Abstract
Nuclear pore complexes (NPC) regulate molecular traffics on nuclear envelope, which plays crucial roles during cell fate specification and diseases. The viral accessory protein NSP9 of SARS-CoV-2 is reported to interact with nucleoporin 62 (NUP62), a structural component of the NPC, but its biological impact on the host cell remain obscure. Here, we established new cell line models with ectopic NSP9 expression and determined the subcellular destination and biological functions of NSP9. Confocal imaging identified NSP9 to be largely localized in close proximity to the endoplasmic reticulum. In agreement with the subcellular distribution of NSP9, association of NSP9 with NUP62 was observed in cytoplasm. Furthermore, the overexpression of NSP9 correlated with a reduction of NUP62 expression on the nuclear envelope, suggesting that attenuating NUP62 expression might have contributed to defective NPC formation. Importantly, the loss of NUP62 impaired translocation of p65, a subunit of NF-κB, upon TNF-α stimulation. Concordantly, NSP9 over-expression blocked p65 nuclear transport. Taken together, these data shed light on the molecular mechanisms underlying the modulation of host cells during SARS-CoV-2 infection.
Keywords: NSP9, SARS-CoV-2, Nucleoporin, NUP62, p65
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease (COVID-19) and is highly transmissible in human population, resulting in a global pandemic in 2020 [1,2]. In order to successfully replicate, viruses employ several strategies to counter antiviral responses in host cells. Recent efforts have identified the suppression of interferons (IFNs) pathway as a major clinical determinant of COVID-19 severity [3], though this insight has yet to be translated into clinical benefits to date. As such, to gain a better understanding of pathophysiological mechanisms remains a pressing need in order to establish effective mitigating strategies to treat COVID-19.
The SARS-CoV-2 encodes 27 proteins with various roles in virus replication and packaging [4]. Those include 4 structural proteins (the nucleocapsid, envelope, membrane and spike), 7 accessory proteins (ORF3a-ORF8), and 16 non-structural proteins (NSP1-NSP16). To delineate the molecular basis of SARS-COV-2 pathogenesis, researchers have sought to comprehensively defined the interactions between SARS-CoV-2 proteins and human proteins or RNAs [4,5]. This approached have identified multiple core biological processes perturbed by SARS-CoV-2, including protein trafficking, translation, transcription, ubiquitin regulation and splicing. Of special note, certain viral proteins such as ORF6 and NSP9 preferably targeted nuclear pore complexes (NPCs) components [4].
NPCs establish nuclear pores studded throughout the nuclear envelope (NE) as portals for bidirectional cytoplasmic-nuclear transport channels. NPCs consist of multiple copies of ∼30 different proteins known as nucleoporins (NUPs). Around one-third of NUPs including NUP62 contain unstructured phenylalanine–glycine (FG) repeats and form a soft and flexible cobweb inside the pore [[6], [7], [8], [9]], which establishes conditional partitions. During nuclear transport, cargos require the aid of nuclear transport receptors called karyopherins to pass through the NPCs. Thus, NPCs establish a selective transport system by collaborating with karyopherins and play a fundamental role in regulating genomic information. Furthermore, disease specific alterations of transport factors result in the pathologic transduction of cellular signaling [[10], [11], [12], [13], [14]]. For example, we and others have reported that the interaction between viral ORF6 protein and NPCs components (NUP98 and Rae1) caused impaired mRNA export as well as nuclear transport [[15], [16], [17]]. On the other hand, while NSP9 was identified to target several NUPs including NUP62 [4], the biological significance of the interaction remains elusive.
Here, we investigated subcellular dynamics between NSP9 and NUP62 using cell line models with ectopic NSP9 expression. The specific localization of NSP9 in close vicinity to endoplasmic reticulum (ER) was observed. These NSP9 interacted with NUP62 in cytoplasm, hence restricting the association of NUP62 to the NPCs. Moreover, NUP62 depletion blocked nuclear transport of p65, a major component of NF-κB. Concordantly, the overexpression of NSP9 in these cells would prevent NF-κB translocation. These findings reveal a hitherto unknown mechanism through which a non-structural protein of SARS-CoV-2 could modulate and restrict NF-κB signaling, a central axis of inflammatory response.
2. Methods
2.1. Cell culture
HeLa cells were obtained from ATCC [18], and maintained in DMEM medium supplemented with 10% (vol/vol) fetal bovine serum (FBS) and 1% (vol/vol) penicillin/streptomycin (P/S). All cells were cultured at 37℃ and 5% CO2 in a humidified incubator.
2.2. DNA vectors and siRNA
In the construction of the DNA vector expressing GFP-fused NSP9, the coding region of NSP9 (YP_009724389.1) was synthesized and inserted into pJET (Thermo Scientific). Subsequently, the DNA sequence of NSP9 was amplified by PCR using primer (F; 5′-CCGCTCGAGGGAATAATGAGCTTAG-3′, R; 5′-CGCGGATCCTCATTGTAGACGTACT-3′) and purified. Lastly, the amplicon containing flanking BamHI/XhoI sites were inserted into pEGFP-C1.
2.3. Transfection
siRNA for NUP62 was from (Sigma-Aldrich, SASI_Hs01_00038069). DNA transfection was performed using Lipofectamine 2000 according to manufacturer's protocol. In brief, cells were seeded on 6-well plate at the density of 4 × 105/well, and incubated for 20 h. Cells were introduced siRNA at 80 nM, and further incubated for 24 h. Those cells were re-seeded on 6-well plate and coverslips to proceed further experiments.
2.4. Reagents
Human TNF-α was purchased from PeproTech, and stored in −20 °C at 0.1 mg/mL. To activate canonical NF-κB signaling in HeLa cells, TNF-α was added to a final concentration of 10ng/ml for all experiments.
2.5. Western blotting
Cells were lysed with RIPA buffer (50 mM Tris-HCL (pH 8.0)), 150 mM sodium chloride, 1% Triton X-100, 0.1% SDS, EDTA free Protease Inhibitor Cocktail (Nacalai tesque). For samples subjected to subcellular localization analysis, nuclear-cytoplasmic fractionation was performed in accordance with the manufacturer's protocol (NE-PER™ Nuclear and Cytoplasmic Extraction Reagents, ThermoFisher). Samples were subjected to SDS-PAGE followed by conventional wet transfer. Primary antibodies were incubated overnight with mixing at 4C. Following washing, membranes are incubated with secondary antibody at room temperature (RT) for 1–2 h. Images were detected by using an C-Digit blot scanner (LI-COR). Information of all antibodies used is shown in Supplementary Table 1.
2.6. Immunofluorescence (IF) microscopic analysis
Cells on coverslips were incubated under indicated conditions. Cells were fixed for 20 min in 4% paraformaldehyde in PBS, then permeabilized with 0.3% Triton X-100 in PBS for 10 min at RT. Subsequently, coverslips with cells were blocked with blocking solution (PBS containing 4% BSA) for 1 h at RT. Coverslips were incubated with the indicated primary antibodies in blocking solution overnight in the cold room. Coverslips were then washed with PBS and incubated with Alexa Fluor-conjugated secondary antibody in blocking solution for 1 h. After PBS washes, samples were mounted onto coverslips using the ProLong™ Gold Antifade reagent (Life Technologies), and examined by confocal microscopy (TCS SP8, Leica, objective × 100/1.4). Images captured on a confocal microscope were processed for deconvolution using Huygens Professional with default parameters, and the intensity of nuclear NF-κB/p65 was quantified with LAS X version 1.8. Information of all antibodies used is shown in Supplementary Table 1.
3. Results
3.1. Ectopic NSP9 is localized on the ER
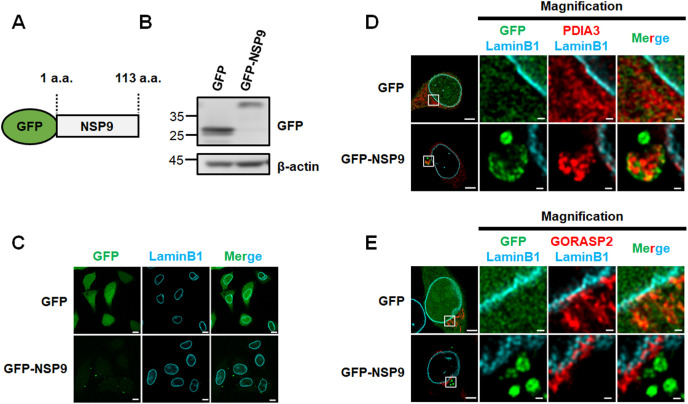
In order to investigate the biological impact of NSP9 in human cells, we established cell line models with ectopic NSP9 expression. The coding region of NSP9 was fused with a N-terminal GFP (GFP-NSP9) and transfected into HeLa cells (Fig. 1 A and B). Most of GFP-NSP9 were observed in cytoplasm by confocal imaging analysis (Fig. 1C). To determine subcellular localization of NSP9, HeLa cells expressing GFP-NSP9 were immunostained with organelle markers (PDIA3 for ER and GORASP2 for Golgi), and subjected to fluorescence confocal microscopic analysis. We found that GFP-NSP9 was mostly localized in PDIA3-stained ER, while such co-localization was rarely observed between GFP and PDIA3 (Fig. 1D). Moreover, no co-localization was observed been GORASP2 and GFP-NSP9 or GFP (Fig. 1E). These data suggested that GFP-NSP9 is preferentially targeted to the ER.
Fig. 1.
NSP9 of SARS-CoV-2 localized in the close vicinity to ER.
(A) Schematic diagram of NSP9-green fluorescent protein fusion protein (GFP-NSP9). (B) Western blot analysis of GFP in HeLa cells transfected with pEGFP-C1 or pEGFP-NSP9. (C) Immunofluorescence confocal microscopic analysis of GFP and LaminB1 in HeLa cells transfected with pEGFP-C1 or pEGFP-NSP9. Scale bars: 10 μm. (D, E) Immunofluorescence confocal microscopic analysis of GFP and ER marker PDIA3 (D) or GORASP2 (E) in HeLa cells transfected with pEGFP-C1 or pEGFP-NSP9. Scale bars: 5 μm. The area within the box is magnified and shown in the right panels. Scale bars: 500 nm.
3.2. NSP9 hampered the localization of NUP62 on the nuclear envelope
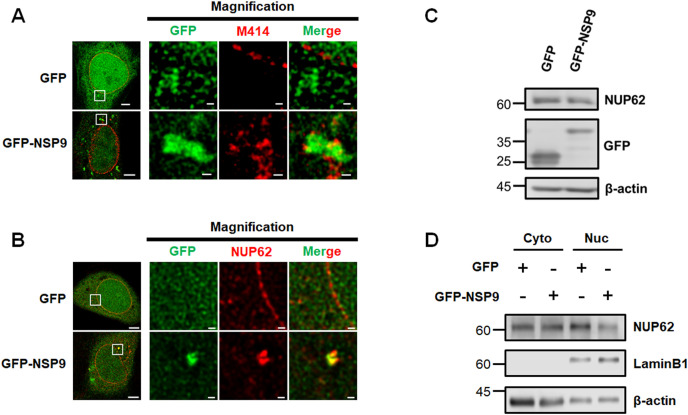
A recent SARS-CoV-2 protein interaction map identified that NSP9 interacted with certain NUPs including NUP62 [4]. We have previously shown that NUP62 plays pivotal roles in nuclear-cytoplasmic traffic as a central channel component of NPCs [8], which prompts us to study the biological significance of the NSP9-NUP62 association. To determine spatial association between NSP9 and NUP62, we first performed immunofluorescence confocal analysis using M414 antibody targeting NUPs harboring FG (FG-NUPs) and determined the co-localization of GFP-NSP9 with FG-NUPs in cytoplasm (Fig. 2 A). Furthermore, immunofluorescence confocal analysis using NUP62 antibody consistently demonstrated the localization of NUP62 and GFP-NSP9 (Fig. 2B). Considering subcellular localization of NSP9 and its previously reported functions [5], we speculated that NSP9 may sequester newly translated NUP62. To determine the effect of ectopic NSP9 on the spatial NUP62 distribution, we investigated the subcellular dynamics of NUP62. We found that total protein levels of NUP62 decreased upon GFP-NSP9 over-expression (Fig. 2C). The WB analysis using fractionated samples demonstrated a reduction of NUP62 in the nuclear fraction (Fig. 2D), which suggests a reduction of NUP62 that could lead to defective NPCs.
Fig. 2.
NSP9 of SARS-CoV-2 altered subcellular dynamics of NUP62.
(A, B) Immunofluorescence confocal microscopic analysis of GFP and M414 (A) or NUP62 (B) in HeLa cells transfected with pEGFP-C1 or pEGFP-NSP9. Scale bars: 5 μm. The area within the box is magnified and shown in the right panels. Scale bars: 500 nm. (C) Western blot analysis of NUP62 in HeLa cells transfected with pEGFP-C1 or pEGFP-NSP9. (D) Western blot analysis of NUP62 in both cytoplasmic and nucleus fraction using HeLa cells expressing either GFP or GFP-NSP9.
3.3. Loss of NUP62 attenuated p65 translocation
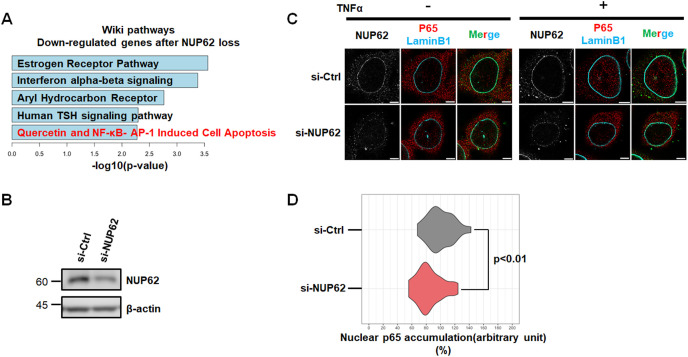
As a fundamental role of NUP62 is in transport regulation, we investigated changes in gene expression upon the RNAi knockdown of NUP62 by reanalyzing microarray data [8]. Remarkably, pathway analysis of down-regulated genes following NUP62 depletion identified several immune response-related pathways such as interferon and NF-κB (Fig. 3 A). Importantly, mounting evidence suggest that SARS-CoV-2 abrogate host immune defenses that includes type I IFN response wherein NF-κB plays a pivotal role to initiate immune response upon viral infections [[19], [20], [21], [22]]. Therefore, we next asked whether NUP62 interference affected the subcellular localization of the p65 (RELA) subunit of NF-κB. In this experiment, we treated HeLa cells with TNF-α to activate the canonical NF-κB pathway. When examined under confocal microscopy, TNF-α-induced p65 nuclear translocation was clearly attenuated in NUP62-depleted cells (Fig. 3B and C). Accordingly, quantification analysis confirmed that nuclear transport of p65 was significantly reduced in NUP62-depleted cells (Fig. 3D), suggesting the role of NUP62 in nuclear transport of p65.
Fig. 3.
Loss of NUP62 caused nuclear transport defects.
(A) Pathway analysis of the down-regulated genes after NUP62 knockdown. (B) Western blot analysis of NUP62 in HeLa cells upon siRNA mediated NUP62 silencing. (C) Immunofluorescence confocal microscopic analysis of p65 in HeLa cells expressing siRNA for NUP62 with TNFα processing for 30 m. Scale bar: 5 μm. (D) Quantification of nuclear p65 intensity in siCtrl (n = 44) and NUP62 silenced HeLa cells (n = 54) after TNFα treatment for 30 m. Relative intensity comparing to siCtrl was shown. Statistical analysis was based on Mann–Whitney U test.
3.4. Impairment of p65 translocation in NSP9 over-expressed cells
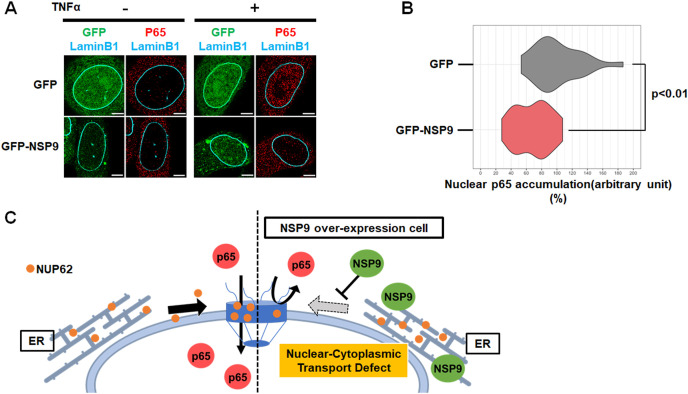
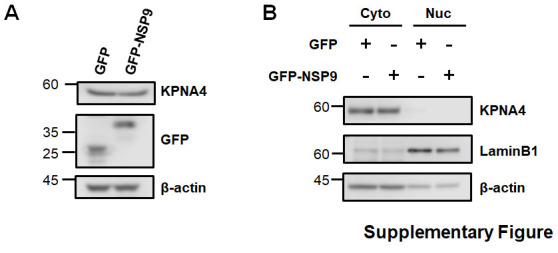
As NSP9 prevented NUP62 subcellular dynamics, we next asked if NSP9 overexpression has an effect on the subcellular localization of p65. In resting GFP or GFP-NSP9 expressing cells, p65 was predominantly located in cytoplasm prior to TNF-α treatment (Fig. 4 A). In the control GFP-expressing cells, the majority of p65 accumulated in the nucleus upon TNF-α stimulation for 30 m (Fig. 4A; Upper panel). In contrast, TNF-α-induced p65 translocation was significantly/markedly impaired in GFP-NSP9 expressing cells (Fig. 4A; Lower panel). Quantitative analysis revealed that p65 is retained in the cytoplasm in cells expressing GFP-NSP9 following TNF-α (Fig. 4B). In contrast, NSP9 over-expression did not alter the total amounts and subcellular localization of KPNA4 (Supplementary Fig. 1), which is determined as a specific transporter of p65 [23]. Collectively, our data demonstrated that NSP9 altered NPC composition and prevented NF-κB nuclear transport (Fig. 4C).
Fig. 4.
NSP9 of SARS-CoV-2 prevented nuclear transport of p65.
(A) Immunofluorescence confocal microscopic analysis of p65 in HeLa cells expressing either GFP or GFP-NSP9 after TNFα treatment for 30 m. Scale bars: 5 μm. (B) Quantification of nuclear p65 intensity in HeLa cells expressing either GFP (n = 23) or GFP-NSP9 (n = 28) after TNFα treatment for 30 m. Relative intensity comparing to GFP control was shown. Statistical analysis was based on Unpaired two-tailed t-test. (C) A model whereby NSP9 attenuates nuclear transport by hampering NUP62 dynamics and functions in host cells.
4. Discussion
The current study focuses on the biological impact of SARS-CoV-2 protein NSP9 on NPC component NUP62, where a novel pathogenic function of NSP9 in altering NPC composition changes and NF-κB nuclear transport was uncovered. At the molecular level, we propose that NSP9 sequesters NUP62 newly translated to the cytoplasm thereby limiting the assembly of NUP62 into NPCs. This in turn perturbs NPC-dependent transportation of molecules relevant to immune response.
Several NSPs are generated in the earliest stage of viral life cycle to interfere with essential cellular processes, so to provide selective advantage for virus propagation [19]. Of special interest, NSP9 has been reported to bind to the 7SL RNA of the signal recognition particle (SPR) cytoplasmic ribonucleoprotein complex to interrupt protein trafficking in host cells [5,24]. Consistent with a recent report, our work demonstrated that NSP9GFP preferably resides in ER. Notably, several NPC components including NUP62 assembled and co-localized with NSP9, which potentially reflects the ability of NSP9 to disturb directing proteins to their correct destination. Therefore, it is important to determine whether and how NSP9 selectively targets NPC components.
Inhibiting host gene expression is a hallmark of virulence factors from SARS-CoV-2 to counteract host defense. We and others recently determined that ORF6, encoded genes by SARS-CoV-2, disrupted mRNA export pathway by interacting with NUP98 and Rae1 of host cells [[15], [16], [17]]. Further, NSP1 also disrupted mRNA export machinery through binding with mRNA export receptor NXF1 while NSP9 is unable to affect mRNA export processes [25]. In agreement with recent interaction maps of viral protein with host factors [25], our present study demonstrated that NSP9 targets NUP62 to render NPCs defective in host cells. Since NPC composition changes are crucial to achieve selective transport underling gene expression processes [8,11,26], interfering NPC functions/components is a promising strategy for viral infections. The current study focuses on the defect of p65 transport upon NSP9 over-expression, and in so doing raises the interesting possibility that NSP9 could prevent the nuclear transportation of other important transcription factors.
The suppression of IFN response is a hallmark strategy of SARS-CoV-2. Several key pathways such as STAT1 and NF-κB are activated in response to IFN stimuli. The activation of NF-κB pathway not only counterbalances antiviral activity but also differentially regulates the expression of several IFN-stimulated genes [20]. In fact, viral protein 4b of MERS-CoV blocked the expression of proinflammatory cytokines by blocking NF-κB transport [22], which further implicates antiviral functions for NF-κB during virus infection. As multiple SARS-CoV-2 derived proteins could differentially or coordinately target immune response, it is therefore important to understand the systemic actions of viral proteins to fully unmask the pathophysiology of SARS-CoV-2.
In conclusion, we demonstrated that SARS-CoV-2 protein NSP9 caused a dysregulated nuclear transport system by altering NPC composition in host cells. Further studies of NSP9 targeting agents could lead to therapeutic regimen to attenuate pathogenesis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank all members of the Wong laboratory. This work was supported by MEXT/JSPS KAKENHI grant numbers 19K07398 (A K), 20K07568 (M H), 20K16262 (S K), 18K07228 (V D) and 21H05744, 21K19043 (R W) from MEXT Japan, Kanazawa University against COVID-19 (R W), the Kobayashi International Scholarship Foundation and the Takeda Science Foundation (R W).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2021.11.046.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
References
- 1.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., Chen J., Wang W., Chen B., Jiang L., Yu S., Lu J., Wang J., Xu M., Yuan Z., Zhang Q., Zhang X., Zhao G., Wang S., Chen S., Lu H. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583:437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 4.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y.F., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G., Goldfarb D., De Lorenzo G., Rihn S.J., Voorhees R.M., Botten J.W., Majumdar D., Guttman M. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339. doi: 10.1016/j.cell.2020.10.004. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamed M.S., Hazawa M., Kobayashi A., Guillaud L., Watanabe-Nakayama T., Nakayama M., Wang H., Kodera N., Oshima M., Ando T., Wong R.W. Spatiotemporally tracking of nano-biofilaments inside the nuclear pore complex core. Biomaterials. 2020;256:120198. doi: 10.1016/j.biomaterials.2020.120198. [DOI] [PubMed] [Google Scholar]
- 7.Mohamed M.S., Kobayashi A., Taoka A., Watanabe-Nakayama T., Kikuchi Y., Hazawa M., Minamoto T., Fukumori Y., Kodera N., Uchihashi T., Ando T., Wong R.W. High-speed atomic force microscopy reveals loss of nuclear pore resilience as a dying code in colorectal cancer cells. ACS Nano. 2017;11:5567–5578. doi: 10.1021/acsnano.7b00906. [DOI] [PubMed] [Google Scholar]
- 8.Hazawa M., Lin D., Kobayashi A., Jiang Y., Xu L., Dewi F.R.P., Mohamed M.S., Hartono, Nakada M., Meguro-Horike M., Horike S., Koeffler H.P., Wong R.W. ROCK -dependent phosphorylation of NUP 62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19:73–88. doi: 10.15252/embr.201744523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sajidah E.S., Lim K., Wong R.W. How SARS-CoV-2 and other viruses build an invasion route to hijack the host nucleocytoplasmic trafficking system. Cells. 2021;10:1–40. doi: 10.3390/cells10061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazawa M., Sakai K., Kobayashi A., Yoshino H., Iga Y., Iwashima Y., Lim K.S., Chih-Cheng Voon D., Jiang Y.Y., ichi Horike S., Lin D.C., Wong R.W. Disease-specific alteration of karyopherin-α subtype establishes feed-forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene. 2020;39:2212–2223. doi: 10.1038/s41388-019-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Bravo V., Pippa R., Song W.M., Carceles-Cordon M., Dominguez-Andres A., Fujiwara N., Woo J., Koh A.P., Ertel A., Lokareddy R.K., Cuesta-Dominguez A., Kim R.S., Rodriguez-Fernandez I., Li P., Gordon R., Hirschfield H., Prats J.M., Reddy E.P., Fatatis A., Petrylak D.P., Gomella L., Kelly W.K., Lowe S.W., Knudsen K.E., Galsky M.D., Cingolani G., Lujambio A., Hoshida Y., Domingo-Domenech J. Nuclear pores promote lethal prostate cancer by increasing pom121-driven E2F1, MYC, and AR nuclear import. Cell. 2018;174:1200–1215. doi: 10.1016/j.cell.2018.07.015. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Guo Y., Lu C., Zhang R., Wang Y., Luo L., Zhang Y., Chu C.H., Wang K.J., Obbad S., Yan W., Li X. Inhibition of Karyopherin beta 1 suppresses prostate cancer growth. Oncogene. 2019;38:4700–4714. doi: 10.1038/s41388-019-0745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang S., Wang Z., Ye Y., Zhang F., Li H., Yang Y., Miao H., Liang H., Zhang Y., Jiang L., Hu Y., Zheng L., Liu X., Liu Y. E2F1 and E2F7 differentially regulate KPNA2 to promote the development of gallbladder cancer. Oncogene. 2019;38:1269–1281. doi: 10.1038/s41388-018-0494-7. [DOI] [PubMed] [Google Scholar]
- 14.Ma A., Tang M., Zhang L., Wang B., Yang Z., Liu Y., Xu G., Wu L., Jing T., Xu X., Yang S., Liu Y. USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene. 2019;38:2405–2419. doi: 10.1038/s41388-018-0590-8. [DOI] [PubMed] [Google Scholar]
- 15.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., García-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U.S.A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato K., Ikliptikawati D.K., Kobayashi A., Kondo H., Lim K., Hazawa M., Wong R.W. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem. Biophys. Res. Commun. 2021;536:59–66. doi: 10.1016/j.bbrc.2020.11.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addetia A., Lieberman N.A.P., Phung Q., Hsiang T.Y., Xie H., Roychoudhury P., Shrestha L., Loprieno M.A., Huang M.L., Gale M., Jerome K.R., Greninger A.L. Sars-cov-2 orf6 disrupts bidirectional nucleocytoplasmic transport through interactions with rae1 and nup98. mBio. 2021;12 doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazawa M., Amemori S., Nishiyama Y., Iga Y., Iwashima Y., Kobayashi A., Nagatani H., Mizuno M., Takahashi K., Wong R.W. A light-switching pyrene probe to detect phase-separated biomolecules. IScience. 2021;24:102865. doi: 10.1016/j.isci.2021.102865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeffer L.M. The role of nuclear factor κb in the interferon response. J. Interferon Cytokine Res. 2011;31:553–559. doi: 10.1089/jir.2011.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhuo Y., Guo Z., Ba T., Zhang C., He L., Zeng C., Dai H. African swine fever virus mgf360-12l inhibits type I interferon production by blocking the interaction of importin α and NF-κB signaling pathway. Virol. Sin. 2021;36:176–186. doi: 10.1007/s12250-020-00304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canton J., Fehr A.R., Fernandez-Delgado R., Gutierrez-Alvarez F.J., Sanchez-Aparicio M.T., García-Sastre A., Perlman S., Enjuanes L., Sola I. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018;14:1–25. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fagerlund R., Kinnunen L., Köhler M., Julkunen I., Melén K. NF-κB is transported into the nucleus by importin α3 and importin α4. J. Biol. Chem. 2005;280:15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 24.Kellogg M.K., Miller S.C., Tikhonova E.B., Karamyshev A.L. Srpassing co-translational targeting: the role of the signal recognition particle in protein targeting and mrna protection. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., Hobman T.C., Ptak C., Gao B., Minna J.D., Chen Z., García-Sastre A., Ren Y., Wozniak R.W., Fontoura B.M.A. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021;7:1–13. doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Angelo M.A., Gomez-Cavazos J.S., Mei A., Lackner D.H., Hetzer M.W. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.