakabe A., Tachiwana H., Takizawa Y., Kagawa W., Kitao H., Dohmae N., Obuse C., Kimura H., Takata M., Kurumizaka H., Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 31, 3524–3536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Childs B. G., Baker D. J., Kirkland J. L., Campisi J., van Deursen J. M., Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 15, 1139–1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takai H., Smogorzewska A., de Lange T., DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., de Lange T., ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Scaffidi P., Misteli T., Bianchi M. E., Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418, 191–195 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Bloom O., Zhang M., Vishnubhakat J. M., Ombrellino M., Che J., Frazier A., Yang H., Ivanova S., Borovikova L., Manogue K. R., Faist E., Abraham E., Andersson J., Andersson U., Molina P. E., Abumrad N. N., Sama A., Tracey K. J., HMG-1 as a late mediator of endotoxin lethality in mice. Science 285, 248–251 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Davalos A. R., Kawahara M., Malhotra G. K., Schaum N., Huang J., Ved U., Beausejour C. M., Coppe J. P., Rodier F., Campisi J., p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 201, 613–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodier F., Coppe J. P., Patil C. K., Hoeijmakers W. A., Munoz D. P., Raza S. R., Freund A., Campeau E., Davalos A. R., Campisi J., Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garinis G. A., Uittenboogaard L. M., Stachelscheid H., Fousteri M., van Ijcken W., Breit T. M., van Steeg H., Mullenders L. H., van der Horst G. T., Bruning J. C., Niessen C. M., Hoeijmakers J. H., Schumacher B., Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat. Cell Biol. 11, 604–615 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I. Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., Al Ghonaium A., Tufenkeji H., Frayha H., Al-Gazlan S., Al-Rayes H., Schreiber R. D., Gresser I., Casanova J. L., Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33, 388–391 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Kowalski D., Kroeker W. D., Laskowski M. Sr., Mung bean nuclease I. Physical, chemical, and catalytic properties. Biochemistry 15, 4457–4463 (1976). [DOI] [PubMed] [Google Scholar]
- 30.Garinis G. A., Mitchell J. R., Moorhouse M. J., Hanada K., de Waard H., Vandeputte D., Jans J., Brand K., Smid M., van der Spek P. J., Hoeijmakers J. H., Kanaar R., van der Horst G. T., Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 24, 3952–3962 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smolka J. A., Sanz L. A., Hartono S. R., Chedin F., Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J. Cell Biol. 220, e202004079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva S., Camino L. P., Aguilera A., Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. U.S.A. 115, 11024–11029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulielmaki E., Tsekrekou M., Batsiotos N., Ascensao-Ferreira M., Ledaki E., Stratigi K., Chatzinikolaou G., Topalis P., Kosteas T., Altmuller J., Demmers J. A., Barbosa-Morais N. L., Garinis G. A., The splicing factor XAB2 interacts with ERCC1-XPF and XPG for R-loop processing. Nat. Commun. 12, 3153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N. J., R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cristini A., Groh M., Kristiansen M. S., Gromak N., RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R. S., Taguchi T., De Cecco M., Leonova K. I., Kogan V., Helfand S. L., Neretti N., Roichman A., Cohen H. Y., Meer M. V., Gladyshev V. N., Antoch M. P., Gudkov A. V., Sedivy J. M., Seluanov A., Gorbunova V., LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuduri S., Crabbe L., Conti C., Tourriere H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C., Pommier Y., Tazi J., Coquelle A., Pasero P., Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Promonet A., Padioleau I., Liu Y., Sanz L., Biernacka A., Schmitz A. L., Skrzypczak M., Sarrazin A., Mettling C., Rowicka M., Ginalski K., Chedin F., Chen C. L., Lin Y. L., Pasero P., Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 11, 3940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Vuuren A. J., Appeldoorn E., Odijk H., Yasui A., Jaspers N. G., Bootsma D., Hoeijmakers J. H., Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J. 12, 3693–3701 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biggerstaff M., Szymkowski D. E., Wood R. D., Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 12, 3685–3692 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harding S. M., Benci J. L., Irianto J., Discher D. E., Minn A. J., Greenberg R. A., Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wold M. S., Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 (1997). [DOI] [PubMed] [Google Scholar]
- 43.Toledo L. I., Altmeyer M., Rask M. B., Lukas C., Larsen D. H., Povlsen L. K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J., ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Baltimore D., Discovering NF-κB. Cold Spring Harb. Perspect. Biol. 1, a000026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterzenbach U., Putz U., Low L. H., Silke J., Tan S. S., Howitt J., Engineered exosomes as vehicles for biologically active proteins. Mol. Ther. 25, 1269–1278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohr J. M., Panic N., Vujasinovic M., Verbeke C. S., The ageing pancreas: A systematic review of the evidence and analysis of the consequences. J. Intern. Med. 283, 446–460 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Skourti-Stathaki K., Proudfoot N. J., Gromak N., Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Manley J. L., Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Huertas P., Aguilera A., Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Sollier J., Stork C. T., Garcia-Rubio M. L., Paulsen R. D., Aguilera A., Cimprich K. A., Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackenzie K. J., Carroll P., Lettice L., Tarnauskaite Z., Reddy K., Dix F., Revuelta A., Abbondati E., Rigby R. E., Rabe B., Kilanowski F., Grimes G., Fluteau A., Devenney P. S., Hill R. E., Reijns M. A., Jackson A. P., Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35, 831–844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coquel F., Silva M. J., Techer H., Zadorozhny K., Sharma S., Nieminuszczy J., Mettling C., Dardillac E., Barthe A., Schmitz A. L., Promonet A., Cribier A., Sarrazin A., Niedzwiedz W., Lopez B., Costanzo V., Krejci L., Chabes A., Benkirane M., Lin Y. L., Pasero P., SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Koo C. X., Kobiyama K., Shen Y. J., LeBert N., Ahmad S., Khatoo M., Aoshi T., Gasser S., Ishii K. J., RNA polymerase III regulates cytosolic RNA:DNA hybrids and intracellular microRNA expression. J. Biol. Chem. 290, 7463–7473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farkash E. A., Luning Prak E. T., DNA damage and L1 retrotransposition. J. Biomed. Biotechnol. 2006, 37285 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ginno P. A., Lott P. L., Christensen H. C., Korf I., Chedin F., R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen W., Sun H., De Hoyos C. L., Bailey J. K., Liang X. H., Crooke S. T., Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Res. 45, 10672–10692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ioannidou A., Goulielmaki E., Garinis G. A., DNA damage: From chronic inflammation to age-related deterioration. Front. Genet. 7, 187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Pluijm I., Garinis G. A., Brandt R. M., Gorgels T. G., Wijnhoven S. W., Diderich K. E., de Wit J., Mitchell J. R., van Oostrom C., Beems R., Niedernhofer L. J., Velasco S., Friedberg E. C., Tanaka K., van Steeg H., Hoeijmakers J. H., van der Horst G. T., Impaired genome maintenance suppresses the growth hormone–insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLOS Biol. 5, e2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta S., Rawat S., Arora V., Kottarath S. K., Dinda A. K., Vaishnav P. K., Nayak B., Mohanty S., An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther 9, 180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cawthon R. M., Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chatzinikolaou G., Apostolou Z., Aid-Pavlidis T., Ioannidou A., Karakasilioti I., Papadopoulos G. L., Aivaliotis M., Tsekrekou M., Strouboulis J., Kosteas T., Garinis G. A., ERCC1-XPF cooperates with CTCF and cohesin to facilitate the developmental silencing of imprinted genes. Nat. Cell Biol. 19, 421–432 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legend for data S1
Data S1