ABSTRACT
Infection by (re-)emerging RNA arboviruses including Chikungunya virus (CHIKV) and Mayaro virus primarily cause acute febrile disease and transient polyarthralgia. However, in a significant subset of infected individuals, debilitating arthralgia persists for weeks over months up to years. The underlying immunopathogenesis of chronification of arthralgia upon primary RNA-viral infection remains unclear. Here, we analysed cell-intrinsic responses to ex vivo arthritogenic alphaviral infection of primary human synovial fibroblasts isolated from knee joints, one the most affected joint types during acute and chronic CHIKV disease. Synovial fibroblasts were susceptible and permissive to alphaviral infection. Base-line and exogenously added type I interferon (IFN) partially and potently restricted infection, respectively. RNA-seq revealed a CHIKV infection-induced transcriptional profile that comprised upregulation of expression of several hundred IFN-stimulated and arthralgia-mediating genes. Single-cell virus-inclusive RNA-seq uncovered a fine-tuned switch from induction to repression of cell-intrinsic immune responses depending on the abundance of viral RNA in an individual cell. Specifically, responses were most pronounced in cells displaying low-to-intermediate amounts of viral RNA and absence of virus-encoded, fluorescent reporter protein expression, arguing for efficient counteraction of innate immunity in cells expressing viral antagonists at sufficient quantities. In summary, cell-intrinsic sensing of viral RNA that potentially persists or replicates at low levels in synovial fibroblasts and other target cell types in vivo may contribute to the chronic arthralgia induced by alphaviral infections. Our findings might advance our understanding of the immunopathophysiology of long-term pathogenesis of RNA-viral infections.
KEYWORDS: Chikungunya, fibroblasts, innate immunity, RNA-seq, transcriptomics
Introduction
Chikungunya virus (CHIKV) and Mayaro virus (MAYV) are arthritogenic alphaviruses of the Togaviridae family, which are transmitted by Aedes sp. mosquitoes and circulate both in urban cycles between vectors and humans, and in sylvatic cycles [1,2]. Hallmarks of the typically relatively short acute disease are febrile illness, rashes, and excruciating pain in multiple joints. Relapsing-remitting arthralgia persists in a subgroup of patients for months to years [3]. The underlying immunopathophysiology of the chronic symptoms remains largely unclear, but appears to associate with circulating IL-6 [4] and IL-12 [5]. Interestingly, a much-discussed hypothesis suggests that it may involve persistence of viral RNA in synovial macrophages, muscle cells, and fibroblasts in vivo [5–7].
Multiple studies on alphaviruses in immortalized model cell lines and in vivo in immunodeficient mice have provided valuable information on key aspects of CHIKV and MAYV tropism and replication, including host factors for entry and replication [8,9], the impact of mutations in the viral glycoproteins on cell entry [10], and cellular restriction factors acting against CHIKV and other alphaviruses [11]. Additionally, studies investigating immune responses to infection have demonstrated that CHIKV nsP2 counteracts host immunity by blocking nuclear translocation of STAT1 [12] and inducing a host transcriptional shutdown [13]. Sensing of infection-induced DNA leakage, as described for RNA viruses before [14], is efficiently subverted by CHIKV through its nonstructural proteins [15]. However, the relevance and consequence of these and potentially additional immunity-subverting mechanisms in infected patients remain unclear. In vivo studies in mice, though recapitulating both innate and adaptive immune responses, require a type I interferon (IFN)-deficient background, neglecting the impact of type I IFN-mediated antiviral responses [16]. Type I IFN induced in and acting on nonhematopoietic cells appears to be essential for the control and early clearance of CHIKV in vivo [17]. Therefore, these models do not fully recapitulate the cellular environment of human primary cells and tissues that are targeted by CHIKV and MAYV in vivo. Primary human cells have been used sporadically for ex vivo infection [18,19], but their unique properties in terms of susceptibility to infection and cell-intrinsic responses remain poorly investigated. Synovial fibroblasts have been described to be a key driver for rheumatoid arthritis by facilitating proinflammatory processes and stimulating the degradation of cartilage [20], and it is plausible that they contribute to CHIKV spread and pathogenesis in vivo. Here, we perform an in depth-characterization of primary human synovial fibroblasts extracted from human knee joint biopsies as an ex vivo model of CHIKV and MAYV infection. We demonstrate that synovial fibroblasts are susceptible and permissive to infection by both arthritogenic alphaviruses. Using bulk and single-cell approaches, we identified cell-intrinsic immune responses that were most pronounced in non-productively infected cells, suggestive of effective viral antagonism of cellular responses in cells undergoing efficient virus replication.
Material and methods
Cells and viruses
Human osteosarcoma U2OS cells (a kind gift from T. Stradal, Hanover), human HEK293 T cells (a kind gift from J. Bohne, Hanover), human foreskin fibroblast HFF-1 cells (ATCC SCRC-1041), human HL116 cells (a kind gift from Sandra Pellegrini, Institut Pasteur, France [21]), and hamster BHK-21 cells (ATCC CCL-10) were grown in Dulbecco’s modified Eagle’s medium – high glucose (DMEM, Sigma-Aldrich D5671) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich F7524), 2 mM L-Glutamine (Gibco 25030081), and 100 units/ml penicillin–streptomycin (Gibco 11548876). HL116 cell received 1X HAT supplement (Gibco 21060017) in addition.
Primary human fibroblasts were obtained from synovial biopsies from knee joins from donors suffering from osteoarthritis (osteroarthrosis synovial fibroblasts, OASF) or a non-arthritic background (healthy donor synovial fibroblasts, HSF), purified, and cultured as described before [22]. Mycoplasma testing was routinely performed and negative in all primary human cell cultures. After 2–4 passages of initial cultivation, cells were expanded and used for experiments in high glucose DMEM supplemented with 20% FBS, 2 mM L-Glutamine, 100 units/ml penicillin–streptomycin, 1% non-essential amino acids (Gibco 11140050), and 1% sodium pyruvate (Gibco 11360070). The CHIKV LR2006-OPY 5′GFP and MAYV TRVL4675 5′GFP infectious clones expressing EGFP under the control of a subgenomic promotor (hereafter referred to as CHIKV and MAYV) have been described previously [23,24]. Virus was produced by in vitro-transcription of and subsequent electroporation of RNA into BHK-21 cells. Virus-containing supernatant was collected, passaged once on BHK-21 cells and viral titers were determined by titration on HEK293T cells.
Infection, treatments, transfections
EGFP expression as surrogate for productive CHIKV or MAYV infection was quantified on a BD FACSCalibur, FACSLyric or Accuri C6. For neutralization assays, virus-containing supernatants were pre-incubated for one hour with anti-CHIKV E2 antibody C9 (Integral Molecular C9, Lot INT MAB-003) at 1 µg/ml or with recombinant MXRA8-Fc (a kind gift from M. Diamond) at 150 ng/ml. Recombinant IFN-α2a (Roferon L03AB04, Roche) and IFN-λ1 (Peprotech 300-02L) was used where indicated. Transfections were performed using Lipofectamine2000 (Thermo Fisher 11668019) for plasmid DNA (pcDNA6 empty vector) or 5′triphosphate dsRNA (InvivoGen tlrl-3prna).
Bulk RNA-Seq analysis
RNA was extracted using the Promega Maxwell 16 with LEV simplyRNA Tissue Kits (Promega AS1270). RNA quality was assessed using the Agilent Bioanalyzer and appropriate samples were used for NGS library preparation with the NEBNext Ultra II Directional RNA kit (NEB E7760) and sequenced with 50 bp paired-end reads and 30 mio reads per sample on the Illumina HiSeq 2500. Data were analysed with CLC Genomics Workbench 12 (QIAGEN) by mapping the human reads onto the hg19 reference genome scaffold (GCA_000001405.28). Unmapped reads not matching the human genome were subsequently mapped onto the CHIKV genome LR2006_OPY (DQ443544.2). For HSF, infection and analysis were performed similarly, but RNA was extracted with the Direct-Zol RNA MiniPrep Kit (Zymo Research R2051), NGS libraries were prepared with the TruSeq stranded mRNA kit (Illumina 20020594) and sequencing was performed on the Illumina NextSeq500 with 65 mio reads per sample. Biological process enrichment was analysed by Gene Ontology [25].
Single-Cell RNA-Seq analysis
Infected cells were trypsinized, debris was removed by filtration, and the suspension was adjusted to a final amount of ∼16,000 cells per lane to achieve the recovery of 10,000 cells per donor after partitioning into Gel-Beads in Emulsion (GEMs) according to the instructions for Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1 provided by the manufacturer (10X Genomics PN-1000121). Polyadenylated mRNAs were tagged with unique 16 bp 10X barcodes and the 10 bp Unique Molecular Identifiers (UMIs), reverse transcribed and resulting cDNAs were bulk amplified. After enzymatic fragmentation and size selection, resulting double-stranded cDNA amplicons optimized for library construction were subjected to adaptor ligation and sample index PCRs needed for Illumina bridge amplification and sequencing. Single-cell libraries were quantified using Qubit (Thermo Fisher) and quality-controlled using the Bioanalyzer System (Agilent). Sequencing was performed on a HiSeq4000 device (Illumina) aiming for 175 mln reads per library (read1: 26 nucleotides, read2: 64 nucleotides). Data were analysed using CellRanger v5.0 (10X Genomics) using human and CHIKV genome scaffolds as described above, and the R packages Seurat v4.0 [26] and DoRothEA v3.12 [27] were used for cell clustering, annotation, and transcription factor activity analysis. Median gene number detected per cell ranged between 2000 and 4400, with 3800–18500 median UMI counts per cell.
Quantitative RT–PCR
RNA was extracted using the Promega Maxwell 16 with the LEV simplyRNA tissue kit (Promega AS1270), the Roche MagNAPure with the Cellular Total RNA Large Volume kit (Roche 05467535001), or the DirectZol RNA Mini kit (Zymo R2051). cDNA was prepared using dNTPs (Thermo Fisher R0181), random hexamers (Jena Bioscience PM-301) and M-MuLV reverse transcriptase (NEB M0253). For quantitative RT–PCR, specific Taqman probes and primers (Thermo Fisher 4331182) were used with TaqMan Universal PCR Master Mix (Applied Biosystems 4305719) or LightCycler© 480 Probes Master (Roche 04887301001). PCRs were performed on the Applied Biosystems ABI 7500 Fast or the Roche LightCycler 480 in technical triplicates.
Flow cytometry, confocal and live cell imaging
For flow cytometric analysis of protein expression, OASF were fixed in 4% PFA (Carl Roth 4235.2), permeabilized in 0.1% Triton-X (Invitrogen HFH10) and immunostained with antibodies against IFIT1 (Origene TA500948, clone OTI3G8), MX1/2 (Santa Cruz sc-47197), and IFITM3 (Abgent AP1153a) in combination with Alexa Fluor-647 conjugated antibodies against mouse- (Thermo Fisher A28181), rabbit- (Thermo Fisher A27040), or goat-IgG (Thermo Fisher A-21447). Flow cytometry was performed on a BD FACSCalibur or FACSLyric and analysed with FlowJo v10. For immunofluorescence microscopy, OASF were seeded in 8-well µ-slides (ibidi 80826), fixed and permeabilized as described above, stained with antibodies against MXRA8 (biorbyt orb221523) with AlexaFluor647-conjugated secondary antibody (Thermo Fisher A28181), and counterstained with DAPI (Invitrogen D1306). For fluorescence microscopy and live cell imaging, cells were infected with CHIKV at an MOI of 10 and imaged with the Zeiss LSM800 Airyscan Confocal Microscope. Images were analysed and merged using Zeiss ZEN Blue 3.0.
Immunoblotting
Cell lysates were separated on 10% acrylamide gels by SDS-PAGE and protein transferred to a 0.45 µm PVDF membrane (GE Healthcare 15259894) using the BioRad TransBlot Turbo system. Expression was detected using primary antibodies detecting MXRA8 (biorbyt orb221523), FHL1 (R&D Systems MAB5938), IFITM3 (Abgent AP1153a), MX2 (Santa Cruz sc-47197), ISG15 (Santa Cruz sc-166755), and α-Tubulin (Cell Signalling Technology 2144S) and appropriate secondary IRDye antibodies. CHIKV proteins were detected using anti-CHIKV antiserum (IBT Bioservices Cat #01-0008 Lot #1703002). Fluorescence was detected and quantified using the LI-COR Odyssey Fc system.
Measurement of IL-6 and bioactive IFN
Anti-IL-6 ELISA (BioLegend 430504) was performed according to manufacturer’s protocols. Briefly, plates were coated with capture antibodies and incubated with diluted supernatant from CHIKV- or mock-infected cell cultures. Detection antibody and substrate were added and the OD measured with the Tecan Sunrise microplate reader. Concentrations were then calculated the concentration according to a standard curve measured on the same plate. Bioactive type I IFN was quantified by incubating supernatant from CHIKV-infected cells on HL116 cells harbouring a firefly luciferase gene under the control of an IFN-sensitive promotor. After six h, cells were lysed, incubated with luciferase substrate solution (Promega E1500), and luciferase activity was quantified with the BioTek Synergy HTX microplate reader.
Data and code availability
RNA-seq and single-cell RNA-seq datasets are available at the NCBI GEO database under the accession number GSE152782 and GSE176361, respectively. All generated code is available at https://github.com/GoffinetLab/CHIKV_scRNAseq-fibroblast.
Statistics
If not stated otherwise, bars and symbols show the arithmetic mean of indicated amount of repetitions. Error bars indicate S.D. from at least three or S.E.M. from the indicated amount of individual experiments. Statistical analysis was performed using CLC Workbench for RNA-seq and GraphPad Prism 8.3.0 for all other analysis. Unpaired, two-sided t-tests were applied with assumed equal standard deviation when comparing results obtained in the same cell line and Mann–Whitney-U-tests when comparing between cell lines or between cell lines and primary cells. For IC50 calculation, nonlinear fit curves with variable slopes were calculated. Differentially expressed genes in RNA-seq data were identified from raw count data, with calculation of false discovery rate correction (FDR) p-values for multiple comparisons. P-values for Gene Ontology analysis were generated after Bonferroni correction for multiple testing. For differential gene expression analysis from single-cell RNA-seq data, Wilcoxon rank sum tests with applied Bonferroni correction were applied. Cumulative distributions between groups were compared using the Kolmogorov–Smirnov (KS) test. Nonparametric Spearman tests with two-tailed p-values were performed to correlate gene expression. P values <0.05 were considered significant (*), <0.01 very significant (**), <0.001 highly significant (***); <0.0001 extremely significant, n.s. = not significant (≥0.05).
Study approval
The local ethic committee (Justus-Liebig-University Giessen) approved the cooperative study (ethical vote IDs 66–08 and 74-05). All patients gave written informed consent prior to inclusion in the study.
Results
Primary human synovial fibroblasts are susceptible and permissive to CHIKV and MAYV infection
First, we examined the ability of primary human synovial fibroblasts to support the entire CHIKV and MAYV replication cycle. Therefore, we infected synovial fibroblasts obtained from osteoarthritic patients (OASF) and from patients with a non-arthritic background (HSF) with CHIKV strain LR2006-OPY or MAYV strain TRVL7546 expressing EGFP under the control of a second subgenomic promotor. 24 h post-infection, the proportion of EGFP-positive cells ranged between 4% and 24.5% for CHIKV and between 8.5 and 39% for MAYV and did not differ between fibroblast types (Figure 1(A)). At the same time point, supernatants of both OASF and HSF displayed CHIKV titers of 1.6–8.8 × 105 infectious particles per ml and MAVV titers of 0.12–2.75 × 105 infection particles per ml, with significantly higher titers produced by OASF. At 48 h post-infection, CHIKV titers produced by HSF did not further increase, whereas the titers produced by OASF reached up to 1.5 × 107 infectious particles per ml (Figure 1(B), left panel), suggesting slightly higher virus production and/or viral spread in OASF as compared to HSF. MAYV titers did not significantly increase in OASF or HSF at 48 h post-infection (Figure 1(B), right panel).
Figure 1.
Primary human synovial fibroblasts are susceptible and permissive to CHIKV and MAYV infection. (A) OASF or HSF were infected with 5′EGFP-CHIKV or -MAYV (MOI 10). 24 h post-infection, the percentage of EGFP-positive cells was quantified by flow cytometry (n = 3–12). (B) Supernatants of CHIKV- and MAYV-infected OASF or HSF were collected at 24 and 48 h post-infection, and titers were determined by analysing EGFP expression at 24 h post-infection of HEK293 T cells (n = 3). (C) Uninfected OASF and HSF were analysed for MXRA8 and FHL1 expression by immunoblotting (n = 4–6) and for MXRA8 expression by immunofluorescence. Scale bar = 50 µm (n = 3, representative images shown). (D) OASF were infected with 5′EGFP-CHIKV at the indicated MOIs upon treatment of the virus with MXRA8-Fc recombinant protein or mock treatment. At 24 h post-infection, cells were analysed for EGFP expression (n = 4). (E) OASF were stimulated with IL-1β at 10 ng/ml for 16 h and subsequently infected with CHIKV (MOI 10) in the presence of IL-1β. At 24 and 48 h post-infection supernatant was collected and analysed for IL-6 secretion by ELISA (n = 3). (F) OASF were infected with 5′EGFP-CHIKV or -MAYV at the indicated MOIs in the presence or absence of 10 µM Ruxolitinib. At 24 and 48 h post-infection, cells were analysed for EGFP expression and (G) for the expression of IFIT1, MX2, and IFNB mRNA. Raw data of EGFP expression is plotted in Fig. S1C. The dotted line in (F) indicates the relative level of EGFP-positive cells (set to 1) in mock-treated, individually infected cell cultures. (H) OASF were infected with 5′EGFP-CHIKV (MOI 10) in the presence or absence of 1 or 10 µM Ruxolitinib or mock-infected. Images were analysed for EGFP intensity using ImageJ (n = 3). Statistical analysis was performed for A, B, D, and E using two-sided, unpaired t-tests, for F and G using ratio paired t-test with assumed equal standard deviation.
Susceptibility of cells to CHIKV infection is enhanced by the attachment factor MXRA8 [8] and the cytosolic protein FHL-1 is essential for CHIKV genome replication [9]. We confirmed expression of these two cellular cofactors in OASF and HSF by immunoblotting and/or immunofluorescence (Figure 1(C)). We assessed the functional relevance of the MXRA8 attachment factor using a soluble MXRA8-Fc fusion protein, which blocks the binding site on the E1-E2 glycoprotein complex on the virus surface [8]. At a low MOI, MXRA8-Fc-preincubated CHIKV was 50% less infectious to synovial fibroblasts, and this inhibition was abolished when saturating amounts of infectious virus particles were used (Figure 1(D)), indicating that endogenous MXRA8 contributes, at least partially, to CHIKV entry in OASF.
Subsequently, we investigated whether IL-1β-mediated activation of synovial fibroblasts, a hallmark of rheumatoid arthritis [28], modulates their susceptibility to CHIKV infection. Treatment with IL-1β did not alter the percentage of EGFP-positive cells upon CHIKV challenge (Figure S1A), while readily inducing IL-6 secretion (Figure 1(E)). Conversely, CHIKV infection induced only low IL-6 secretion and mildly, if at all, enhanced IL-1β-induced IL-6 secretion (Figure 1(E)). Overall, these data suggest that IL-6 secretion is not driven by the infection in a direct manner, but may be enhanced through external stimuli.
To determine the importance of IFN-mediated antiviral immunity in this primary cell system, we analysed the secretion of type I IFN upon CHIKV and MAYV infection, which was detectable in cultures infected with CHIKV and MAYV at an MOI of 10, while detectable IFN secretion was mostly absent in cultures infected at an MOI of 1 (Figure S1B). Additionally, we monitored efficiency of CHIKV and MAYV infection in the absence or presence of the JAK/STAT inhibitor Ruxolitinib. Infection efficiency was increased 1.3–4.7-fold and 1.3–7.7-fold in infected, Ruxolitinib-treated cells at 24 and 48 h post-infection, respectively, as compared to mock-treated, infected cultures (Figure 1(F), Figure S1C). In line with the enhanced infection efficiency, ISG expression was dampened in Ruxolitinib-treated cells, with a significant reduction of IFIT1 expression at 48 h post-infection and a complete suppression of induction of the IFN-dependent gene MX2 at both time points (Figure 1(G)). Interestingly, MAYV infection was only mildly enhanced by Ruxolitinib treatment (Figure 1(F)), potentially due to a higher base-line infection rate (Figure S1C) and a stronger JAK/STAT-independent induction of IFIT1 by MAYV infection (Figure 1(G)). Using live-cell imaging, we documented the increase in EGFP-positive, CHIKV-infected cells between ten and 48 h post-infection, which progressed faster in Ruxolitinib-treated cultures, with an onset of cytopathic effects observed after 24 h in all infected cultures (Figure S1D, Suppl. Mov. 1). Analysis of the EGFP intensity in each frame over time confirmed the higher expression of EGFP in Ruxolitinib-treated cultures (Figure 1(H), Suppl. Mov. 2). Overall, these experiments establish the susceptibility and permissiveness of synovial fibroblasts to CHIKV and MAYV infection and their expression of important cellular cofactors. Furthermore, we show that the restriction of infection in this system is, to a large extent, dependent on JAK/STAT-mediated IFN signalling and secretion, and demonstrate an absence of interconnection between IL1-β activation and susceptibility to CHIKV infection.
CHIKV infection provokes a strong cell-intrinsic immune response in synovial fibroblasts
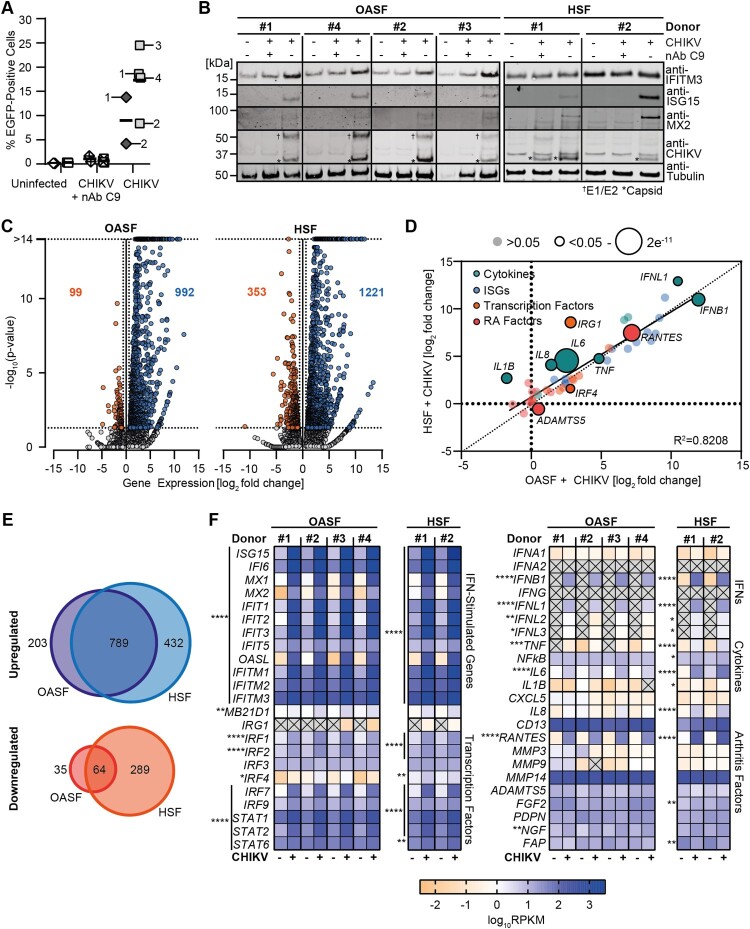
Next, we performed RNA-seq analysis on OASF and HSF that had been infected with CHIKV in the presence or absence of the glycoprotein E2-binding, neutralizing antibody C9 [29], and on mock-infected cells. C9 pre-treatment resulted in potent inhibition of the infection by on average 16-fold (Figure 2(A)). Upon infection, expression of numerous IFN-stimulated genes (ISGs) was induced at the protein level in a C9 treatment-sensitive manner, including IFITM3, ISG15, and MX2. As expected, production of the viral E1-E2 and capsid proteins was detectable specifically in CHIKV-infected, but not in cells exposed to C9-pretreated virus (Figure 2(B)). Global transcriptional profiling by RNA-seq revealed 992 (OASF) and 1221 (HSF) upregulated genes as well as 99 (OASF) and 353 (HSF) downregulated genes in CHIKV-infected cells 24 h post-infection as compared to uninfected cells (Figure 2(C)). Uninfected cells and cells exposed to C9-treated virus shared a similar profile (data not shown). A high similarity of the gene expression profile of uninfected OASF and HSF (R2 = 0.9086) argues against a potential transcriptional predisposition that could have exerted a rheumatoid arthritis-related gene expression profile or a broad proinflammatory activation (Figure S2A). Uninfected OASF and HSF differed in genes involved in organ development and cellular regulatory processes, and not inflammatory or antiviral processes (Figure S2B). Additionally, the transcriptional profile in infected OASF and HSF was very similar (R2 = 0.9085, Figure S2C-D), with an equivalently strong upregulation of a set of prototypic inflammation and arthritis-related genes which we defined for further analysis (R2 = 0.8202, Figure 2(D)). Interestingly, the number of genes significantly up- and downregulated upon infection was 1.23-fold and 3.57-fold higher in HSF compared to OASF, respectively, but 55.4% of upregulated genes from both groups overlapped (Figure 2(E)). Most of the prototypic antiviral and proinflammatory genes were highly upregulated in infected cultures, demonstrating a broad and strong activation of antiviral immune responses in cells from four different donors with no statistically significant deviation in the magnitude of induction (Figure 2(F), left panel). Upregulation of IFNB and IFNL1, IFNL2, and IFNL3 expression was statistically significant but low in magnitude, with almost no IFNA mRNA detectable. Expression of arthritis-associated genes, including genes encoding immune cell chemoattractants (CXCL5, IL8, CD13, RANTES/CCL5), matrix-metalloproteases (MMP3, −9, −14, ADAMTS5) and genes commonly expressed by fibroblasts in rheumatoid arthritis (FGF2, PDPN, NGF, FAP), was not grossly altered in CHIKV-infected cells. Exceptions were a strong CHIKV-induced upregulation of RANTES/CCL5 in both OASF and HSF and IL8 in HSF (Figure 2(F), right panel). mRNAs for all IFN receptors were detectable and stable with exception of IFNLR1, whose expression was upregulated upon CHIKV infection (Figure S2E). Established host factors for CHIKV as well as fibroblast marker genes and cellular housekeeping genes were not quantitatively altered in their expression. Virtual absence of expression of monocyte/macrophage lineage-specific genes excluded the possibility of a contamination of the fibroblast culture with macrophages, which occasionally has been reported in early passages of ex vivo-cultured synovial fibroblasts [22] (Figure S2E). Conclusively, OASF and HSF share similar basal and CHIKV infection-induced transcriptional profiles. Overall, CHIKV-infected synovial fibroblasts react to CHIKV infection by extensive upregulation of antiviral and proinflammatory ISGs. IFN expression itself was low at 24 h post-infection, not excluding the possibility that it peaked transiently at earlier time points.
Figure 2.
CHIKV infection provokes a strong cell-intrinsic immune response in synovial fibroblasts. (A) OASF were infected with 5′EGFP-CHIKV at an MOI of 10 in the presence or absence of the anti-E2 antibody C9 and the percentage of EGFP-positive cells was measured by flow cytometry (OASF: squares, n = 4; HSF: diamonds, n = 2. The infected samples are marked with their respective donor number). (B) Selected proteins of cells infected in A were analysed by immunoblotting (n = 4). (C-F) RNA from cells infected in A was extracted and subjected to RNA-seq (n = 4). Differentially expressed genes were identified by comparison of raw count data, with calculation of false-discovery rate (FDR) p-value for multiple comparisons. (C) Analysis of up- and downregulated genes in CHIKV-infected samples compared to mock. Dotted lines indicate cutoff for <1.5 fold regulation and a p-value of >0.05. (D) Visualization of the fold change induction of indicated genes in CHIKV-infected OASF and HSF. Average fold change (log2) values for infected OASF are plotted on the x-axis, with corresponding values from infected HSF plotted on the y-axis. R2 value and regression line for the comparison are inset, dot sizes indicate significance. (E) Overlap of significantly (FDR-p <0.05) up- and downregulated genes in infected OASF and HSF. Numbers of genes up- or downregulated in either OASF or HSF only, or in both cell-types, are indicated. (F) Heatmaps of selected gene expression profiles related to innate immune responses (left) or to secreted proinflammatory mediators and arthritis-connected genes (right) in uninfected or CHIKV-infected cells.
The CHIKV genome replicates to a high degree with a bias towards the structural subgenome
We noticed very little inter-donor variation regarding the distribution of identified viral reads along the viral genome. The 5´ region of the genome, encoding the non-structural CHIKV proteins, replicated to a lower extent than the 3´, 26S subgenomic promoter-driven, structural protein-encoding genomic region. Interestingly, this differential abundance of 5´and 3´ reads was also detected in cultures inoculated with C9-neutralized virus, suggesting infection of a small number of cells (Figure 3(A)). Overall, the 26S subgenomic viral RNA was 5.3-fold more abundant than nonstructural subgenomes (Figure 3(B)). 18–54% and 17–44% of the total reads in productively infected OASF and HSF, respectively, were attributed to the CHIKV genome (Figure 3(C,D)). In summary, our analysis revealed efficient replication of the CHIKV genome in infected fibroblasts with an excess of structural protein-encoding subgenomic RNA.
Figure 3.
The CHIKV genome replicates to a high degree with a bias towards the structural subgenome. (A) NGS reads attributed to each individual position in the CHIKV genome plotted for synovial fibroblasts infected with CHIKV in the presence or absence of neutralizing antibody (nAb). SGP: subgenomic promotor. (B) Normalized amount of reads attributed to the structural and nonstructural part of the CHIKV genome in CHIKV-infected OASF and HSF. (C) Number of NGS reads attributed to the human or CHIKV reference genome in CHIKV or neutralizing antibody-treated CHIKV infected OASF or (D) HSF (n = 4).
Exogenous IFN administration provokes higher immune responses and leads to improved protection from infection in primary fibroblasts than in commonly used cell lines
CHIKV and MAYV infection rates in OASF did not increase after 24 h post-infection (Figure 4(A)), and we suspected this to be the result of the strong immune activation and subsequent IFN signalling. The commonly used osteosarcoma cell line U2OS was more susceptible, while the immortalized fibroblast cell line HFF-1 displayed reduced susceptibility to alphaviral infection (Figure 4(A)). OASF exhibited strong induction of IFIT1 and MX2 CHIKV infection, which exceeded those mounted by U2OS and HFF-1 cells at both 24 and 48 h post-infection by 15- to 150-fold. MAYV infection-provoked ISG responses in OASF were inferior to those induced by CHIKV, despite similar percentages of infected cells (Figure 4(B)). Contrasting the cell system-specific magnitude of gene expression upon CHIKV infection, both OASF and cell lines shared similar responsiveness to 5′-triphosphate dsRNA (5´-ppp-RNA) transfection, which exclusively stimulates the RNA sensor RIG-I [30], the main sensor of CHIKV RNA in infected cells [31], and plasmid DNA transfection (Figure S3A).
Figure 4.
Exogenous IFN administration provokes higher immune responses and leads to improved protection from infection in primary fibroblasts than in commonly used cell lines. (A) OASF and HFF-1 cells were infected with 5′EGFP-CHIKV at an MOI of 10, U2OS cells were infected at an MOI of 0.5. EGFP-positive cells were quantified at 24 and 48 h post-infection by flow cytometry (n = 3-6). (B) Cells infected in (A) were analysed for expression of IFIT1 and MX2 mRNA at 24 and 48 h post-infection by quantitative RT-PCR (n = 3-6). Statistical analysis between samples of the same time point was performed using Mann-Whitney tests. (C) Cells were treated with IFN-α or -λ for 48 h before infection with 5′EGFP-CHIKV (OASF and HFF-1: MOI 10; U2OS: MOI 0.5) in the continuous presence of IFN. Inset numbers indicate IC50 values for each time point. (D) Cells were infected with 5′-EGFP CHIKV (OASF and HFF-1: MOI 10; U2OS: MOI 0.5) and IFN-α was added four hours post-infection. 24 and 48 h post-infection, EGFP-positive cells were quantified by flow cytometry. Inset numbers indicate IC50 values for each time point. UT: untreated, IU: international units (n = 3 for all experiments). For C and D, nonlinear fit curves with variable slopes for IC50 calculation were used.
Next, we tested the cells´ ability to respond to exogenous type I and III IFNs, which play a crucial role in limiting virus infection and protecting the host [32,33]. We stimulated OASF individually with a range of IFN-α2 and -λ concentrations at 48 h prior to infection. At all investigated concentrations, even at the lowest dose, IFN-α induced a potent upregulation of IFIT1 and MX2 (Figure S3B), and almost completely inhibited CHIKV infection (Figure 4(C)). In contrast, IFN-λ induced lower ISG expression levels (Figure S3B), and inhibited infection less efficiently (Figure 4(C)). Although less effective than in OASF, IFN-α restricted CHIKV infection both in U2OS and HFF-1 cells, while IFN-λ pre-treatment was more potent in U2OS cells than in OASF, and ineffective in HFF-1 cells (Figure 4(C)). These antiviral activities were largely consistent with the respective degree of ISG expression at the time point of infection (Figure S3B). IFN-α and -λ induced expression of IFIT1 and MX2 was higher in U2OS cells than in HFF-1. We next investigated the sensitivity of CHIKV infection to IFN when applied four hours post-infection. In this set-up, IFN-α still displayed a clear, though less potent antiviral activity when compared to the pre-treatment setting (Figure 4(D)). In contrast, treatment of both immortalized cell lines with IFN-α post-infection was very ineffective (Figure 4(D)).
Interestingly, in all three cells systems, a preceding CHIKV infection did not antagonize IFN-mediated induction of ISGs, and led to expression levels of IFIT1 and MX2 exceeding those induced by IFN-α alone (Figure S3C). Overall, the data suggest a stronger sensitivity of OASF to IFN-α-induced immunity compared to commonly used immortalized cell lines. Most interestingly, and in striking contrast to the immortalized cell lines, OASF were unique in their ability to transform a post-infection treatment of IFN-α into a relatively potent antiviral programme. Collectively, these data uncover crucial differences between primary synovial fibroblasts and widely used immortalized cell lines regarding their cell-intrinsic innate response to infection and their sensitivity to exogenous IFNs.
Strong immune activation In EGFP protein-negative and viral RNA-low cells
Finally, we asked how the cell-intrinsic defenses correlate with the infection status within individual cells of a given infected culture by analysing virus-exposed OASF for their expression of antiviral proteins using flow cytometry. As expected, expression of IFIT1, IFITM3 and MX1/2 was enhanced in OASF upon IFN-α treatment (Figure 5(A)). Interestingly, these proteins were expressed at even higher levels in EGFP-negative cells of CHIKV-infected cultures, while the productively infected, EGFP-positive cells displayed markedly reduced expression levels of these factors (Figure 5(A)).
Figure 5.
Strong immune activation in EGFP protein-negative and viral RNA-low cells. (A) OASF were infected with 5′EGFP-CHIKV (MOI 10) or treated with 100 IU/ml IFN-α and immunostained for IFIT1, IFITM3, and MX1/2 24 h post-infection. Numbers in the dot plots indicate mean fluorescence intensities (MFI) of one representative experiment, and the bar diagram shows quantification of three individual experiments with statistical analysis by two-sided unpaired t-tests with assumed equal standard deviation (n = 3). (B) OASF were infected with 5′EGFP-CHIKV at indicated MOIs. Six and 24 h post-infection, EGFP-positive cells were quantified by flow cytometry (left panel), and cells were analysed for expression of IFIT1 and MX2 mRNA (right panel, n = 6). (C) Using OASF infected with 5′EGFP-CHIKV, single-cell RNA-sequencing was conducted and UMAP visualizations for sample overlapping after integration are shown (n = 2). (D) UMAP projections from infected OASF (24 h post-infection) indicate the abundance of CHIKV 3′ end reads, EGFP 3′ end reads, and IFN signalling gene expression as calculated by IMS (n = 2).
Since absence of EGFP expression does not necessarily exclude the presence of viral, potentially abortive RNA, we performed virus-inclusive single-cell RNA-seq to establish potential correlations of the quantity of viral RNA and a specific cellular transcriptional profile. To this end, we analysed OASF infected at escalating MOIs. No EGFP-positive cells were detectable at six hours post-infection by flow cytometry (Figure 5(B), left panel). In contrast, 24 h post-infection, the reporter was expressed in an MOI-dependent fashion, ranging from virtually 0% to 15% (Figure 5(B), left panel). IFIT1 and MX2 mRNA expression was largely proportional to EGFP expression (Figure 5(B), right panel).
Single-cell (sc) RNA-seq of the very same cells showed very little inter-donor variability, and we merged data from both donors throughout the rest of the analysis (Figure 5(C)). In order to identify potential correlations of viral RNA abundance and the cellular transcription profile, we compared the expression of CHIKV RNA to expression of 203 IFN signalling genes listed in the REACTOME database (identifier R-HSA-913531, Table 1). For each cell, the expression of this collection of genes was summarized using Seurat’s AddModuleScore function. Briefly, this summarizes the expression of a select group of genes by normalizing the aggregate expression to a randomly selected, non-overlapping subset of genes and scores each cell based on its expression of genes in this module, creating a module score (IFN Module Score, IMS). 24 h post-infection, most identified CHIKV reads corresponded to the 3′ end of the genome, along with a minor number of reads mapping to the 3′ end of EGFP, which is expressed as a subgenomic RNA in infected cells (Figure S4A). As expected for mock-infected cells, CHIKV reads were undetectable, and IFN signalling genes were expressed at basal levels, as calculated by the IMS. CHIKV RNA abundance per cell increased in an MOI-dependent manner, however susceptibility to infection was unequally distributed over individual cells, and a subset of cells displayed a higher susceptibility than others, as reflected by a high percentage of reads attributed to the viral genome (Figure S4B). Strikingly, IFN signalling genes appeared to be induced predominantly in cells displaying low CHIKV gene expression. Vice versa, clearly CHIKV RNA-positive cells maintained basal or reduced expression of IFN signalling related genes (Figure 5(D)). Of note, six hours post-infection, CHIKV expression was low and antiviral responses as presented by the IMS was were largely absent at low MOIs, while individual ISGs were induced at higher MOIs (Figure S4C-D). As opposed to the induction of IFN signalling genes, known CHIKV cofactors MXRA8, FHL1, and the fibroblast marker genes VIM and COL3A1 were broadly and stably expressed under all experimental conditions. Surprisingly, FURIN, encoding the cellular protease considered important for viral polyprotein cleavage, was detectable only in a minority of cells (Figure S5).
Table 1.
Interferon signalling genes identified by the REACTOME database.
| AAAS | HLA-DQA1 | IFNGR1 | NUP37 | STAT1 |
| ABCE1 | HLA-DQA2 | IFNGR2 | NUP42 | STAT2 |
| ADAR | HLA-DQB1 | IP6K2 | NUP43 | SUMO1 |
| ARIH1 | HLA-DQB2 | IRF1 | NUP50 | TPR |
| B2M | HLA-DRA | IRF2 | NUP54 | TRIM10 |
| BST2 | HLA-DRB1 | IRF3 | NUP58 | TRIM14 |
| CAMK2A | HLA-DRB3 | IRF4 | NUP62 | TRIM17 |
| CAMK2B | HLA-DRB4 | IRF5 | NUP85 | TRIM2 |
| CAMK2D | HLA-DRB5 | IRF6 | NUP88 | TRIM21 |
| CAMK2G | HLA-E | IRF7 | NUP93 | TRIM22 |
| CD44 | HLA-F | IRF8 | NUP98 | TRIM25 |
| CIITA | HLA-G | IRF9 | OAS1 | TRIM26 |
| DDX58 | HLA-H | ISG15 | OAS2 | TRIM29 |
| EGR1 | ICAM1 | ISG20 | OAS3 | TRIM3 |
| EIF2AK2 | IFI27 | JAK1 | OASL | TRIM31 |
| EIF4A1 | IFI30 | JAK2 | PDE12 | TRIM34 |
| EIF4A2 | IFI35 | KPNA1 | PIAS1 | TRIM35 |
| EIF4A3 | IFI6 | KPNA2 | PIN1 | TRIM38 |
| EIF4E | IFIT1 | KPNA3 | PLCG1 | TRIM45 |
| EIF4E2 | IFIT2 | KPNA4 | PML | TRIM46 |
| EIF4E3 | IFIT3 | KPNA5 | POM121 | TRIM48 |
| EIF4G1 | IFITM1 | KPNA7 | POM121C | TRIM5 |
| EIF4G2 | IFITM2 | KPNB1 | PPM1B | TRIM6 |
| EIF4G3 | IFITM3 | MAPK3 | PRKCD | TRIM62 |
| FCGR1A | IFNA1 | MID1 | PSMB8 | TRIM68 |
| FCGR1B | IFNA10 | MT2A | PTAFR | TRIM8 |
| FLNA | IFNA13 | MX1 | PTPN1 | TYK2 |
| FLNB | IFNA14 | MX2 | PTPN11 | UBA52 |
| GBP1 | IFNA16 | NCAM1 | PTPN2 | UBA7 |
| GBP2 | IFNA17 | NDC1 | PTPN6 | UBB |
| GBP3 | IFNA2 | NEDD4 | RAE1 | UBC |
| GBP4 | IFNA21 | NUP107 | RANBP2 | UBE2E1 |
| GBP5 | IFNA4 | NUP133 | RNASEL | UBE2L6 |
| GBP6 | IFNA5 | NUP153 | RPS27A | UBE2N |
| GBP7 | IFNA6 | NUP155 | RSAD2 | USP18 |
| HERC5 | IFNA7 | NUP160 | SAMHD1 | USP41 |
| HLA-A | IFNA8 | NUP188 | SEC13 | VCAM1 |
| HLA-B | IFNAR1 | NUP205 | SEH1L | XAF1 |
| HLA-C | IFNAR2 | NUP210 | SOCS1 | |
| HLA-DPA1 | IFNB1 | NUP214 | SOCS3 | |
| HLA-DPB1 | IFNG | NUP35 | SP100 |
Correlation analysis of viral and cellular gene expression reveals a switch from induction to suppression of transcription factor and ISG expression
In order to quantify expression of IFN signalling genes according to viral RNA abundance, we divided cells into three groups: cells without detectable viral RNA expression (bystander), cells displaying low amounts of viral RNA (low) and cells displaying high levels of viral RNA (high) (Figure 6(A)). Mirroring our initial observations (Figure 5), we detected a significantly lower IMS in high cells when compared to low or bystander cells of the identical culture (Figure S6A). Six hours post-infection, differential expression of non-ISGs was very modest between bystander and viral RNA-positive cells, while it was more pronounced 24 h post-infection (Figure S6B). In contrast, over 250 ISGs, including ISG15, IFIT1, MX2, IFITM3, MX1, and IFI6, were upregulated in viral RNA-positive cells as compared to bystander cells at both investigated time points. Individual comparisons of either low or high cells with bystander cells gave similar overall observations. However, at both investigated time points, no further upregulation of ISGs was detected in the high cells as compared to low cells, but rather a significant downregulation of three ISGs at 24 h post-infection and six ISGs at six hours post-infection. This suggests either a loss of cellular transcription activity or a lowered stability of cellular RNA in cells containing high loads of CHIKV RNA (Figure S6B).
Figure 6.
Correlation analysis of viral and cellular gene expression reveals a switch from induction to suppression of transcription factor and ISG expression. (A) Visualization of the viral RNA content of infected OASF from Figure 5 at six and 24 h post-infection. Line indicates the cutoff dividing cells displaying low and high content of viral RNA. Bystander cells were defined as cells with no detectable viral RNA. (B) Infected OASF were sorted into digital bins of 1000 cells displaying a gradual increase of the amount of viral reads per cell. Viral reads and the IMS at six and 24 h post-infection are plotted. Coloured bins indicate selected representative cells for low and high content of viral RNA. (C) Expression of selected genes within mock, bystander, representative low cell bins (bin 26–29) and high cell bins (bin 34–36) defined in A and B at six and 24 h post-infection. Arrows indicate a statistically significant (p < 0.05, fold change >1.5) up- or downregulation (depending on the arrow direction) in low or high CHIKV bins versus bystander (inside the boxes) or in high CHIKV bins versus low (next to the boxes). Differential gene expression was tested by Wilcoxon rank sum tests with applied Bonferroni correction. (D) Correlation of CHIKV RNA expression with expression of IFN signalling genes in high and low CHIKV RNA groups calculated by non-parametric Spearman’s test. Transcription factors are plotted in white, with selected genes in red. (E) Activity of transcription factor regulons within groups defined in A at six and 24 h post-infection. Arrows indicate a significant up- or downregulation between bystander and low or high CHIKV groups (inside the boxes) or between low and high CHIKV groups (next to the boxes). Cumulative distributions between groups were compared using non-parametric Kolmogorov-Smirnov tests.
To increase resolution, we calculated the average CHIKV and EGFP RNA expression and the average IMS in bins of 1000 cells for a total of 36 bins, sorted by their expression level of CHIKV RNA. At both time points, while the first 7–10 bins represented cells expressing no or virtually no CHIKV RNA, the following 18–21 bins represented cells displaying (according to the cut-off defined in Figure 6(A)) low, but gradually increasing levels of CHIKV RNA, and largely undetectable EGFP RNA. We considered the latter cells to represent unproductively infected cells due to their lack of subgenomic transcripts. The last eight bins displayed cells with overall high, starkly increasing levels of CHIKV RNA and with significant levels of EGFP mRNA. We hypothesize that these cells represent productively infected cells. Strikingly, in unproductively infected cells, IMS values increased proportionally to the abundance of viral RNA per cell, whereas in productively infected cells, an inverse proportionality was observed (Figure 6(B)). This dataset suggests that expression of IFN signalling genes is upregulated in cells harbouring low-to-intermediate levels of viral RNA, which, however, do not or have not yet progressed to a productive infection. In contrast, cells that exceed a certain threshold of viral RNA show a prevention or downregulation of the expression of IFN signalling genes. The analysis of expression of selected genes confirmed this observation. Expression of individual ISGs, including ISG15, IFI6, MX1, OASL, IFITs, and IFITMs, and transcription factors, including STAT1 and IRF7, was low in mock-infected and bystander cells, and more pronounced in representative low cells (six hours post-infection: bin 27–29, 24 h post-infection: bin 26–28) than in high cells (bins 34–36). Furthermore, expression of genes mediating arthralgia, including as RANTES, IL1B, IL6, and IL8 was lower in high cells as compared to low cells (Figure 6(C)). To identify further putative targets of viral antagonism, we correlated the expression of all 203 genes of the IMS to the viral RNA expression in infected cells at 24 h post-infection. We identified 13 genes displaying a significant positive correlation (r > 0.3) in the low CHIKV group, and a significant negative correlation (r < −0.3) in the high CHIKV group (IFITM3, IFIT3, OAS1, XAF1, GBP1, EIF4A1, EIF2AK2, STAT1, GBP3, UBC, PSMB8, UBA52). Strikingly, the only transcription factor present in both groups, STAT1, was also negatively correlated at six hours post-infection in the high viral RNA group. We additionally identified transcription factors JAK1 and IRF7 to switch from weak correlation in the low CHIKV group to a negative correlation in the high CHIKV group at 24 h post-infection (Figure 6(D)). We confirmed this finding using a transcription factor activity score analysis using the DoRotheEa database, which scores cells based on the activity of transcription factors inferred from the expression of the associated target genes in regulons. The regulon of STAT1 was strongly induced in bystander and low CHIKV groups at high MOIs, yet highly susceptible to viral antagonism in the high CHIKV group. The same observation was obtained for multiple other transcription factors, including IRFs and STATs as well as NFκB and c-JUN, at higher MOIs, indicating a strong and sensitive induction that is counteracted in high cells (Figure 6(E)). Taken together, our analyses at single-cell resolution specifically uncovered a spectrum of cellular states, displaying a range from induced to repressed cell-intrinsic immune responses depending on a threshold abundance of viral RNA per individual cell. Efficacy of mounting innate immune responses was highest in cells displaying low-to-intermediate quantities of viral RNA. Furthermore, viral antagonism manifested itself specifically in the relatively low percentage of cells presenting efficient virus replication, consistent with expression and functionality of virally encoded IFN antagonists.
Discussion
Given that the joints are the primarily affected body compartments and that fibroblasts are susceptible to arthritogenic alphavirus infection in general [6,34], we hypothesized that cells of the joint synovium are directly implicated in the pathophysiology of CHIKV-induced arthralgia. Cells of the synovial tissue and synovial fluid contain CHIKV RNA and protein upon CHIKV infection in vivo in humans [5], experimentally infected macaques and mice [7,35]. Additionally, infectious virus was recovered from the joints of infected macaques at six days post-infection, indicating the synovium as an active replication site [7]. The main cell types composing the synovium are macrophages and fibroblasts. The latter have been identified to be susceptible to CHIKV infection ex vivo [8]. However, the corresponding basal innate immune state of primary synovial fibroblasts and their ability to exert IFN-mediated antiviral restriction is unknown. Here, we establish that the widely available OASF and less available HSF share susceptibility and permissiveness to CHIKV infection, and describe their basal and infection-induced transcriptional programmes. We confirmed that the two cell types share a highly similar overall transcriptional profile, except in some signalling pathways unrelated to immunity. CHIKV infection provoked a striking cellular response that involves upregulation of multiple ISGs in a JAK/STAT-dependent manner, many of them exerting antiviral activity, and rheumatoid arthritis-mediating genes. Although we did not define the PAMP(s) that trigger responses in synovial macrophages, ISG induction occurred specifically in cells containing virus genome-, but not EGFP-specific reads, indicating the absence of de novo synthesized subgenomic RNA and therefore suggesting that these cells represent an early or unproductive infection state. Infection by alphaviruses typically raises RIG-I-mediated responses through exposure of dsRNA intermediates and provokes mitochondrial DNA leakage that is sensed via cGAS/STING [31]. Indeed, experimental ligands of both sensors were highly reactive in OASF, as was IFN-α treatment. Surprisingly, also IFN-λ pre-treatment translated into an antiviral state, indicating that synovial fibroblasts may represent an exception to the notion of otherwise IFN-λ-nonresponsive fibroblasts [36]. Finally, CHIKV infection of synovial fibroblasts was sensitive to IFN-α applied after inoculation with virus. These findings appear to contrast with potent virus-mediated antagonism of IFN in U2OS and HFF-1 cell lines, which has been suggested to involve counteraction of nuclear translocation of STAT1 [12]. CHIKV was unable to suppress ISG expression upon exogenous IFN treatment in any cell type, indicating that the proposed antagonistic functions may not be strong enough to be detectable at the bulk level. Also, unaltered levels of expression of housekeeping genes and genes encoding fibroblast markers in primary synovial fibroblasts did not generate evidence for a general virus-mediated host transcriptional shut-off that has been reported for several cell lines [13]. Overall, synovial fibroblasts appear to respond differently to CHIKV infection as commonly used cell lines. The underlying reason for this difference is unknown, but may involve a different intracellular milieu that is hyper-responsive to CHIKV infection.
While our single-cell RNA-seq approach is based on 3′ end capture and does not allow to discriminate between full-length and subgenomic viral RNA, we identified a relative excess of subgenomic RNA in infected cultures by bulk RNA sequencing, in analogy to reports for Sindbis virus-infected cells [37]. Enhanced replication of the subgenomic RNA, which is mediated by the four cleaved nonstructural proteins forming a replication complex, ensures the rapid production of viral structural proteins and the formation of new virions [38]. While packaging of subgenomic RNA into virions has been described so far solely for Aura virus among alphaviruses [39], CHIKV bears a packaging signal in the nsP2-encoding region of its genome, selecting specifically full genomic RNA to be packaged into virions [40]. Therefore, we assume that the different abundance is based on de novo produced subgenomic RNA rather than on incoming viral RNA.
Our attempts to identify correlations of cellular gene expression with CHIKV RNA abundance in individual infected cells revealed that a certain threshold of viral RNA is required to initiate viral RNA sensing and eventually trigger ISG expression. However, expression of most ISGs is negatively regulated in the presence of a high viral RNA burden per cell. This is consistent with the idea that productive infection involves the synthesis of viral antagonists that hamper the induction and/or evade the function of ISGs, resulting in efficient virus propagation. Along these lines, West Nile virus infection also results in lowered ISG expression levels in cells harbouring high viral RNA quantities [41]. In vivo, actively SARS-CoV-2 infected monocytes of COVID-19 patients expressed lower levels of ISGs than non-infected bystander cells [42]. Monocytes of Ebola-infected rhesus monkeys display similar dynamics, with an additional downregulation of STAT1 mRNA in infected cells [43]. On the contrary, cells that undergo abortive infection, or alternatively have not yet reached sufficient levels of virus replication fail to mount or repress a strong antiviral profile. Depending on the longevity of cells harbouring abortive genomic replication products and intermediates, their presence may influence the immunopathogenesis of chronic aspects of RNA-viral infection. Owing to genetic recombination and low fidelity of the alphaviral RNA-dependent polymerase, defective alphaviral genomes (DVGs) and defective alphaviral particles arise during virus replication, but are themselves replication-incompetent [44]. Of note, our virus-inclusive sequencing approach does not have the power to distinguish between full-length viral genomes and defective or otherwise dead-end genomes. It will be interesting to test the contribution of the latter to triggering the strong cell-intrinsic innate recognition that we linked here to low intracellular viral RNA quantities in general. Strikingly, we find indication that the expression of some genes, such as proinflammatory transcription factors, may be actively targeted by CHIKV.
Particularly interesting in the context of an a priori acute RNA virus infection, arthritogenic alphavirus infection has been suspected to result in the generation of long-lived cellular reservoirs that may maintain low-levels of viral RNA. In mice, fibroblasts survive a CHIKV infection and joint-associated tissue can harbour viral RNA in the absence of infectious virus production for at least 16 weeks [6,45]. Prolonged shedding of infectious virus for up to 35 days from ex vivo-infected human synovial fibroblasts has been observed for Ross River virus (RRV), another arthritogenic alphavirus [46]. In infected macaques, infectious CHIKV particles were not shed for extended periods in the joints, but was detected in other tissues, including the liver and the spleen, at 44 days post-infection [7]. In addition to fibroblasts, other discussed cell types for long-term persistence are (synovial) macrophages and, to a lesser extent, dendritic cells. Macrophages harbour persistent viral RNA in a nonhuman primate infection model [7] and in human patients, where joint biopsies found synovial macrophages to be CHIKV RNA- and antigen-positive for as long as 18 months post-infection [5]. In vitro, CHIKV has been proposed to productively infect primary human macrophages [34]. Additionally, murine macrophages have been observed to periodically relapse from undetectable RRV production to spontaneous or inducible viral shedding [47]. On the other hand, persistent infection with MAYV has so far only be observed in RAG-/- mice, suggesting an efficient clearance of infection by adaptive immune responses [48]. In the absence of viral proteins antagonizing host antiviral responses, replication sites in the cytoplasm separated by single- or double membranes may shield the viral RNA from cellular detection and degradation, as has been demonstrated for flaviviruses [49] and coronaviruses [50]. Continuous sensing of viral RNA, which is not necessarily replicated to a level that would suffice for de novo virus production and virus spread, could thereby resonate in a state of chronic inflammation in the joints of infected patients. It is tempting to speculate that those cell-intrinsic responses that we identified to be most pronounced in cells displaying low levels of viral RNA could be identical to those driving the pain- and inflammation-related joint immunopathology linked to chronic alphavirus infection-induced arthritis. Other suggested mechanisms for alphavirus-mediated arthralgia include, similar to Parvovirus B19- or Epstein–Barr virus-induced arthralgia [51,52], the formation of immunogenic autoantibodies through molecular mimicry [53].
Furthermore, human synovial fibroblasts secrete cytokines such as IL-6, IL1β, and RANTES, stimulating monocyte migration upon CHIKV infection, and drive them towards an osteoclast-like phenotype [4]. Interestingly, we identify a similar pattern in infected fibroblasts with upregulation and/or secretion of IL-6 and RANTES, but not matrix-metalloproteases (MMPs), as described before [4]. MMP expression and secretion by synovial fibroblasts can, similar to IL-6 secretion, be induced through external stimulation with IL-1 and TNF and by activated immune cells [54]. A paracrine stimulation of MMP expression by infiltrating immune cells has not been addressed in this model, but is likely to contribute to the direct induction of rheumatoid arthritis-like symptoms. Early IFN-mediated bystander cell activation and death has been reported in La Crosse virus infected cultures [55], processes which we do not observe, suggesting a limited role of bystander cell responses in this model system. On the other hand, at six hours post-infection in cultures where we expect almost all cells to have made contact with virus particles, we observe a strong activation of the RNA-negative cells. Additionally, infected synovial fibroblasts mount an immune response in the absence of JAK/STAT signalling, although low and without the induction of the strictly IFN-dependent ISG MX2 in this model. This indicates that incoming viral RNA in the absence of a productive infection is sufficient to trigger pattern-recognition receptors such as RIG-I in an IFN-independent manner, as shown before [56], which may lead to the suppression of an active infection. Nevertheless, IFN-mediated immunity is majorly responsible for the suppression of viral spread in infected synovial fibroblasts. While paracrine and autocrine activation of expression by secreted IFN would lead to a basic level of immunity in all cells of the culture, we observed a reduced ISG activation in highly infected cells, again supporting the hypothesis that CHIKV actively counteracts JAK/STAT signalling. On the other hand, uninfected bystander cells were partially ISG-positive, which may protect them to a certain degree against infection or progression to highly infected cells. The marginal expression of IFN genes in most cells in the presented single-cell RNA-seq dataset indicates that at a given time, only a small number of cells contribute to the secretion of IFN, which may explain the overall low amount of type I IFN detected here.
Finally, an interesting hypothesis foresees that pharmacological interference with the synovial fibroblast-specific hyperreactivity represents a feasible intervention approach towards the alleviation of long-term arthralgia. In rheumatoid arthritis, hyperactivated synovial fibroblasts invade the joint matrix, destroying/disrupting the cartilage and causing long-term inflammation [57,58]. This and the subsequent attraction of immune cells, including monocyte-derived macrophages to the damaged sites, may represent important events in the progression to long-term morbidity [59]. Indeed, data obtained in recent clinical studies suggests that treatment of chikungunya-induced arthritis with the immunosuppressant methotrexate may be beneficial [60]. The data presented here support the hypothesis that infected synovial fibroblasts display a phenotype that is reminiscent of those in rheumatoid arthritis, and that they are a driver of the typical symptoms in interplay with infiltrating immune cells. Key features such as the IL-1β-mediated IL-6 release, the aggressive proinflammatory gene expression in productively infected cells, and the strong expression of important cofactors make them likely to contribute to viral replication and disease progression in vivo.
Supplementary Material
Acknowledgements
We thank the sequencing core of the Helmholtz Centre for Infection Research (HZI) in Braunschweig and the Genomics platform of the Berlin Institute of Health (BIH) for preparation of the Illumina sequencing libraries and the next generation sequencing. Additionally, we thank the sequencing facility of the Max Delbrück Center for Molecular Medicine for the next generation sequencing and bioinformatic support. We thank M. Diamond for providing the MXRA8-Fc proteins. We thank Theresia Stradal, Jens Bohne, and Sandra Pellegrini for providing the U2OS cells, HEK293T cells, and HL116 cells, respectively. We thank Thomas Pietschmann, Institute for Experimental Virology, TWINCORE, and Christian Drosten for constant support.
Funding Statement
This work was supported by funding from Deutsche Forschungsgemeinschaft (DFG) to CG (GO2153/3-1; GO2153/6-1), by the Impulse and Networking Fund of the Helmholtz Association through the HGFEU partnering grant PIE-008 to CG, and by funding from the Helmholtz Center for Infection Research (HZI) and Berlin Institute of Health (BIH) to CG.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Matusali G, Colavita F, Bordi L, et al. . Tropism of the Chikungunya virus. Viruses. 2019 Feb 20;11(2):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diagne CT, Bengue M, Choumet V, et al. . Mayaro virus pathogenesis and transmission mechanisms. Pathogens. 2020 Sep 8;9(9):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santiago FW, Halsey ES, Siles C, et al. . Long-term arthralgia after Mayaro virus infection correlates with sustained pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2015;9(10):e0004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phuklia W, Kasisith J, Modhiran N, et al. . Osteoclastogenesis induced by CHIKV-infected fibroblast-like synoviocytes: a possible interplay between synoviocytes and monocytes/macrophages in CHIKV-induced arthralgia/arthritis. Virus Res. 2013 Nov 6;177(2):179–188. [DOI] [PubMed] [Google Scholar]
- 5.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, et al. . Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010 May 15;184(10):5914–5927. [DOI] [PubMed] [Google Scholar]
- 6.Young AR, Locke MC, Cook LE, et al. . Dermal and muscle fibroblasts and skeletal myofibers survive chikungunya virus infection and harbor persistent RNA. PLoS Pathog. 2019 Aug;15(8):e1007993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labadie K, Larcher T, Joubert C, et al. . Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010 Mar;120(3):894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R, Kim AS, Fox JM, et al. . Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature. 2018 May;557(7706):570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meertens L, Hafirassou ML, Couderc T, et al. . FHL1 is a major host factor for chikungunya virus infection. Nature. 2019 Oct;574:(7777):259–263. [DOI] [PubMed] [Google Scholar]
- 10.Salvador B, Zhou Y, Michault A, et al. . Characterization of Chikungunya pseudotyped viruses: identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology. 2009 Oct 10;393(1):33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynaud JM, Kim DY, Atasheva S, et al. . IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog. 2015 Apr;11(4):e1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goertz GP, McNally KL, Robertson SJ, et al. . The methyltransferase-like domain of Chikungunya virus nsP2 inhibits the interferon response by promoting the nuclear export of STAT1. J Virol. 2018 Aug 16;92(17):e01008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhrymuk I, Lukash T, Frolov I, et al. . Novel mutations in nsP2 abolish Chikungunya virus-induced transcriptional shutoff and make the virus less cytopathic without affecting its replication rates. J Virol. 2019 Feb 5;93(4):e02062–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW, MacDuff DA, Imanaka N, et al. . Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014 Jan 30;505(7485):691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb LG, Veloz J, Pintado-Silva J, et al. . Chikungunya virus antagonizes cGAS-STING mediated type-I interferon responses by degrading cGAS. PLoS Pathog. 2020 Oct 15;16(10):e1008999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haese NN, Broeckel RM, Hawman DW, et al. . Animal models of Chikungunya virus infection and disease. J Infect Dis. 2016 Dec 15;214(suppl 5):S482–s487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simarmata D, Ng DC, Kam YW, et al. . Early clearance of Chikungunya virus In children is associated with a strong innate immune response. Sci Rep. 2016 May 16;6:26097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain KM, Lee RC, Ng MM, et al. . Establishment of a novel primary human skeletal myoblast cellular model for Chikungunya virus infection and pathogenesis. Sci Rep. 2016 Feb 19;6:21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard E, Hamel R, Neyret A, et al. . Human keratinocytes restrict chikungunya virus replication at a post-fusion step. Virology. 2015 Feb;476:1–10. [DOI] [PubMed] [Google Scholar]
- 20.Lefevre S, Meier FM, Neumann E, et al. . Role of synovial fibroblasts in rheumatoid arthritis. Curr Pharm Des. 2015;21(2):130–141. [DOI] [PubMed] [Google Scholar]
- 21.Uzé G, Di Marco S, Mouchel-Vielh E, et al. . Domains of interaction between alpha interferon and its receptor components. J Mol Biol. 1994 Oct 21;243(2):245–257. [DOI] [PubMed] [Google Scholar]
- 22.Neumann E, Riepl B, Knedla A, et al. . Cell culture and passaging alters gene expression pattern and proliferation rate in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2010;12(3):R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsetsarkin K, Higgs S, McGee CE, et al. . Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006 Winter;6(4):325–337. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Zhang H, Zhang Y, et al. . Development of a rapid antiviral screening assay based on eGFP reporter virus of Mayaro virus. Antiviral Res. 2019 Aug;168:82–90. [DOI] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, et al. . Gene ontology: tool for the unification of biology. Nat Genet. 2000 May;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. bioRxiv. 2020:2020.10.12.335331.
- 27.Holland CH, Tanevski J, Perales-Patón J, et al. . Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020 Feb 12;21(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georganas C, Liu H, Perlman H, et al. . Regulation of IL-6 and IL-8 expression in rheumatoid arthritis synovial fibroblasts: the dominant role for NF-κB But Not C/EBPβ or c-Jun. J Immunol. 2000 Dec 15;165(12):7199–7206. [DOI] [PubMed] [Google Scholar]
- 29.Selvarajah S, Sexton NR, Kahle KM, et al. . A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. 2013;7(9):e2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornung V, Ellegast J, Kim S, et al. . 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006 Nov 10;314(5801):994–997. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez David RY, Combredet C, Sismeiro O, et al. . Comparative analysis of viral RNA signatures on different RIG-I-like receptors. Elife. 2016 Mar 24;5:e11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoggins JW, Wilson SJ, Panis M, et al. . A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011 Apr 28;472(7344):481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou JH, Wang YN, Chang QY, et al. . Type III interferons in viral infection and antiviral immunity. Cell Physiol Biochem. 2018;51(1):173–185. [DOI] [PubMed] [Google Scholar]
- 34.Sourisseau M, Schilte C, Casartelli N, et al. . Characterization of reemerging chikungunya virus. PLoS Pathog. 2007 Jun;3(6):e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couderc T, Chrétien F, Schilte C, et al. . A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008 Feb 8;4(2):e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommereyns C, Paul S, Staeheli P, et al. . IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008 Mar 14;4(3):e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemm JA, Rümenapf T, Strauss EG, et al. . Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. Embo J. 1994 Jun 15;13(12):2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupp JC, Sokoloski KJ, Gebhart NN, et al. . Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol. 2015 Sep;96(9):2483–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rümenapf T, Strauss EG, Strauss JH.. Subgenomic mRNA of Aura alphavirus is packaged into virions. J Virol. 1994 Jan;68(1):56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim DY, Firth AE, Atasheva S, et al. . Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J Virol. 2011 Aug;85(16):8022–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Neal JT, Upadhyay AA, Wolabaugh A, et al. . West Nile virus-inclusive single-cell RNA sequencing reveals heterogeneity in the type I interferon response within single cells. J Virol. 2019 Mar 5;93(6):e01778–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bost P, Giladi A, Liu Y, et al. . Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020 Jun 25;181(7):1475–1488.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotliar D, Lin AE, Logue J, et al. . Single-cell profiling of Ebola virus disease in vivo reveals viral and host dynamics. Cell. 2020 Nov 25;183(5):1383–1401.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poirier EZ, Mounce BC, Rozen-Gagnon K, et al. . Low-fidelity polymerases of alphaviruses recombine at higher rates to overproduce defective interfering particles. J Virol. 2016;90(5):2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawman DW, Stoermer KA, Montgomery SA, et al. . Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J Virol. 2013 Dec;87(24):13878–13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Journeaux SF, Brown WG, Aaskov JG.. Prolonged infection of human synovial cells with Ross River virus. J Gen Virol. 1987 Dec;68(Pt 12):3165–3169. [DOI] [PubMed] [Google Scholar]
- 47.Way SJ, Lidbury BA, Banyer JL.. Persistent Ross River virus infection of murine macrophages: an in vitro model for the study of viral relapse and immune modulation during long-term infection. Virology. 2002 Sep 30;301(2):281–292. [DOI] [PubMed] [Google Scholar]
- 48.Figueiredo CM, Neris R, Gavino-Leopoldino D, et al. . Mayaro virus replication restriction and induction of muscular inflammation in mice are dependent on age, type-I interferon response, and adaptive immunity. Front Microbiol. 2019;10:2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Garcia MD, Mazzon M, Jacobs M, et al. . Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe. 2009 Apr 23;5(4):318–328. [DOI] [PubMed] [Google Scholar]
- 50.Wolff G, Melia CE, Snijder EJ, et al. . Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020 Dec;28(12):1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerr JR. The role of parvovirus B19 in the pathogenesis of autoimmunity and autoimmune disease. J Clin Pathol. 2016 Apr;69(4):279–291. [DOI] [PubMed] [Google Scholar]
- 52.Houen G, Trier NH.. Epstein-Barr virus and systemic autoimmune diseases. Front Immunol. 2020;11:587380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venigalla SSK, Premakumar S, Janakiraman V.. A possible role for autoimmunity through molecular mimicry in alphavirus mediated arthritis. Sci Rep. 2020 Jan 22;10(1):938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuchs S, Skwara A, Bloch M, et al. . Differential induction and regulation of matrix metalloproteinases in osteoarthritic tissue and fluid synovial fibroblasts. Osteoarthritis Cartilage. 2004 May;12(5):409–418. [DOI] [PubMed] [Google Scholar]
- 55.Cruz MA, Parks GD.. La Crosse virus infection of human keratinocytes leads to interferon-dependent apoptosis of bystander non-infected cells in vitro. Viruses. 2020 Feb 25;12(3):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber M, Gawanbacht A, Habjan M, et al. . Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013 Mar 13;13(3):336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumann E, Lefevre S, Zimmermann B, et al. . Rheumatoid arthritis progression mediated by activated synovial fibroblasts. Trends Mol Med. 2010 Oct;16(10):458–468. [DOI] [PubMed] [Google Scholar]
- 58.Hillen J, Geyer C, Heitzmann M, et al. . Structural cartilage damage attracts circulating rheumatoid arthritis synovial fibroblasts into affected joints. Arthritis Res Ther. 2017 Feb 28;19(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falconer J, Murphy AN, Young SP, et al. . Review: synovial cell metabolism and chronic inflammation In rheumatoid arthritis. Arthritis Rheumatol. 2018 Jul;70(7):984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amaral JK, Sutaria R, Schoen RT.. Treatment of chronic Chikungunya arthritis with methotrexate: A systematic review. Arthritis Care Res. 2018 Oct;70(10):1501–1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and single-cell RNA-seq datasets are available at the NCBI GEO database under the accession number GSE152782 and GSE176361, respectively. All generated code is available at https://github.com/GoffinetLab/CHIKV_scRNAseq-fibroblast.