Abstract
Drug-induced liver injury (DILI) is a challenging clinical problem with respect to both diagnosis and management. As a newly emerging biomarker of liver injury, miR122 shows great potential in early and sensitive in situ detection of DILI. Glycyrrhetinic acid (GA) possesses desirable therapeutic effect on DILI, but its certain dose-dependent side effects after long-term and/or high-dose administration limit its clinical application. In this study, in order to improve the precise diagnosis and effective treatment of DILI, GA loaded all-in-one theranostic nanoplatform was designed by assembling of upconversion nanoparticles and gold nanocages. As a proof of concept, we demonstrated the applicability of this single-wavelength laser-triggered theranostic nanoplatform for the spatiotemporally controllable in situ imaging of DILI and miR122-controlled on-demand drug release in vitro and in vivo. This novel nanoplatform opens a promising avenue for the clinical diagnosis and treatment of DILI.
Keywords: miR122, All-in-one theranostic nanoplatform, Drug-induced liver injury, In situ detection, On-demand drug release
Graphical abstract

1. Introduction
Drug-induced liver injury (DILI) has become a major clinical issue of global concern due to its relatively high incidence [1]. It is estimated that 25% of safety issues in the clinic are caused by DILI, among which nearly 40% are related with herbs [2,3]. For instance, Dioscorea bulbifera rhizome (DBR), a herb with the therapeutic function to treat goiters and cancer, could trigger liver injury [[4], [5], [6]]. Currently, the diagnosis and treatment of DILI are still difficult in clinical practice, because common clinical diagnostic indicators, such as aminotransferase aspartate aminotransferase (AST), alanine aminotransferase (ALT), and/or alkaline phosphatase (ALP) are proven to be lack of sensitivity and specificity [7]. miR122, a non-coding RNA, shows high sensitivity in DILI diagnosis, several clinical studies have demonstrated that miR122, with a shorter half-life than ALT and AST, changes at least 8 h earlier than ALT after the initial toxic invasion [[8], [9], [10]], and its abnormal expression is closely related to the degree of liver injury [11]. Besides, miR122 is liver specific, its level in liver is much higher than in other organs; for instance, the level of miR122 in heart is as low as negligible [12,13]. These unique properties make miR122 more suitable for serving as a biomarker for in situ detection of liver injury.
Glycyrrhetinic acid (GA), one of the bioactive natural products from Glycyrrhiza uralensis, has been extensively applied in the treatment of various liver diseases [14,15]. However, its clinical application is limited due to dose-dependent side effects, such as pseudoaldosteronism, hypersalicin corticosteroid action and rhabdomyolysis, after long-term and/or high dose administration [[16], [17], [18]]. Therefore, it is urgent to design a multifunctional nanoplatform that can realize not only the real-time detection of DILI in response to miR122, but also target-delivery and on-demand release of GA according to the status of DILI. To achieve these aims, photo-regulation is becoming a highly sought-after means owing to its spatiotemporal precision and non-invasion [[19], [20], [21], [22], [23]].
At present, the majority of existing light-controlled systems are driven by ultraviolet visible (UV–vis) light, however, the weak points of UV–vis light including poor penetration, phototoxicity and low photoactivation efficiency in biological tissues hamper their application in vivo [24,25]. Upconversion nanoparticles (UCNPs) have been discovered as a practicable photo-responsive nanoplatform in disease theranostics since UCNPs can absorb near-infrared (NIR) light and emit tuneable UV–vis or NIR light, which significantly overcomes the abovementioned drawbacks of UV–vis-driven systems [[26], [27], [28]].
In this study, UCNPs of a strong emission at 800 nm were employed as donor, while gold nanocages (Au NCs) with the localized surface plasmon resonance (LSPR) peak at 780 nm were chosen as acceptor. Meanwhile, Au NCs also functioned as drug carrier due to their porous walls and hollow interiors. Based on the chain replacement reaction involving miR122 aptamer and the luminescence resonance energy transfer (LRET) between UCNPs and Au NCs, viz. UCNPs-GA@Au NCs, was thus designed. In the nanoplatform (Scheme 1), Au NCs are loaded with GA and conjugated with photosensitizer-modified G-quadruplex DNA (Gate) to regulate GA release, which is then assembled with UCNPs by the complementary base pairs between two nucleic acid chains L1 and L2. miR122 acts as the start and regulation switch that can bind with L1 in UCNPs-GA@Au NCs, resulting in L2 replacement and UCNPs-GA@Au NC disassembly. Under 980 nm laser irradiation, the LRET between UCNPs and Au NCs ceases, while the upconversion luminescence (UCL) of UCNPs at 800 nm recoveries. At this point, the UV light from UCNPs cannot be transmitted to the Gate on Au NCs; as a result, the Gate remains intact and no GA is released. In contrast, in the absence of miR122, UCNPs-GA@Au NCs remains stable, at this moment, the UCL of UCNPs at 800 nm is quenched and GA is released. This proposed nanoplatform could detect miR122 with highly sensitive and release GA on-demand under single wavelength laser irradiation, which provides a basis for DILI in situ theranostic.
Scheme 1.
Schematic illustration of the fabrication of UCNPs-GA@Au NCs (a) and its application in DILI in situ detection and on-demand drug release therapy under single-wavelength laser stimulation (b).
2. Materials and methods
2.1. Instruments
The TEM measurements were performed on an HT7700 (Hitachi, Japan) transmission electron microscope operating at an acceleration voltage of 100 kV. X-ray powder diffraction (XRD) measurements were carried out on a Bruker D8 Advance diffractometer. Energy dispersive spectroscopy (EDS) was performed on an S–3400 N (Hitachi, Japan) scanning electron microscope. Zeta potentials and dynamic light scattering (DLS) measurements were performed on a Zetasizer Nano ZS901 (Malvern, UK) instrument. UV–vis spectra were acquired on a microplate reader (U-3900, BioTeK Synergy, USA). Fourier transform infrared (FT-IR) spectroscopy was conducted on an IR Affinity-1S1 spectrometer (Shimadzu, Japan). Fluorescence imaging in vitro was performed on an Olympus FV3000 confocal microscope (Japan). UCL spectra were recorded on a fluorescence spectrometer (RF-5301PC, Shimadzu, Japan) using an external 980 nm laser (Ningbo Yuanming Laser Technology Co. Ltd, China) as the excitation source. UCL imaging in vitro was acquired using an Olympus microscope equipped with an additional 980 nm external laser and CCD camera (Andor, UK). UCL imaging in vivo was performed with a modified Maestro in vivo imaging system (CRi Inc) from EINST Technology equipped with a 980 nm optical laser (Hi-Tech Optoelectronics Co., Ltd, China) as the excitation source.
2.2. Fabrication of UCNPs-GA@Au NCs
First was the preparation of UCNPs-L1. UCNPs-L1 was synthesized as previously reported methods with some adjustments [29]. Briefly, UCNPs (1 mg mL−1) were mixed with L1 (20 nM) in 2 mL PBS (pH 7.4). The mixture was ultrasonicated for 30 min, and then incubated at 37 °C for 12 h. After centrifugation at 6654 g for 10 min, the precipitates were washed with PBS thrice and mixed with disodium glycerol phosphate (GDSH, 100 × 10−6 M) in 2 mL PBS. Subsequently, the mixture was stirred at room temperature for 24 h. The obtained UCNPS-L1/GDSH was collected by centrifugation at 6654 g for 10 min, washed with PBS thrice, and re-dispersed in 1 mL PBS.
Next was the preparation of GA@Au NCs-L2/Gate/PEG. Au NCs (0.6 mg) were dispersed in 1 mL ethanol containing 1.95 mg mL−1 GA and stirred in dark at room temperature for 24 h. Free GA was removed by rapid centrifugation (9600 g, 15 min) and the resultant products were washed with ethanol (2 mL) thrice. Subsequently, the precipitates were re-dispersed in 2 mL PBS (pH 7.4) containing 40 nM L2 and 40 nM Gate, and the mixture was incubated at 37 °C for 3 h 10 μL of NaCl (1.67 M) was added drop wise to the above solution every 30 min and repeated 6 times, then the mixture was incubated for another 16 h. Centrifugation was performed at 9600 g for 15 min and the precipitates were washed with PBS thrice. Upon the precipitates were incubated with HS-PEG (4 μg mL−1, Sigma) at 37 °C for 1 h, the products were precipitated by centrifugation at 9600 g for 15 min and washed with PBS thrice, and finally re-dispersed in 1 mL PBS.
The synthesis of UCNPs-GA@Au NCs was as follows. UCNPs-L1/GDSH were mixed with GA@Au NCs-L2/Gate/PEG in 3 mL PBS and incubated at 37 °C for 2 h. The products were obtained by centrifugation at 6000 g for 15 min, washed with PBS for three times, and re-dispersed in 20 mL PBS. To calculate the conjugation efficiency of L1, L2 and Gate on the nanoplatform, fluorescent group-modified nucleic acids (L1-FAM, L2-ROX, Gate-Cy5) were used to fabricate the nanoplatform. The fluorescence was measured by an Envision®Multimode Plate Reader (PerkinElmer). The loading efficiency of GA was calculated by high-performance liquid chromatography (HPLC), the mobile phase was methanol-0.1% phosphoric acid aqueous solution (95:5), the UV detection wavelength was set at 250 nm, the flow rate of the mobile phase was 1.0 mL min−1, and the column temperature was 30 °C.
The loading efficiency of GA was calculated as follows:
| (1) |
where MGA (original) is the mass of initial GA, MGA (supernatant) is the mass of GA in the supernatant, and MUCNPs-Au NCs is the mass of null UCNPs-Au NCs without GA.
2.3. UCL measurements
UCNPs-GA@Au NCs were mixed with different amounts of miRNA, where the equivalent concentration of UCNPs was 0.1 mg mL−1 and miRNA ranged from 1.0 × 10−12 nM to 1.0 × 10−7 nM. After being incubated at 37 °C for 15–90 min, the UCL spectra of these mixtures were recorded on a fluorescence spectrometer with an external 980 nm laser (1 W) as the excitation source. UCL ratio (UCL-UCL0)/UCL0 was applied to measure the diagnosis characteristic of the nanoplatform, where UCL0 is the UCL intensity of the control group and UCL is the UCL intensity of the experimental group. The standard deviation produced from three measurements was depicted as error bars.
2.4. NIR-responsive drug release in vitro
UCNPs-GA@Au NCs and miR122 were mixed in 40 mL of 1% PBST-80, where the equivalent concentration of GA was 0.3 mg mL−1 and miR122 was 10−9 M. The mixture was incubated at 37 °C for 1 h and irradiated at 980 nm (0.5 W) for 1 min at an interval of 30 s for 5 consecutive times. The mixture was continuously incubated and 1 mL solution was collected at different time point within 0.1–48 h, meanwhile, the same volume of fresh medium was added. These solutions were centrifuged at 4000 g for 5 min and the concentration of GA was calculated by HPLC. The UCNPs-GA@Au NCs prepared by photo-insensitive Gate (in-Gate), incubated without miR122, or irradiated with 365 nm light were as control.
2.5. UCL imaging in vitro
HL7702 cells or RAW264.7 cells were seeded in a 24-well plate (5 × 103 cells well−1) containing cell slides. After the cells were cultured for 12 h and adhered to the slides, UCNPs-GA@Au NCs were added with a concentration of 100 μg mL−1 4 h later, the medium was removed and the cells were washed with cold PBS thrice. The cells were fixed with 4% paraformaldehyde (Beyotime Biotechnology Co., Ltd., Shanghai) and stained with DAPI (Beyotime Biotechnology Co., Ltd., Shanghai). The UCL images (800 ± 25 nm) were acquired by a 60 × objective on a microscope equipped with an additional 980 nm external laser (0.5 W) and CCD camera.
2.6. UCL imaging in vivo
Male ICR mice (20 ± 2 g) were purchased from Laboratory Animal Centre of Yangzhou University (Yangzhou, China) and housed under standard laboratory conditions with free feeding of standard diet and tap water ad libitum. The extraction procedure of DBR and the establishment of DBR-induced liver injury mouse model were performed as previously reported [30]. The mice were intragastrically (i.g.) administered with DBR extract (25 g kg−1) for 3 days, the control group was i.g. administered with 200 μL of 0.5% carboxymethylcellulose sodium. Different nanoplatform (25 mg kg−1) and saline (China Resources Double-crane Pharmaceutical) were first intravenously (i.v.) injected into mice for 3 days via the tail vein when they were given DBR, and then continued to be given without additional DBR for another 3 days. After the mice were administered with the nanoplatform for 2 h, the UCL imaging (800 ± 40 nm) was performed, or the mice were irradiated with or without a 980 nm laser (1 W for 2 min time−1,1 min interval, 5 times). Seven days later, the serum and organs of mice were collected for biochemical index detection and haematoxylin and eosin (H&E) staining.
3. Results and discussion
3.1. Characterizations of NaYF4:Yb, Tm and NaYF4:Yb,Tm@NaYF4
Because the doped ions are homogeneously distributed in the host lattice, the conventional UCNPs are prone to be quenched by surface quenchers and lose their excitation energy, eventually leading to weak and vulnerable emission. Conversely, the upconversion emission of the UCNPs with core-shell structure and doped with lanthanide ions is greatly enhanced, and the optical integrity is largely preserved [31]. Therefore, in order to improve the detection sensitivity and therapy efficiency of the nanoplatform, core–shell UCNPs were adopted in this research. The morphologies of the UCNPs were characterized by transmission electron microscopy (TEM), and it illustrated that the UCNPs exhibited high uniformity and monodispersity, the core nanoparticles (NaYF4:Yb,Tm) were approximately 25 nm, and the core–shell UCNPs (NaYF4: Yb,Tm@NaYF4, OA-UCNPs) were about 27 nm (Fig. 1a and b, Fig. S1a-b). DLS measurement further verified the particle size of the nanoparticle (Fig. S1c-d). The XRD spectra showed that the peaks of NaYF4:Yb, Tm and NaYF4:Yb,Tm@NaYF4 corresponded well to the pure hexagonal of NaYF4 (JCPDS card: No.16-0334) (Fig. 1c), combining with high-resolution TEM (Fig. 1d) and EDS analysis (Fig. S1e), it could be concluded that NaYF4:Yb,Tm@NaYF4 have been successfully synthesized and have an integrity hexagonal crystal structure. Under 980 nm single laser irradiation, the fabricated UCNPs could emit 800 nm NIR light and 365 nm UV light, thus enabling in situ detection and allowing release drug without exogenous UV light (Fig. 1e). Subsequently, the OA-UCNPs were transformed into hydrophilic UCNPs to prepare the nanoplatform, the results indicated that after modification, the morphology (Fig. S2a), size (Fig. S2b-c) and UCL spectrum (Fig. S2d) of UCNPs were all not affected.
Fig. 1.
TEM images of core: NaYF4:Yb, Tm (a) and core-shell: NaYF4:Yb,Tm@NaYF4 (OA-UCNPs) (b). (c) XRD pattern of NaYF4:Yb, Tm and OA-UCNPs. (d) High resolution TEM image of OA-UCNPs. (e) UCL spectrum of OA-UCNPs. (f) TEM image of Ag NCs. (g) LSPR of Ag NCs. (h) LSPR of Au NCs. (i) TEM image of Au NCs. (j) Scheme illustration of miR122 activated chain substitution detection principle. (k) SDS-PAGE image of M: Marker, lane 1: miR122, lane 2: L1, lane 3: L2, lane 4: L1+miR122, lane 5: L1+L2, lane 6: L1+L2+miR122. The concentration of H1 was 1 × 10−8 M, H2 was 1 × 10−8 M, miR122 was 1 × 10−8 M. Each team was cultured in PBS at 37 °C for 1 h. Gels conditions: 20% PAGE, 100 V of separated voltage, 20 μL loading amount of each sample, 10 μL of 2 × loading buffer.
3.2. Characterizations of Ag NCs and Au NCs
Au NCs were synthesized by galvanic replacement, in this reaction, preparation of Ag NCs with high-quality was the critical step [32]. As could be seen, the Ag NCs were cube shaped with good dispersibility and relatively uniform size at about 43 nm (Fig. 1f). DLS measurement further validated the size of Ag NCs (Fig. S2e). The UV–vis spectrum of Ag NCs revealed that the LSPR peak of Ag NCs was about 420 nm, and the small peak around 354 nm corresponded to the angular position of Ag NCs (Fig. 1g). With addition of HAuCl4, Ag NCs were converted into Au NCs with different void/pore sizes and the LSPR peak was continuously shifted from visible to NIR region [32]. Fig. 1h shows that the LSPR peak of Au NCs was 780 nm, which satisfied the need of LRET between Au NCs and UCNPs. The DLS measurement of Au NCs shows that the synthesized Au NCs were about 45 nm (Fig. 1i, Fig. S2f).
3.3. Validation of the chain substitution detection feasibility of miR122
In this study, the nucleic acid chains L1 and L2 (Table S1) were involved in the detection of miR122 by chain substitution detection method in addition to being the linkers connecting UCNPs and Au NCs. Fig. 1j shows an overview of the chain substitution detection method triggered by miR122. As can be seen, L1 and L2 can be partially connected to form the complex of L1-L2. As the aptamer of miR122, L1 can specifically recognize miR122 with stronger binding ability than L2. When miR122 is present, L2 in the complex will be replaced, causing L1-L2 broken and forming L1-miR122. 20% PAGE was applied to verify the feasibility of the chain substitution detection assay. As illustrated in Fig. 1k, three bands due to miR122, L1 and L2 were detected in Lane 1, Lane 2 and Lane 3, respectively. When L1 was mixed with miR122 (Lane 4) or L2 (Lane 5), an extra band due to L1-miR122 or L1-L2 appeared, respectively. When miR122 was mixed with L1-L2 complex and incubated at 37 °C for 1 h (Lane 6), a band corresponding to L2 emerged. These results indicated that the chain substitution detection assay is feasible.
3.4. Construction and characterization of UCNPs-GA@Au NCs
First was the preparation of UCNPs-L1/GDSH. L1 was modified to UCNPs through the high affinity between polycytosine (poly-C) and UCNPs [29], and GDSH was modified to improve the stability and biocompatibility of the nanoplatform. As shown in Fig. S3a, after modifying the negatively charged L1, the charge of UCNPs changed from +31 ± 1.6 mV to −13.6 ± 1.8 mV, and the electronegativity increased (−21.3 ± 2.6 mV) after the modification of GDSH, suggesting the successful preparation of UCNP-L1/GDSH. The particle size was measured as approximately 37 nm (DPI = 0.25, Fig. S3b) by DLS method.
The following was the construction of GA@Au NCs-L2/Gate/PEG. Circular dichroism spectroscopy (CD) was applied to investigate the structure of the Gate, as shown in Fig. S3c, the positive peak at 260 nm and the negative peak between 240 and 245 nm indicated that the Gate had G-quadruplex parallel structure. Subsequently, L2, Gate and PEG were modified to Au NCs via gold-sulfur bonds. As Fig. S3d shown, compared with Au NCs (−18.2 ± 1.4 mV), the negative potential of Au NCs-L2/Gate (−30.2 ± 1.4 mV) increased due to L2 and Gate were negatively charged; after modification with PEG, the zeta potential of Au NCs-L2/Gate/PEG increased (−16.5 ± 0.6 mV). Fig. S3e shows that the prepared Au NCs-L2/Gate/PEG was about 78 nm (DPI = 0.37).
Finally, UCNPs-GA@Au NCs were constructed by UCNPs-L1/GDSH and GA@Au NCs-L2/Gate/PEG through the complementary base pair between L1 and L2. The TEM (Fig. 2a) and DLS (Fig. S3f) results depicted that UCNPs-GA@Au NCs were successfully fabricated with a size of about 102 nm. The synthesis of UCNPs-GA@Au NCs was further studied by FT-IR, as shown in Fig. S3g, the characteristic vibrations of thymine at 1714 cm−1 (υC O) and sugar phosphate at 1250 - 1000 cm−1 confirmed the successful preparation of UCNPs-GA@Au NCs. L1-FAM, L2-ROX and Gate-Cy5 were applied to roughly quantify the modification amount of L1, L2 and Gate on UCNPs-GA@Au NCs. It was calculated that the modification amounts of L1, L2 and Gate on UCNPs-GA@Au NCs were approximately 12.9 ± 1.7 nmol mg−1 (Fig. S4a,b), 48.9 ± 3.6 nmol mg−1 (Fig. S4c,d) and 34.8 ± 0.6 nmol mg−1 (Fig. S4e,f), respectively. Driven by the concentration-gradient diffusion, GA was encapsulated in the Au NCs, and the loading efficiency of this nanoplatform was calculated by HPLC. As Fig. S5 and Fig. 2b shown, when the mass ratio of GA to Au NCs was 1.5, UCNPs-GA@Au NCs had the highest drug-loading efficiency of about 22 wt%.
Fig. 2.
(a) TEM images of UCNPs-GA@Au NCs. (b) Drug loading efficiency of GA with different ratios of GA to UCNPs–Au NCs. The weight of UCNPs-Au NCs was 1.3 mg. (c) UCL spectra of UCNPs-GA@Au NCs when incubated with different concentrations of miR122 in the range of 0 M – 1 × 10−7 M. The equivalent concentration of UCNPs was 0.1 mg mL−1. (d) Relationship between quenching efficiency UCL-UCL0)/UCL0 and logarithm of miR122 ranging from 1 × 10−12 M to 1 × 10−7 M. The calibration equation was with a correlation coefficient of . The equivalent concentration of UCNPs was 0.1 mg mL−1. (e) Quenching efficiency of UCNPs-GA@Au NCs with different miRNAs. The equivalent concentration of UCNPs was 0.1 mg mL−1, and miRNAs were all 1 × 10−8 M. (f) Drug release behaviours of UCNPs-GA@Au NCs or in-Gate modified UCNPs-GA@Au NCs (in-Gate) with or without miR122 and a 980 nm laser or 365 nm UV–vis light irradiation (0.5 W for 1 min, 30 s interval, 5 times in total). The equivalent concentration of UCNPs was 0.1 mg mL−1. The concentration of the miR122 was 1 × 10−8 M (Data are medians ± quartiles, n = 3).
3.5. Performance of UCNPs-GA@Au NCs in miR122 detection and on-demand drug release
Coupling amount of UCNPs and Au NCs affects the performance of the nanoplatform. Therefore, the ratio of UCNPs to Au NCs was optimized and set as 0.3 (Fig. S6a). The incubation time of UCNPs-GA@Au NCs and biomarker also plays an important role in improving the detection efficiency of the nanoplatform. As Fig. S6b shown, (UCL-UCL0)/UCL0 increased rapidly as incubation time went on, and the curve entered a plateau stage after 60 min. Thus, the incubation time of UCNPs-GA@Au NCs was optimized as 60 min.
Subsequently, the detection feasibility of the nanoplatform was studied by comparing the normalized UCL of different nanoparticles. As shown in Fig. S7a, compared with UCNPs, the UCL of UCNPs-GA@Au NCs distinctly decreased due to the LRET among UCNPs and Au NCs; what's more, this phenomenon indicated again that UCNPs-GA@Au NCs were successfully fabricated. However, the UCL of the mixture of UCNPs and GA@Au NCs (UCNPs + GA@Au NCs group) did not change, indicating that the free Au NCs had little effect on the UCL of UCNPs. When miR122 was added to UCNPs-GA@Au NCs (UCNPs-GA@Au NCs + miR122 group), the UCL restored due to the structure destruction of UCNPs-GA@Au NCs. This result indicated that UCNPs-GA@Au NCs could be applied for miR122 detection.
To evaluate the performance of the nanoprobe, the sensitivity and specificity of UCNPs-GA@Au NCs were investigated. It could be seen that with the increase of miR122, the UCL intensity of UCNPs-GA@AuNCs enhanced (Fig. 2c). When miR122 ranged from 1 × 10−12 M to 1 × 10−7 M, the calibration curve of (UCL-UCL0)/UCL0 corresponding to the logarithm relative value of miR122 exhibited a well-fitted linear relationship (Fig. 2d), the calibration equation was with a correlation coefficient of , and the detection limit was 5 × 10−13 M. The specificity of the nanoplatform was demonstrated in the potential interferences such as miR155, miR192, and bases mismatched miR122 (mis-miR122). Compared with miR122, the (UCL-UCL0)/UCL0 of these nonspecific miRNAs had nearly no noticeable change (Fig. 2e), suggesting that this nanoprobe had favourable selectivity for miR122.
The following was the investigation of on-demand drug release of UCNPs-GA@AuNCs. As shown in Fig. 2f, once UCNPs-GA@Au NCs were irradiated with 365 nm UV–vis light, almost all GA was released 12 h later due to the Gate had no selectivity for 365 nm UV–vis light, and the cumulative release of GA reached about 90% (miR122 + 365 nm group, and 365 nm group). A similar effect occurred when UCNPs-GA@Au NCs were irradiated with 980 laser (980 nm group). When miR122 was present, less drug was released under irradiation of 980 nm laser (miR122 + 980 nm group), which could be explained by the fact that after the emergence of miR122, the structure of UCNPs-GA@Au NCs was destroyed and the UV emitted from UCNPs could not transfer to the Gate on Au NCs. On the contrary, without 980 nm laser irradiation, UCNPs-GA@Au NCs did not undergo drug release (miR122 group). However, the UCNPs-GA@Au NCs fabricated by photo-insensitive Gate (in-Gate) had no drug release no matter irradiated with 365 nm UV light (in-Gate+365 nm group) or 980 nm laser (in-Gate+980 nm group). In conclusion, 980 nm laser and photosensitive Gate were the necessary conditions for the drug release of UCNPs-GA@Au NCs, in which miR22 played a regulatory role.
UCNPs-GA@Au NCs will eventually be used for in situ detection in vivo; therefore, the stability of the nanoplatform should not be ignored. The stability of the nanoplatform was studied in the serum diluted 8-fold with PBS and 0.1 U μL−1 DNase Ⅰ (Sangon). The UCNPs-GA@Au NCs were diluted in above medium and incubated at 37 °C for 24 h, and the UCL signal and the drug release were measured every 2 h. It could be seen that after 24 h of incubation, UCL (Fig. S7b), cumulative drug release (Fig. S7c) and size of the nanoplatform (Fig. S7d-e) did not change significantly. Long-term stability studies showed that when the nanoplatform was stored at 4 °C for 28 d, the UCL (Fig. S7f) and size (Fig. S7g) showed no significant changes, but a small amount of drug leakage occurred at 21st days (Fig. S7f). These results demonstrated that the UCNPs-GA@Au NCs had favourable optical stability and can be stored for a long time.
3.6. Performance of UCNPs-GA@Au NCs in vitro
After determining the feasibility and optimal operating conditions of the nanoplatform, its imaging detection capability and on-demand drug release performance in vitro were studied. HL7702 cell line was applied in this research unless otherwise stated. First was the cytotoxicity study of UCNPs-Au NCs, methyl thiazolyl tetrazolium (MTT) assay was adopted in this research. HL7702 cells were exposed to different amounts of UCNPs-Au NCs and irradiated with or without 980 nm laser (0.5 W for 1 min time−1, 30 s interval, 5 times). As shown in Fig. S8, the cell viability was higher than 90%, indicating that the synthesized nanoplatform exhibited good biocompatibility and had little effect on cell viability.
To evaluate the cellular uptake of the nanoplatform, L1-FAM and L2-ROX fabricated nanoplatform (UCNPs(L1-FAM)-GA@Au NCs(L2-ROX)) was co-incubated with hepatocytes (HL7702 cells) and macrophages (RAW264.7 cells), respectively. Confocal fluorescence images of the cells pre-treated with different nanoplatform showed that compared with the normal nanoplatform (UCNPs-GA@Au NCs), the HL7702 cells pre-treated with UCNPs(L1-FAM)-GA@Au NCs(L2-ROX) exhibited a significant fluorescence signal (Fig. S9a). The nanoplatform incubated with RAW264.7 cells had the same performance (Fig. S9b). These results suggested that both hepatocytes and macrophages could internalize the nanoplatform. The prevailing hypothesis is that most of the particles are absorbed or internalized by macrophages and further transferred to hepatocytes, where internalization of macrophages facilitates the insertion of nanoparticles into hepatocytes [33,34]. To better demonstrate this result, darkfield images of UCNPs-GA@Au NCs and UCNPs-GA@AuNCs pre-treated HL7702 cells were obtained. It could be seen that some bright green points appeared when the UCNPs-GA@Au NCs were placed in the darkfield; in contrast, there were no bright points in the images of the HL7702 cells; after 3 h co-incubation of UCNPs-GA@AuNCs and HL7702 cells, some bright points appeared (Fig. S10). This result further proved that the nanoplatform could be endocytosed by HL7702 cells.
To study the lysosome escape ability of UCNPs-GA@AuNCs, L1-FAM fabricated nanoplatform (UCNPs(L1-FAM)-GA@Au NCs) were used in this research. As could be seen from the time-dependent colocalization (Fig. S11), the fluorescence of red (LysoTracker deep red stained endo/lysosome) and green (UCNPs(L1-FAM)-GA@AuNCs) were merged at 1st h, which indicated that the nanoplatform was engulfed by endo/lysosomes. 3 h later, the merged fluorescence separated in the cytoplasm, which illustrated that the nanoplatform could escape from the endo/lysosome entrapment. Accordingly, the UCNPs-GA@Au NCs could enter the cell and escape from the endo/lysosome to fulfil their detection and drug delivery functions.
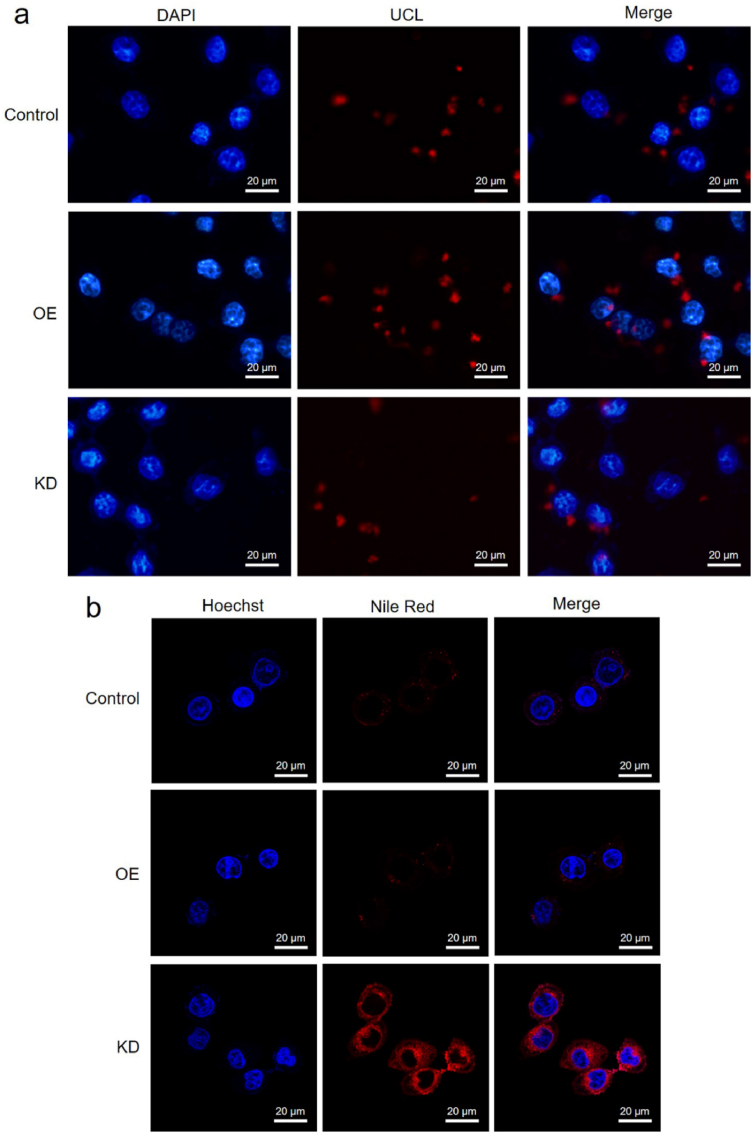
In order to explore the performance of the nanoplatform in DILI detection and treatment, HL7702 cells with miR122 overexpression (OE) or knockdown (KD) were applied to simulate different states of DILI. The RT-qPCR analysis is shown in Fig. S12, it could be seen that the cell model with miR122 OE or KD was successfully constructed. The UCL imaging of miR122 OE or KD cells treated by UCNPs-GA@Au NCs are shown in Fig. 3a, as could be seen, compared with normal cells, the UCL of miR122 OE cells was markedly increased; conversely, the UCL of miR122 KD cells was significantly decreased. These results confirmed that the nanoplatform could be used for intracellular visual imaging of miR122.
Fig. 3.
(a) UCL images of HL7702 cells with miR122 OE or KD incubated with UCNPs-GA@Au NCs for 3 h. Wild-type HL7702 cells were used as a control. The power of the 980 nm laser was 0.5 W. (b) Confocal images of HL7702 cells with miR122 OE or KD incubated with UCNPs-Nile red@Au NCs. These cells were irradiated with a 980 nm laser (0.5 W for 1 min, 30 s interval, 5 times) after 3 h of incubation and then continued for another 4 h incubation. The wild-type HL7702 cells were used as control.
The feasibility of on-demand drug release of this nanoplatform in vitro was studied by Nile red-loaded nanoplatform (UCNPs-Nile red@Au NCs). UCNPs-Nile red@Au NCs were incubated with HL7702 cells containing different amounts of miR122 for 3 h, and then irradiated with 980 nm laser (0.5 W for 1 min time−1, 30 s interval, 5 times), after another 4 h incubation, the release of Nile red was observed by confocal microscopy. As shown in Fig. 3b, compared with the control group, almost no Nile red was released in miR122 OE cells, while a large amount of Nile red was released in miR122 KD cells. Subsequently, the in-Gate modified UCNPs-Nile red@Au NCs were set as the negative control to further investigate the role of 980 nm laser and photosensitive Gate in on-demand drug release. As Fig. S13 shown, the in-Gate modified UCNPs-Nile red@Au NCs did not engage in drug release when incubated with miR122 KD cells and irradiated with 980 nm laser or 365 nm UV light. This result was consistent with the study of drug release in buffer solution (Fig. 2f), which further demonstrated that under 980 nm laser irradiation, the nanoplatform can release drugs on-demand.
3.7. Performance of UCNPs-GA@Au NCs in vivo
The following described the investigation of the diagnostic and therapeutic performance of UCNPs-GA@AuNCs in vivo. The first was to study the in vivo distribution of the nanoplatform. UCNPs-GA@Au NCs (25 mg kg−1) were i. v. administered to the mice pre-treated with DBR (200 mg kg−1). As shown in Fig. 4a and b, after the administration of the nanoplatform, the UCL in the liver was significantly enhanced and reached to the plateau 2 h later, and it kept stronger luminance even 6 h later, which clarified that the nanoplatform had good stability in vivo. Subsequently, the mice were killed to investigate the distribution of the nanoplatform in various organs after administered with UCNPs-GA@Au NCs for 2 h. As Fig. 4c shown, the nanoplatform mainly accumulated in the liver, which stated that the nanoplatform exhibited good liver specificity. The UCL imaging of liver slices (Fig. 4d) showed that both hepatocytes and liver macrophage-Kupffer cells could take up the nanoplatform. It was speculated that Kupffer cells contributed to the liver-targeting property of nanoparticles.
Fig. 4.
(a) Representative UCL images of mice pretreated with DBR and i. v. with UCNPs-GA@Au NCs (25 mg kg−1) at different times. (b) Quantification of the UCL intensity from (a). (Data are medians ± quartiles, n = 5). (c) Distribution of UCNPs-GA@Au NCs in heart, liver, spleen, lung, kidney after the mice was i. v with UCNPs-GA@Au NCs (25 mg kg−1) for 2 h. (d) UCL imaging of liver slice.
Our previous study showed that the variation of miR122 in liver were related to the degree of hepatotoxicity [30]. To investigate the role of the nanoplatform in guiding the treatment of DILI, the UCL of the mice that treated with this nanoplatform for different times was explored. The treatment timeline is shown in Fig. 5a. As can be seen, the UCL enhanced as time went on and basically returned to normal after 6 days of treatment (Fig. 5b and c). To explore the diagnostic and therapeutic performance of UCNPs-GA@AuNCs on DILI, the mice were i. v. injected with different nanoparticles and irradiated with 980 nm laser or not, and then the UCLs were compared. As shown in Fig. 5d and e, the UCL of the DILI mice (DBR group) was lower than the wild mice; however, the UCLs of the mice irradiated alone with 980 nm laser (DBR + Laser group), treated with the nanoplatform (DBR + NPs group), or administrated with in-Gate modified nanoplatform (DBR+in-GateNPs + Laser group) were same to that of DBR treated mice; conversely, when the mice gained 6 days of effective treatment (DBR + NPs + Laser group), their UCL returned to a normal level as the control group. On the last day, the relative expression of miR122 in liver, serum biochemistry (ALT, AST), and liver ratio were investigated to validate the efficiency of the nanoplatform, and it showed that only the DBR + NPs + laser group got recovered (Fig. S14). The histopathologic evaluation (H&E staining) of the liver (Fig. S15) demonstrated that except the DBR + NPs + laser group, the liver of other four groups still had necrotic lesions. Subsequently, UCNPs-GA@Au NCs (25 mg kg−1) were given to the mice that pre-treated with different doses of DBR (0, 12.5, 25, 50 g kg−1) to verify their diagnosis and treatment effectiveness of DILI in different degree. After 6 days of treatment, the UCL imaging of the mice in the low- and medium-dose groups was similar to the control group; while the UCL images of the mice in the high-dose group was lower (Fig. S16a-b). The serum biochemistry (ALT, AST), liver ratio, and liver H&E staining supported above results (Fig. S16c-f). These studies indicated that UCNPs-GA@AuNCs played a positive role in guiding the treatment of DILI, and could be used for the effective treatment of DILI.
Fig. 5.
(a) Treatment Timeline. (b) Representative in vivo UCL images of mice pretreated with UCNPs-GA@Au NCs for 3 d, 4 d, 5 d, 6 d. (c) Corresponding UCL intensity of mice treated with UCNPs-GA@Au NCs for different time in (b). (n = 5 mice per group). (d) Representative UCL images of DILI mice with different treatments. DBR: the mice were only given DBR and without other treatment; DBR + Laser: the mice were treated with DBR and irradiated with a 980 nm laser; DBR + NPs: the mice were treated with DBR and injected with UCNPs-GA@Au NCs; DBR+in-GateNPs + Laser: the mice were treated with DBR and injected with in-Gate modified UCNPs-GA@Au NCs, and then irradiated with a 980 nm laser; DBR + NPs + Laser: the mice were treated with DBR and injected with UCNPs-GA@Au NCs and irradiated with a 980 nm laser. WT ICR mice were as control. (e) Corresponding UCL intensity of mice with different treatments in (d). The power of the 980 nm laser was 0.5 W, and n = 6 mice per group.
Finally, the hepatorenal toxicity of the nanoplatform was studied by i. v. injecting GA unloaded nanoplatform (eNPs, 25 mg kg−1) to the wild mice for different days, and the serum biochemical indices were observed. It could be seen that compared with the control group, even the mice were given eNPs for 14 days consecutively, their hepatic function indexes (ALT, AST, TBA, Fig. S17a-c) and renal function indexes (CRE, BUN, UA, Fig. S17d-f) had no significant changes. H&E staining of the liver and kidney agreed with the above measurements (Fig. S17g). These results suggested that UCNP–Au NCs were safe and had no significant hepatorenal side effects on mice.
4. Conclusion
In summary, a novel theranostic nanoplatform was designed. Under a single 980 nm laser irradiation, it could not only achieve the visually sensitive detection of miR122 in vitro and in vivo, but also intelligently release drug on demand, enabling accurate in situ detection and efficient treatment of DILI. The use of a 980 nm laser as the activation switch could avoid the photodamage and phototoxicity caused by exogenous UV–vis light. Undoubtedly, this smartly assembled nanoplatform shows great promise for the in situ diagnosis and treatment of other diseases.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant No. 81773993).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100157.
Contributor Information
Yan Jiang, Email: jiangyancpu@126.com.
Hui-Jun Li, Email: cpuli@163.com.
Credit author statement
Lingchang Meng and Qiaolei Wang designed and performed the experiment, and wrote the manuscript. Lai Wang, Zhen Zhao, Ping Zhou and Zuguo Zheng analysed the data. Gui-Zhong Xin, Yan Jiang and Ping Li provided reagents, Yan Jiang and Hui-Jun Li were the guarantor of this research work.
Authors’ contributions
LCM and QLW designed and performed the experiment, and wrote the manuscript. LW, ZZ (Zhen Zhao), PZ and ZZ (Zuguo Zheng) analysed the data. GZX, YJ and PL provided reagents, YJ and HJL were the guarantor of this research work. HJL had the full right to obtain the research data and was responsible for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Andrade R.J., Chalasani N., Björnsson E.S., Suzuki A., Kullak-Ublick G.A., Watkins P.B., Devarbhavi H., Merz M., Lucena M.I., Kaplowitz N., Aithal G.P. Drug-induced liver injury. Nat Rev Dis Primers. 2019;5:58. doi: 10.1038/s41572-019-0105-0. [DOI] [PubMed] [Google Scholar]
- 2.Hou F.Q., Zeng Z., Wang G.Q. Hospital admissions for drug-induced liver injury: clinical features, therapy, and outcomes. Cell Biochem. Biophys. 2012;64:77–83. doi: 10.1007/s12013-012-9373-y. [DOI] [PubMed] [Google Scholar]
- 3.Watkins P.B. Drug safety sciences and the bottleneck in drug development. Clin. Pharmacol. Ther. 2011;89:788–790. doi: 10.1038/clpt.2011.63. [DOI] [PubMed] [Google Scholar]
- 4.Wang J.M., Ji L.L., Branford-White C.J., Wang Z.Y., Shen K.K., Liu H., Wang Z.T. Antitumor activity of Dioscorea bulbifera L. rhizome in vivo. Fitoterapia. 2012;83:388–394. doi: 10.1016/j.fitote.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhao D.S., Wu Z.T., Li Z.Q., Wang L.L., Jiang L.L., Shi W., Li P., Li H.J. Liver-specific metabolomics characterizes the hepatotoxicity of Dioscorea bulbifera rhizome in rats by integration of GC-MS and (1)H-NMR. J. Ethnopharmacol. 2018;226:111–119. doi: 10.1016/j.jep.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Li H., Peng Y., Zheng J. Dioscorea bulbifera L.-induced hepatotoxicity and involvement of metabolic activation of furanoterpenoids. Drug Metab. Rev. 2020;52:568–584. doi: 10.1080/03602532.2020.1800724. [DOI] [PubMed] [Google Scholar]
- 7.Church R.J., Watkins P.B. The transformation in biomarker detection and management of drug-induced liver injury. Liver Int. 2017;37:1582–1590. doi: 10.1111/liv.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starlinger P., Hackl H., Pereyra D., Skalicky S., Geiger E., Finsterbusch M., Tamandl D., Brostjan C., Grünberger T., Hackl M., Assinger A. Predicting postoperative liver dysfunction based on blood-derived MicroRNA signatures. Hepatology. 2019;69:2636–2651. doi: 10.1002/hep.30572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell L.S., Ireland L., Park B.K., Goldring C.E. MiR-122 and other microRNAs as potential circulating biomarkers of drug-induced liver injury. Expert Rev. Mol. Diagn. 2018;18:47–54. doi: 10.1080/14737159.2018.1415145. [DOI] [PubMed] [Google Scholar]
- 10.Dear J.W., Clarke J.I., Francis B., Allen L., Wraight J., Shen J., Dargan P.I., Wood D., Cooper J., Thomas S.H.L., Jorgensen A.L., Pirmohamed M., Park B.K., Antoine D.J. Risk stratification after paracetamol overdose using mechanistic biomarkers: results from two prospective cohort studies. Lancet Gastroenterol Hepatol. 2018;3:104–113. doi: 10.1016/s2468-1253(17)30266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q., Liu H.O., Liu Y.D., Liu W.S., Pan D., Zhang W.J., Yang L., Fu Q., Xu J.J., Gu J.X. Decreased expression of hepatocyte nuclear factor 4alpha (Hnf4alpha)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J., Xu Y.X., Hao J.L., Wang S.F., Li C.F., Meng S.D. MiR-122 in hepatic function and liver diseases. Protein & Cell. 2012;3:364–371. doi: 10.1007/s13238-012-2036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. https://doi.org/Pii-S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Coon J.T., Ernst E. Complementary and alternative therapies in the treatment of chronic hepatitis C: a systematic review. J. Hepatol. 2004;40:491–500. doi: 10.1016/j.jhep.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Wu S.Y., Cui S.C., Wang L., Zhang Y.T., Yan X.X., Lu H.L., Xing G.Z., Ren J., Gong L.K. 18β-Glycyrrhetinic acid protects against alpha-naphthylisothiocyanate-induced cholestasis through activation of the Sirt1/FXR signaling pathway. Acta Pharmacol. Sin. 2018;39:1865–1873. doi: 10.1038/s41401-018-0110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y., Li J., Wang G. In vitro and in vivo evaluation of mPEG-PLA modified liposomes loaded glycyrrhetinic acid. Int. J. Pharm. 2008;356:274–281. doi: 10.1016/j.ijpharm.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Yu H., Li S., Wang G.J. Enhanced distribution and extended elimination of glycyrrhetinic acid in mice liver by mPEG-PLA modified (mPEGylated) liposome. J. Pharmaceut. Biomed. Anal. 2010;51:1147–1153. doi: 10.1016/j.jpba.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Yoshino T., Yanagawa T., Watanabe K. Risk factors for pseudoaldosteronism with rhabdomyolysis caused by consumption of drugs containing licorice and differences between incidence of these conditions in Japan and other countries: case report and literature review. J. Alternative Compl. Med. 2014;20:516–520. doi: 10.1089/acm.2013.0454. [DOI] [PubMed] [Google Scholar]
- 19.Nelidova D., Morikawa R.K., Cowan C.S., Raics Z., Goldblum D., Scholl H.P.N., Szikra T., Szabo A., Hillier D., Roska B. Restoring light sensitivity using tunable near-infrared sensors. Science. 2020;368:1108–1113. doi: 10.1126/science.aaz5887. [DOI] [PubMed] [Google Scholar]
- 20.Franke K., Vlasits A. Unblinding with infrared nanosensors. Science. 2020;368:1057–1058. doi: 10.1126/science.abc2294. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y., Zhang J., Yang Z., Mou Q., Ma Y., Xiong Y., Lu Y. Functional DNA regulated CRISPR-cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J. Am. Chem. Soc. 2020;142:207–213. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Zhang Y., Zhang X., Li Y., He Y., Liu Y., Ju H. A photo zipper locked DNA nanomachine with an internal standard for precise miRNA imaging in living cells. Chem. Sci. 2020;11:6289–6296. doi: 10.1039/d0sc00394h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu L., Wu C., You M., Han D., Chen T., Zhu G., Jiang J., Yu R., Tan W. A targeted, self-delivered, and photocontrolled molecular beacon for mRNA detection in living cells. J. Am. Chem. Soc. 2013;135:12952–12955. doi: 10.1021/ja406252w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ankenbruck N., Courtney T., Naro Y., Deiters A. Optochemical control of biological processes in cells and animals. Angew Chem. Int. Ed. Engl. 2018;57:2768–2798. doi: 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velema W.A., Szymanski W., Feringa B.L. Photopharmacology: beyond proof of principle. J. Am. Chem. Soc. 2014;136:2178–2191. doi: 10.1021/ja413063e. [DOI] [PubMed] [Google Scholar]
- 26.Jayakumar M.K., Idris N.M., Zhang Y. Remote activation of biomolecules in deep tissues using near-infrared-to-UV upconversion nanotransducers. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8483–8488. doi: 10.1073/pnas.1114551109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Weitemier A.Z., Zeng X., He L., Wang X., Tao Y., Huang A.J.Y., Hashimotodani Y., Kano M., Iwasaki H., Parajuli L.K., Okabe S., Teh D.B.L., All A.H., Tsutsui-Kimura I., Tanaka K.F., Liu X., McHugh T.J. Near-infrared deep brain stimulation via upconversion nanoparticle-mediated optogenetics. Science. 2018;359:679–684. doi: 10.1126/science.aaq1144. [DOI] [PubMed] [Google Scholar]
- 28.Teh D.B.L., Bansal A., Chai C., Toh T.B., Tucker R.A.J., Gammad G.G.L., Yeo Y., Lei Z., Zheng X., Yang F., Ho J.S., Bolem N., Wu B.C., Gnanasammandhan M.K., Hooi L., Dawe G.S., Libedinsky C., Ong W.Y., Halliwell B., Chow E.K., Lim K.L., Zhang Y., Kennedy B.K. A flexi-PEGDA upconversion implant for wireless brain photodynamic therapy. Adv. Mater. 2020;32 doi: 10.1002/adma.202001459. [DOI] [PubMed] [Google Scholar]
- 29.Ge H., Wang D., Pan Y., Guo Y., Li H., Zhang F., Zhu X., Li Y., Zhang C., Huang L. Sequence-dependent DNA functionalization of upconversion nanoparticles and their programmable assemblies. Angew Chem. Int. Ed. Engl. 2020;59:8133–8137. doi: 10.1002/anie.202000831. [DOI] [PubMed] [Google Scholar]
- 30.Meng L., Zheng X., Zheng Z., Zhao Z., Wang L., Zhou P., Xin G.Z., Li P., Li H.J. A sensitive upconverting nanoprobe based on signal amplification technology for real-time in situ monitoring of drug-induced liver injury. Nanoscale. 2020;12:15325–15335. doi: 10.1039/d0nr01493a. [DOI] [PubMed] [Google Scholar]
- 31.Chen X., Peng D., Ju Q., Wang F. Photon upconversion in core-shell nanoparticles. Chem. Soc. Rev. 2015;44:1318–1330. doi: 10.1039/c4cs00151f. [DOI] [PubMed] [Google Scholar]
- 32.Skrabalak S.E., Au L., Li X., Xia Y. Facile synthesis of Ag nanocubes and Au nanocages. Nat. Protoc. 2007;2:2182–2190. doi: 10.1038/nprot.2007.326. [DOI] [PubMed] [Google Scholar]
- 33.Tsoi K.M., MacParland S.A., Ma X.Z., Spetzler V.N., Echeverri J., Ouyang B., Fadel S.M., Sykes E.A., Goldaracena N., Kaths J.M., Conneely J.B., Alman B.A., Selzner M., Ostrowski M.A., Adeyi O.A., Zilman A., McGilvray I.D., Chan W.C. Mechanism of hard-nanomaterial clearance by the liver. Nat. Mater. 2016;15:1212–1221. doi: 10.1038/nmat4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z., Li Y., Li W., Xiao C., Liu D., Dong C., Zhang M., Mäkilä E., Kemell M., Salonen J., Hirvonen J.T., Zhang H., Zhou D., Deng X., Santos H.A. Multifunctional nanohybrid based on porous silicon nanoparticles, gold nanoparticles, and acetalated dextran for liver regeneration and acute liver failure theranostics. Adv. Mater. 2018;30 doi: 10.1002/adma.201703393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.