Abstract
Responsible for more than 4.9 million deaths so far, COVID-19, caused by SARS-CoV-2, is instigating devastating effects on the global health care system whose impacts could be longer for the years to come. Acquiring a comprehensive knowledge of host-virus interaction is critical for designing effective vaccines and/or drugs. Understanding the evolution of the virus and the impact of genetic variability on host immune evasion and vaccine efficacy is helpful to design novel strategies to minimize the effects of the emerging variants of concern (VOC). Most vaccines under development and/or in current use target the spike protein owning to its unique function of host receptor binding, relatively conserved nature, potent immunogenicity in inducing neutralizing antibodies, and being a good target of T cell responses. However, emerging SARS-CoV-2 strains are exhibiting variability on the spike protein which could affect the efficacy of vaccines and antibody-based therapies in addition to enhancing viral immune evasion mechanisms. Currently, the degree to which mutations on the spike protein affect immunity and vaccination, and the ability of the current vaccines to confer protection against the emerging variants attracts much attention. This review discusses the implications of SARS-CoV-2 spike protein mutations on immune evasion and vaccine-induced immunity and forward directions which could contribute to future studies focusing on designing effective vaccines and/or immunotherapies to consider viral evolution. Combining vaccines derived from different regions of the spike protein that boost both the humoral and cellular wings of adaptive immunity could be the best options to cope with the emerging VOC.
Keywords: Spike protein, RBD, Mutation, Immunity, Vaccine, SARS-CoV-2
1. Introduction
According to the worldometer data (https://www.worldometers.info/coronavirus/), COVID-19 cases surpass 244 million with more than 4.9 million deaths reported as of October 24, 2021. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for COVID-19, is the seventh coronavirus (CoV) identified infecting humans and causing the most lethal cases so far [[1], [2], [3]]. Although some vaccines are currently under clinical use, their effectiveness on emerging VOC could be compromised. Besides, several neutralizing antibodies are under development or approved as treatment options; however, their clinical efficacy especially in severely ill patients and new viral strains is controversial. The current primary COVID-19 treatment relies on symptomatic and oxygen therapy to manage respiratory impairment [[4], [5], [6], [7]].
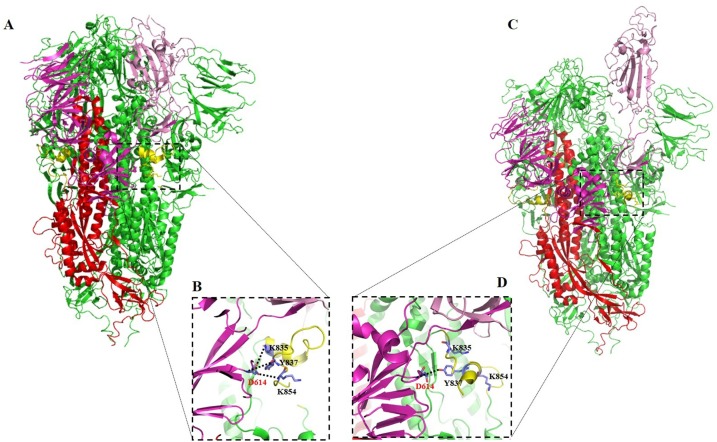
Understanding the biology of SARS-CoV-2 is critical in developing effective drugs, vaccines, and immunotherapies. As a member of the Betacoronaviridae family, SARS-CoV-2 is an enveloped virus with a large positive-sense RNA genome (about 30 kb). SARS-CoV-2 has a similar entry receptor, the human angiotensin-converting enzyme 2 (hACE2), with SARS-CoV except for differences in some amino acid residues [5,[8], [9], [10], [11], [12]]. Additionally, a recent study showed that tyrosine-protein kinase receptor UFO (AXL) is a candidate receptor for the spike protein of SARS-CoV-2 [13]. Despite its large size, the SARS-CoV-2 genome encodes for only a few proteins, including 4 structural proteins (spike (S), nucleoprotein (N), envelope (E), and membrane (M)) and 16–17 nonstructural proteins (nsp) [14,15]. The entry of CoVs including SARS-CoV-2 is mediated by the envelope anchored spike (S) protein [16,17] which, in most viruses, is cleaved into S1 and S2 subunits by viral proteases. The S1 subunit recognizes receptors while the S2 subunit is important for fusion [18]. A portion of the spike protein at the S1 subunit, the receptor-binding domain (RBD) encompassing a core structure and receptor binding motif (RBM), recognizes and binds to ACE2 during the entry process of CoVs [5,[19], [20], [21]]. When CoVs spike protein binds to ACE2 receptor, it is activated by transmembrane (TM) protease serine 2 (TMPRSS2) promoting virus entry [22]. Fig. 1 describes the overall trimeric structure of the SARS-CoV-2 S protein and its subunits.
Fig. 1.
Structure of trimeric SARS-CoV-2 spike protein (PDB entry: 7JWY) [274]. The SARS-CoV-2 S1 subunit (with S1 core domain in magenta and RBD in light purple) and S2 subunit (with S2 core domain in red and fusion peptide fragment in yellow) are presented. (A-B) Pre-fusion stabilized closed conformation of Spike trimer (7JWY), with a close-up view of the interaction between G614 from one SD2 Spike monomer and K835, Y837, and K854 from the neighbor FP fragment (yellow). (C-D) Partial open conformation of Spike trimer (6XM4), highlighting the loss of the binding lead by conformational changed, which allows the open conformation and favors the binding with ligand (ACE2). D614 G is involved in the stabilization of FP and the tight pre-fusion closed conformation of SARS-Co-2 S protein. Any mutation of D614 destabilizes the intradomain interaction, favors an open conformation, and increase the viral infectivity.
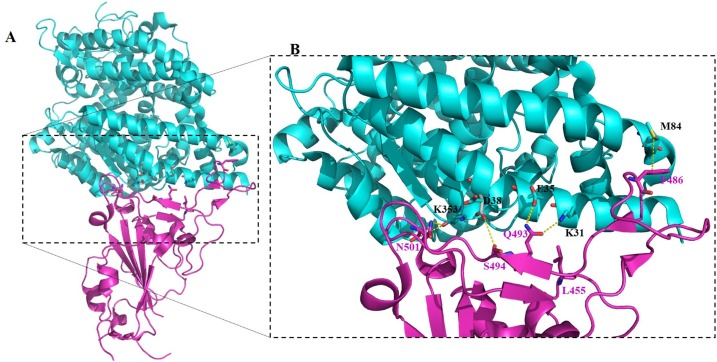
Encoded by a 1273 long amino acids sequence, SARS-CoV-2 S protein measures about 180−200 kDa. The spike protein undergoes structural rearrangement upon interacting with host receptors facilitating viral fusion. Residues 14-685 constitute the S1 subunit that carries the receptor-binding domain (RBD) or C-terminal domain (CTD) (residues 319-541) [[23], [24], [25]]. In addition to the RBD, the SARS-CoV-2 S1 subunit contains an N-terminal domain (NTD) (residues 14-305) [1,8,26,27]. The known function of the NTD includes allosteric regulation of spike protein conformation and some of the mutations on the NTD resulting in increased cell entry that are correlated with the greater presentation of RBDs to ACE2 receptors [28]. Wang et al. [1] identified SARS-CoV-2 CTD as a key region interacting with hACE2 indicating an overall similarity in receptor recognition between SARS-CoV and SARS-CoV-2. SARS-CoV-2 RBD-hACE2 binding is illustrated in Fig. 2 .
Fig. 2.
Complex structure of SARS-CoV-2 RBD with human ACE2 (PDB entry: 6VW1) [111]. (A) Structure showing the binding of RBD (magentas) with human ACE2 (cyan) and (B) the binding interface of the RBD (magentas) and human ACE2 (cyan). The key residues at the binding interface between the RBD (red highlighted N501, Q493, S494, L455, and F486) and human ACE2 (blue highlighted K353, D38, E35, K31, and M82) are presented by red highlighted sticks.
The S2 subunit, composed of the fusion protein (FP), heptapeptide repeat sequences 1 and 2 (HR1 and HR2), transmembrane domain (TM), and the cytoplasmic domain (CT), is responsible for viral fusion and entry. The FP is conserved and relatively short composed of 15–20 main hydrophobic residues used to anchor to the target receptor when the S protein assumes the pre-hairpin conformation. This protein facilitates virus entry by disrupting the lipid bilayers of the target membrane. HR1 and HR2, composed of repetitive heptapeptides, are crucial for the fusion and entry of the S2 subunit through forming six helical bundle structures. HR1 is located in the C-terminus of the FP while HR2 is located at the N-terminus of the TM domain. Structurally, the TM domain anchors the S protein to the viral M protein and the S2 subunit at the tail of the CT [[29], [30], [31], [32], [33], [34]].
Studies reported that SARS-CoV-2 RBD has a higher affinity for the ACE2 receptor than the RBD of SARS-CoV has due to key amino acid differences [1,8,26,27]. Besides its conserved nature, the S protein is crucial in the life cycle of CoVs as it contributes to receptor recognition, viral attachment, and entry [9,[25], [26], [27],[35], [36], [37]]. Other studies showed that S protein is involved in immune evasion [38] making it the primary target for vaccine and therapeutic studies. Several vaccines are under clinical trial and/or being delivered to the targeted community. Vaccination is aimed to boost the immune response of individuals similar to natural infection. However, SARS-CoV-2 mutations at key residues especially in the S protein evade the immune responses and thus reduce the immunogenicity and efficacy of the vaccines posing a significant challenge to the prevention and control strategies [39]. This review summarizes the impact of SARS-CoV-2 spike protein mutations on immune evasion and vaccine-induced immunity which could have a potential contribution for future studies focusing on vaccine design and immunotherapy.
2. Immune response to SARS-CoV-2 infection
2.1. Innate immunity
The innate immune system is an important first-line defense during infection. Innate immunity produces pro-inflammatory cytokines which can inhibit viral replication, stimulate the adaptive immune system and recruit immune cells to the site of infection. Granulocytes, monocytes, macrophages, and dendritic cells contribute to the fighting against infections through releasing toxins, enzymes and presenting antigens to T cells. Also, natural killer (NK) cells contribute to immunity through killing virus-infected cells, inducing apoptosis and antibody-dependent cell-mediated cytotoxicity (ADCC) [[40], [41], [42], [43], [44]]. Besides, interferons and the complement system (through recruiting immune cells, activating cells, and killing pathogens) have crucial roles in preventing viral infection and propagation [45]. Lung epithelial cells also produce inflammatory cytokines to combat SARS-CoV-2 infection [46,47]. Provided, SARS-CoV-2 infection is known to evade innate immunity by inducing a cytokine storm, impairing interferon responses, and suppressing antigen presentation on both MHC class I and class II cells [48].
Innate immunity plays a crucial role in fighting against SARS-CoV-2 infection. The presence of unique gene signature and variability of the innate immune response among individuals is the cause of disease heterogeneity observed among COVID-19 cases [49,50]. A combination of innate immune cells (granulocytes, monocytes, and macrophages) and adaptive immune cells (B cells and T cells) are important to tackle the pandemic [51]. Surprisingly, there is no evidence whether innate immunity has role in preventing SARS-CoV-2 infection in the retina and the brain [52,53]. Findings reported that in vitro exposure of SARS-CoV-2 and its spike protein results in platelet aggregation, up-regulation of activation markers, the release of coagulation factors, and secretion of inflammatory cytokines [[54], [55], [56]] indicating innate immunity-based complications in severe COVID-19 cases. During viral entry, pattern recognition receptors (PRR) including toll-like receptor 3 (TLR3) and TLR7 are assumed to be involved in preventing SARS-CoV-2 infection as proved in other CoVs [57]. In addition, viral RNA is suggested to be recognized by cytosolic sensors like retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) [58].
The other innate immune component involved in viral defense is the interferon system. Infection with SARS-CoV-2 induces unique pro-inflammatory cytokines such as IL-1β, IL-6, and TNF, as well as many chemokines (CCL20, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL16) [46]. However, some interferons like IFN-β were not detected regardless of disease severity [59]. Induction of cytokine production results in cytokine storm in severe disease due to failure of the immune system to resolve the inflammation [60]. Innate immune cells have been reported to be affected by COVID-19. Reduction of NK cells was observed in severe COVID-19 cases [61] and recovery from COVID-19 was associated with an increased level of cytotoxicity, DNA replication, and decreased inhibition of NF-κB in NK cells [62]. This reveals the role of NK cells in preventing SARS-CoV-2 infection as an increased level of NK cells was observed in COVID-19 convalescent patients [63,64]. Elevation of eosinophils [65], reduction of basophils [66], and dysregulation of monocytes and dendritic cells [67,68] were observed in different disease stages of COVID-19 and recovered patients.
According to Huang et al. [69], compared to healthy controls, both severe and mild COVID-19 patients showed elevated plasma concentration of cytokines and chemokines (IL-1β, IL-1Rα, IL-7, IL-8, IL-9, IL-10, basic FGF, GCSF, GM-CSF, IFN-ɣ, CXCL10, CCL2, CCL3, CCL4, PDGF, TNFα and VEGF). Similarly, severe COVID-19 patients showed elevated levels of proinflammatory cytokines compared to non-COVID-19 respiratory tract patients [46]. Although there is no evidence supporting whether immune cells are targets of SARS-CoV-2 infection [70], both adaptive and innate immunity are the gears leading to the way out of the pandemic. Besides, mucosal antibodies to SARS-CoV-2 antigens have been shown [71] indicating the potential role of mucosal immunity and/or mucosal vaccination in disease prevention. The spike protein of SARS-CoV-2 is recognized by multiple innate immune receptors including the mannose receptors MR/CD206, DC-SIGN/CD209, L-SIGN/CD209L, and MGL/CLEC10A/CD301 [72] indicating the important role of spike induced innate immune response in controlling SARS-CoV-2 infection. Since innate immunity is triggered by infections with a complex heterogeneous response, specific innate immune response to SARS-CoV-2 infection is not well-established [49] requiring future scrutiny.
2.2. Adaptive immunity
In the meantime, adaptive immunity is also important for controlling viral infections. It is suggested that the upper respiratory tract is protected by secretory IgA while IgG guards the lower respiratory tract [73,74]. Studies reported that spike-specific B cell subsets were also identified [75,76] and SARS-CoV-2 specific T-cell immune memory was found in convalescent COVID-19 patients [77] suggesting the role of adaptive immunity in combating SARS-CoV-2 infection. Emerging pieces of evidence reveal that neutralizing antibodies, CD4+ and CD8+ T cells are contributing to the control of SARS-CoV-2 infection in both hospitalized and non-hospitalized COVID-19 patients [51,[78], [79], [80]]. A longitudinal study reported increasing titers of neutralizing anti-spike IgG antibodies exhibiting a positive correlation with disease severity in COVID-19 patients [81]. Our study also revealed serum RBD-specific IgA levels were positively correlated with COVID-19 disease severity [82]. The role of CD4+ T cells is prominent than CD8+ T cells [78,83] in responding to SARS-CoV-2 infection through control of primary infection [84]. It is also reported that CD8+ T cells result in better COVID-19 outcomes; however, their consistent expression was limited compared to CD4+ T cells [78,[83], [84], [85]]. On the contrary, decreased percentage of lymphocytes and decrement in CD4+ and CD8+ T cells count was observed in COVID-19 patients [86] revealing the immune evasion ability of the virus. A study reported that rapid induction of humoral responses were associated with disease severity while early induction of IFN-ɣ secreting SARS-CoV-2-specific T cells was present in patients with a mild disease which accelerated viral clearance [87].
SARS-CoV-2 spike protein specifically the RBD region is a critical target for neutralizing antibodies and vaccine design. Above 90 % of neutralizing antibodies target the RBD of the S protein [[88], [89], [90], [91], [92], [93], [94], [95], [96]] and most vaccine discoveries for COVID-19 are particularly interested in the spike protein. T cell responses, besides antibody responses, to the spike protein, are believed to be important [79] and SARS-CoV-2 S protein is a strong target for CD4+ T cells [78]. In addition, the induction of anti-spike protein antibodies is dependent on spike-specific CD4+ T cells [97,98]. It is noted that most residues in the RBM of the SARS-CoV-2 RBD that directly interact with the ACE2 receptor are conserved in CoVs [99]; however, differences in the antigenicity of SARS-CoV and SARS-CoV-2 S protein are observed. This is evidenced by the failure of some murine SARS-CoV monoclonal and polyclonal antibodies to bind on the S protein of SARS-CoV-2 [1,25,100] indicating the necessity of updating previously designed SARS-CoV RBD based vaccines.
It is noted that the majority of neutralizing antibodies target the RBD and thus halt virus entry through disrupting ligand-receptor engagement [101,102]. However, despite structural similarities between the two proteins, antibodies specific for SARS-CoV RBD were unable to bind on SARS-CoV-2 RBD [1] entailing differences in immunogenicity secondary to differences in key residues. Although antibodies that potentially neutralize SARS-CoV-2 infection through inhibiting the binding of RBD to ACE2 are discovered, these antibodies are found to be largely virus-species-specific inhibitors [103]. The emergence of potent neutralizing antibodies targeting multiple epitopes on the different regions of SARS-CoV-2 S protein has been reported [99,104] making the vaccine discovery efforts promising. Interestingly, cross-reactive anti-spike antibodies and memory B cells are rare [87,92,[105], [106], [107]]. However, studies showed the presence of significant cross-reactive T cells [78,108,109] with a majority of CD4+ T cells being involved while limited involvement of CD8+ T cells was observed [78].
In a study by Yang et al. [41], immunization of mice, rabbit, and other non-human primates with recombinant SARS-CoV-2 RBD induced a dose-dependent antibody (IgG and IgM) production within 1–2 weeks where adjuvants enhanced its immunogenicity. Sera from the immunized animals blocked the binding of RBD to ACE2 with subsequent inhibition of infection. Here, CD4+ T cells were found to contribute to the induction of antibody responses. Similarly, superior diagnostic test accuracy was achieved by antibodies (IgG/IgM/IgA) produced against the RBD [110] making RBD the optimal antigen important for more efficient diagnostic, therapeutic, and vaccine formulations. Studies recommended that neutralizing monoclonal antibodies (mAbs) against the RBM of the RBD could be promising antiviral drugs in preventing viral attachment [111].
Adaptive immune responses to mild and severe SARS-CoV-2 infections are reported to be different and heterogeneous [112]. Zhang et al. [113] reported a positive correlation between disease severity and vigorous adaptive immune response (both humoral and T cell responses) provided that the response of three major clusters of memory T cells (CD8+ TEM, CD8+ TTE, and CD4+ TTE) appear to be independent of disease severity. Monoclonal antibodies targeting the RBD-ACE2 interaction are important to block viral attachment. Accordingly, C8 IgG mAb neutralized SARS-CoV-2 pseudovirus infection in ACE2 expressing 293 cells in vitro [114]. In addition, linear epitopes of SARS-CoV-2 spike protein showed strong induction of neutralizing antibodies in COVID-19 patients [115].
2.3. Vaccine induced-immunity
The human immune system mostly launches an immunological response to a particular pathogen when a natural infection occurs. This immunological response protects the body from the associated diseases, and ideally, forever. Antibodies and cytotoxic cells combat the disease-related pathogen while memory cells are formed and persist for a long time then become active when the same type of antigenic material enters the body on a later occasion. The principle of immunization emanates from this fact [116]. Since the discovery of the first vaccine, Variolae vaccinae, for smallpox by Edward Jenner in 1798 [117], vaccines have long been used in the prevention of many infectious diseases and have achieved great success. Vaccines activate an antigen-specific adaptive immune response by initiating the innate immune response. They induce cell-mediated immunity by activating highly specific subsets of T lymphocytes and humoral immunity by stimulating B lymphocytes to produce specific antibodies. The adaptive immune system then establishes immunological memory following the elimination of the pathogen [118,119].
Most COVID-19 vaccines induce disease-preventing/attenuating immunity than disease sterilizing immunity [79]. The development and application of vaccines is quite a complex, lengthy and tedious process. Moreover, it needs assessment of vaccine efficacy, safety, and use with or without major side effects. Spike protein and specifically RBD-based vaccine designing is so far the commonest strategy to combat the COVID-19 pandemic [120]. After three months of the emergence of COVID-19, a clinical trial of the first vaccine candidate (NCT04283461) was started in March 2020. Currently, different types of COVID-19 vaccines are under development or in clinical use. Among these, inactivated virus vaccines (CoronaVac) [121,122], live attenuated vaccines like a vaccine under development by Codagenix and Serum Institute of India, recombinant protein vaccines like RBD and spike protein-based vaccines, vector-based vaccines, inactivated vector virus vaccines, DNA and RNA vaccines [123] are amongst the current COVID-19 vaccines. Several vaccines including Sinovac (inactivated virion) [121], Adenovirus vector vaccines (AstraZeneca [124], Cansino, and Johnson & Johnson), Moderna (mRNA vaccine) [125], Novavax (recombinant spike protein vaccine) [126], and Pfizer (mRNA expressing trimeric RBD vaccine) [127,128] are licensed; many of which are in clinical use. All these vaccines have their pros and cons, especially regarding their safety and ability to prevent VOC. Besides, some of these vaccines are being developed by new companies who lack experience in vaccine production, thus making the application of anticipated vaccines delayed [79].
Several COVID-19 vaccines have been evaluated in a macaque model to verify whether they pass clinical trials. Sinovac exhibited protection of the lower respiratory tract by inducing low to moderate antibody titers [121]. ADV26 vaccine (a vector-based vaccine produced by Janssen) showed sterilizing immunity without significant induction of antibody titers [129]. An mRNA vaccine, Moderna, elicited CD4+ and follicular helper T cell responses in addition to boosting antibody production [125]. A recombinant spike protein-based vaccine, Novavax, increased antibody titers and prevent infection both in the lower and upper respiratory tract [130]. AstraZeneca (produced by Oxford) is a replication-incompetent adenovirus vector vaccine encoding the full length of SARS-CoV-2 spike protein. In a study, this vaccine boosted both humoral and cellular immunity with some side effects [94]. Pfizer vaccine elicits short-term neutralizing antibodies better than natural SARS-CoV-2 infection in phase 1 clinical trial [131] and this RBD-based vaccine boosted CD4+ and CD8+ T cell responses where Th1 CD4+ T cells were mostly expressed [132]. Most COVID-19 candidate vaccines showed comparable immunogenicity, specifically, mRNA-based vaccines exhibit similar (or even better) neutralizing antibody titers and/or CD4+ T counts [[94], [95], [96]] as the natural infection.
The different forms of vaccines have their mechanisms of eliciting immunity with corresponding advantages and drawbacks. It is noted that recombinant virus vector-based vaccines work similarly to natural endogenous pathogens where a processed antigen is presented to CD8+ T cells with subsequent activation of cytotoxic T lymphocytes (CTL) [133]. Whereas, pathogenicity deficient adenoviral-based vector vaccines result in exponential induction of CD8+ T cells and antibody production [134]. DNA vaccines elicit both humoral and cellular immunity through activating both CD4+ and CD8+ T cells. Although the exact mechanism of action of DNA vaccines is not well established, signaling pathways including STING/TBK1/IRF3 and the AIM2 inflammasome play crucial roles [[135], [136], [137]]. Subunit and inactivated vaccines are stable and non-pathogenic, but they elicit short memory immunity and require booster doses. Live attenuated vaccines elicit strong immunity similar to natural infection; however, they are inconvenient for immunocompromised patients and may be reverted to pathogenic form. Although recombinant viruses induce strong immunity, pre-existing immunity can affect their efficiency. On the other hand, RNA vaccines can be easily modified and are considered to be safe; though, their instability and low immunogenicity could result in short immune memory [39,138]. In addition, their instability makes them unsuitable in resource-limited settings where there is a lack of adequate storage facilities.
3. Spike protein mutations: Impacts on immune evasion and vaccine-induced immunity
RNA viruses have higher mutation rates than DNA viruses [139,140] where SARS-CoV-2 is not an exception. Amino acid changes, especially on the surface proteins of viruses, affect viral pathogenicity, immune response, and thus vaccine efficacy as observed in Chikungunya, Ebola, and Influenza viruses resulting in increased transmissibility, infectivity, and resistance to neutralizing antibodies [[141], [142], [143], [144], [145]]. There are continuous genetic variations of the SARS-CoV-2 genome and mutations in the S protein are unceasingly reported [[146], [147], [148], [149], [150]]. Unfortunately, studies reported that 65 % of viral C > T mutations are imposed by host immune responses [151] which could result in attenuation of the virus owing to reduced immunogenicity [152]. The SARS-CoV-2 genome alterations are estimated to be 1–2 mutations every month [153] and are highly glycosylated [95]. Evolutionarily essential mutations have also been reported in SARS-CoV-2 genes encoding for immunologically important proteins [154].
Emerging SARS-CoV-2 variants, importantly the VOC, are demonstrating reduced sensitivity to convalescent sera and monoclonal antibodies [155] posing a significant concern on the ongoing prevention strategies [156]. Reports showed that a reduced anti-RBD humoral response is associated with viral persistence and shading in the gastrointestinal tract [157]. Surprisingly, earlier studies reported more than 329 naturally occurring S protein variants deposited in the public domain [158] where these mutations are known to affect the glycosylation of viral proteins resulting in differences in virulence and host-pathogen interaction [[159], [160], [161]]. More importantly, the receptor binding motif (RBM) of the RBD is a primary target for neutralizing antibodies on one hand and is a highly variable region of the spike protein on the other hand [89]. Evolvability of SARS-CoV-2 S protein causes a significant alert on possible evasion of immunity and vaccinations [162]. Overall, these reports warn the scientific world to cautiously design and/or update vaccines and therapeutics considering the impact of prospective genetic variations on the transmissibility, infectivity, pathogenicity, immune evasion, and antigenicity of SARS-CoV-2.
SARS-CoV-2 has different spike protein variants categorized based on their spreading ability, disease severity, immunity, and treatment response. As of 21 October 2021, the European Centre for Disease Prevention and Control (ECDPC) classified three variants as VOC, variants of interest (VOI), and variants under monitoring. Accordingly, VOC includes Beta or B.1.351 (K417N, E484K, N501Y, D614G, A701V)), Gamma or P.1 (K417T, E484K, N501Y, D614G, H655Y), and Delta or B.1.617.2 (L452R, T478K, D614G, P681R). VOI encompass Mu or B.1.621 (R346K, E484K, N501Y, D614G, and P681H) and Lambda or C.37 (L452Q, F490S, and D614G) whereas variants under monitoring include various spike protein mutations circulating in different parts of the world.
3.1. Evasion of adaptive immunity
3.1.1. Mutations compromise humoral responses
The adaptive immune response is of utmost importance to get rid of the virus, as it is specifically well known that antibodies are effective in preventing severe complications from SARS-CoV2 infection. However, they effectiveness are reduced by mutations affecting spike protein [163], and currently circulating SARS-CoV-2 S variants are reported to successfully evade the humoral immune response. Although SARS-CoV-2 VOCs get more attention to study, emerging variants from patients under convalescent plasma (CP) therapy are also escaping neutralization to existing CP neutralizing antibodies [164]. In a study, sera from RBD nanoparticle vaccinated rhesus macaques showed moderate to low efficiency for different variants. The sera efficiently neutralized B.1.1.7 pseudotyped virus but exhibited weak to no inhibition effect against 501Y.V2 variant pseudotyped virus [165].
Numerous studies identified and reported SARS-CoV-2 spike mutations that are involved and compromise humoral response [120,163,165]. Specifically, these studies revealed that the most mutations on SARS-CoV-2 spike protein occur within RBD fragment, especially in the – RBM (S438 to Q506 and K417) – residues involved in ACE2 binding (Fig. 2) [103,[166], [167], [168]]. These results suggest that the RBD is the substantial immune-dominance region of SARS-CoV-2 spike protein. Among all those reported, the common RBD mutations of concern (MOCs) (present in the common VOCs) which compromise immune response against antibodies, identified in FDA Emergency Use Authorization (EUA) Fact sheets, include P337H/L/R/T, E340A/K/G, K417E/N, D420N, N439K, K444Q, V445A, N450D, L452R, Y453F, L455F, N460K/S/T, V483A, E484K/Q/P/D, F486V, F490S, Q493K/R, S494P, and N501Y/T (https://www.fda.gov/media/149534/download).
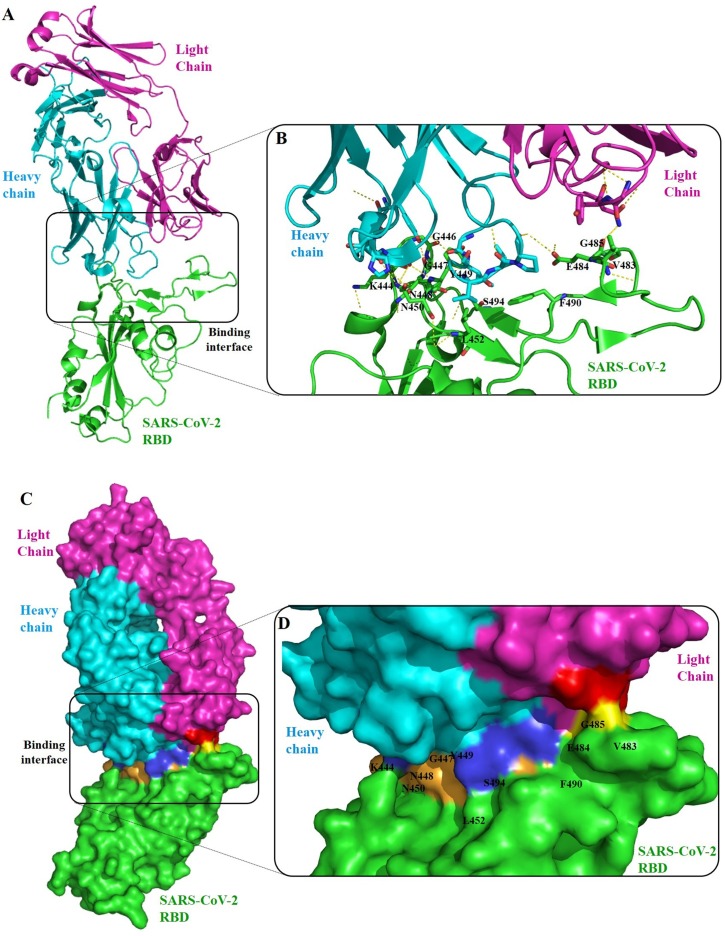
Mutations E484GK/Q/P, present in variants B.1.617.1 and B.1.1.7 (the highest contagious variants) have been reported to be the most important mutations in reducing antibody binding and neutralization titer of polyclonal serum antibodies by more than 10 times compare with other mutations [169,170]. E484K for instance, has the ability to escape the known high neutralizing antibodies, including the two antibody cocktails formed by mAbs C121 and C144, and REGN10989 and REGN10934, respectively [171]. Moreover, in cell culture, the B.I.617 (which carries mutation E484Q) showed enhanced entry and pathogenesis, and in treatment showed resistance to neutralization by bamlanivimab (an antibody used in COVID-19 treatment), COVID-19 CPs, and plasma from BNT162b2 vaccinated individuals indicating enhanced evasion of humoral immunity induced by both infection and vaccination [172]. Interestingly, at the molecular level, it has been shown that E484 forms attraction with most antibodies, while mutations of this residue has repulsion from most antibodies (demonstrated for the K484), explaining the low binding affinity of E484 mutants to most antibodies [173]. A typical antibody-RBD complex structure is illustrated in Fig. 3 . In the represented structure, residue E484 is highly engaged in the complex formation of RBD with P2B-2F6 antibody. A mutation of this residue abolishes the binding and destabilizes the complex, favoring the virus immune escape.
Fig. 3.
Cartoon (upper) and surface (lower) representation of a structure of a typical antibody (P2B-2F6 mAb) bound onto the RBD of SARS-CoV-2(PDB entry: 7BWJ) [275]. (A) Overall structure of the antibody (light chain (magentas) and heavy chain (cyan)) binding with RBD (green). (B) The epitope residues are all in the RBD receptor-binding motif, including residues K444, G446, G447, N448, Y449, N450, L452, V483, E484, G485, F490 and S494. P2B-2F6 attachment uses hydrophobic interactions around RBD residues Y449, L452 and F490 and hydrophilic interactions at the interface. RBD residues at the binding interface engaged in the complex are colored in yellow (bind with light chain residues in red) and in bright orange (bind with heavy chain residues in dark blue) [276]. Mutations on these epitopes are reported to affect the antigenicity and susceptibility of RBD to neutralizing antibodies. Specially A475 V and F490 L substitution mutations resulted in increased resistance to mAbs and COVID-19 convalescent sera [95].
Another crucial mutation that compromises humoral response with limited neutralizing activity includes N439K [174], the second commonest RBD and the sixth commonest S protein mutation. N439K mutation is reported to favor spike flexibility allowing an open conformation where a free RBD can bind to ACE2 and therefore enhancing infectivity and escaping humoral immune response [174,175]. Robertson & Snell's group reported that the N439K variant-associated increased resistance to neutralizing mAbs and COVID-19 sera, is not followed by any effect on viral replication, indicating RBM-based measures to be taken during vaccine and drug formulation.
Mutations in residues K444, G446, L452, and F490 have also been associated with immune escape of serum polyclonal antibodies [170], and mutations K444Q, V445A, and N450D have also been reported with high immune escape ability. Allison et al. [169] demonstrated that mutations in the fragment spanning residues 443–450 (also targeted by REGN10989 and REGN10934 cocktail [171]) strongly reduced neutralization effect of convalescent plasma, besides mutations in F456 and E484 which have the same immune escape molecular mechanism.
In their investigation, Li et al. [176] reported that mutations A475KV and F490L confer resistance to neutralizing polyclonal antibodies, monoclonal antibodies, and CPs. Interestingly, changing A475 by V475 leads to formation of weaker hydrogen bonds and hydrophobic interactions between RBD and neutralizing antibodies, and changing F490 by L490 leads to the disturbance of the hydrophobic interaction of molecules involved in antibody-RBD integration [95].
One of the famous non-RBM S protein mutants is the D614G which has a central role in viral infectivity. As presented in Fig. 1A & B, D614 of SD2 fragment is involved in the stabilization of the close state conformation of SARS-CoV-2, by engaging four hydrogen bonds/salt bridges with residues K835, Y837, and K854 of fusion peptide (FP) from the neighbor S promotor. So, mutations of D614 abolished SD2-FP interaction, which may lead to the destabilization of the close conformation, and favoring the change to open conformation which exposes RBD for engaging interaction with ACE2, strengthening affinity and enhancing the infectivity (Fig. 1C & D) [177].
These previous show the negative effect of single or point mutations in RBD on humoral immunity. When multiple mutations occur in RBD, the humoral immune response in highly compromised, facilitating the immune escape. Multiple substitutions occurring simultaneously at the same time, such as P384A, K417V, and L452K are highly resistant to neutralizing monoclonal antibodies COVA1-16, COVA2-04, and COVA2-29 which respectively recognize different binding/neutralizing sites without affecting each other. Similarly, using a computational approach, Shah and colleagues [178] demonstrated that the V417K mutation enhanced RBD affinity to ACE2, associated with high infectivity, while that P475A and G482 result in resistance to neutralization by mAbs. These multiple mutations simultaneously increase RBD-ACE2 affinity and resist to antibodies, which can greatly hinder the immunogenicity of most vaccines [179]. This multiple substitutions on spike are observed in the VOCs, including B.1.617.1 and B.1.1.7, known to have high infectivity and ability to resist to pre-existing antibodies, CPs, or vaccine-induced immunity.
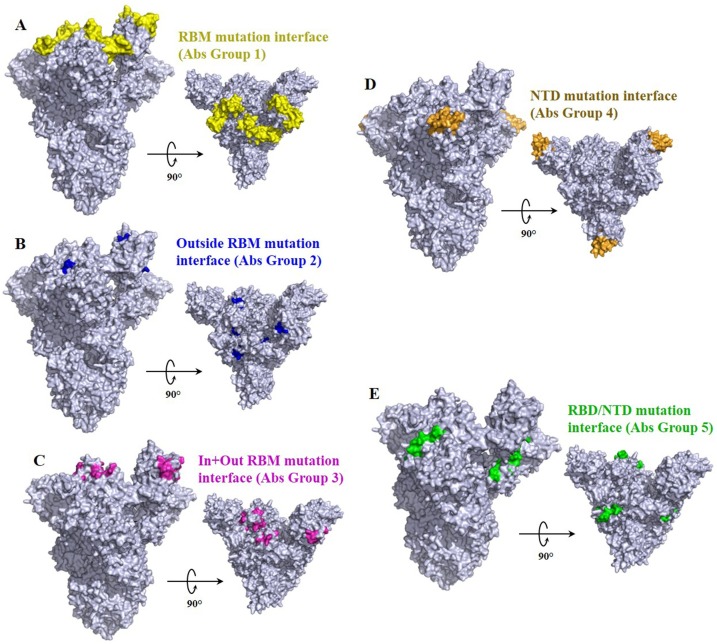
Besides RBD mutations, mutations occurring in NTD are also been associated with immune escape. And most of the evidence of immune escape associated with mutations in NTD have been centered in loop N3 (140-156) and loop N5 (246-260), which overlap epitope of antibody 4A8 [99] (Fig. 4 ). Deletion in the two following segment 141–144 and 146, and Δ243–244 abolished binding with 4A8 [180].
Fig. 4.
Representation of the antibody binding interface susceptible for mutations compromising immune response (modified from PDB: 7DK3) with one RBD (RBD Chain C) in ‘’open’’ conformation [177]. RBD interface engaged in the complex with common antibodies are colored regarding specific antibody groups described in [99].
Several studies have raised the fact that even though some SARS-CoV neutralizing mAbs recognize conserved epitopes between SARS-CoV RBD and SARS-CoV-2 RBD, they are unable to neutralize SARS-CoV-2 (reviewed in [99]). For instance, SARS-CoV neutralizing antibody CR3022 is unable to neutralize SARS-CoV-2, although CR3022 display high binding affinity with SARS-CoV-2 RBD [38]. Reciprocally, another study showed that anti-SARS-CoV-2-RBD Antibody, HA001, failed to recognize SARS-CoV RBD representing novel binding sites of neutralizing antibodies [181]. These studies revealed that there are key amino acid differences on the RBD epitopes of the two viruses (SARS-CoV and SARS-CoV-2) affecting the neutralizing ability of common mAbs. Furthermore, these studies also suggest that emerging SARS-CoV-2 RBD variants could affect the current vaccine and therapeutic candidates requiring continuous optimization. Overall, further studies are critically important to reconcile the inconsistencies observed across these studies.
Unlike previously explained, all mutations do not necessarily confer a local effect with direct resistance to antibodies, vaccine or convalescent plasma. For example, a computational analysis revealed that Y449 F and N501D result in reduced RBD affinity to ACE2, but most mutations are tolerated [105,182]. The K417N mutation also showed a decrease in binding affinity to ACE2, but this mutation evades neutralization by CB6 nAb through eliminating an interfacial salt bridge between the RBD and the antibody [183]. Mutation in 452 and 484 in RBDs are involved in evading the immune response, though they are not in direct contact with interfacial residues in ACE2. Overall, these mutations display an immune escape mechanism unknown. Amino acid substitutions causing significant changes in the structure and stability of epitopes in the RBD could be more significant as these mutations could result in reduced binding affinity and energy between antibodies and the target protein. Therefore, antibodies disrupting important epitopes in the RBD are crucial in eroding the binding of the RBD to the ACE2 receptor.
In summary, the mechanism of some mutations on RBD is unveiled, many of them resulted the reduction of effectiveness of preexisting neutralization antibodies, these mutation sites are presented in Fig. 4.
3.1.2. Mutations compromise cellular immunity
In addition to antibodies, CD4+ and CD8 + T cells are suggested to play critical roles in resolving SARS-CoV-2 infection and COVID-19 [184]. Studies claimed that the ability of SARS-CoV-2 variants to escape from T cell immunity is unlikely due to recognition of a broad array of epitopes by the adaptive immunity [78]. Intriguingly, CD4+ and CD8 + T cells recognize more than 10 epitopes in the SARS-CoV-2 genome [185]. In addition, it is reported that some SARS-CoV-2 positive CD8 + T cell epitopes are conserved in other coronaviruses suggesting the presence of pre-existing immunity against coronavirus in some people infected with other coronaviruses [186]. Although more studies are required, the D614 G mutation is suggested not to escape T cell responses as computational mapping studies showed this mutation is not included in T cell epitopes [187,188]. However, a study reported that two recently emerged RBD mutations (L452R in B.1.427/429 and Y453 F in B.1.298) can escape from the HLA-24-restricted cellular immunity where the authors suggested these mutants be future threats [189].
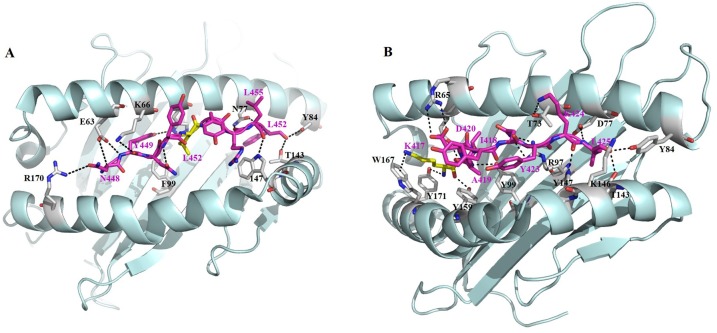
Reduced cellular immune responses to SARS-CoV-2 variants were also suggested in a CD8+ T cell epitope profiling study from COVID-19 convalescents [186]. Mechanistically, some mutations in MHC-I-restricted epitopes (in the S protein and other proteins) evade in vitro CD8+ T cell responses through abolishing MHC-I binding [190]. Indeed, our recent data showed that the K417 N mutation found in B.1.1.7, B.1.351. and P.1 variants and in the Y155- mutation found in B.1.1.7, B.1.351, B.1.525 variants significantly abolished the capability of the peptide to be loaded onto the relevant HLA-A class I molecules [186]. In the crystal structure of K417 containing epitope, KIADYNYKL derived from spike protein of SARA-CoV-2, in complexed with a dominant HLA-I subtype A*02:01 and a β2 m microglobulin, the positive-charged side chain amino group of the N-terminal lysine (K1) of this epitope adopts a Pi-cation interaction with the indole ring of W167 of HLA-A*02:01 (Fig. 5 A). This structure provides direct evidence that if mutation the K417 to any other residues, such interaction would be abolished. While from the complex structure of another spike epitope, NYNYLYRLF that harbors recent mutation sites of L452R or Y453 F in the B.1.1.7 and B.1.1.298 variants, with the HLA-A*24:02, we found hydrophobic side chain of L452 is buried in a pocket on the surface of HLA-A*24:02. The mutation to a hydrophilic large side chain may change the overall conformation of this HLA-peptide complex, thus change the corresponding TCR recognition and T-cell activation (Fig. 5B).
Fig. 5.
Structural basis of variant related SARS-CoV-2 T-cell epitope presentation by HLA-I. (A) The KIADYNYKL epitope was presented by a HLA-A*02:01 HLA (PDB 7KEU). (B) NYN epitope in complex with HLA-A*24:02 (PDB 7F4W).
These pieces of evidence are; therefore, providing insights that emerging SARS-CoV-2 variants are evading cellular immunity and some variants rapidly become dominating variants in certain countries and spread globally.
3.2. Evasion of vaccine-induced immunity
Vaccines are potential weapons in the fight against COVID-19 [191]. Several vaccines are under use to control the pandemic but the emergence of VOC poses a big concern on vaccines efficacy. In addition to mutations at the RBD, mutations at the NTD also exhibited reduced neutralization in vaccinated individuals [192]. Although Alpha variants caused no significant impact, Delta, Beta, Gamma, and Lambda variants raise a worry on the efficacy of subunit vaccines [39].
A meta-analysis study found that full vaccination, but not the partial one, with the current vaccines was effective (88.3 % efficacy) against Alpha, Beta/Gamma, and Delta variants with mRNA-based vaccines exhibiting higher efficacy [193]. This is supported by a progress study revealing the effectiveness of the current vaccines especially in preventing severe disease [194]. Sera from Sputnik V vaccinated people retained good neutralization activity against VOC including B.1.1.7, B.1.351, P.1, B.1.617.2, and B.1.617.3 [195]. In a phase 1 trial study, GRAd (gorilla adenovirus-based vaccines) induced Th1 cells in younger and older adults [196]. However, the results of studies reporting the effectiveness of vaccines against VOC are mixed requiring ongoing efforts.
The B.1.351 variant or the beta variant contains nine spike protein mutations and the D614G substitution (five mutations in the NTD, three mutations in the RBD, and one mutation near the furin cleavage site) [197] posing a growing concern that this variant could halt vaccines and mAb-based therapies [198,199]. The B.1.1.7 variant didn’t resist neutralization by sera from Moderna vaccinated individuals while the B.1.351 variant does [200]. Similarly, B.1.1.7 variant failed to escape from neutralization by Pfizer mRNA vaccine-elicited sera [201]. On the contrary, BNT162b2 vaccine-elicited antibody neutralization was modestly resisted by the D614 G variant [202] causing another threat to available vaccines. However, neutralization of the B.1.351 variant using the BNT162b2 vaccine was diminished by 7.6-fold, and both the B.1.1.7 and B.1.351 (with more resistance) variants showed strong resistance to neutralization by sera from AstraZeneca vaccinated individuals [203,204]. Also, the B.1.351 variant moderately affected the neutralization ability of sera from individuals vaccinated with inactivated BBIBP-CorV and ZF2001 RBD recombinant vaccines [155]. These results suggest that mRNA-based vaccines have better tolerance to genetic alterations compared to other types of vaccines.
SARS-CoV-2 spike 69/70 deletion, E484K, and N501Y variants exhibited a small impact on the neutralization activity of two doses of BNT162b2 vaccine-elicited sera [205]. AZD1222 vaccine was also effective against one of the VOC, B.1.1.7 in the United Kingdom [206] and emerging variants (Zeta, Gamma, and B.1.1.28) in Brazil [207]. But sera from mRNA-1273 vaccinated humans/non-human primates showed reduced neutralization against B.1.351 variant [200]. B.1.351 variant is known to be more resistant to convalescent plasma and sera from vaccinated people [208,209] and in macaques [210]. In addition, the B.1.617.2 Delta variant showed significant resistance to neutralization by BNT162b2 [211] and AZD1222 [212] vaccines induced sera. Of note, a single dose of BNT162b2 and AZD 1222 vaccines showed low efficacy against the Delta variant [213]. Comparatively, the B.1.617.2 Delta variant showed resistance to vaccines in mild infections while vaccine efficacy was sustainable in severe COVID-19 cases [214]. Therefore, important mutations in these VOC should be given extra attention and a full dose vaccination should be strictly implemented to prevent the emergence of new variants.
E484 K in VOC is regarded as an immune escape mutation since it is used to bypass the body’s immune system. This mutation is reported to require increased serum antibodies to prevent infection [215,216]. Also, this mutation evaded antibody neutralization elicited by infection or vaccination [217]. Novavax and Johnson & Johnson vaccines exhibited poor efficacy in clinical trials in South Africa possibly due to the high circulation of this variant in the country [218,219]. This variant also demonstrated enhanced resistance to vaccine-elicited antibodies and mAbs resulting in a threat to the efficacy of the Pfizer mRNA-based BNT162b2 vaccine [216] as this variant resulted in a 3.4-fold reduction of neutralization by sera from individuals who received two doses of the vaccine [220].
While there is no consensus on the optimal antigen sequence used in next generation vaccines to cope with VOCs, results from booster dose are exciting. Several new studies on some mRNA vaccines and inactivated vaccines both supported increased effectiveness against emerging Delta (B1617.2) variant after a booster dose [221]. As a result, several countries including the United States, China and UK are implanting the booster dose starting late 2020.
According to a modeling study [222], vaccines reduce infections but may not avoid a new wave. Although updating vaccines based on variants is recommended [223], the safety and efficacy of the prospective vaccines are under question. Overall, the currently available vaccines have good efficacy against the VOC including the Delta variant, specifically these vaccines significantly reduced severe disease [224]. But, worth-attention efficacy drop-off is observed among vaccines against the emerging VOC. Therefore, the production of multivalent vaccines, massive vaccine deployment to the community, and reactivating current vaccines are pivotal in addition to vaccine updating [225,226]. The impact of SARS-CoV-2 VOC on immunity is summarized in Table 1 .
Table 1.
Summary of the impact of SARS-CoV-2 VOC on immunity.
| Mutation/variant | Virus type | Control strain | Location | Impact on natural and vaccine-induced immunity |
|---|---|---|---|---|
| L452R and Y453F [189] | Pseudovirus | Parental S protein | RBD | Escape from the HLA-24-restricted cellular immunity |
| B.1.1.7, B.1.351, P.1, or B.1.617.2 [186] | Lentivirus (plasmid) | Among the variants | S protein | This resulted in diminished activation of T cells mediated by major HLA alleles from COVID-19 convalescents. Mutations at the K417, L452, and Y144 of the spike protein evade T cells via weakening epitope ligand interaction |
| 501Y.V2 [165] | Pseudovirus | Wuhan-Hu-1 | RBD | Resulted in weak neutralization by sera from pseudotyped virus RBD nanoparticle vaccinated macaques |
| P384A [277] | Pseudovirus | Wild type RBD (P384) | S protein CR3022 IgG epitope | This resulted in a reduced affinity of the antibody to the RBD. Increased affinity of SARS-CoV CR3022 to the RBD of SARS-CoV-2 after the substitution mutation on the epitope |
| N234Q, A475V, L452R, V483A, F490L [103,168] | Pseudovirus | Beta/Shenzhen/SZTH-003/2020 | Other regions of S protein and RBD | Resisted neutralizing antibodies from patient convalescent sera and rhesus monkeys. SARS-CoV-2 antibodies didn’t cross-react with RBD of SARS-Co-V |
| A475V and F490L [95] | Pseudovirus | Natural variants | RBD | Exhibited reduced sensitivity to monoclonal antibodies and altered sensitivity to convalescent COVID-19 sera |
| N439K [174] | Live virus | Wild type RBD (N439) | RBD | Exhibited increased resistance to neutralizing mAbs and COVID-19 sera |
| P384A | Pseudovirus | Wuhan reference strain | RBD | Resisted neutralization by a cluster III RBD specific mAb (COVA1-16). Showed poor neutralization by RBD-specific mAb (COVA2-04). Resisted neutralization by a cluster I RBD specific mAb (COVA2-29). These mutations resulted in reduced potency of serum neutralization |
| K417V | ||||
| L452K [179] | ||||
| E484K [215,216,220] | Infectious cDNA clone | Wuhan reference strain | RBD | Showed enhanced resistance to vaccine-elicited antibodies and monoclonal antibodies. It also showed resistance to Pfizer elicited antibody neutralization from individuals who received two doses of the vaccine. This substitution is considered as host defense escaping mutation |
| D614G [202] | Pseudovirus | Wild type S protein (D614) | S protein | Resisted Pfizer vaccine-elicited antibody neutralization by mouse, rhesus, and human sera. |
| B.1.351 variant [155,200,201,203,204] | Live virus | Wuhan reference strain | S protein | Showed reduced neutralization by sera from AstraZeneca, Moderna and Pfizer vaccinated individuals. It also moderately resisted neutralization by sera from individuals vaccinated with inactivated BBIBP-CorV and ZF2001 RBD recombinant vaccines |
| B.1.1.7 variant [203,204] | Live virus | Wuhan reference strain | S protein | Showed heightened resistance to neutralization by sera from AstraZeneca vaccinated individuals |
| P475A and G482 insertion on SARS-CoV-2 RBD [178] | n/a | SARS-Co-V RBD | RBD | Predicted to escape from recognition by SARS-CoV mAbs (m396, 80R, s230, and CR3014) and thus resist SARS-CoV mAb cross-neutralization |
| L452R and Y453F [278] | Pseudovirus | NF9 (parental) | RBM | Escaped HLA-A24-restricted cellular immunity (CD8 + T cells). L452R increases viral infectivity by enhancing spike stability, viral replication, and fusogenicity in Vero cells |
n/a: not applicable for the computational study.
4. Challenges and future opportunities
SARS-CoV-2 genetic alterations would be the most critical challenge to the prevention and control of the pandemic as mutations could hamper the ongoing efforts on therapeutics, diagnostic, and vaccination efforts. Genetic alterations in VOC help the virus evade immunity through different mechanisms of which include altered interaction with immune regulatory genes [227], losing epitopes [228], escaping T-cell killing and low affinity to neutralizing antibodies [229]. On the other hand, the circulating VOC showed enhanced transmissibility, infectivity, and immune evasion owing to structural rearrangements resulting in increased accessibility and binding affinity of the RBD to ACE2 [230]. Due to differences in key amino acids, the RBM of SARS-CoV-2 forms a larger binding interface and higher affinity to ACE2 than SARS-CoV RBM does [111] which is a lesson that future virus evolutions will affect immunity and vaccinations. Unfortunately, studies showed that the SARS-CoV-2 S protein has lower stability than the SARS-CoV S protein [231] and the S2 fusion subunit is more conserved than the S1 subunit [26]. Therefore, a combination of mAbs that can identify various epitopes is recommended to combat this problem [232].
The mutations in VOC (D614 G and E484 K) improve the stability of S protein in its open state [233] which in turn increases virulence [234]. For example, a mutant SARS-CoV-2 showed effective zoonotic transmission [235] and the famous D614 G spike protein variant demonstrated more efficient entry and receptor binding affinity compared to the wild type [236]. The D614 G mutation is reported to stabilize the structure of the S1/S2 complex through destabilizing the free S1 structure [237] providing insights into designing potent vaccines/antibodies targeting this mutation. A study by Zhang et al. [238] reported that the D614 G substitution stabilizes the S trimer which could have an impact on vaccines and immunity. Besides, the D614 G mutation is reported to increase the up state of the RBD and enhance S1/S2 junction protease cleavage [239]. Therefore, targeting multiple sites in the spike protein could be a promising strategy to cope with VOC as mutations in the NTD and other regions are suggested to affect the structure and thus the immunogenicity RBD epitopes [240]. In this regard, Cho et al. [241] designed bispecific antibodies which potently neutralize VOC (Alpha, Beta, Gamma, and Delta variants) and the wild-type virus. In addition, combining RBD and NTD neutralizing antibodies is suggested to be another means of fighting with VOC [[242], [243], [244]]. Compared to the wild-type virus, the currently circulating VOC have significantly altered NTD, but the presence of only local changes on the RBD brightens the hope of coping VOC with RBD-based vaccines and therapeutics [245].
Intriguingly, recombined antibodies showed potent neutralization of S protein variants [246]. This indicates the use of a combination of vaccines derived from different domains of the spike protein could be more effective. However, prediction of mutational pathways, mapping of immunogenic epitopes in the S protein, and structure-based analysis of Ab-epitope binding should be the primary concern to design potent vaccines and/or antibody-based therapies [170,243,247,248]. The application of small proteins like nanobodies could also be an interesting approach. In this regard, Jin’s group identified potent hetero-bivalent alpaca nanobodies readily blocking RBD-ACE2 interaction with high binding affinity and stability [249].
P.1 variant showed resistance to neutralization by several mAbs, but antibodies with epitopes outside the RBD can readily neutralize the virus. For example, P.1 from Brazil, B.1.351 from South Africa, and B.1.1.7 from the UK are neutralized by a modified mAb (222) providing evidence of coping genetic variations through modifying antibody epitopes [250]. Modification of the spike protein is important for designing better vaccines for COVID-19. Notably, deletion of polybasic cleavage sites, inclusion of stabilizing mutations, and inclusion of trimerization domains in producing recombinant spike-related vaccines greatly influence their antigenicity [79,96,[251], [252], [253], [254], [255]]. In this regard, Amanat et al. [253] modified the recombinant spike protein of SARS-CoV-2 and tested its immunogenicity in a mouse model transiently expressing hACE2. The authors found that the spike protein with two Prolines introduced and the polybasic cleavage site removed exhibited better immunogenicity with robust induction of anti-RBD antibodies. Moreover, recurrent mutations other than VOC are speculated to reduce disease severity [256]. This reveals that all mutations on the spike protein are not always in favor of the virus bearing hopes in tackling new variants of SARS-CoV-2.
Even though there are growing efforts in producing different forms of effective vaccines against SARS-CoV-2 infection, there are still challenges hindering these efforts. These challenges are the limitations and drawbacks of the different forms of COVID-19 vaccines [79]. While inactivated vaccines are good to boost immunity, it is difficult to produce them in large quantities. Live attenuated vaccines have safety concerns in addition to their time-consuming production process. Moreover, some proteins like the spike protein are hard to express making the production of recombinant vaccines limited. While RBD is highly immunogenic, the protein is small enough to be prone to antigenic drift. Whereas, vector-based vaccines might be neutralized by pre-existing immunity which requires utilization of animal strains or medically rare strains. DNA vaccines are poorly immunogenic, in contrast, RNA vaccines are highly immunogenic. However, RNA vaccines are not stable, requiring specialized storage facilities and they also induce poor mucosal immunity. Ultimately, virus evolution is a big challenge requiring continuous update of vaccines which is a resource, technical, and time demanding contrary to urgency. Moreover, predicting the pathways of SARS-CoV-2 mutations is critically challenging which also pause threats to the treatment and vaccination efforts. Systematic genetic surveillance and modeling approaches are important to predict the evolutionary routes of variants and identify their phenotypic effects [257]. Further, identifying a universal antigen to deal with mutations is a critical point of intervention. Therefore, designing vaccines and immunotherapy considering these drawbacks are ultimately needed to curb the ongoing COVID-19 pandemic.
5. Conclusion
Although it has a comparatively low mutation rate, SARS-CoV-2 is evolving antigenically to escape from neutralizing antibodies [258] and cellular immunity, thus requiring timely updating of vaccines and immunotherapy. Although their circulation is low, SARS-CoV-2 spike protein variants (in the RBD and NTD) escaped from neutralizing antibodies and convalescent sera [259]. Immune responses due to infection and/or vaccination could also result in mutants. Notably, three spike protein mutations were observed after growing SARS-CoV-2 in the presence of neutralizing convalescent plasma where the new variants developed resistance to neutralization [260]. Multiple mutations in the RBD and NTD are reported to resist neutralization [261]. Besides, chronic SARS-CoV-2 infection in immunocompromised individuals is suggested to increase the mutation rate [262]. A low frequency of Delta variant sublineages harboring new and wild mutations has been reported even in vaccinated individuals [263]. Therefore, continuous molecular surveillance and monitoring are necessary to direct the development and effective application of vaccines and immunotherapy [174,264] to tackle virus transmission [265].
A deep understanding of the immune evasion mechanisms of SARS-CoV-2 and host immune responses help design better vaccines and antibody-based therapies [266]. Unlike HIV, HBV, and HCV [267], the ability of SARS-CoV-2 variants to escape T cell immunity is less well understood. The role of T cells in fighting against SARS-CoV-2 infection is also not well documented, but the polyclonal nature of T cell response might resist the effect of some viral mutations [268]. Also, since T cell epitopes are located along the genome in addition to the spike protein [269], designing peptide vaccines targeting T cells could be an attractive direction for future vaccine design.
Having escalating safety and efficacy concerns, it is encouraged to scale up immunization using the current vaccines provided together with periodic updating of vaccines matching circulating strains is recommended [268]. It is suggested that different response strategies including increasing vaccination doses, designing next-generation vaccines against VOC, improving the immunogenicity of vaccines, and massive vaccination could help tackle the ongoing COVID-19 caused especially by the VOC [39]. More studies are required to better understand the correlation between immune evasion, disease severity, and immunotherapy associated with the VOC [270]. The emergence of VOC is unavoidable requiring appropriate countermeasures. Thus, a focus should be given to co-circulating VOC in the same region as these variants could pause another challenge upon recombination [271]. Targeting mutation hotspots and synergic mutations for therapy, diagnostics and vaccination is of utmost recommended [217]. Public education about the importance of vaccination should also be a priority agenda as negative attitude towards vaccination and vaccine hesitancy is observed [272,273].
Updated structure-based studies are important to decipher the immune escaping mechanisms of S protein variants to design structurally suitable vaccines/antibodies. Looking for conserved epitopes in the spike protein is also another potential means of developing effective antibodies and vaccines that can resist future virus genetic alterations. Mucosal targeting vaccines using potent adjuvants could also be a new approach to prevent initial virus entry; however, current vaccine studies focus majorly on neutralizing antibodies in blood. More importantly, timely reformulating vaccines and antibodies in parallel with viral evolution are at most crucial. Overall, the protective efficacy of most COVID-19 vaccines is achieved through boosting neutralizing antibodies targeting the spike protein that could be greatly affected by virus evolution and deserves conscious follow-up.
Data availability
No data was used for the research described in the article.
Author contributions
T.J. provided funding, conceived the topic, and edited the paper. HMM conceived the topic and drafted the manuscript and the figures. DM, AA, AK, and MG helped with the write-up and manuscript editing and figures.
Declaration of Competing Interest
All authors declare that there is no competing interests.
Acknowledgments
The authors acknowledge their funding institutions. TJ is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29030104), the National Natural Science Fund (Grant Nos.: 31870731 and 31971129), the Fundamental Research Funds for the Central Universities, the 100 Talents Program of the Chinese Academy of Sciences and USTC new medicine joint fund training program (WK9110000136). HMM is supported by the University of Science and Technology of China scholarship program. DM is supported by ANSO scholarship. AKK is supported by the Chinese Government Scholarship. The funder has no role in designing, writing, preparation, and submission of this article.
References
- 1.Wang Q., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4) doi: 10.1016/j.cell.2020.03.045. p. 894-904. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gates B. Responding to Covid-19—a once-in-a-century pandemic? N. Engl. J. Med. 2020;382(18):1677–1679. doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., et al. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascella M., et al. Statpearls [internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- 5.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beigel J.H., et al. Remdesivir for the treatment of Covid-19—preliminary report. N. Engl. J. Med. 2020;383(10):993–994. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 7.Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9(7) doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. p. 271-280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dömling A., Gao L. Chemistry and biology of SARS-CoV-2. Chem. 2020;6(6):1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan J.F.-W., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23(8):468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai M.M.C., Perlman S., Anderson L.J. In: Fields Virology. Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; 2007. Coronaviridae; pp. 1305–1335. [Google Scholar]
- 19.Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 22.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch B.J., et al. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y., et al. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y., et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.058. p. 281-292. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 28.Qing E., et al. Dynamics of SARS-CoV-2 spike proteins in cell entry: control elements in the amino-terminal domains. Mbio. 2021;12(4):e01590–21. doi: 10.1128/mBio.01590-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang T., et al. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71(12):3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 33.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020;119:103670. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia S., et al. Peptide-based membrane fusion inhibitors targeting HCoV-229E spike protein HR1 and HR2 domains. Int. J. Mol. Sci. 2018;19(2):487. doi: 10.3390/ijms19020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gui M., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hulswit R., De Haan C., Bosch B.-J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan M., et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021;25 doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao T., et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv. 2020 p. 2020.03.29.20041962. [Google Scholar]
- 41.Liao M., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 42.Zheng M., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6) doi: 10.1016/j.chom.2020.04.017. p. 883-890. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong Y., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibabaw T., et al. Role of IFN and complements system: innate immunity in SARS-CoV-2. J. Inflamm. Res. 2020;13:507. doi: 10.2147/JIR.S267280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5) doi: 10.1016/j.cell.2020.04.026. p. 1036-1045. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taefehshokr N., et al. Covid-19: perspectives on innate immune evasion. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. Carapito, et al., Identification of driver genes for critical forms of COVID-19 in a deeply phenotyped young patient cohort. Sci. Transl. Med. p. eabj7521. 2021. [DOI] [PubMed]
- 51.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Figueiredo C.S., Raony Í., Giestal-de-Araujo E. SARS-CoV-2 targeting the retina: host–virus interaction and possible mechanisms of viral tropism. Ocul. Immunol. Inflamm. 2020;28(8):1301–1304. doi: 10.1080/09273948.2020.1799037. [DOI] [PubMed] [Google Scholar]
- 53.Song E., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218(3):e20202135. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackermann M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta A., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S., et al. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J. Hematol. Oncol. 2020;13(1):1–22. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazaleuskaya L., et al. Protective role of Toll-like receptor 3-induced type I interferon in murine coronavirus infection of macrophages. Viruses. 2012;4(5):901–923. doi: 10.3390/v4050901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sa Ribero M., et al. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16(7):e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hadjadj J., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fajgenbaum D.C., June C.H. Cytokine storm. N. Engl. J. Med. 2020;383(23):2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Y., et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11(10):740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su Y., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6) doi: 10.1016/j.cell.2020.10.037. p. 1479-1495. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez L., et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep. Med. 2020;1(5):100078. doi: 10.1016/j.xcrm.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni L., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6) doi: 10.1016/j.immuni.2020.04.023. p. 971-977. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laing A.G., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 67.Files J.K., et al. Sustained cellular immune dysregulation in individuals recovering from SARS-CoV-2 infection. J. Clin. Invest. 2021;131(1) doi: 10.1172/JCI140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou R., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53(4) doi: 10.1016/j.immuni.2020.07.026. p. 864-877. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilk A.J., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Isho B., et al. Mucosal versus systemic antibody responses to SARS-CoV-2 antigens in COVID-19 patients. medRxiv. 2020 p. 2020.08.01.20166553. [Google Scholar]
- 72.Gao C., et al. SARS-CoV-2 spike protein interacts with multiple innate immune receptors. bioRxiv: the preprint server for biology. 2020 p. 2020.07.29.227462. [Google Scholar]
- 73.Reynolds H.Y. Immunoglobulin G and its function in the human respiratory tract. Mayo Clinic Proceedings. 1988 doi: 10.1016/s0025-6196(12)64949-0. Elsevier. [DOI] [PubMed] [Google Scholar]
- 74.Pakkanen S.H., et al. Expression of homing receptors on IgA1 and IgA2 plasmablasts in blood reflects differential distribution of IgA1 and IgA2 in various body fluids. Clin. Vaccine Immunol. 2010;17(3):393–401. doi: 10.1128/CVI.00475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson P. 2020. Distinct B Cell Subsets Give Rise to Antigen-Specific Antibody Responses against SARS-CoV-2. [Google Scholar]
- 76.Guthmiller J.J., et al. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. bioRxiv. 2020 doi: 10.1128/mBio.02940-20. p. 2020.09.12.294066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang F., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020;5(1):156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grifoni A., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7) doi: 10.1016/j.cell.2020.05.015. p. 1489-1501. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 80.Stephens D.S., McElrath M.J. COVID-19 and the path to immunity. JAMA. 2020;324(13):1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Legros V., et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell. Mol. Immunol. 2021;18(2):318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma H., et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sekine T., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1) doi: 10.1016/j.cell.2020.08.017. p. 158-168. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moderbacher C.R., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4) doi: 10.1016/j.cell.2020.09.038. p. 996-1012. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng Y., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Z., et al. Clinical and laboratory profiles of 75 hospitalized patients with novel coronavirus disease 2019 in Hefei, China. medRxiv. 2020 p. 2020.03.01.20029785. [Google Scholar]
- 87.Tan A.T., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34(6):108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brouwer P.J., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piccoli L., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4) doi: 10.1016/j.cell.2020.09.037. p. 1024-1042. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robbiani D.F., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rogers T.F., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suthar M.S., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Reports Medicine. 2020;1(3):100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41(5):355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Folegatti P.M., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jackson L.A., et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. Nov 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Keech C., et al. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15(3):185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elsayed H., et al. Intrastructural help: harnessing T helper cells induced by licensed vaccines for improvement of HIV Env antibody responses to virus-like particle vaccines. J. Virol. 2018;92(14) doi: 10.1128/JVI.00141-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kombe Kombe A.J., et al. Potent molecular feature-based neutralizing monoclonal antibodies as promising therapeutics against SARS-CoV-2 infection. Front. Mol. Biosci. 2021;8(461) doi: 10.3389/fmolb.2021.670815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xia S., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du L., et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Q., et al. MERS-CoV spike protein: targets for vaccines and therapeutics. Antiviral Res. 2016;133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 104.Liu L., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 105.Amanat F., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Okba N.M., et al. Severe acute respiratory syndrome coronavirus 2− specific antibody responses in coronavirus disease patients. Emerging Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wec A.Z., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369(6504):731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Le Bert N., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 109.Braun J., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 110.Mekonnen D., Mengist H.M., Derbie A., Nibret E., Munshea A., He H., Li B., Jin T. Diagnostic accuracy of serological tests and kinetics of severe acute respiratory syndrome coronavirus 2 antibody: A systematic review and meta-analysis. Rev Med Virol. 2021 doi: 10.1002/rmv.2181. May;31(3):e2181. Epub 2020 Nov 5. PMID: 33152146. [DOI] [PubMed] [Google Scholar]
- 111.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilk A.J., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang F., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020;5(1):1–11. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan M., et al. Identification and characterization of a monoclonal antibody blocking the SARS-CoV-2 spike protein–ACE2 interaction. Cell. Mol. Immunol. 2021;18(6):1562–1564. doi: 10.1038/s41423-021-00684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y., et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020;17(10):1095–1097. doi: 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Canouï E., Launay O. Histoire et principes de la vaccination. Rev. Mal. Respir. 2019;36(1):74–81. doi: 10.1016/j.rmr.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 117.Rroux D.Pl., Rhodes W. An enquiry into the causes and effects of variolae vaccinae. South Afr. Med. J. 1921;19(20):374–376. [PubMed] [Google Scholar]
- 118.Clem A. Fundamentals of vaccine immunology. J. Glob. Infect. Dis. 2011;3:73–78. doi: 10.4103/0974-777X.77299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mort M., et al. World Health Organization; 2013. Vaccine Safety Basics: Learning Manual. [Google Scholar]
- 120.Lusvarghi S., et al. Key substitutions in the spike protein of SARS-CoV-2 variants can predict resistance to monoclonal antibodies, but other substitutions can modify the effects. J. Virol. 2021:JVI0111021. doi: 10.1128/JVI.01110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao Q., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bala P.C., Eisenreich B.R., Yoo S.B.M., Hayden B.Y., Park H.S., Zimmermann J. Automated markerless pose estimation in freely moving macaques with OpenMonkeyStudio. Nat Commun. 2020;11(1):4560. doi: 10.1038/s41467-020-18441-5. Sep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.(WHO) 2020. W.H.o. Draft Landscape of COVID-19 Candidate Vaccines.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [cited 2021 21/04/2021]; Available from: [Google Scholar]
- 124.van Doremalen N., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586(7830):578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Corbett K.S., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guebre-Xabier M., et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine. 2020;38(50):7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mulligan M.J., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 128.Walsh E.E., et al. RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv. 2020 p. 2020.08.17.20176651. [Google Scholar]
- 129.Logunov D.Y., et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396(10255):887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guebre-Xabier M., et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine. 2020;38(50):7892–7896. doi: 10.1016/j.vaccine.2020.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Walsh E.E., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sahin U., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH 1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 133.Bonavia A., et al. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Humphreys I.R., Sebastian S. Novel viral vectors in infectious diseases. Immunology. 2018;153(1):1–9. doi: 10.1111/imm.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines. 2016;15(3):313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang Z.-y., et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pandey S.C., et al. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020;256:117956. doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rodriguez-Coira J., Sokolowska M. SARS-CoV -2 candidate vaccines - composition, mechanisms of action and stages of clinical development. Allergy. 2021 doi: 10.1111/all.14714. [DOI] [PubMed] [Google Scholar]
- 139.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8):e3000003. doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lauring A.S., Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6(7):e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tsetsarkin K.A., et al. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3(12):e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Diehl W.E., et al. Ebola virus glycoprotein with increased infectivity dominated the 2013–2016 epidemic. Cell. 2016;167(4) doi: 10.1016/j.cell.2016.10.014. p. 1088-1098. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Urbanowicz R.A., et al. Human adaptation of Ebola virus during the West African outbreak. Cell. 2016;167(4) doi: 10.1016/j.cell.2016.10.013. p. 1079-1087. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ning T., et al. Antigenic drift of influenza A (H7N9) virus hemagglutinin. J. Infect. Dis. 2019;219(1):19–25. doi: 10.1093/infdis/jiy408. [DOI] [PubMed] [Google Scholar]
- 145.Petrie J.G., Lauring A.S. Influenza a(H7N9) virus evolution: which genetic mutations are antigenically important? J. Infect. Dis. 2018;219(1):3–5. doi: 10.1093/infdis/jiy409. [DOI] [PubMed] [Google Scholar]
- 146.Dawood A.A. Mutated COVID-19 may foretell a great risk for mankind in the future. New Microbes New Infect. 2020;35:100673. doi: 10.1016/j.nmni.2020.100673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Korber B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4) doi: 10.1016/j.cell.2020.06.043. p. 812-827. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saha P., et al. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40(5) doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sheikh J.A., et al. Emerging genetic diversity among clinical isolates of SARS-CoV-2: lessons for today. Infect. Genet. Evol. 2020;84:104330. doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.van Dorp L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang R., et al. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12(10):1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rice A.M., et al. Evidence for strong mutation Bias toward, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 2021;38(1):67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Duchene S., et al. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6(2):veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Banoun H. Evolution of SARS-CoV-2: review of mutations, role of the host immune system. Nephron. 2021:1–12. doi: 10.1159/000515417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hu J., et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18(4):1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sharun K., et al. Emerging SARS-CoV-2 variants: impact on vaccine efficacy and neutralizing antibodies. Hum. Vaccin. Immunother. 2021:1–4. doi: 10.1080/21645515.2021.1923350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hu F., et al. A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract. Cell. Mol. Immunol. 2020;17(11):1119–1125. doi: 10.1038/s41423-020-00550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Becerra‐Flores M., Cardozo T. SARS‐CoV‐2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020;74(8):e13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.François K.O., Balzarini J. The highly conserved glycan at asparagine 260 of HIV-1 gp120 is indispensable for viral entry. J. Biol. Chem. 2011;286(50):42900–42910. doi: 10.1074/jbc.M111.274456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kong L., Wilson I.A., Kwong P.D. Crystal structure of a fully glycosylated HIV‐1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins Struct. Funct. Bioinform. 2015;83(3):590–596. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang W., et al. N463 glycosylation site on V5 loop of a mutant gp120 regulates the sensitivity of HIV-1 to neutralizing monoclonal antibodies VRC01/03. J. Acquir. Immune Defic. Syndr. 2015;69(3):270. doi: 10.1097/QAI.0000000000000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Van Egeren D., et al. Risk of rapid evolutionary escape from biomedical interventions targeting SARS-CoV-2 spike protein. PLoS One. 2021;16(4):e0250780. doi: 10.1371/journal.pone.0250780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu Z., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3) doi: 10.1016/j.chom.2021.01.014. p. 477-488 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Müller K., et al. Sensitivity of two SARS-CoV-2 variants with spike protein mutations to neutralising antibodies. Virus Genes. 2021:1–8. doi: 10.1007/s11262-021-01871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu Z., et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2021 doi: 10.1101/2020.11.06.372037. Jan 11:2020.11.06.372037. Update in: Cell Host Microbe. 2021 Mar 10;29(3):477-488.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cao Y., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1) doi: 10.1016/j.cell.2020.05.025. p. 73-84. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lv Z., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369(6510):1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 169.Greaney A.J., et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3) doi: 10.1016/j.chom.2021.02.003. p. 463-476 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Harvey W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baum A., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506) doi: 10.1126/science.abd0831. p. 1014-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hoffmann M., et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv. 2021 doi: 10.1101/2021.05.04.442663. p. 2021.05.04.442663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wu L., et al. Exploring the immune evasion of SARS-CoV-2 variant harboring E484K by molecular dynamics simulations. Brief Bioinform. 2021 doi: 10.1093/bib/bbab383. Sep 22:bbab383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Thomson E.C., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5) doi: 10.1016/j.cell.2021.01.037. p. 1171-1187. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Thomson E.C., et al. The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. bioRxiv. 2021 doi: 10.1016/j.cell.2021.01.037. Mar 4;184(5):1171-1187.e20. Epub 2021 Jan 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li Q., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5) doi: 10.1016/j.cell.2020.07.012. p. 1284-1294. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu C., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7(1):eabe5575. doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shah M., et al. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 2020;18:3402–3414. doi: 10.1016/j.csbj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rees-Spear C., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34(12):108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.McCarthy K.R., et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yi C., et al. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17(6):621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bozdaganyan M.E., et al. Effects of mutations in the receptor-binding domain of SARS-CoV-2 spike on its binding affinity to ACE2 and neutralizing antibodies revealed by computational analysis. bioRxiv. 2021 p. 2021.03.14.435322. [Google Scholar]
- 183.Luan B., Huynh T. Insights into SARS-CoV-2’s mutations for evading human antibodies: sacrifice and survival. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00311. [DOI] [PubMed] [Google Scholar]
- 184.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tarke A., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2(2):100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang H., et al. Profiling CD8+ T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants. Cell Rep. 2021;36(11):109708. doi: 10.1016/j.celrep.2021.109708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Peng Y., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Groves D.C., Rowland-Jones S.L., Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem. Biophys. Res. Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Motozono C., et al. An emerging SARS-CoV-2 mutant evading cellular immunity and increasing viral infectivity. bioRxiv. 2021 doi: 10.1101/2021.04.02.438288. p. 2021.04.02.438288. [DOI] [Google Scholar]
- 190.Agerer B., et al. SARS-CoV-2 mutations in MHC-I–restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021;6(57):eabg6461. doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mekonnen D., Mengist H.M., Jin T. SARS-CoV-2 subunit vaccine adjuvants and their signaling pathways. Expert Rev. Vaccines. 2021 doi: 10.1080/14760584.2021.1991794. p. null–null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Singh Y., et al. N‐terminal domain of SARS CoV‐2 spike protein mutation associated reduction in effectivity of neutralizing antibody with vaccinated individuals. J. Med. Virol. 2021;93(10):5726. doi: 10.1002/jmv.27181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zeng B., et al. Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern: a systematic review and meta-analysis. medRxiv. 2021;26(41):2100920. doi: 10.2807/1560-7917.ES.2021.26.41.2100920. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tregoning J.S., et al. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021:1–11. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gushchin V.A., et al. Neutralizing activity of sera from Sputnik V-vaccinated people against variants of concern (VOC: B. 1.1. 7, B. 1.351, P. 1, B. 1.617. 2, B. 1.617. 3) and Moscow endemic SARS-CoV-2 variants. Vaccines. 2021;9(7):779. doi: 10.3390/vaccines9070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.S. Lanini, et al., GRAd-COV2, a gorilla adenovirus-based candidate vaccine against COVID-19, is safe and immunogenic in younger and older adults. Sci. Transl. Med. 2021 p. eabj1996. [DOI] [PubMed]
- 197.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature. 2021:1–6. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 198.McCallum M., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–2347. doi: 10.1016/j.cell.2021.03.028. Apr 29 .e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cerutti G., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29(5):819–833. doi: 10.1016/j.chom.2021.03.005. May 12. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wu K., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 p. 2021.01.25.427948. [Google Scholar]
- 201.Muik A., et al. Neutralization of SARS-CoV-2 lineage B. 1.1. 7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science. 2021;371(6534):1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zou J., et al. The effect of SARS-CoV-2 D614G mutation on BNT162b2 vaccine-elicited neutralization. NPJ Vaccines. 2021;6(1):1–4. doi: 10.1038/s41541-021-00313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Supasa P., et al. Reduced neutralization of SARS-CoV-2 B. 1.1. 7 variant by convalescent and vaccine sera. Cell. 2021;184(8):2201–2211. doi: 10.1016/j.cell.2021.02.033. Apr 15.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou D., et al. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361. doi: 10.1016/j.cell.2021.02.037. Apr 29.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xie X., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 206.Emary K.R.W., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397(10282):1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Clemens S.A.C., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 lineages circulating in Brazil. Nat. Commun. 2021;12(1):5861. doi: 10.1038/s41467-021-25982-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 209.Caniels T.G., et al. Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination. medRxiv. 2021 doi: 10.1101/2021.05.26.21257441. Jun 1:2021.05.26.21257441. Update in: Sci Adv. 2021 Sep 3;7(36):eabj5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.King H.A., et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. Proc Natl Acad Sci U S A. 2021;118(38) doi: 10.1073/pnas.2106433118. Sep 21, e2106433118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang B., et al. Resistance of SARS-CoV-2 Delta variant to neutralization by BNT162b2-elicited antibodies in Asians. Lancet Regional Health–Western Pacific. 2021;15 doi: 10.1016/j.lanwpc.2021.100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mlcochova P., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lopez Bernal J., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Harder T., et al. Effectiveness of COVID-19 vaccines against SARS-CoV-2 infection with the Delta (B. 1.617. 2) variant: second interim results of a living systematic review and meta-analysis, 1 January to 25 August 2021. Eurosurveillance. 2021;26(41):2100920. doi: 10.2807/1560-7917.ES.2021.26.41.2100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. May. [DOI] [PubMed] [Google Scholar]
- 216.Collier D.A., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593(7857):136–141. doi: 10.1038/s41586-021-03412-7. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Alenquer M., et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021;17(8):e1009772. doi: 10.1371/journal.ppat.1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mahase E. Covid-19: novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 219.Wise J. Covid-19: the E484K mutation and the risks it poses. BMJ. 2021;372:n359. doi: 10.1136/bmj.n359. [DOI] [PubMed] [Google Scholar]
- 220.Jangra S., et al. The E484K mutation in the SARS-CoV-2 spike protein reduces but does not abolish neutralizing activity of human convalescent and post-vaccination sera. medRxiv. 2021 p. 2021.01.26.21250543. [Google Scholar]
- 221.Bar-On Y.M., et al. Protection of BNT162b2 vaccine booster against Covid-19 in Israel. N. Engl. J. Med. 2021;385(15):1393–1400. doi: 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ramos A.M., Vela-Pérez M., Ferrández M.R., Kubik A.B., Ivorra B. Modeling the impact of SARS-CoV-2 variants and vaccines on the spread of COVID -19. Commun Nonlinear Sci Numer Simul. 2021 doi: 10.1016/j.cnsns.2021.105937. Nov;102:105937. Epub 2021 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang Z., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Boyle K. Technology Networks-Biopharma; 2021. Vaccine Efficacy Against Different SARS-CoV-2 Variants. August 12. [Google Scholar]
- 225.Zhou W., Wang W. Fast-spreading SARS-CoV-2 variants: challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021;6(1):226. doi: 10.1038/s41392-021-00644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cevik M., et al. COVID-19 vaccines: keeping pace with SARS-CoV-2 variants. Cell. 2021;184(20):5077–5081. doi: 10.1016/j.cell.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rashid F., et al. Mutations in SARS-CoV-2 ORF8 altered the bonding network with interferon regulatory factor 3 to evade host immune system. Front. Microbiol. 2021:1811. doi: 10.3389/fmicb.2021.703145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Triveri A., et al. SARS-CoV-2 spike protein mutations and escape from antibodies: a computational model of epitope loss in variants of concern. J. Chem. Inf. Model. 2021;61(9):4687–4700. doi: 10.1021/acs.jcim.1c00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ferraz M.V., et al. Immune evasion of SARS-CoV-2 variants of concern is driven by low affinity to neutralizing antibodies. Chem. Commun. 2021;57(49):6094–6097. doi: 10.1039/d1cc01747k. Jun 21. [DOI] [PubMed] [Google Scholar]
- 230.Cai Y., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.04.13.439709. Apr 14:2021.04.13.439709. Update in: Science. 2021 Jun 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rabaan A., et al. Exploring the genetics, ecology of SARS-CoV-2 and climatic factors as possible control strategies against COVID-19. Infez. Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- 232.Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16(10):1718. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang Z., et al. SARS-CoV-2 variants exhibit increased kinetic stability of open spike conformations as an evolutionary strategy. bioRxiv. 2021 doi: 10.1128/mbio.03227-21. p. 2021.10.11.463956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Giron C.C., Laaksonen A., Barroso da Silva F.L. Up state of the SARS-COV-2 spike homotrimer favors an increased virulence for new variants. Front. Med. Technol. 2021;3(29) doi: 10.3389/fmedt.2021.694347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lesté-Lasserre C. Mutant coronaviruses found in mink spark massive culls and doom a Danish group’s research. Science (New York, N.Y.) 2020 doi: 10.1126/science.abf6565. [DOI] [Google Scholar]
- 236.Ozono S., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12(1):848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Fernández A. Structural impact of mutation D614G in SARS-CoV-2 spike protein: enhanced infectivity and therapeutic opportunity. ACS Med. Chem. Lett. 2020;11(9):1667–1670. doi: 10.1021/acsmedchemlett.0c00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang J., et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372(6541):525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gobeil S.M.C., et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34(2):108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Klinakis A., Cournia Z., Rampias T. N-terminal domain mutations of the spike protein are structurally implicated in epitope recognition in emerging SARS-CoV-2 strains. Comput. Struct. Biotechnol. J. 2021;4(19):5556–5567. doi: 10.1016/j.csbj.2021.10.004. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cho H., et al. Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021:eabj5413. doi: 10.1126/scitranslmed.abj5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Haslwanter D., et al. A combination of receptor-binding domain and N-terminal domain neutralizing antibodies limits the generation of SARS-CoV-2 spike neutralization-escape mutants. Mbio. 2021;12(5):e02473–21. doi: 10.1128/mBio.02473-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Liu L., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 244.Suryadevara N., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184(9) doi: 10.1016/j.cell.2021.03.029. p. 2316-2331. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.J. Zhang, et al., Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021. p. eabl9463. [DOI] [PMC free article] [PubMed]
- 246.Xie J., et al. Novel monoclonal antibodies and recombined antibodies against variant SARS-CoV-2. Front. Immunol. 2021:3430. doi: 10.3389/fimmu.2021.715464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.McCallum M., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9) doi: 10.1016/j.cell.2021.03.028. p. 2332-2347. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Barnes C.O., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ma H., et al. Potent neutralization of sars-cov-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. J. Virol. 2021;95(10):e02438–20. doi: 10.1128/JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dejnirattisai W., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184(11) doi: 10.1016/j.cell.2021.03.055. p. 2939-2954.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mercado N.B., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Pallesen J., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. PNAS. 2017;114(35):E7348–e7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Amanat F., et al. Introduction of two prolines and removal of the polybasic cleavage site lead to higher efficacy of a recombinant spike-based SARS-CoV-2 vaccine in the mouse model. mBio. 2021;12(2):e02648–20. doi: 10.1128/mBio.02648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Corbett K.S., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hsieh C.-L., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369(6510):1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Singh J., et al. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021;18(1):1–21. doi: 10.1186/s12985-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Harvey W.T., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Eguia R.T., et al. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17(4):e1009453. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Weisblum Y., Schmidt F. vol. 9. 2020. (Escape from Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Andreano E., et al. SARS-CoV-2 escape <em>in vitro</em> from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1073/pnas.2103154118. p. 2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wibmer C.K., et al. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 262.Choi B., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Baj A., et al. Spike protein evolution in the SARS-CoV-2 delta variant of concern: a case series from Northern Lombardy. Emerg. Microbes Infect. 2021:1–10. doi: 10.1080/22221751.2021.1994356. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Rolland M., Gilbert P.B. Sieve analysis to understand how SARS-CoV-2 diversity can impact vaccine protection. PLoS Pathog. 2021;17(3):e1009406. doi: 10.1371/journal.ppat.1009406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Meredith L.W., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020;20(11):1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yang H., Lyu Y., Hou F. SARS-CoV-2 infection and the antiviral innate immune response. J. Mol. Cell Biol. 2020;12(12):963–967. doi: 10.1093/jmcb/mjaa071. Nov 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.McMichael A. T cell responses and viral escape. Cell. 1998;93(5):673–676. doi: 10.1016/s0092-8674(00)81428-2. [DOI] [PubMed] [Google Scholar]
- 268.Williams T.C., Burgers W.A. SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir. Med. 2021;9(4):333–335. doi: 10.1016/S2213-2600(21)00075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mateus J., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Schenten D., Bhattacharya D. Immunology of SARS-CoV-2 infections and vaccines. Adv. Immunol. 2021 doi: 10.1016/bs.ai.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lazarevic I., et al. Immune evasion of SARS-CoV-2 emerging variants: what have we learnt so far? Viruses. 2021;13(7):1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Paul E., Steptoe A., Fancourt D. Attitudes towards vaccines and intention to vaccinate against COVID-19: implications for public health communications. Lancet Regional Health - Europe. 2021;1:100012. doi: 10.1016/j.lanepe.2020.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Danabal K.G.M., et al. Attitude towards COVID 19 vaccines and vaccine hesitancy in urban and rural communities in Tamil Nadu, India – a community based survey. BMC Health Serv. Res. 2021;21(1):994. doi: 10.1186/s12913-021-07037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhou T., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28(6) doi: 10.1016/j.chom.2020.11.004. p. 867-879. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ju B., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 276.Kombe Kombe A.J., et al. Potent molecular feature-based neutralizing monoclonal antibodies as promising therapeutics against SARS-CoV-2 infection. Front. Mol. Biosci. 2021;8:461. doi: 10.3389/fmolb.2021.670815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wu N.C., et al. A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. PLoS Pathog. 2020;16(12):e1009089. doi: 10.1371/journal.ppat.1009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Motozono C., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29(7) doi: 10.1016/j.chom.2021.06.006. p. 1124-1136. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.