Abstract
Human endogenous retroviruses (HERVs) represent ∼8% of human genome, deriving from exogenous retroviral infections of germ line cells occurred millions of years ago and being inherited by the offspring in a Mendelian fashion. Most of HERVs are nonprotein-coding because of the accumulation of mutations, insertions, deletions, and/or truncations. It has been long thought that HERVs were “junk DNA”. However, it is now known that HERVs are involved in various biological processes through encoding proteins, acting as promoters/enhancers, or lncRNAs to affect human health and disease. In this review, we summarized recent findings about HERVs, with implications in embryonic development, pluripotency, cancer, aging, and neurodegenerative diseases.
Keywords: Human endogenous retroviruses (HERVs), Embryonic development and pluripotency, Cancer, Aging, Neurodegenerative diseases
1. Introduction
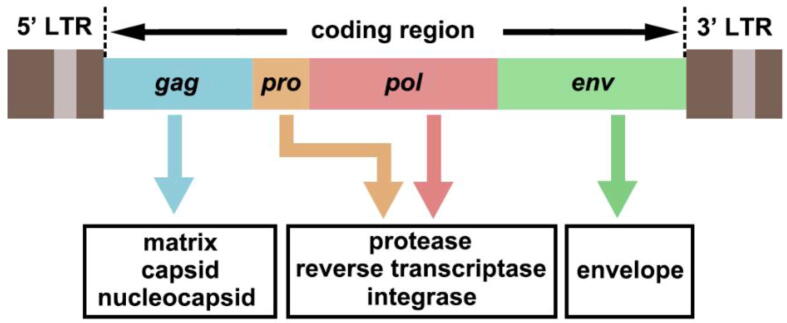
Reverse-transcribing viruses, which synthesize a copy of genomic DNA from an RNA template, are widespread in animals, plants, algae, and fungi. Among reverse-transcribing viruses, belpaoviruses, metaviruses, pseudoviruses [better known as Bel/Pao, Ty3/Gypsy, and Ty1/Copia long terminal repeat (LTR) retrotransposons, respectively], and retroviruses typically encapsidate single-stranded RNA (ssRNA) genomes and frequently integrate into the host genomes as part of their replication cycles (Baltimore class VI) [1]. In fact, endogenous retroviruses (ERVs) and LTR retrotransposons represent a considerable part of their host genomes [2]. Human ERVs (HERVs), which integrated into the human genome via exogenous retroviral infections of germ line cells over millions of years ago [3], account for ∼8% of the human genome [4]. The complete genomic structure of HERVs is composed of gag, pro, pol, and env, flanked by two LTRs (Fig. 1). Among them, gag encodes capsid, nuclecapsid, and matrix protein, pro encodes protease, pol encodes reverse transcriptase and integrase, and env encodes envelope protein [5]. LTR is non-coding region that comprises many regulatory functions [promoter, enhancer, primer-binding site (PBS) for reverse transcription, polyA signal, and others]. However, most of HERVs are nonprotein-coding because of the accumulation of mutations, insertions, deletions, and/or truncations [6], [7].
Fig. 1.
The genomic structure of HERVs. The general structure of a full-length HERV provirus is represented: the two long terminal repeats (LTRs) flank the gag, pro, pol, and env genes that encoded matrix, capsid, nucleocapsid; proteinase, reverse transcriptase, integrase; and envelop, respectively. Although full-length HERVs exist, HERVs DNA sequences are often truncated, and contain mutations, insertions, or deletions.
HERVs are divided into three main classes based on sequence similarity with the exogenous members: class I (Gammaretrovirus- and Epsilonretrovirus-like), class II (Betaretrovirus-like) and class III (Spumaretrovirus-like) while no Alpha-, Deltaretrovirus- or Lentivirus-like elements were detectable in human genome [7]. In other species, endogenous Alpharetrovirus, avian leukosis virus (ALV) in the domestic fowl (Gallus gallus), is one of the first discovered ERVs [8]. An endogenous Deltaretrovirus, MINERVa, was identified in the germline of long-fingered bats (Miniopteridae) [9] and the first endogenous lentivirus, RELIK, was discovered and characterized in the European rabbit (Oryctolagus cuniculus) [10]. Notably, Chong et al. revealed a new class of ERV, ERV4, which appears to be common to crocodilians and turtles, is phylogenetically distinct from the other ERV classes and currently has no characterized exogenous counterpart [11]. Recently, Vargiu et al. classified the HERVs by analyzing the phylogenetic relationships among all the well-conserved HERV sequences in human genome and provided a comprehensive HERV classification and characterization approach, which should be applicable for classification of all ERVs [7]. Based on the tRNA binding to the viral PBS to prime reverse transcription, HERVs are further classified into several groups. For instance, HERV-K implies a group of proviruses that utilize a lysine (K) tRNA, no matter their relationship to one another [12]. In some cases, the PBS sequence was not available when novel elements were first discovered, leading to the names based on neighboring genes (e.g. HERV-ADP), clone number (e.g. HERV-S71), or amino acid motifs (e.g. HERV-FRD) [13]. Over the span of millions of years, the genomes of vertebrates have accumulated thousands, even hundreds of thousands of ERV loci [3]. Thus, a unified system for naming ERV loci is needed to assist genome annotation and facilitate the research on ERVs. Recently, a nomenclature both providing unique identifiers for individual loci and denoting orthologous relationships between ERVs in different species has been proposed [13]. In this nomenclature, an ERV loci is assigned standard, unique IDs composed of three elements, each separated by a hyphen, in a “Category – Taxonomic group. Numeric ID – Species ID” format [13]. For example, ERV-K(HML2).4352-Hsa refers to human copy HERV-K (HML2) at chromosome 4q35.2. Currently, several databases containing all the transposable elements (TEs, HERVs included) coordinates in human genome, including RepBase [14], RepeatMasker [15], and Dfam [16], have been established. Software tools have also been developed specifically to assist in the identification and characterization of ERVs, such as RetroTector [17], [18]. Benefit from the new resources in genetics, several works collected and analyzed the coordinates of specific HERV families, including HERV-K (HML-2) [19], HERV-K (HML-6) [20], HERV-K (HML-7) [21], HERV-K (HML-10) [22], and HERV-W [23]. Furthermore, a recent study provided a reference of global population diversity in HERV-K proviruses at all currently known polymorphic loci in the human genome and revealed notable differences in the prevalence of HERV-Ks in different global populations and in the total number of HERV-Ks currently known to be polymorphic within a person’s genome [24].
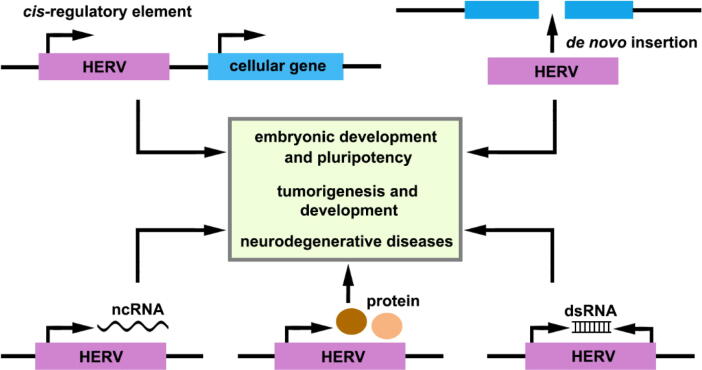
HERVs have been considered as “junk DNA sequences” for a long time. Accumulating evidence now reveals that HERVs play a role in normal physiologic processes and have been implicated in several diseases (Fig. 2). In this review, we will discuss the physiological and pathological functions of these long-misunderstood elements.
Fig. 2.
Mechanistic actions of HERVs in human development and diseases. HERVs are involved in physiological and pathological processes through several mechanisms: HERVs may act as promoters or enhance cellular gene expression through LTR cis-regulatory elements; insertion of HERV sequences can induce chromosomal rearrangements and genome instability; HERVs can encode ncRNAs, proteins, and dsRNAs.
2. HERVs in embryonic development and pluripotency
The envelope proteins encoded by env are involved in the morphogenesis of placenta in mammals, which is one of the best characterized biological functions [25]. Syncytin-1 and syncytin-2, which are envelope proteins encoded by HERV-W and HERV-FRD respectively, are specifically expressed in the placenta and with cell–cell fusogenic activities, contributing to the formation of the placenta syncytiotrophoblast layer at the materno-fetal interface [26], [27]. Likewise, the envelope proteins syncytin-A and syncytin-B with similar expression specificity and function are found in mouse [28]. Therefore, capture of retroviral envelope genes may have played a role in the emergence of placental mammals.
ERVs and other types of TEs have been revealed as regulatory sequences for many host genes in a wide range of cell types throughout mammalian evolution by their cis-regulatory element activities [29]. As LTRs harbor the regulatory regions required for proviral transcription, generally including various transcription factor binding sites (TFBSs), they are able to recruit cellular transcription factors and enhance the transcription of their neighboring genes in specific cell types [30]. TEs contribute ∼20% of TFBSs across the genome [31]. Among them, LTR elements tend to contribute more than other TE types, probably because LTRs are more likely to possess and to retain their ancestral cisregulatory activities [32]. The cis-regulatory element activities of ERVs during embryonic development have been deeply studied in mouse cleavage and 2-cell stage. Briefly, after fertilization, the 1-cell zygote undergoes a series of cleavage cell divisions, during which the embryo increases cell numbers while maintaining overall size [33]. The initial phases of cleavage rely on maternally inherited mRNAs and proteins, but cannot be accomplished without transcription from the embryonic genome. Thus a process known as zygotic genome activation (ZGA) occurs [33]. Mouse ZGA is prominent at 2-cell stage [34] and activation of MuERVs (e.g., murine endogenous retrovirus with leucine tRNA primer [MuERV-L]) [35] and stage-specific genes (e.g., Zscan4, Tcstv1/3) [36], [37] are unique features of ZGA [34]. In fact, MuERV-L are transiently activated and produce ∼ 3% of the transcribed messenger RNAs at the 2-cell stage [35]. The pattern of MuERV-L expression overlaps with hundreds of 2-cell-specific genes, which have co-opted regulatory elements from these retroviruses to initiate transcription, indicating that MuERV-L serve as functional promoters in the activation of their transcription [35]. For example, MT2, an LTR derived from MuERV-L family, acts as promoters for over five hundred 2-cell-specific gene transcripts [30]. Although activated MuERVs and other non-LTR TEs have been found at mouse 2-cell stage, their regulatory mechanism and biological function remain largely unknown.
In human, HERVs are systematically transcribed during human early embryogenesis in a stage-specific manner, and HERVs expression is a hallmark of cellular identity and cell potency that characterizes the cell populations in early human embryos [38]. Specifically, HERV-K (HML-2) has been reported to be transcribed during normal human embryogenesis, beginning with embryonic genome activation at the eight-cell stage, and continuing through the emergence of epiblast cells in preimplantation blastocysts [39]. Such proviral RNAs lead to the production of viral-like particles and Gag proteins in human blastocysts, indicating that early human development proceeds in the presence of retroviral products [39]. Interestingly, the envelope protein of HERV-K (HML-2) from loci in chromosomes 12 and 19 is highly expressed on the cell membrane of human pluripotent stem cells (hPSCs), and critical in maintaining the stemness via mammalian target of rapamycin (mTOR) pathway [40]. Down-regulation or epigenetic silencing of HML-2 Env resulted in dissociation of the stem cell colonies and enhanced differentiation along neuronal pathways, suggesting that HML-2 regulation is critical for human embryonic development and neural differentiation [40]. HERV-H, which is a primate-specific ERV and accounts for 2% of all poly-A RNAs in human embryonic stem cells (hESCs), is essential for maintenance of pluripotency in human stem cells [41]. Based on evidence observed so far, HERV-H has several roles, including as functional long noncoding RNAs (lncRNAs), enhancers or alternative promoters, and markers of topologically associating domain (TAD) boundaries [41]. Considering the length and transcriptional activity, HERV-H themselves can be considered as lncRNAs [41], and depletion of HERV-H RNA in hESCs by short hairpin RNA (shRNA) leads to loss of self-renewal, downregulation of hallmark pluripotency transcription factors OCT4, SOX2, and NANOG, and upregulation of differentiation markers [42], [43]. Although a general knockdown of HERV-H is sufficient to impair pluripotency, individual HERVH-derived lncRNAs with unique chimeric sequences also play specific roles in hPSCs, such as ESRG [42], linc-ROR [44], LINC00458 [45], and HPAT [46]. However, a recent study showed that ESRG is dispensable for pluripotency maintenance by a complete excision of the entire ESRG region [47]. The disagreement between these studies could be attributed to the different methods, Wang et al. using an RNAi knockdown [42] while Takahashi et al. using CRISPR/Cas9 knockout [47]. In addition, HERV-H LTRs contain binding sites for the LTR-binding protein 9 (LBP9) [42], OCT4 [43], and NANOG [48] to recruit these pluripotency factors. Moreover, HERV-H is involved in establishing TAD boundaries in hPSCs [49]. TADs represent a physical compartmentalization of the genome, and two regions within a TAD associate on average more frequently with each other than with regions outside of the TAD [50]. HERV-H sequences affect gene regulatory programs by creating new hPSC-specific TAD boundaries that shape the chromatin architecture, providing a new insight into how these elements may impact gene regulatory networks [49]. Recently, an eutherian-specific multicopy retrogene, human DUX4 and its mouse ortholog, Dux, were identified to share central roles in driving cleavage-specific gene expression (including ZSCAN4, KDM4E and PRAMEF-family genes), ERVL-family retrotransposon transcription, and chromatin remodeling, suggesting that DUX4 resides at the top of a transcriptional hierarchy initiated at embryonic genome activation (EGA) and is implicated in developmental events during mammalian embryogenesis [51].
3. HERVs in cancer
The transcriptional activation of HERVs is a common feature in human cancers, involving HERVs as causative elements or cofactors contributing to the onset and progression of human cancer [52]. So far, several studies have demonstrated the presence of HERVs transcripts, proteins, and viral-like particles in various human cancers (Table 1).
Table 1.
HERVs expression in tumors.
| Tumor type | HERVs Detected | HERVs-Products/Oncogenic Mechanisms |
|---|---|---|
| Breast | HERV-K (HML-2) | Env induces EMT and activates the ERK pathway [61] |
| Env [81], [111] | ||
| Env splice variants [112] | ||
| Reverse Transcriptase [113] | ||
| HERV-H | Env [111] | |
| LTR7, etc. contribute to lncRNA linc-ROR [75] | ||
| HERV-R | Env [111] | |
| HERV-P | Env [111] | |
| ERVW-1/syncytin-1 | Syncytin-1-mediated cancer-endothelial cell fusions [60] | |
| ERVMER34-1/HEMO | Env [114] | |
| Cervical | HERV-H | LTR as alternative promoter for GSDML[115], [116] |
| Colon | HERV-H | Non-coding spliced transcripts [117] |
| More active HERV-H loci [117] | ||
| HERV-H is associated with the microsatellite and nodal status [118] | ||
| Head and Neck | HERV-H | Transcripts [83] |
| Hepatocellular | HERV-K (HML-2) | Transcripts [119] |
| HERV-H | LTR7, etc. contribute to lncRNA linc-ROR [77] | |
| ERV-9 | LTR12C contributes to lncRNA PRLH1 [120] | |
| Kidney | HERV-E | Env [84] |
| HERV-H | HHLA2 in tumor immune suppression [121] | |
| Leukemia | HERV-K (HML-2) | Np9 as a critical molecular switch of multiple signaling pathways [70] |
| Lung | HERV-R | Env [122], [123] |
| HERV-P | Env [123] | |
| HERV-H | Env [123] | |
| HERV-K (HML-2) | Env [123] | |
| Novel insertion polymorphisms [124] | ||
| Lymphoma | HERV-K (HML-2) | Env [125] |
| Gag [126] | ||
| Transcripts [127] | ||
| HERV-W | Transcripts [127] | |
| MaLR | LTR as alternative promoter for CSF1R[54] | |
| Melanoma | HERV-K (HML-2) | Env-mediated intercellular fusion [58] |
| Rec [128] | ||
| Virus-like particles [129] | ||
| anti-HERV-K antibodies in patient [129] | ||
| Maintaining CD133+ melanoma cells with stemness features [130] | ||
| Ovarian | ERV3 | Env [131] |
| HERV-K (HML-2) | Env [131] | |
| Gag [132] | ||
| Hypomethylation [133] | ||
| HERV-W | Transcripts [134] | |
| HERV-E | Env [131] | |
| ERVMER34-1/HEMO | Env [114] | |
| Pancreatic | HERV-K (HML-2) | Env [62] |
| Reverse transcriptase [62] | ||
| Virus-like particles [62] | ||
| anti-HERV-K antibodies in patient [62] | ||
| HERV-H | LTR7, etc. contribute to lncRNA linc-ROR [76] | |
| Prostate | HERV-K (HML-2) | Env [82] |
| Gag [82] | ||
| Transcripts [135] | ||
| HERV-E | Env [136] | |
| ERV3 | Env [136] | |
| Sarcoma | HERV-K (HML-2) | Transcripts [137] |
| HERV-F | Transcripts [137] | |
| Seminoma | ERVW-1/syncytin-1 | Env [138] |
| HERV-K (HML-2) | HERV-K-specific T cells in patients [139] | |
| Teratocarcinoma | HERV-K (HML-2) | Virus-like particles [140] |
| Np9 [141] | ||
| Urothelial | HERV-K (HML-2) | Hypomethylation [142] |
| HERV-H | LTR7Y and HERV-H contribute to lncRNA UCA1 [72] | |
| HHLA2 in tumor immune suppression [143] | ||
| HERV-E | Modulating PLA2G4A transcription [144] | |
| ERVW-1/Syncytin-1 | Env [145] |
Given to the potential transposable ability of retrotransposon, it is believed that the tumorigenicity of HERVs could depend on retroviruses movement and consequently disrupting the stability of the host genome [52]. Actually, new insertions of TEs, especially HERVs, have been described in several tumors [53]. LTRs can act as alternative promoter or enhancer, leading to the deregulation of proto-oncogenes or tumor suppressor genes. In Hodgkin’s lymphoma, CSF1R transcription does not originate from its own promoter, but instead initiates at an LTR element of the MaLR THE1B family, located ∼ 6.2 kb upstream of the normal promoter [54]. An HERV type C inserted in Pleiotrophin (PTN) between its 5′ untranslated and coding regions generates an additional promoter with trophoblast-specific activity, resulting in fusion transcripts between HERV and PTN (HERV-PTN) specifically expressed in human trophoblast cell cultures and trophoblast-derived choriocarcinoma cell lines, suggesting that the tissue-specific expression of PTN due to the HERV insertion supports the highly aggressive growth of human choriocarcinoma and possibly of the human trophoblast [55]. However, it should be noted that the LTRs activity can have an anti-oncogenic effect by driving the expression of tumor suppressor genes. For example, TP63 and TNFRSF10B, encoding the p63 homologue of the tumour suppressor p53 and death receptor 5 (DR5), respectively, are regulated by upstream LTRs belonging to the ERV9 group of HERVs [56], [57].
HERVs can exert a direct action through their own proteins in cancer. The envelop proteins of HERVs contribute to tumorigenesis and development by inducing cell–cell fusion or epithelial to mesenchymal transition (EMT) in melanoma [58], endometrial carcinoma [59], and breast cancer [60], [61]. In pancreatic cancer, HERV-K (HML-2) Env protein was reported to play an integral role in pancreatic cancer proliferation, tumorigenesis, and metastasis formation by both regulating KRAS through hyperactivation of the RAS/MEK/ERK pathway and upregulating p70S6 Kinase/JNK/C-Jun signalling [62]. Another mechanism of Env in tumor progression is involved in the promotion of the immune escape, as the immunosuppressive domain (ISD) of Env can abolish the anti-oncogenic cytolytic immune response [63]. For example, tumor cells expressing Env protein of HERV-H are able to escape the immune rejection and to proliferate when injected into immunocompetent mice [64], and the knockdown of HERV-H Env in tumor cells improves immune responses in vitro and in vivo accompanied by tumor regression [65], suggesting that HERV-H is a critical determinant of immune escape in cancer. In addition, HERV proteins Rec and Np9 encoded by HERV-K (HML-2) may act oncogenically by derepressing c-MYC through the inhibition of promyelocytic leukemia zinc finger protein (PLZF) [66], and Rec alone is able to overcome the direct transcriptional repression by testicular zinc-finger protein (TZFP) of the c-MYC gene promoter [67]. In addition, Rec can form a trimeric complex with TZFP and androgen receptor (AR) [67], or bind to the human small glutamine-rich tetratricopeptide repeat protein (hSGT) [68], relieving the repression of AR activity. Np9 is also a tumor-specific biomarker, it can interact with ligand of Numb protein X (LNX, a RING-type E3 ubiquitin ligase), affecting tumorigenesis through the LNX/Numb/Notch pathway [69], and act as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1, promoting the growth of human leukemia stem/progenitor cells [70].
Long noncoding RNAs (lncRNAs) have critical roles in various biological processes, such as cancer progression. Strikingly, 75–83% of lncRNAs contain TE sequences, especially ERVs [71]. Several HERVs-derived lncRNAs have been described. UCA1, a lncRNA consists of LTR7Y and HERV-H, promotes proliferation, motility, and invasion of bladder cancer [72]. The HERVs-derived lncRNAs SAMMSON and BANCR are involved in melanoma progression [73], [74]. Additionally, linc-ROR derived from LTR7, HERVH-int, etc., has been reported to contribute to the progression, metastasis or chemoresistance in breast cancer [75], pancreatic cancer [76] and hepatocellular carcinoma [77].
It has been reported that the expression of HERVs is associated with the modulation of innate immune response in different physiological and pathological conditions [52]. In the context of tumorigenesis and development, HERVs may have opposite effects on modulating immune response. They may be involved in both anti-tumor defense and oncogenic process. On the one hand, DNA methyltransferase inhibitors (DNMTis) upregulate HERVs in tumor cells to induce a growth-inhibiting immune response. High expression of the genes associated with the anti-viral response seems to potentiate a response to immune checkpoint therapy [78], [79]. On the other hand, the double-stranded RNA (dsRNA) transcribed from HERVs can induce immune-suppressed microenvironment of tumors, with important implications for cancer immunotherapy [80].
Considering the activation of HERVs in many human cancers, HERVs can be used as biomarkers for tumor diagnosis and/or prognosis [52]. For example, in breast cancer patients, HERV-K (HML-2) Env expression is closely associated with tumor size, TNM stage, and lymph node metastases [81]. Moreover, breast cancer patients with high expression of HERV-K (HML-2) have a decreased overall survival, indicating that HERV-K (HML-2) Env protein is a novel candidate prognostic marker for breast cancer [81]. A study in prostate cancer reported that the combination testing of HERV-K (HML-2) with traditional prostate-specific antigen may improve the efficacy of prostate cancer detection, specifically among older men and smokers who tend to develop a more aggressive disease [82]. Recently, Kolbe et al. performed the first locus-specific analysis of HERVs expression in head and neck cancer and found that members of the HERV-H family are highly expressed in tumor tissues and may serve as biomarkers for disease progression [83]. In this view, HERVs have the potential to be targets for new cancer therapeutic opportunity. A recent study revealed that CT-RCC HERV-E envelope-encoding peptides, which are expressed in the majority of ccRCC tumors but not in normal tissues, are immunogenic and capable of stimulating CD8+ T cells in vitro that recognize CT-RCC HERV-E-expressing ccRCC tumors, suggesting that HERV-E Env can represent an excellent target for T cell-based immunotherapy for kidney cancer [84]. Another study reported that anti-HERV-K (HML-2) Env antibodies inhibited growth and induced apoptosis of breast cancer cells in vitro and reduced growth of xenograft tumors in mice [85]. Consistently, HERV-K Env-specific CAR+ T cells are able to lyse melanoma tumor cells in an antigen-specific manner in vitro and in vivo [86]. In addition, DNMTis activate the viral recognition and interferon response pathway by inducing dsRNAs transcribed by HERVs, which can potentiate the response to immune checkpoint therapy [78], [79].
4. HERVs in aging and neurodegenerative diseases
In general, ERVs are largely transcriptionally silenced through a heterochromatic structure, which is established by histone modification and DNA methylation mechanisms [87]. However, there may be a net loss of heterochromatin with ageing, resulting to the abnormal activation of retrotransposons, including ERVs, in aging individuals [88], [89]. In aging mouse, the MuERV intracisternal A-particle (IAP) becomes active [90], and an increase in the RNA transcription and copy number of MusD is detected [91]. In human, HERV-K (HML-2) and HERV-W exhibit different expression patterns in young and old individuals [92]. The expression of HERV-H and HERV-W in human peripheral blood mononuclear cells (PBMCs) shows a significant positive correlation with age over 30 years old [93]. Notably, the expression of HERV-W shows a significant increase in individuals over 40 years old, and neurodegenerative diseases such as multiple sclerosis (MS) also occur around this range of age [93]. It has been recognized that the reactivation of HERVs is related to the occurrence and development of human aging-related diseases [94], [95].
Currently, abnormal activation of HERVs has been detected in certain neurodegenerative diseases [5]. For example, elevated HERV-W envelope protein expression is detected in muscle cells of amyotrophic lateral sclerosis (ALS) patients [96]. HERV-K (HML-2) is expressed in cortical and spinal neurons of ALS patients, but not in neurons from control healthy individuals, and its envelope protein may contribute to neurodegeneration [97]. MS is another neurodegenerative disease associated with HERVs abnormal expression. HERV-W envelope protein was detected in MS brains and specially in lesions [98], and HERV-W Env-mediated microglial polarization contributes to neurodegeneration in MS [99]. In fact, a recently completed clinical study in MS patients, which used monoclonal neutralizing antibodies against the HERV-W envelope protein, showed that the antibody-mediated neutralization exerts neuroprotective effects [99]. In neurodevelopmental disorder autistic spectrum disorder (ASD), HERV-H and HERV-W are differentially expressed in ASD patients and healthy controls, with HERV-H being more abundantly expressed while HERV-W having lower abundance in PBMCs from ASD patients compared to healthy controls [100]. Furthermore, HERV-H exhibits a statistically significant higher expression in ASD patients with severe disease development [100]. Moreover, it has been reported that expression of HEMO (ERVMER34-1) is altered in ASD patients and may be useful for the disease diagnosis [101]. Therefore, HERVs expression could be considered as a biomarker, which is easily detectable in blood and potentially contributing to an early diagnosis. In addition, several HERVs and non-LTR TEs are activated in Alzheimer’s disease (AD) [102], and some HERV-W and HERV-H loci are transcriptionally upregulated in postmortem brain samples from schizophrenic and bipolar patients [103]. Taken together, these studies suggest that the activation of HERVs may be a mechanistic link between aging and neurodegeneration.
5. Conclusion and perspective
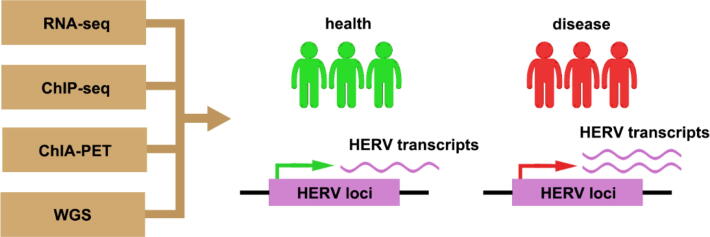
HERVs are involved in various biological processes through encoding proteins, acting as promoters/enhancers, or lncRNAs to affect human health and disease. In addition to the discoveries mentioned above, HERVs are also involved in neurodevelopment [104], [105], HIV-1 infection [106], chronic hepatitis C virus (HCV) infection [107], and type 1 diabetes [108], [109]. Although there are a large number of studies focusing on the expression of whole HERVs families, most of these works did not detect the individual coordinates of the expressed loci, or measure the expression of a single locus. Recently, the application of omics approaches has allowed to solve these problems, for instance, high-throughput sequencing, which allows the obtaining of important information about the individual HERV loci, can be used to investigate the contribution of HERVs to human development and diseases [110] (Fig 3). Previously, due to the technical difficulties in analyzing these highly repetitive elements, HERVs have not received enough attention. With the development of novel technologies, such as high-throughput sequencing, single-cell RNA-seq, genome editing technology, and integration of big data, etc., the mystery of HERVs is being further uncovered. Nonetheless, there are still more questions about these ancient remnants than definite answers, such as the identification of specific HERV loci associated with specific diseases. This provides a promising opportunity to enrich our knowledge for HERVs, the complex genomic elements that make up ∼ 8% of human genome.
Fig. 3.
Advanced technologies provide opportunities to explore HERVs in health and disease. The application of advanced technologies such as high-throughput sequencing provides opportunities for identifying functional HERV loci in patients versus healthy individuals.
CRediT authorship contribution statement
Jian Mao: Conceptualization, Writing – review & editing. Qian Zhang: Writing – review & editing. Yu-Sheng Cong: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31730020, 32000512, 31801155), and the Hangzhou Science and Technology Bureau (20182014B01).
Contributor Information
Jian Mao, Email: maojian@hznu.edu.cn.
Yu-Sheng Cong, Email: yscong@hznu.edu.cn.
References
- 1.Krupovic M., Blomberg J., Coffin J.M., Dasgupta I., Fan H., Geering A.D., et al. New virus order unifying five families of reverse-transcribing viruses. J Virol. 2018;92(12):e00515–18. doi: 10.1128/JVI.00515-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodonova S.O., Prinz S., Bilanchone V., Sandmeyer S., Briggs J.A.G. Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc Natl Acad Sci U S A. 2019;116(20):10048–10057. doi: 10.1073/pnas.1900931116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat Rev Microbiol. 2019;17(6):355–370. doi: 10.1038/s41579-019-0189-2. [DOI] [PubMed] [Google Scholar]
- 4.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Kury P., Nath A., Creange A., Dolei A., Marche P., Gold J., et al. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol Med. 2018;24(4):379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Parseval N., Heidmann T. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res. 2005;110(1–4):318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 7.Vargiu L., Rodriguez-Tomé P., Sperber G.O., Cadeddu M., Grandi N., Blikstad V., et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss R.A. The discovery of endogenous retroviruses. Retrovirology. 2006;3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farkašová H., Hron T., Pačes J., Hulva P., Benda P., Gifford R.J., et al. Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: Miniopteridae) Proc Natl Acad Sci USA. 2017;114(12):3145–3150. doi: 10.1073/pnas.1621224114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzourakis A., Tristem M., Pybus O.G., Gifford R.J. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104(15):6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong A.Y., Kojima K.K., Jurka J., Ray D.A., Smit A.F.A., Isberg S.R., et al. Evolution and gene capture in ancient endogenous retroviruses - insights from the crocodilian genomes. Retrovirology. 2014;11:71. doi: 10.1186/s12977-014-0071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mager D.L., Stoye J.P. Mammalian Endogenous Retroviruses. Microbiol Spectr. 2015;3(1) doi: 10.1128/microbiolspec.MDNA3-0009-2014. MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 13.Gifford R.J., Blomberg J., Coffin J.M., Fan H., Heidmann T., Mayer J., et al. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology. 2018;15(1):59. doi: 10.1186/s12977-018-0442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9(6):657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 16.Storer J., Hubley R., Rosen J., Wheeler T.J., Smit A.F. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA. 2021;12(1):2. doi: 10.1186/s13100-020-00230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperber G.O., Airola T., Jern P., Blomberg J. Automated recognition of retroviral sequences in genomic data–RetroTector. Nucleic Acids Res. 2007;35(15):4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sperber G., Lövgren A., Eriksson N.-E., Benachenhou F., Blomberg J. RetroTector online, a rational tool for analysis of retroviral elements in small and medium size vertebrate genomic sequences. BMC Bioinf. 2009;10(Suppl 6):S4. doi: 10.1186/1471-2105-10-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian R.P., Wildschutte J.H., Russo C., Coffin J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisano M.P., Grandi N., Cadeddu M., Blomberg J., Tramontano E. Comprehensive characterization of the human endogenous retrovirus HERV-K(HML-6) group: overview of structure, phylogeny, and contribution to the human genome. J Virol. 2019;93(16):e00110–19. doi: 10.1128/JVI.00110-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandi N., Pisano M.P., Pessiu E., Scognamiglio S., Tramontano E. HERV-K(HML7) integrations in the human genome: comprehensive characterization and comparative analysis in non-human primates. Biology (Basel) 2021;10(5):439. doi: 10.3390/biology10050439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandi N., Cadeddu M., Pisano M.P., Esposito F., Blomberg J., Tramontano E. Identification of a novel HERV-K(HML10): comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob DNA. 2017;8:15. doi: 10.1186/s13100-017-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandi N., Cadeddu M., Blomberg J., Tramontano E. Contribution of type W human endogenous retroviruses to the human genome: characterization of HERV-W proviral insertions and processed pseudogenes. Retrovirology. 2016;13(1):67. doi: 10.1186/s12977-016-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Lin L., Malhotra R., Yang L., Acharya R., Poss M. A computational framework to assess genome-wide distribution of polymorphic human endogenous retrovirus-K In human populations. PLoS Comput Biol. 2019;15(3) doi: 10.1371/journal.pcbi.1006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., et al. Paleovirology of 'syncytins', retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 27.Blaise S., de Parseval N., Bénit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A. 2003;100(22):13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dupressoir A., Marceau G., Vernochet C., Bénit L., Kanellopoulos C., Sapin V., et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson P.J., Macfarlan T.S., Lorincz M.C. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Mol Cell. 2016;62(5):766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24(12):1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chuong E.B., Elde N.C., Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18(2):71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossant J., Tam P.P.L. New insights into early human development: lessons for stem cell derivation and differentiation. Cell Stem Cell. 2017;20(1):18–28. doi: 10.1016/j.stem.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Lu F., Zhang Y. Cell totipotency: molecular features, induction, and maintenance. Natl Sci Rev. 2015;2(2):217–225. doi: 10.1093/nsr/nwv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falco G., Lee S.-L., Stanghellini I., Bassey U.C., Hamatani T., Ko M.S.H. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307(2):539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q., Dan J., Wang H., Guo R., Mao J., Fu H., et al. Tcstv1 and Tcstv3 elongate telomeres of mouse ES cells. Sci Rep. 2016;6:19852. doi: 10.1038/srep19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Göke J., Lu X., Chan Y.-S., Ng H.-H., Ly L.-H., Sachs F., et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16(2):135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Grow E.J., Flynn R.A., Chavez S.L., Bayless N.L., Wossidlo M., Wesche D.J., et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T., Medynets M., Johnson K.R., Doucet-O'Hare T.T., DiSanza B., Li W., et al. Regulation of stem cell function and neuronal differentiation by HERV-K via mTOR pathway. Proc Natl Acad Sci USA. 2020;117(30):17842–17853. doi: 10.1073/pnas.2002427117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sexton C.E., Tillett R.L., Han M.V. The essential but enigmatic regulatory role of HERVH in pluripotency. Trends Genet. 2021 doi: 10.1016/j.tig.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Xie G., Singh M., Ghanbarian A.T., Raskó T., Szvetnik A., et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516(7531):405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 43.Lu X., Sachs F., Ramsay L., Jacques P.-É., Göke J., Bourque G., et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21(4):423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Ng S.-Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glinsky G., Durruthy-Durruthy J., Wossidlo M., Grow E.J., Weirather J.L., Au K.F., et al. Single cell expression analysis of primate-specific retroviruses-derived HPAT lincRNAs in viable human blastocysts identifies embryonic cells co-expressing genetic markers of multiple lineages. Heliyon. 2018;4(6) doi: 10.1016/j.heliyon.2018.e00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi K., Nakamura M., Okubo C., Kliesmete Z., Ohnuki M., Narita M., et al. The pluripotent stem cell-specific transcript ESRG is dispensable for human pluripotency. PLoS Genet. 2021;17(5) doi: 10.1371/journal.pgen.1009587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoni F.A., Guerra J., Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., Li T., Preissl S., Amaral M.L., Grinstein J.D., Farah E.N., et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet. 2019;51(9):1380–1388. doi: 10.1038/s41588-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixon J.R., Gorkin D.U., Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62(5):668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.-W., Wike C.L., et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matteucci C., Balestrieri E., Argaw-Denboba A., Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018;53:17–30. doi: 10.1016/j.semcancer.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J., 3rd, et al. Cancer genome atlas research N. landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamprecht B., Walter K., Kreher S., Kumar R., Hummel M., Lenze D., et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16(5):571–579. doi: 10.1038/nm.2129. [DOI] [PubMed] [Google Scholar]
- 55.Schulte A.M., Lai S., Kurtz A., Czubayko F., Riegel A.T., Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc Natl Acad Sci U S A. 1996;93(25):14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beyer U., Moll-Rocek J., Moll U.M., Dobbelstein M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci U S A. 2011;108(9):3624–3629. doi: 10.1073/pnas.1016201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beyer U., Kronung S.K., Leha A., Walter L., Dobbelstein M. Comprehensive identification of genes driven by ERV9-LTRs reveals TNFRSF10B as a re-activatable mediator of testicular cancer cell death. Cell Death Differ. 2016;23(1):64–75. doi: 10.1038/cdd.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang G., Li Z., Wan X., Wang Y., Dong J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J Carcinog. 2013;12:5. doi: 10.4103/1477-3163.109032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strick R., Ackermann S., Langbein M., Swiatek J., Schubert S.W., Hashemolhosseini S., et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J Mol Med (Berl) 2007;85(1):23–38. doi: 10.1007/s00109-006-0104-y. [DOI] [PubMed] [Google Scholar]
- 60.Bjerregaard B., Holck S., Christensen I.J., Larsson L.I. Syncytin is involved in breast cancer-endothelial cell fusions. Cell Mol Life Sci. 2006;63(16):1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemaitre C., Tsang J., Bireau C., Heidmann T., Dewannieux M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017;13(6) doi: 10.1371/journal.ppat.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li M., Radvanyi L., Yin B., Rycaj K., Li J., Chivukula R., et al. Downregulation of human endogenous retrovirus type K (HERV-K) viral RNA in pancreatic cancer cells decreases cell proliferation and tumor growth. Clin Cancer Res. 2017;23(19):5892–5911. doi: 10.1158/1078-0432.CCR-17-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grandi N., Tramontano E. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol. 2018;9:462. doi: 10.3389/fmicb.2018.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mangeney M., de Parseval N., Thomas G., Heidmann T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J Gen Virol. 2001;82(Pt 10):2515–2518. doi: 10.1099/0022-1317-82-10-2515. [DOI] [PubMed] [Google Scholar]
- 65.Kudo-Saito C., Yura M., Yamamoto R., Kawakami Y. Induction of immunoregulatory CD271+ cells by metastatic tumor cells that express human endogenous retrovirus H. Cancer Res. 2014;74(5):1361–1370. doi: 10.1158/0008-5472.CAN-13-1349. [DOI] [PubMed] [Google Scholar]
- 66.Denne M., Sauter M., Armbruester V., Licht J.D., Roemer K., Mueller-Lantzsch N. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol. 2007;81(11):5607–5616. doi: 10.1128/JVI.02771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaufmann S., Sauter M., Schmitt M., Baumert B., Best B., Boese A., et al. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol. 2010;91(Pt 6):1494–1502. doi: 10.1099/vir.0.014241-0. [DOI] [PubMed] [Google Scholar]
- 68.Hanke K., Chudak C., Kurth R., Bannert N. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT) Int J Cancer. 2013;132(3):556–567. doi: 10.1002/ijc.27693. [DOI] [PubMed] [Google Scholar]
- 69.Armbruester V., Sauter M., Roemer K., Best B., Hahn S., Nty A., et al. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J Virol. 2004;78(19):10310–10319. doi: 10.1128/JVI.78.19.10310-10319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen T., Meng Z., Gan Y., Wang X., Xu F., Gu Y., et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia. 2013;27(7):1469–1478. doi: 10.1038/leu.2013.8. [DOI] [PubMed] [Google Scholar]
- 71.Goke J., Ng H.H. CTRL+INSERT: retrotransposons and their contribution to regulation and innovation of the transcriptome. EMBO Rep. 2016;17(8):1131–1144. doi: 10.15252/embr.201642743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang F., Li X., Xie X., Zhao L., Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 73.Leucci E., Vendramin R., Spinazzi M., Laurette P., Fiers M., Wouters J., et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531(7595):518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 74.Flockhart R.J., Webster D.E., Qu K., Mascarenhas N., Kovalski J., Kretz M., et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hou P., Zhao Y., Li Z., Yao R., Ma M., Gao Y., et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhan H.X., Wang Y., Li C., Xu J.W., Zhou B., Zhu J.K., et al. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374(2):261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi K., Yan I.K., Kogure T., Haga H., Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cañadas I., Thummalapalli R., Kim J.W., Kitajima S., Jenkins R.W., Christensen C.L., et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24(8):1143–1150. doi: 10.1038/s41591-018-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J., Rycaj K., Geng S., Li M., Plummer J.B., Yin B., et al. Expression of human endogenous retrovirus type K envelope protein is a novel candidate prognostic marker for human breast cancer. Genes Cancer. 2011;2(9):914–922. doi: 10.1177/1947601911431841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wallace T.A., Downey R.F., Seufert C.J., Schetter A., Dorsey T.H., Johnson C.A., et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis. 2014;35(9):2074–2083. doi: 10.1093/carcin/bgu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolbe A.R., Bendall M.L., Pearson A.T., Paul D., Nixon D.F., Pérez-Losada M., et al. Human endogenous retrovirus expression is associated with head and neck cancer and differential survival. Viruses. 2020;12(9):956. doi: 10.3390/v12090956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cherkasova E., Scrivani C., Doh S., Weisman Q., Takahashi Y., Harashima N., et al. Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res. 2016;76(8):2177–2185. doi: 10.1158/0008-5472.CAN-15-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang-Johanning F., Rycaj K., Plummer J.B., Li M., Yin B., Frerich K., et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J Natl Cancer Inst. 2012;104(3):189–210. doi: 10.1093/jnci/djr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krishnamurthy J., Rabinovich B.A., Mi T., Switzer K.C., Olivares S., Maiti S.N., et al. Genetic engineering of T cells to target HERV-K, an ancient retrovirus on melanoma. Clin Cancer Res. 2015;21(14):3241–3251. doi: 10.1158/1078-0432.CCR-14-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Groh S., Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci. 2017;74(11):2055–2065. doi: 10.1007/s00018-017-2454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol. 1997;32(4–5):383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 89.Pal S., Tyler J.K. Epigenetics and aging. Sci Adv. 2016;2(7) doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barbot W., Dupressoir A., Lazar V., Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30(11):2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De Cecco M., Criscione S.W., Peterson A.L., Neretti N., Sedivy J.M., Kreiling J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013;5(12):867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nevalainen T., Autio A., Mishra B.H., Marttila S., Jylha M., Hurme M. Aging-associated patterns in the expression of human endogenous retroviruses. PLoS ONE. 2018;13(12) doi: 10.1371/journal.pone.0207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balestrieri E., Pica F., Matteucci C., Zenobi R., Sorrentino R., Argaw-Denboba A., et al. Transcriptional activity of human endogenous retroviruses in human peripheral blood mononuclear cells. Biomed Res Int. 2015;2015 doi: 10.1155/2015/164529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hohn O., Hanke K., Bannert N. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front Oncol. 2013;3:246. doi: 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Volkman H.E., Stetson D.B. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. 2014;15(5):415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oluwole S.O.A., Yao Y., Conradi S., Kristensson K., Karlsson H. Elevated levels of transcripts encoding a human retroviral envelope protein (syncytin) in muscles from patients with motor neuron disease. Amyotroph Lateral Scler. 2007;8(2):67–72. doi: 10.1080/17482960600864207. [DOI] [PubMed] [Google Scholar]
- 97.Li W., Lee M.-H., Henderson L., Tyagi R., Bachani M., Steiner J., et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7(307) doi: 10.1126/scitranslmed.aac8201. 307ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Horssen J., van der Pol S., Nijland P., Amor S., Perron H. Human endogenous retrovirus W in brain lesions: Rationale for targeted therapy in multiple sclerosis. Mult Scler Relat Disord. 2016;8:11–18. doi: 10.1016/j.msard.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 99.Kremer D., Gruchot J., Weyers V., Oldemeier L., Göttle P., Healy L., et al. pHERV-W envelope protein fuels microglial cell-dependent damage of myelinated axons in multiple sclerosis. Proc Natl Acad Sci USA. 2019;116(30):15216–15225. doi: 10.1073/pnas.1901283116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Balestrieri E., Arpino C., Matteucci C., Sorrentino R., Pica F., Alessandrelli R., et al. HERVs expression in Autism spectrum disorders. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0048831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Balestrieri E., Cipriani C., Matteucci C., Benvenuto A., Coniglio A., Argaw-Denboba A., et al. Children With autism spectrum disorder and their mothers share abnormal expression of selected endogenous retroviruses families and cytokines. Front Immunol. 2019;10:2244. doi: 10.3389/fimmu.2019.02244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo C., Jeong H.H., Hsieh Y.C., Klein H.U., Bennett D.A., De Jager P.L., et al. Tau activates transposable elements in Alzheimer's disease. Cell Rep. 2018;23(10):2874–2880. doi: 10.1016/j.celrep.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li F., Sabunciyan S., Yolken R.H., Lee D., Kim S., Karlsson H. Transcription of human endogenous retroviruses in human brain by RNA-seq analysis. PLoS ONE. 2019;14(1) doi: 10.1371/journal.pone.0207353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brattås P.L., Jönsson M.E., Fasching L., Nelander Wahlestedt J., Shahsavani M., Falk R., et al. TRIM28 controls a gene regulatory network based on endogenous retroviruses in human neural progenitor cells. Cell Rep. 2017;18(1):1–11. doi: 10.1016/j.celrep.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 105.Padmanabhan Nair V., Liu H., Ciceri G., Jungverdorben J., Frishman G., Tchieu J., et al. Activation of HERV-K(HML-2) disrupts cortical patterning and neuronal differentiation by increasing NTRK3. Cell Stem Cell. 2021;28(9):1566–1581. doi: 10.1016/j.stem.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vincendeau M., Göttesdorfer I., Schreml J.M.H., Wetie A.G.N., Mayer J., Greenwood A.D., et al. Modulation of human endogenous retrovirus (HERV) transcription during persistent and de novo HIV-1 infection. Retrovirology. 2015;12:27. doi: 10.1186/s12977-015-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tovo P.-A., Garazzino S., Daprà V., Alliaudi C., Silvestro E., Calvi C., et al. Chronic HCV infection Is associated with overexpression of human endogenous retroviruses that persists after drug-induced viral clearance. Int J Mol Sci. 2020;21(11):3980. doi: 10.3390/ijms21113980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levet S., Charvet B., Bertin A., Deschaumes A., Perron H., Hober D. Human endogenous retroviruses and type 1 diabetes. Curr Diab Rep. 2019;19(12):141. doi: 10.1007/s11892-019-1256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tovo P.-A., Rabbone I., Tinti D., Galliano I., Trada M., Daprà V., et al. Enhanced expression of human endogenous retroviruses in new-onset type 1 diabetes: potential pathogenetic and therapeutic implications. Autoimmunity. 2020;53(5):283–288. doi: 10.1080/08916934.2020.1777281. [DOI] [PubMed] [Google Scholar]
- 110.Pisano M.P., Grandi N., Tramontano E. High-throughput sequencing is a crucial tool to investigate the contribution of human endogenous retroviruses (HERVs) to human biology and development. Viruses. 2020;12(6):633. doi: 10.3390/v12060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rhyu D.-W., Kang Y.-J., Ock M.-S., Eo J.-W., Choi Y.-H., Kim W.-J., et al. Expression of human endogenous retrovirus env genes in the blood of breast cancer patients. Int J Mol Sci. 2014;15(6):9173–9183. doi: 10.3390/ijms15069173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang-Johanning F., Frost A.R., Jian B., Epp L., Lu D.W., Johanning G.L. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22(10):1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 113.Golan M., Hizi A., Resau J.H., Yaal-Hahoshen N., Reichman H., Keydar I., et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10(6):521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heidmann O., Béguin A., Paternina J., Berthier R., Deloger M., Bawa O., et al. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci USA. 2017;114(32):E6642–E6651. doi: 10.1073/pnas.1702204114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sin H.S., Huh J.W., Kim D.S., Kang D.W., Min D.S., Kim T.H., et al. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch Virol. 2006;151(10):1985–1994. doi: 10.1007/s00705-006-0764-5. [DOI] [PubMed] [Google Scholar]
- 116.Sun Q., Yang J., Xing G., Sun Q., Zhang L., He F. Expression of GSDML associates with tumor progression in uterine cervix cancer. Transl Oncol. 2008;1(2):73–83. doi: 10.1593/tlo.08112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang Q., Xu Z., Xu R., Wu L., Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0029950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pérot P., Mullins C.S., Naville M., Bressan C., Hühns M., Gock M., et al. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget. 2015;6(37):40095–40111. doi: 10.18632/oncotarget.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma W., Hong Z., Liu H., Chen X., Ding L., Liu Z., et al. Human endogenous retroviruses-K (HML-2) expression is correlated with prognosis and progress of hepatocellular carcinoma. Biomed Res Int. 2016;2016:8201642. doi: 10.1155/2016/8201642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deng B., Xu W., Wang Z., Liu C., Lin P., Li B., et al. An LTR retrotransposon-derived lncRNA interacts with RNF169 to promote homologous recombination. EMBO Rep. 2019;20(11) doi: 10.15252/embr.201847650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen D., Chen W., Xu Y., Zhu M., Xiao Y., Shen Y., et al. Upregulated immune checkpoint HHLA2 in clear cell renal cell carcinoma: a novel prognostic biomarker and potential therapeutic target. J Med Genet. 2019;56(1):43–49. doi: 10.1136/jmedgenet-2018-105454. [DOI] [PubMed] [Google Scholar]
- 122.Andersson A.C., Svensson A.C., Rolny C., Andersson G., Larsson E. Expression of human endogenous retrovirus ERV3 (HERV-R) mRNA in normal and neoplastic tissues. Int J Oncol. 1998;12(2):309–313. doi: 10.3892/ijo.12.2.309. [DOI] [PubMed] [Google Scholar]
- 123.Zare M., Mostafaei S., Ahmadi A., Azimzadeh Jamalkandi S., Abedini A., Esfahani-Monfared Z., et al. Human endogenous retrovirus env genes: Potential blood biomarkers in lung cancer. Microb Pathog. 2018;115:189–193. doi: 10.1016/j.micpath.2017.12.040. [DOI] [PubMed] [Google Scholar]
- 124.Kahyo T., Tao H., Shinmura K., Yamada H., Mori H., Funai K., et al. Identification and association study with lung cancer for novel insertion polymorphisms of human endogenous retrovirus. Carcinogenesis. 2013;34(11):2531–2538. doi: 10.1093/carcin/bgt253. [DOI] [PubMed] [Google Scholar]
- 125.Januszkiewicz-Lewandowska D., Nowicka K., Rembowska J., Fichna M., Żurawek M., Derwich K., et al. Env gene expression of human endogenous retrovirus-k and human endogenous retrovirus-w in childhood acute leukemia cells. Acta Haematol. 2013;129(4):232–237. doi: 10.1159/000345407. [DOI] [PubMed] [Google Scholar]
- 126.Depil S., Roche C., Dussart P., Prin L. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia. 2002;16(2):254–259. doi: 10.1038/sj.leu.2402355. [DOI] [PubMed] [Google Scholar]
- 127.Fava P., Bergallo M., Astrua C., Brizio M., Galliano I., Montanari P., et al. Human Endogenous Retrovirus Expression in Primary Cutaneous T-Cell Lymphomas. Dermatology. 2016;232(1):38–43. doi: 10.1159/000438669. [DOI] [PubMed] [Google Scholar]
- 128.Singh S., Kaye S., Francis N., Peston D., Gore M., McClure M., et al. Human endogenous retrovirus K (HERV-K) rec mRNA is expressed in primary melanoma but not in benign naevi or normal skin. Pigment Cell Melanoma Res. 2013;26(3):426–428. doi: 10.1111/pcmr.12066. [DOI] [PubMed] [Google Scholar]
- 129.Büscher K., Trefzer U., Hofmann M., Sterry W., Kurth R., Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65(10):4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 130.Argaw-Denboba A., Balestrieri E., Serafino A., Cipriani C., Bucci I., Sorrentino R., et al. HERV-K activation is strictly required to sustain CD133+ melanoma cells with stemness features. J Exp Clin Cancer Res. 2017;36(1):20. doi: 10.1186/s13046-016-0485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang-Johanning F., Liu J., Rycaj K., Huang M., Tsai K., Rosen D.G., et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 2007;120(1):81–90. doi: 10.1002/ijc.22256. [DOI] [PubMed] [Google Scholar]
- 132.Götzinger N., Sauter M., Roemer K., Mueller-Lantzsch N. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J Gen Virol. 1996;77(Pt 12):2983–2990. doi: 10.1099/0022-1317-77-12-2983. [DOI] [PubMed] [Google Scholar]
- 133.Iramaneerat K., Rattanatunyong P., Khemapech N., Triratanachat S., Mutirangura A. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer. 2011;21(1):51–57. doi: 10.1097/IGC.0b013e3182021c1a. [DOI] [PubMed] [Google Scholar]
- 134.Menendez L., Benigno B.B., McDonald J.F. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Agoni L., Guha C., Lenz J. Detection of Human Endogenous Retrovirus K (HERV-K) Transcripts in Human Prostate Cancer Cell Lines. Front Oncol. 2013;3:180. doi: 10.3389/fonc.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang-Johanning F., Frost A.R., Jian B., Azerou R., Lu D.W., Chen D.-T., et al. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98(1):187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 137.Giebler M., Staege M.S., Blauschmidt S., Ohm L.I., Kraus M., Würl P., et al. Elevated HERV-K expression in soft tissue sarcoma is associated with worsened relapse-free survival. Front Microbiol. 2018;9:211. doi: 10.3389/fmicb.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Benešová M., Trejbalová K., Kovářová D., Vernerová Z., Hron T., Kučerová D., et al. DNA hypomethylation and aberrant expression of the human endogenous retrovirus ERVWE1/syncytin-1 in seminomas. Retrovirology. 2017;14(1):20. doi: 10.1186/s12977-017-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rakoff-Nahoum S., Kuebler P.J., Heymann J.J., Sheehy E.M., Ortiz G.M., Ogg S.G., Barbour J.D., Lenz J., Steinfeld A.D., Nixon D.F. Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res Hum Retroviruses. 2006;22(1):52–56. doi: 10.1089/aid.2006.22.52. [DOI] [PubMed] [Google Scholar]
- 140.Bieda K., Hoffmann A., Boller K. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol. 2001;82(Pt 3):591–596. doi: 10.1099/0022-1317-82-3-591. [DOI] [PubMed] [Google Scholar]
- 141.Chan S.M., Sapir T., Park S.-S., Rual J.-F., Contreras-Galindo R., Reiner O., et al. The HERV-K accessory protein Np9 controls viability and migration of teratocarcinoma cells. PLoS ONE. 2019;14(2) doi: 10.1371/journal.pone.0212970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Florl A.R., Löwer R., Schmitz-Dräger B.J., Schulz W.A. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80(9):1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin G., Ye H., Wang J., Chen S., Chen X., Zhang C. Immune checkpoint human endogenous retrovirus-H Long terminal repeat-associating protein 2 is upregulated and independently predicts unfavorable prognosis in bladder urothelial carcinoma. Nephron. 2019;141(4):256–264. doi: 10.1159/000495887. [DOI] [PubMed] [Google Scholar]
- 144.Gosenca D., Gabriel U., Steidler A., Mayer J., Diem O., Erben P., et al. HERV-E-mediated modulation of PLA2G4A transcription in urothelial carcinoma. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu H., Liu T., Zhao Z., Chen Y., Zeng J., Liu S., et al. Mutations in 3'-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene. 2014;33(30):3947–3958. doi: 10.1038/onc.2013.366. [DOI] [PubMed] [Google Scholar]