Abstract
A method utilizing PCR-restriction fragment length polymorphism (RFLP) in the Helicobacter pylori genes is widely used to differentiate strains. However, with this typing method only a single base change at a specific restriction site can be detected. In addition, it is unclear whether the nucleotide base change recognized by RFLP is related to a substitution of encoded amino acid. To examine the validity of the PCR-RFLP method, 933-bp PCR products were obtained from 41 different clinical H. pylori isolates and were digested with Sau3A restriction endonuclease. Furthermore, the nucleotides of the same region in the ureB gene were directly sequenced and compared. PCR-RFLP confirmed that there was genetic diversity within the ureB gene with three distinct types, one being well conserved and the other two being variations. However, the direct sequencing method revealed that there was no difference at the nucleotide level among these RFLP types. Base substitutions recognized by Sau3A occurred in the third-base position and did not change the encoded amino acid. In addition, many nucleotide mutations, which could not be recognized by Sau3A, were frequently found. These results suggest that the PCR-RFLP method provides for an easy typing scheme of isolates, but does not reveal the true extent of genetic diversity. It is proposed that careful observation is required for the interpretation of results when clinical isolates are differentiated.
Helicobacter pylori is a gram-negative, microaerophilic organism that colonizes the human gastric mucosa. It has been shown that H. pylori is not only the causative agent of chronic gastritis and peptic ulcer disease (15) but also a risk factor for gastric cancer (16, 19), and it has been designated a class I carcinogen by the World Health Organization (10). Indeed, the eradication of this organism by antibacterial therapy has led to the normalization of chronic gastritis and to lower rates of peptic ulcer relapse (9). H. pylori infection occurs worldwide at a high prevalence rate, and an accurate method for the differentiation of H. pylori strains is of great importance for diagnosis and monitoring after treatment.
To differentiate H. pylori isolates, many approaches have been presented, but no typing scheme for precise strain identification is generally available. Several molecular techniques have been applied to separate clinical isolates from different patients. PCR-restriction fragment length polymorphism (RFLP) analysis has been widely developed for the typing of clinical isolates, with several genes within H. pylori having been targets for this method (1, 4, 5, 7, 8, 13, 14, 17, 18, 23). However, this method is limited when it comes to differentiating isolates because it detects only a single base change at a specific restriction site. Therefore, it is unable to evaluate the amino acid alignment and reveal whether the base change affected the amino acid itself.
Recently, a PCR-direct sequencing method has been applied for the typing of H. pylori clinical isolates (12, 25). This method has the clear advantage of revealing not only the full-length nucleotide sequence but the amino acid sequence, showing the genomic differentiation among the strains.
In the present study, we tested the validity of PCR-RFLP analysis of the H. pylori ureB gene by examining whether the nucleotide variation as determined by PCR-direct sequencing is related to the restriction sites.
MATERIALS AND METHODS
H. pylori strains and patients.
Forty-one H. pylori isolates from 22 patients were obtained from gastric tissue during gastroduodenal endoscopy in the Third Department of Internal Medicine, Kyoto Prefectural University of Medicine, Kyoto, Japan. All patients gave informed consent for the biopsy samples, and this study was approved by the Human Research Committee of the university. Endoscopic diagnoses were recorded for all patients by two trained endoscopists. The diagnosis was classified according to the guidelines of the Sydney System (24). The endoscopic diagnoses in the patients were as follows: 6 patients had duodenal ulcer, 3 had gastric ulcer, and 13 had chronic gastritis without peptic ulcer.
Culture of H. pylori from gastric biopsy specimens.
Two biopsy specimens were taken from both the antrum (pyloric gland area) and the body (fundic gland area) of the stomach with a sterilized endoscope. The biopsy specimens were initially spread out with an applicator and placed in a Helicobacter-selective agar plate (Eiken Chemical Co., Ltd., Tokyo, Japan) containing 7% (vol/vol) horse serum, vancomycin (10 μg/ml), polymyxin B (2.5 U/ml), trimethoprim (5 μg/ml), and amphotericin B (2 μg/ml). The plates were incubated at 37°C under microaerophilic conditions (AnaeroPack Systems; Mitsubishi Gas Chemical Co., Inc., Osaka, Japan) for up to 7 days. The organisms were identified as H. pylori by Gram staining, colony morphology, and positivity for oxidase and catalase.
Extraction of genomic DNA from clinical isolates.
Chromosomal DNA was extracted and purified from the H. pylori strains with the use of Instagene Matrix (Bio-Rad Laboratories, Richmond, Calif.). Briefly, the isolated bacterial colony was suspended in 1 ml of distilled water. The suspension was centrifuged at 10,000 × g for 1 min, and the supernatant was removed. The Instagene Matrix was added to the pellets and boiled for 8 min after preincubation at 56°C. The supernatants were stored at −20°C until used as PCR templates.
PCR amplification.
Oligonucleotide primers were synthesized by using a DNA synthesizer. The oligonucleotides used as PCR primers were derived from the known sequence of ureB, which encodes the urease structural gene (3). The amplification product of the forward (5′-GAACATGACTACACCAT-3′) and reverse (5′-TGGTTTGAGGGCGAATC-3′) primers was a 933-bp nucleotide. Bacterial DNA (5 μl) was added to 50-μl reaction mixtures containing 5 μl 10× PCR buffer, which consisted of 100 mmol of KCl, 20 mmol of Tris-HCl (pH 7.5), 15 mmol of MgCl2, 1 mmol of dithiothreitol, and 0.1 mmol of EDTA per liter, 200 μl of each deoxynucleotide (Pharmacia Biotech AB), 200 nmol of each primer per liter, 1.0 U of Taq DNA polymerase (included in the Expand High Fidelity PCR System; (Boehringer Mannheim, Mannheim, Germany), and H2O. The PCR was performed with an automatic thermal cycler (TP-3000; Takara Biomedicals, Otsu, Japan). The amplification cycle consisted of an initial denaturation of target DNA at 95°C for 5 min and then denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1 min. The final cycle included an extension step for 5 min at 72°C to ensure full extension of the product. Samples were amplified through 35 consecutive cycles.
Enzymatic digestion of amplified DNA.
We selected Sau3A (Takara Biomedicals, Otsu, Japan) as a representative restriction enzyme, which has been widely used to differentiate H. pylori strains, and whose usefulness has been indicated (4, 5, 23). The site recognized by this enzyme is GATC. A 10-μl sample of the PCR product was digested with 10 U of Sau3A for 3 h at 37°C in buffer recommended by the manufacturer. The digested samples were analyzed by electrophoresis by using 5% NuSieve agarose (3:1; NuSieve; FMC BioProducts, Rockland, Maine) containing ethidium bromide. The restriction fragments were separated at 50 V in 1× Tris-borate-EDTA buffer for 60 min and examined by transillumination before being photographed. A 100-bp DNA ladder (Takara Biomedicals) was used as the standard for the molecular size marker.
Nucleotide sequencing.
The PCR products were purified with Centricon-100 Concentrator columns (Amicon, Beverly, Mass.). DNA sequencing was performed by the dideoxynucleotide primer method with a Thermo Sequenase premixed cycle sequencing kit (Amersham Pharmacia Biotech) in an automated DNA sequencer, model SQ-5500 (Hitachi Co., Ltd., Tokyo, Japan). According to the manufacturer's protocol, reagent mixtures containing 1 μl of purified PCR product, 1 pmol of primer labeled by Texas Red, 6 μl of Thermo Sequenase reagent, and 22 μl of sterilized distilled water were prepared. Reaction tubes were placed in the thermal cycler, and the thermal sequencing cycle was started under the following conditions: first heating at 94°C for 5 min and then 25 cycles consisting of 94°C for 30 s and 60°C for 30 s. Cycle sequencing reactions were performed for both DNA strands by using two primers (sense, 5′-GAACATGACTACACCAT-3′; antisense, 5′-TGGTTTGAGGGCGAATC-3′) and the 933-bp product as a template. Any sequences that were difficult to read were also resequenced.
Sequence data analysis.
The nucleotide sequence and the deduced amino acid sequence within the ureB gene were analyzed with Genetyx-Mac, version 9.0, software (Software Development Co., Ltd., Tokyo, Japan). The sequence of one H. pylori strain (85P) previously reported was used as a reference. The sequence was taken from the GenBank sequence data library (accession number M60398). The nucleotide and the deduced amino acid sequence identities between each strain were determined as the mean ± the standard deviation (SD). The recognition sites of the enzyme within the sequence region were analyzed with the same software.
Nucleotide sequence accession numbers.
The nucleotide sequence of KP48a, KP48b (strain from a duodenal ulcer patient), KP72b (from a chronic gastritis patient), KP96a, and KP96b (from a chronic gastritis patient) are in the DDBJ, EMBL, and GenBank nucleotide sequence databases under the accession numbers AB028034, AB028035, AB028036, AB028037, and AB028038.
RESULTS
Isolation of H. pylori from biopsy specimens.
A total of 41 strains were isolated from biopsy specimens: 12 strains from patients with duodenal ulcer, 6 strains from patients with gastric ulcer, and 23 strains from patients with chronic gastritis. In 19 of the 22 patients, two isolates were obtained from both the antrum and the body of the stomach.
PCR amplification and RFLP types.
PCR was performed for each of the 41 strains. In all strains, the PCR products were successfully amplified to the expected 933-bp fragment within ureB gene (corresponding to nucleotides 96 to 1,029 of strain 85P).
We examined whether PCR-RFLP analysis could differentiate H. pylori strains. The digestion fragments of the PCR products obtained with restriction endonuclease indicated that the H. pylori strains could be separated into three types (types A, B, and C) on the basis of the presence of two, one, and three recognition sites for the enzyme, respectively (Table 1 and Fig. 1). The precise fragment size was determined by using the known locations of Sau3A sites in a linear map of previously reported H. pylori ureB gene sequence (3), which was classified as type A, in spite of the presence of three Sau3A sites. The B and C types were considered variants.
TABLE 1.
Typing by PCR-RFLP and comparison of the fragments by nucleotide sequencing
| Strain group (no. of strains) | RFLP type | Fragment lengths (bp) | Sum (bp) | Calculated froma: |
|---|---|---|---|---|
| Reference strain 85P | 389, 281, 246, 17 | 933 | Nuc Seq | |
| 1 (24) | A | 390, 280, 250 | 920 | Gel |
| 2 (3) | B | 530, 390 | 920 | Gel |
| 3 (14) | C | 390, 250, 170, 110 | 920 | Gel |
Nuc Seq, nucleotide sequence; Gel, gel electrophoresis.
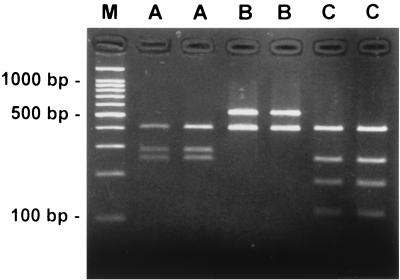
FIG. 1.
Restriction digest types of the 933-bp PCR product from the H. pylori ureB gene of six representative clinical isolates. Amplified DNA was digested with Sau3A and separated by electrophoresis on a 5% gel. Lane M is the molecular mass standard of 100 bp. Lanes show type A (A), type B (B), and type C (C) by PCR-RFLP analysis. The B and C types were considered variants.
As shown in Table 1, the results of RFLP analysis resolved by agarose gel electrophoresis were nearly identical to the predicted fragments based on the nucleotide sequence data. However, the 17-bp Sau3A DNA fragment predicted from the nucleotide sequence was too small to be detected by the PCR-RFLP method as used here (Fig. 1).
Most of the strains isolated from the patients with duodenal ulcer or gastric ulcer were type A, with ratios of 10/12 (83.3%) and 6/6 (100%), respectively. In contrast, 15 of 23 (65.2%) strains isolated from patients with chronic gastritis were of the B or C type, each of which was considered a variant (Table 2).
TABLE 2.
PCR-RFLP types from the different gastroduodenal diseases
| Strain group (no. of strains) | RFLP type | No. of isolates from gastroduodenal diseasea
|
||
|---|---|---|---|---|
| DU | GU | CG | ||
| 1 (24) | A | 10 | 6 | 8 |
| 2 (3) | B | 0 | 0 | 3 |
| 3 (14) | C | 2 | 0 | 12 |
DU, duodenal ulcer; GU, gastric ulcer; CG, chronic gastritis.
Comparison of RFLP types by nucleotide and deduced amino acid sequence determined by PCR-direct sequencing.
To examine the nature of PCR-RFLP analysis, the 933-bp PCR products were directly sequenced. From the primary sequencing data, the nucleotide sequences of the 660-bp ureB gene (corresponding to nucleotides 226 to 886 of strain 85P) were determined. No insertions or deletions were found within this 660-bp ureB region, and the deduced amino acid sequences could be determined without a stop codon (Fig. 2).
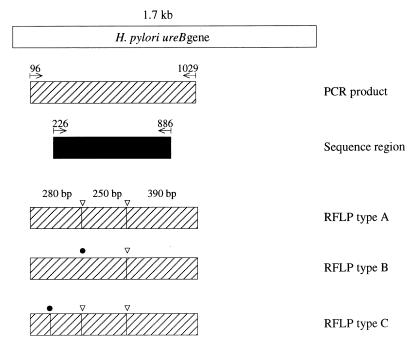
FIG. 2.
The region of the H. pylori ureB gene PCR was amplified and sequenced. The PCR-amplified region corresponding to nucleotides 96 to 1029 of strain 85P is indicated as a hatched bar. The sequenced region corresponding to nucleotides 226 to 886 is indicated as a black bar. White arrowheads show the recognition sites for Sau3A. Closed circles indicate the nucleotide mutation site found by Sau3A. The B and C PCR-RFLP types were considered to be variants.
Analysis of the nucleotide sequences indicated that the base substitution events within the ureB gene did not relate to the observed PCR-RFLP types (Table 3). Although the B and C types were judged as variants by PCR-RFLP, their nucleotide sequence identities were the same as for type A. There was no difference in the nucleotide and the amino acid sequences among the three RFLP types.
TABLE 3.
Comparison of PCR-RFLP types by nucleotide and deduced amino acid sequence by PCR-direct sequencing
| Strain group (no. of strains) | RFLP type | % Identity of nucleotide sequencea | % Identity of amino acid sequencea |
|---|---|---|---|
| 1 (24) | A | 94.4 ± 2.6 | 97.7 ± 3.2 |
| 2 (3) | B | 95.2 ± 0.4 | 98.3 ± 0.5 |
| 3 (14) | C | 95.3 ± 0.3 | 99.3 ± 0.4 |
Nucleotide and amino acid sequence identities were calculated by comparison with that of 85P, as previously reported. The results are given as the mean ± the SD.
Mutational events of the ureB gene explained by PCR-RFLP analysis.
Two regions were recognized by Sau3A within the nucleotide sequence data obtained in the present study (Fig. 2). In the sequence data, the base substitutions were detected at one position in three strains of type B. These mutations included one G-to-A transition and two C-to-T transitions. They occurred in the third-base position and did not change the encoded amino acid. Furthermore, nucleotide mutations of the third-base position were frequently found at other sites, which could not be recognized by Sau3A. Most of the nucleotide mutations did not affect the encoded amino acid alignment (Fig. 3).
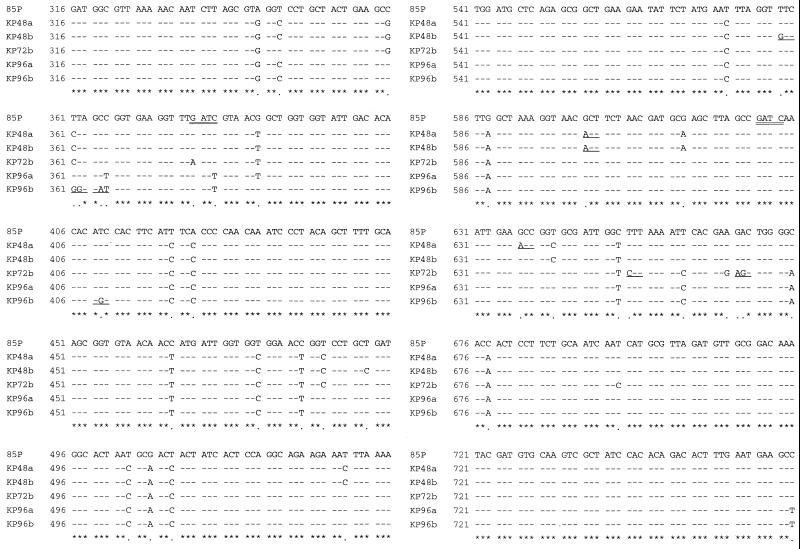
FIG. 3.
Partial nucleotide sequences of the 933-bp ureB gene PCR product obtained from five H. pylori strains representing two different PCR-RFLP types and one reference strain (85P), as previously reported. PCR-RFLP of KP48a and KP48b strains showed them to be type A. KP72b, KP96a, and KP96b were considered to be variant strains showing type B by PCR-RFLP. Numbers on the left indicate the base positions corresponding to nucleotides 316 to 765 of strain 85P. Bases included in the Sau3A restriction site (GATC) are double underlined. Asterisks indicate complete identity of the nucleotides, and dots indicate base mutations. The positions of the encoded amino acid substitution are underlined.
DISCUSSION
In recent years, many investigators applying the molecular techniques have revealed that H. pylori possesses a remarkable degree of genetic diversity, which closely relates with its epidemiological and pathological characteristics and dynamics of transmission. Various typing methods have been tried, including PCR-based randomly amplified polymorphic DNA fingerprinting (6), pulsed-field gel electrophoresis (PFGE) (21), and hybridization with specific probes (20).
In addition to the above techniques, a PCR-RFLP analysis has been widely developed for the typing and the differentiation of H. pylori strains from clinical isolates. This method has been used to analyze H. pylori genes, especially those encoding urease structural and accessory proteins (1, 4, 7, 8, 13, 14, 17, 18, 23). These results have indicated that PCR-RFLP method was an effective tool and that a diversity of H. pylori urease genes existed among clinical isolates. However, its use in identification is limited because the method detects mutations only at the restriction sites of the enzymes even if many other regions differed throughout the entire genome.
On the other hand, the sequencing method has provided a means of examining the nucleotide alignment within a gene and thus has advantages over other methods that examine only restriction site changes in a single gene, such as PFGE and PCR-RFLP. Therefore, we examined the nucleotide sequence of H. pylori ureB genes with different RFLP types and compared the results.
Seventeen strains were judged as variants by RFLP analysis, but their ureB gene sequences were shown to be well conserved by direct sequencing (>95% identity at the nucleotide level and >98% identity at the deduced amino acid level). In addition, small fragments (17 bp) were not detected by the RFLP method, as supported by another study (5). The present study confirmed that there is diversity in the ureB genes of isolates (1, 4, 7, 11, 18). However, the nucleotide mutations within the ureB gene occurred randomly and were unrelated to the restriction sites used here. By comparing the direct sequencing results to the results obtained by PCR-RFLP, it was concluded that the nucleotide sequence variations detected in this gene were base substitutions that conserve the amino acid alignment. It could be speculated that the nucleotide sequence of a virulence factor as important as urease should be conserved among strains (22), a notion supported by our findings.
The present study showed that there is a relation between the RFLP type of the ureB gene and the clinical outcome, a finding in agreement with a previous report (11). However, this association seems doubtful given that the sequencing method indicated there was no difference at the nucleotide level among the RFLP types. The differences in RFLP types are due mainly to the silent nucleotide variation within the gene. Thus, the results obtained with lower-resolution techniques, such as PCR-RFLP or PFGE, have probably led to an overestimation of the true extent of genetic diversity in H. pylori (2).
Versalovic et al. recently reported that an A-to-G mutation at position 2143 or 2144 in domain V of the 23S rRNA gene of H. pylori was closely associated with resistance to clarithromycin (26). In addition, they also established a PCR-RFLP system to detect these base mutations precisely. In this case, RFLP analysis was useful for identifying the mutations, because the assay was able to catch the nucleotide mutation directly, and these mutations were closely related to the amino acid associated with antimicrobial resistance.
In conclusion, the present study suggests that PCR-RFLP analysis of a portion of the H. pylori ureB gene does not provide for an accurate identification of bacteria. Although the PCR-RFLP technique will continue to be useful for simple strain typing, a more careful examination may be required when differentiating clinical isolates of H. pylori.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Berg D E. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20:6221–6225. doi: 10.1093/nar/20.23.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Millis D M, Ives C, Gibson R, Merberg D, Millis S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Clayton C L, Pallen M, Kleanthous H, Wren B, Tabaqchali S. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res. 1990;18:362. doi: 10.1093/nar/18.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton C L, Kleanthous H, Morgan D D, Puckey L, Tabaqchali S. Rapid fingerprinting of Helicobacter pylori by polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1420–1425. doi: 10.1128/jcm.31.6.1420-1425.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans D G, Evans D J, Lampert H C, Graham D Y. Restriction fragment length polymorphism in the adhesin gene hpaA of Helicobacter pylori. Am J Gastroenterol. 1995;90:1282–1288. [PubMed] [Google Scholar]
- 6.Fantry G T, Zheng Q X, Darwin P E, Rosenstein A H, James S P. Mixed infection with cagA-positive and cagA-negative strains of Helicobacter pylori. Helicobacter. 1996;1:98–106. doi: 10.1111/j.1523-5378.1996.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 7.Foxall P A, Hu L T, Mobley H L. Use of polymerase chain reaction-amplified Helicobacter pylori urease structural genes for differentiation of isolates. J Clin Microbiol. 1992;30:739–741. doi: 10.1128/jcm.30.3.739-741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto S, Marshall B, Blaser M J. PCR-based restriction fragment length polymorphism typing of Helicobacter pylori. J Clin Microbiol. 1994;32:331–334. doi: 10.1128/jcm.32.2.331-334.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentschel E, Brandstatter G, Dragosics B, Hirschl A M, Nemec H, Schutze K, Taufer M, Wurzer H. Effect of ranitidine and amoxicillin plus metronidazole on the eradication of Helicobacter pylori and the recurrence of duodenal ulcer. N Engl J Med. 1993;328:308–312. doi: 10.1056/NEJM199302043280503. [DOI] [PubMed] [Google Scholar]
- 10.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 11.Ito A, Fujioka T, Kubota T, Nasu M. Molecular typing of Helicobacter pylori: differences in pathogenicity among diverse strains. J Gastroenterol. 1996;31:1–5. doi: 10.1007/BF01211179. [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazakai Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Ha T, Chi D S, Ferguson D A, Jr, Jiang C, Laffan J J, Thomas E. Differentiation of Helicobacter pylori strains directly from gastric biopsy specimens by PCR-based restriction fragment length polymorphism analysis without culture. J Clin Microbiol. 1997;35:3021–3025. doi: 10.1128/jcm.35.12.3021-3025.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore R A, Kureishi A, Wong S, Bryan L. Categorization of clinical isolates of Helicobacter pylori on the basis of restriction digest analyses of polymerase chain reaction-amplified ureC genes. J Clin Microbiol. 1993;31:1334–1355. doi: 10.1128/jcm.31.5.1334-1335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- 16.Nomura A, Stemmermann G, Chyou P, Kato I, Perez-Perez G, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 17.Owen R J, Bickley J, Hurtado A, Fraser A, Pounder R E. Comparison of PCR-based restriction length polymorphism analysis of urease genes with rRNA gene profiling for monitoring Helicobacter pylori infections in patients on triple therapy. J Clin Microbiol. 1994;32:1203–1210. doi: 10.1128/jcm.32.5.1203-1210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owen R J, Slater E R, Xerry J, Peters T M, Teare E L, Grant A. Development of a scheme for genotyping Helicobacter pylori based on allelic variation in urease subunit genes. J Clin Microbiol. 1998;36:3710–3712. doi: 10.1128/jcm.36.12.3710-3712.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsonnet J, Friedman G, Vandersteen D, Chang Y, Vogelman J, Orentreich N, Sibley R. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 20.Pina M, Occhialini A, Monteiro L, Doermann H P, Megraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prewett E J, Bickley J, Owen R J, Pounder R E. DNA patterns of Helicobacter pylori isolated from gastric antrum, body, and duodenum. Gastroenterology. 1992;102:829–833. doi: 10.1016/0016-5085(92)90165-u. [DOI] [PubMed] [Google Scholar]
- 22.Shen Z, Schauer D B, Mobley H L, Fox J G. Development of a PCR-restriction fragment length polymorphism assay using the nucleotide sequence of the Helicobacter hepaticus urease structural genes ureAB. J Clin Microbiol. 1998;36:2447–2453. doi: 10.1128/jcm.36.9.2447-2453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shortridge V D, Stone G G, Flamm R K, Beyer J, Versalovic J, Graham D W, Tanaka S K. Molecular typing of Helicobacter pylori isolates from a multicenter U.S. clinical trial by ureC restriction fragment length polymorphism. J Clin Microbiol. 1997;35:471–473. doi: 10.1128/jcm.35.2.471-473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tytgat G. The Sydney System: endoscopic division. Endoscopic appearances in gastritis/duodenitis. J Gastroenterol Hepatol. 1991;6:223–234. doi: 10.1111/j.1440-1746.1991.tb01469.x. [DOI] [PubMed] [Google Scholar]
- 25.van der Ende A, Pan Z, Bart A, van der Hulst R, Feller M, Xiao S, Tytgat D, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]