Highlights
-
•
The inoculated cefotaxime-resistant E. coli was a good pig gut colonizer.
-
•
Probiotics could not reduce faecal excretion of resistant E. coli in inoculated pigs.
-
•
Resistant E. coli titers were lower in digestive tracts of the probiotic-treated pigs.
-
•
No transfer of the blaCTX−M-1 gene was detected.
Keywords: E. coli, Probiotic, Cephalosporin resistance, Pig, Microbiota
Abstract
We evaluated the impact of the administration of two Escherichia coli probiotic strains (ED1a and Nissle 1917) to pigs on the gut carriage or shedding of extended-spectrum beta-lactamase-producing E. coli. The probiotics were given to four sows from 12 days before farrowing to the weaning day, and to the 23 piglets (infected treated group (IPro)) from birth to the age of 49 days. Four other sows and their 24 piglets (infected non-treated group (INT)) did not receive the probiotics. IPro and INT piglets (n = 47) were orally inoculated with the strain E. coli 17–348F-RifR carrying the blaCTX−M-1 gene and resistant to rifampicin. Cefotaxime-resistant (CTXR) E. coli and rifampicin-resistant (RifR) E. coli were cultured and excretion of probiotics was studied using PCR on individual faecal and post-mortem samples, and from manure collected after the challenge with resistant E. coli. CTXR and RifRE.coli isolates were characterized to detect transfer of the blaCTX−M-1 to other strains.. Overall, there was no significant reduction in faecal excretion of CTXR and RifRE. coli in IPro pigs compared with INT pigs, although the CTXR and RifRE. coli titres were slightly, but significantly lower in the colon, caecum and rectum at post mortem. Excretion of the probiotics decreased with age, but Nissle 1917 was detected in most pigs at post-mortem. No transfer of the blaCTX−M-1 gene to probiotic and other E. coli strains was detected. In conclusion, in our experimental conditions, the used probiotics did not reduce shedding of the challenge strain.
Introduction
Extended-spectrum cephalosporins (ESC) are critically important antibiotics for human health (WHO, 2019). Enterobacterales resistant to these antimicrobials are isolated from human and animal infections, and carriage of such ESC-resistant (ESCR) bacteria in food-producing animals is suspected to be a source of human contamination (EFSA, 2011). The 2018–2019 European monitoring of antimicrobial resistance in the caeca of animals at the slaughterhouse showed that the median levels of resistance to cefotaxime and ceftazidime are lower than 1.5% in isolates from pigs, calves, broilers and turkeys, and amongst the reporting Member States, the occurrence of ESC resistance varies from 0% to 5.9% in fattening pigs, from 0% to 5.6% in calves and from 0% to 30.1% in broilers (EFSA/ECDC, 2020). The use of selective media containing cefotaxime made it possible to evaluate the prevalence of presumptive E. coli producing extended-spectrum beta-lactamases (ESBLs) and/or AmpC beta-lactamases in samples of pig caeca, and revealed that 42.7% of 6792 samples from 28 Member States contained ESCR E. coli. amongst these ESCR isolates, according to phenotypic tests, presumptive ESBL producers were more common than AmpC producers (EFSA/ECDC, 2021). In France, the analysis of commensal or pathogenic ESCR E. coli isolates from pigs showed that resistance is — as in other food animals in France — mainly carried by highly similar blaCTX−M-1 IncI1/ST3 plasmids (Lucas et al., 2018).
The high prevalence of ESCR E. coli carriage in pigs constitutes a hazard for human health due to the possible contamination of farmers, carcasses and the environment via manure. In France, an efficient cephalosporin stewardship programme deployed in swine production units has helped reduce cephalosporin use and contain resistance (Verliat et al., 2021), but strategies to reduce the number of ESCR Enterobacterales in pig faeces are still needed. In a previous study (Mourand et al., 2017), we tested the possibility of competition between ESCR Enterobacterales and E. coli probiotics (Mourand et al., 2017). Thus, the possibility of preventing the colonization of weaned specific-pathogen-free (SPF) piglets via an oral inoculation of ESCR E. coli was explored by comparing the ESCR E. coli titres of piglets born to E. coli probiotic (ED1a)-treated sows and given the probiotic from birth to weaning, with non-treated piglets born to non-treated sows. The results showed that, in the conditions of the experiments, E. coli ED1a (Clermont et al., 2008) had a limited impact on the gut carriage of ESBL-producing E. coli. In the present study, we modified the protocols with the hope of increasing the impact of the administration of probiotics. We tested the simultaneous administration of two probiotics (E. coli ED1a and E. coli Nissle 1917 (EcN)), increased the doses and duration of the period of administration, and modified the challenging ESCR E. coli strain and inoculation dose. We selected the ED1a and EcN probiotics, because ED1a is a good colonizer, avirulent and its numerous isolates are susceptible to antimicrobials (Clermont et al., 2008), and EcN has been used for decades as a probiotic for human consumption (Hancock, Dahl & Klemm, 2010). Both probiotic strains belong to the B2 phylogroup, but to distinct sequence types (STs): ST452 for ED1a and ST127 for EcN (Denamur, Clermont, Bonacorsi & Gordon, 2021). They have different metabolic properties (Bouvet, Bourdelier, Glodt, Clermont & Denamur, 2017) and EcN is highly virulent in a mouse model of sepsis, whereas ED1a is avirulent in this model (Clermont et al., 2008). The administration period was extended after the ESCR E. coli inoculation day, to favour the development of the probiotic strains. Moreover, to better simulate field situations, we used a lower inoculum dose prepared with a pig commensal strain, E. coli 17–348F, containing a blaCTX−M-1 IncI1/ST3 plasmid, because such plasmids are the most common in French ESCR E. coli isolates (Lucas et al., 2018). Our hypothesis was that the use of these two probiotics, with possibly different niches in the digestive tract, and given during an extended period, increases the probability of preventing ESCR E. coli colonization resulting from a moderate inoculum dose.
Materials and methods
Preparation of the probiotics
Suspensions of the two probiotics were prepared as follows. Each week, the E. coli ED1a and EcN strains were grown separately at 37 °C ± 1 °C in Mueller-Hinton (MH) broth. For each strain, a suspension of approximately 9.30 log10 colony-forming units (CFU)/mL was obtained by centrifuging cultures in MH broth. Equal volumes of the two suspensions were mixed and stored at 5+/−3 °C for up to one week, as well as small aliquots of the non-mixed suspensions. The mean titres of the non-mixed suspensions determined, on the first and the last day of storage, by plating decimal dilutions on agar plates were 9.32 log10 CFU/mL [9.07–9.49 log10 CFU/mL] for E. coli ED1a, and 9.39 log10 CFU/mL [8.96–9.60 log10 CFU/mL] for E. coli EcN.
A mixed-culture assay was performed to check the absence of competition between the two probiotic strains. Separate cultures of each probiotic strain were prepared, and diluted suspensions (0.5 McFarland) were mixed and grown in MH broth at 37 °C ± 1 °C for 24 h. The initial and final titres of the culture were measured by plating decimal dilutions on agar plates. Using plates containing well-isolated colonies, the percentage of colonies of E. coli ED1a and EcN in the initial mixture and after 24 h were determined using PCR tests specific to each strain as described below.
Preparation and whole genome sequencing (WGS) of the challenge strain
For the challenge, we used a mutant of the previously described ESCR E. coli 17–348F strain Lucas et al. (2018). E. coli 17–348F was isolated from pig caeca at the slaughterhouse; it belongs to the B1 phylogenetic group and contains an IncI1/ST3 plasmid with the resistance genes blaCTX−M-1, sul2 and tetA. A mutant resistant to rifampicin (17–348F-RifR) was obtained from a culture of a concentrated suspension of E. coli 17–348F in MH media supplemented with rifampicin (250 mg/L). The inoculum prepared for pigs consisted of suspensions of the E. coli 17–348F-RifR cultures obtained on cefotaxime-supplemented MH agar plates, and diluted. The titre determined by plating serial dilutions was 6.40 log10 CFU per pig.
DNA from the 17–348F strain was prepared using QIAmp DNA mini kits according to the manufacturer's instructions. WGS was performed with the Ion Proton system (Ion Torrent™). We cleaned reads with Trimmomatic (Bolger, Lohse & Usadel, 2014) and the following parameters: ILLUMINACLIP: oligos.fasta: 2:30:5:1: true; LEADING: 3; TRAILING: 3; MAXINFO: 40:0.2; MINLEN: 36). Then used BWA-MEM (Li, 2013) to align them with the E. coli KV7 strain (NCBI Reference Sequence LT795502.1), unmapped reads were extracted. The cleaned reads were down-sampled to fit a global coverage depth estimation of 80 x and were assembled using the SPAdes (3.10.0) de novo assembler (Bankevich et al., 2012). The de novo contigs were then screened in Megablast (Chen, Ye, Zhang & Xu, 2015) on a local nt database. The unmapped reads were also assembled in the SPAdes de novo assembler and the de novo contigs were aligned against a local nt database with Megablast.
Sequences were analysed using the web tool (https://cge.cbs.dtu.dk/services) to detect antimicrobial resistance genes and virulence genes, and to determine the multi-locus sequence type (MLST), the fumC and fimH alleles and O serogroup of E. coli 17–348F (Thomsen et al., 2016).
Animals and experiment design
The experiments were performed in accordance with French animal welfare regulations and the protocol was approved by the ANSES/ENVA/UPEC ethical committee and the French Ministry for Higher Education, Research and Innovation (ComEth authorization 2019/01/17–5, no. APAFIS: 2,018,121,812,512,422 (# 18,136)). The experiments were conducted at the ANSES Ploufragan animal facilities. Strict biosecurity measures were implemented in order to avoid contamination of the pigs, people or the environment, including the use of an air filtration system and airlocks for each unit, unit-specific clothes and compulsory showering before and after visiting the pigs.
Eight SPF Large White pregnant sows and 54 of their piglets were used. Four sows were housed in two animal rooms, and after farrowing, the piglets were left with their mother for 4 weeks before weaning. Then, 24 piglets (infected non-treated (INT) group), were housed in two other rooms (six piglets per pen, two pens per room) (Figure S1).
The infected treated (IPro) piglets were borne to four other pregnant SPF sows and were housed in two different animal rooms. From 12 days before the expected day of farrowing and up to the weaning day (Day 28), these four sows were daily given a 10 mL suspension of both E. coli ED1a and E. coli EcN. The mixed probiotic suspension was deposited in a small quantity of each animal's food before the full meal was offered, to ensure that the sows ate all the probiotic treatment. During the suckling period, the piglets from the treated sows were given the probiotic suspension daily by oral gavage from the first day of life up to the weaning day, with increasing doses (1 mL the first week, 2 mL week 2, 3 mL week 3 and 4 mL week 4). After weaning, at four weeks of age, 23 of these probiotic-treated piglets were moved to two other rooms (five or six piglets per pen, two pens per room). Then, from weaning to the age of 49 days, they received the probiotics deposited in the food, with doses of 5, 6 and 7 mL/pig/day for week 5, 6 and 7 respectively.
On the day after weaning (day 29), the 47 INT and IPro piglets received the 5 mL inoculum of the suspension prepared from E. coli 17–348F-RifR. Clinical signs and rectal temperatures were recorded, and the infected piglets were weighed once a week. Faecal samples were collected from the sows before farrowing (days −2 and −12) and up to the weaning of their piglets (days 5, 12, 19 and 28), and from the INT and IPro piglets on days 30, 31, 33, 36, 42 and 49. The faecal samples were diluted 1:10 in peptone buffer containing 20% glycerol and stored at <−18 °C.
All piglets were sacrificed from 55 to 58 days of age (IPro piglets on day 55 and day 57, and INT piglets on day 56 and day 58). Samples from the jejunum, ileum, colon, caecum and rectum were collected. All these post-mortem (PM) samples contained both digesta and scraped mucosa.
During the experiment, after E. coli 17–348F-RifR inoculation, one large plastic box was placed under each piglet pen to collect the mixture of all faeces and urine produced by the piglets. These mixtures were collected weekly. The faeces and urine were vigorously mixed and the resulting liquid manure samples were diluted and stored at <−18 °C.
Bacteriological analysis of faecal, post-mortem and manure samples
The titres of ESCR E. coli for the individual faecal, PM and manure samples were determined by spreading 100 µL of three tenfold dilutions on MacConkey (MC) agar plates containing 2 mg/L cefotaxime (MC—CTX) and on MC agar plates containing 250 mg/L rifampicin (MC-Rif). After incubation at 37 °C, the ESCR or RifR colonies on supplemented MC plates were enumerated and the titres were calculated for each pig per day. When no colony was detected, 0.1 mL of the 1:10 faecal suspension was inoculated into 0.9 mL of MH broth containing 2 mg/L cefotaxime and MH broth containing 250 mg/L rifampicin. After 24 h of incubation at 37 °C ± 2 °C under agitation, 10 µL of the cultures were streaked onto MC—CTX and MC-Rif plates. The detection limit was 2 log10 CFU/g of faeces. For each faecal, PM and manure sample, one colony from each medium was characterized: colonies obtained on MC-Rif were streaked on MC—CTX plates, and those obtained on MC—CTX were streaked on MC-Rif plates. They were identified with an E. coli-specific PCR (Furet et al., 2009); the presence of the blaCTX−M-1 gene was screened for using PCR (Woodford, Fagan & Ellington, 2006). Moreover, the isolates obtained from pigs were also tested by PCRs specific to the E. coli ED1a or EcN strains (Mourand et al., 2017). Finally, for all faecal samples collected on day 49 and rectum samples on day 58, a cell lysate was prepared from all the pooled colonies obtained on non-supplemented MC plates, and for manure samples, a suspension was prepared from all the pooled colonies of the MC—CTX plates for PCR analysis, as described below.
Molecular analysis
DNA extracts were prepared from 0.25 g of each faecal, PM or manure sample using a protocol, including bead beating with high concentrations of sodium dodecyl sulfate (SDS), salt and EDTA, using a TissueLyser (Qiagen) for 2 min. After centrifugation, (2 min, 1500 g) the supernatants were digested with proteinase K for 1 h at 70 °C, before DNA extraction with the NucleoMag Tissue kit (Macherey-Nagel) and the KingFisher Duo Prime System (Thermofisher). Each DNA extract was quantified using the NanoDrop 2000 spectrophotometer (Thermo Scientific) then adjusted to a concentration of 10 ng/µL, before storage at −20 °C. E. coli, E. coli ED1a and E. coli EcN were quantified in faeces, PM and manure samples from piglets according to Mourand et al. (2017). For each sample, results were expressed in log10 copies of DNA per gram of faecal material or per 10 ng of DNA.
PCR was also used to detect E. coli ED1a from the lysates prepared from the suspensions of all colonies grown on non-supplemented MC from faecal samples collected on day 58 or from rectum samples. In addition, PCR was used for detection of E. coli ED1a or EcN amongst colonies grown on MC—CTX from manure samples.
In vitro conjugation
In vitro conjugation tests were performed to determine whether the IncI1 plasmid of E. coli 17–348F-RifR could be transferred to commensal E. coli. Recipient E. coli cells were prepared from two commensal isolates (C-V1 and C2–6F, ANSES collection) and the reference strain E. coli CIP 7224. These commensal strains were chosen because they are susceptible to beta-lactams, tetracycline, trimethoprim-sulfamethoxazole and colistin, but resistant to nalidixic acid and ciprofloxacin. Broth cultures of E. coli 17–348F-RifR and of each recipient strain were mixed and incubated for 6 h at 37 °C in MH broth containing ciprofloxacin (0.25 or 1 mg/L) and cefotaxime (1 mg/L). The cells were pelleted and the re-suspended pellet was inoculated onto ciprofloxacin- and cefotaxime-MH agar plates. After incubation, colonies were re-streaked on ciprofloxacin- and cefotaxime-MH agar plates.
Similarly, several assays of conjugation between E. coli 17–348F-RifR and each probiotic strain were performed. To do so, fosfomycin-resistant mutants of E. coli ED1a and EcN were prepared by culturing in brain heart infusion broth supplemented with fosfomycin (64 mg/L). Then, the occurrence of conjugation was tested using different proportions of recipient (fosfomycin-resistant E. coli ED1a or EcN) and donor (E. coli 17–348F-RifR) strains. The potential transconjugants were cultured on MH media supplemented with fosfomycin (64 mg/L) and cefotaxime (2 mg/L).
Statistical analysis
The individual titres of ESCR and RifR E. coli and gene copy numbers were log10-transformed. When only the enrichment was positive, the titre was arbitrarily fixed at 2 log10/mL. For faecal samples, a mixed analysis of variance explaining the results of culture on medium with rifampicin/cefotaxime by considering two fixed factors (i.e., Treatment and Time) and 'Animal' as a random one. An auto-correlation structure of rank 1 was applied (i.e., the results of time T depend on those of time T-1). The parametric hypothesis were checked on the model's residuals, i.e., (normality (Shapiro test), equality of variances (Bartlett test) and independence (Durbin-Watson-Test). The associated pairwise mean comparisons are adjusted with the Tukey method. The mixed model is performed by means of the 'geeglm' function of the R (version 4.1.1) package 'geepack'(version 1.3–2) and the mean comparisons are performed by the ‘emmeans’ R package (version 1.6.3). Significant differences between individual body weight, body weight gain, ESCR E. coli in PM samples and gene copy numbers of INT and IPro groups were tested using Mann-Whitney-Wilcoxon tests. The distribution of the number of positive tests was compared using a Chi2 test or a Fisher exact test (n ≤ 5). The results were considered significant when p < 0.05.
Accession numbers
The sequence of E. coli 17–348F has been deposited at DDBJ/ENA/GenBank under the accession SAMN23005185 (PRJNA778926).
Results
Characterization of the E. coli 17–348F strain
According to WGS results, E. coli 17–348F belongs to serogroup O8 and to ST 767, and carries the fumC_4 and fimH_32 alleles. We also detected the following virulence genes: astA (heat stable toxin), cea (colicin E1), cib (colicin Ib), cvaC (microcin C), etsC (putative type I secretion outer membrane protein), fyuA (siderophore receptor), hlyF (hemolysin F), iha (adherence protein), iroN (enterobactin siderophore receptor protein), iss (increased serum survival), lpfA (long polar fimbriae), mchC (MchC protein), mchF (ABC transporter protein MchF), ompT (outer membrane protease), sitA (iron transport protein), terC (tellurium ion resistance protein) and traT (outer membrane protein complement resistance). The strain did not harbour any other resistance genes than those previously detected on its IncI1/ST3 plasmid, i.e. blaCTX−M-1, sul2 and tetA (Mourand et al., 2017). Several attempts of in vitro conjugation between E. coli 17–348F-RifR and recipient E. coli cells (the two commensal C-V1 and C2–6F E. coli strains, the reference E. coli CIP 7224 strain, and the two probiotic strains) were unsuccessful.
Comptetion between the probiotics
Regarding the competition assay between the two probiotics, the initial and final titres were respectively 5.3 and 9.0 log10 CFU/mL. The PCR analysis of 100 colonies from the initial suspension and the 24 h culture showed that the initial mixture contained 56% ED1a and 44% EcN and similar percentages were obtained after 24 h (57% ED1a and 43% EcN).
Clinical signs and body weight gains
Mild diarrhoea with no signs of dehydration were observed from day 35 to day 37 in IPro pigs and from day 51 to day 55 in INT pigs. Temperatures above 40.0 °C were rarely detected in IPro pigs, but recorded on 16 occasions in INT pigs (3/357 observations vs 16/312, p = 0.0008), mainly during the first week after weaning and inoculation. The maximum observed temperature was 41.6 °C. The mean body weights of the INT piglets were significantly higher than those of the IPro piglets on all weeks (data not shown), and their mean body weight gains from day 25 to day 51 were significantly higher (respectively 12.1 kg ± 2.0 kg vs 10.3 kg ± 1.4 kg, p = 0.0008).
ESCR and rifrE. coli in faeces, PM and manure samples
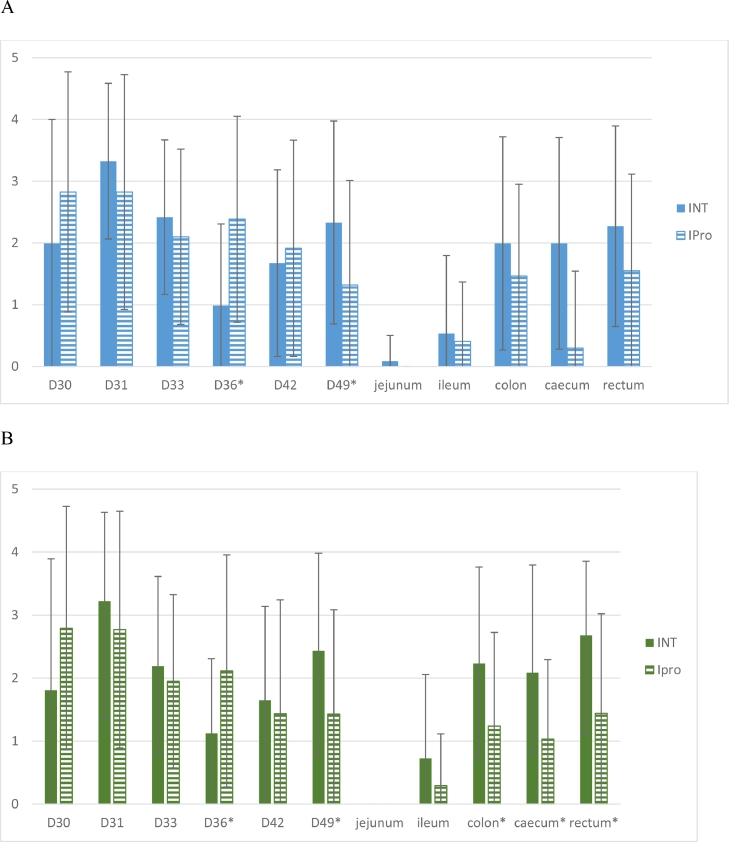
No ESCR E. coli were detected in the 29 faeces samples collected from sows from 12 days before the expected day of farrowing and up to the weaning day (day 28). No ESCR and no RifR E.coli were detected in any of the piglet faecal samples before E. coli 17–348F-RifR inoculation. The mean numbers of CFU/g of faecal samples collected from day 30 to day 49 and PM samples obtained on MC—CTX and on MC-Rif are shown in Fig. 1, and the numbers of MC—CTX- and MC-Rif- positive samples are given in Table 1. Concerning the titers obtained on MC—CTX, the overall impact of probiotics was not significant (p = 0.14) whereas the one of the interaction between ‘Treatment’ and ‘Time’ was significant. More precisely, the time evolution of the titers was significantly different between groups on day 36 (p = 0.0010, marked decrease for the INT group) and day 49 (p = 0.0340, with a global decrease and a lower titre for the Ipro group). Significantly more positive faecal samples were detected in the IPro group compared with the INT group on day 36 (p = 0.007). For PM samples, there were no significant differences between titres or ratios of positive samples of the INT and IPro groups. Considering all PM samples, there was no significant difference between the ratios of animals yielding at least one positive sample on MC—CTX plates (19/24 for INT pigs vs 12/23 for IPro pigs, (p > 0.05)). All tested faecal and PM isolates (n = 264) were identified as E. coli, grew on MC-Rif agar, and had the blaCTX−M-1 gene. According to PCR, none of these isolates carried ED1a- or EcN-specific genes.
Fig. 1.
Titres (in log10 CFU/g) obtained for faecal samples from day 30 to day 49 and post-mortem samples (mean titres +/- standard deviation) on MC—CTX (A) or on MC-Rif (B) selective agar.*Asterisks indicate significant differences between groups.All samples collected on day 28 (before inoculation of E. coli 17–348F-RifR) gave negative results. .
Table 1.
Numbers of positive faecal samples obtained on MC—CTX and MC-Rif agar.
| Faecal samples | PM samples | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Agar |
Group |
D28a (−1b) |
D30 (+1) |
D31 (+2) |
D33 (+4) |
D36 (+7) |
D42 (+13) |
D49 (+20) |
Jejunum | Ileum | Colon | Caecum | Rectum | ||
| MC—CTX | INT | 0/24c | 13/24 | 22/24 | 20/24 | 9/24* | 14/24 | 17/24 | 1/24 | 4/24 | 15/24 | 16/24 | 17/24 | ||
| IPro | 0/24 | 17/23 | 17/23 | 17/23 | 18/23* | 14/23 | 10/23 | 0/23 | 4/23 | 12/23 | 11/23 | 12/23 | |||
| MC-Rif | INT | 0/24 | 11/24* | 21/24 | 18/24 | 12/24 | 14/24 | 18/24 | 0/24 | 6/24 | 18/24* | 16/24 | 22/24* | ||
| IPro | 0/24 | 17/23* | 17/23 | 17/23 | 15/23 | 10/23 | 11/23 | 0/23 | 3/23 | 10/23* | 10/23 | 11/23* | |||
Age of piglets in days;.
Day before or after inoculation of E. coli 17–348F-RifR.
number of positive samples/number of tested samples.
For each agar medium, significant differences between groups are indicated with an asterisk.
Concerning the titers obtained on MC-Rif, there was only a tendency (p = 0.09) for the probiotic administration to yield higher titers. The time evolution model showed significant differences between groups on day 36 (p = 0.0257) and on day 49 (p = 0.0286), with more fluctuant titers in the INT group compared to the rather regular decrease for the Ipro group. The numbers of MC-Rif-positive faecal samples were significantly different on day 36 (more positive samples in the IPro group, p = 0.049) and day 49 (more positive samples in the INT group, p = 0.001). For PM samples, significant differences were detected for the titres of caecum, colon and rectum samples (p = 0.02, p = 0.04 and p = 0.02, respectively), with lower titres in the IPro group, and the number of positive colon and rectum samples were significantly higher in the INT pigs than in the IPro group (p = 0.04 and p = 0.02, respectively). Considering all PM samples, significantly more INT pigs were positive than IPro pigs (respectively 23/24 vs 11/23, p = 0.0003). All tested isolates (n = 257) were E. coli; they were able to grow on MC—CTX and carried the blaCTX−M-1 gene.
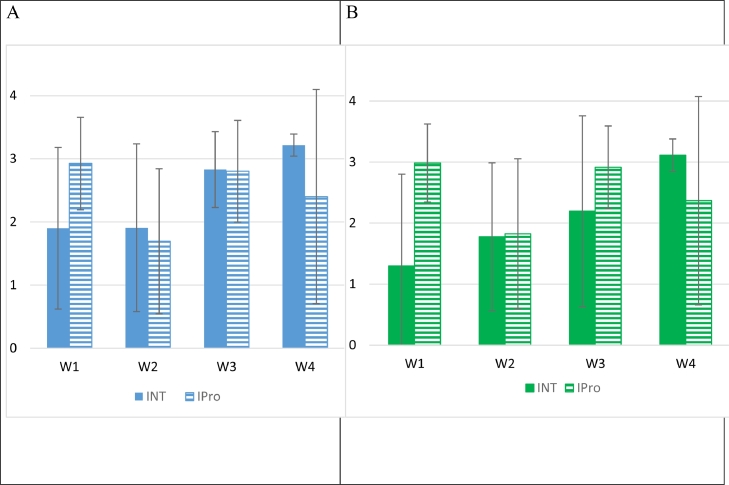
Titres obtained for manure on MC—CTX and MC-Rif are given in Fig. 2. No significant differences were detected between the INT and IPro groups (p > 0.05). The 24 lysates prepared from colonies grown on MC—CTX gave negative results when tested with the ED1a- and EcN-specific PCR.
Fig. 2.
Titres (in log10 CFU/g) obtained for manure samples (mean titres +/- standard deviation) collected each week (W) after E. coli 17–348F-RifR inoculation, on MC—CTX (A) or on Rif-CTX (B) selective agar.
Quantification of the probiotic strains and total E. coli
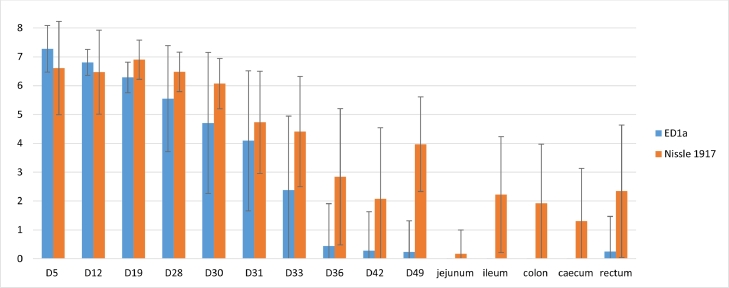
The results of qPCR for E. coli ED1a, E. coli EcN and total E. coli in faecal and PM samples are shown in Figs. 3, S2 and S3 and Table 2.
Fig. 3.
Quantification using qPCR of E. coli ED1a and E. coli Nissle 1917 in faecal and post-mortem samples from infected treated (IPro) pigs.Titres are expressed in log10 copies per g of sample (mean titre +/- standard deviation) Piglets were weaned on day 28 (D28) and inoculated on day 29. They were given the probiotics from birth to day 49.
Table 2.
Numbers of faecal and PM samples positive for E. coli ED1a and Nissle 1917 (EcN) in the infected treated (IPro) group.
| Faecal samples | PM samples | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day or sample | D5a | D12 | D19 | D28 | D30 | D31 | D33 | D36 | D42 | D49 | Jejunum | Ileum | Colon | Caecum | Rectum | |
| ED1a | 23/23b | 23/23 | 23/23 | 20/23 | 13/16 | 16/21 | 10/21* | 2/23* | 1/23* | 1/23* | 0/23 | 0/23* | 0/23* | 0/23* | 1/23* | |
| EcN | 23/23 | 23/23 | 23/23 | 23/23 | 16/16 | 19/21 | 18/21* | 14/23* | 10/23* | 20/23* | 1/23 | 13/23* | 11/23* | 8/23* | 12/23* | |
Age of piglets;.
number of positive samples/number of tested samples.All samples from the infected non-treated (INT) group were negative.
For each day, significant differences are indicated with an asterisk.
Regarding total E. coli, all faecal samples were positive, as were colon, caecum and rectum samples. Concerning PM samples, only 1/24 INT and 4/23 IPro jejunum samples were positive (p > 0.05), but 17 out of 24 INT ileum samples and 19/23 IPro ileum samples were positive (p > 0.05).
All samples from non-treated sows and piglets were negative for E. coli ED1a and EcN. The mean titres obtained for E. coli ED1a for treated sows were higher than 5 log10/g from day-12 to day 19, and decreased to 3.51+/−3.16 on day 28. For E. coli EcN, the mean titres remained higher than 5.62+/- 1.83 (Figure S3). Before weaning and during the first days after inoculation, the two probiotics were detected in most faecal samples collected from piglets. Thereafter, the mean titres and the numbers of positive samples for ED1a decreased (Fig. 3) and only 1 PM sample — a rectum sample — was positive. E. coli EcN was significantly more frequently detected than E. coli ED1a in faecal samples from day 33 until the end of the experiment, and in ileum, colon, caecum and rectum samples (p < 0.05 each). PM samples from 19 out of 23 pigs were detected positive for E. coli EcN, but only 1 pig tested positive for ED1a. Overall, EcN was more frequently detected in faecal and PM samples than was ED1a (234 vs 133 positive samples out of 334, p < 0.001).
Because very few faecal samples tested positive for ED1a, cell lysates from all pooled colonies grown on MC non-supplemented plates were prepared from faecal samples collected on day 49 and rectum samples at PM. All 48 suspensions prepared from INT pigs gave negative results, and 16 out of 23 faecal samples on day 49 and 9 rectum out of 23 IPro samples were positive, respectively. Thus, the sensitivity of the PCR on lysates of pooled E. coli colonies was better than that of the PCR on DNA from faecal samples (25 vs 2 positive samples out of 46 samples, p < 0.001).
Results obtained for manure samples are presented in Figure S4. The mean numbers of total E. coli copies were not significantly different between INT and IPro samples. E. coli ED1a and EcN were not detected in the manure from INT pigs. The comparison between the numbers of copies for the different weeks showed that the total E. coli and EcN copies numbers were stable over time, whereas the ED1a copies numbers were significantly lower during the fourth week, after the end of the probiotic administration (p < 0.03).
Discussion
Overall, there was no significant reduction in faecal excretion of CTXR and RifR E. coli in IPro pigs compared with INT pigs, although the CTXR and RifR E. coli titres were slightly, but significantly lower in the colon, caecum and rectum at post mortem. Excretion of ESCR E. coli by animals is a public health issue. In a previous study (Mourand et al., 2017), we showed that the reduction in the level of faecal excretion of CTXR- E. coli in E. coli ED1a-treated pigs compared with that in non-treated pigs was usually less than 1 log10 CFU and was mainly observed during the probiotic administration period. The aim of the present trial was to determine whether an extended administration of two probiotics (E. coli ED1a and EcN) can reduce the shedding or carriage of an ESCR E. coli strain. In comparison with the fourth trial described in (Mourand et al., 2017), two probiotics were used and given together to piglets before and after challenge with ESCR E. coli. We also used a different ESCR E. coli strain and the challenge inoculum dose was 10 times lower.
In the very first days following inoculation, we observed temperature increases in the INT group, but not in the IPro group and several episodes of mild diarrhoea were observed. Different events (stress of weaning, dietary changes…) may lead to such clinical signs which might also indicate a moderate pathogenic effect of the E. coli 17–348F-RifR inoculation, probably associated with the detected virulence genes of E. coli 17–348F-RifR including extra-intestinal pathogenic E. coli (ExPEC) genes. The frequent presence of ExPEC-associated genes, such as iss, iha, tsh or iroN, in commensal E. coli isolated from pigs has previously been reported (Li, Ma, Li, Dai, & Zhang, 2020). E. coli 17–348F-RifR carries four genes (hlyF, iroN, iss and ompT) of the five plasmid genes that have been shown to be predictors of pathogenicity of avian pathogenic E. coli (Johnson et al., 2008a), as well as other genes such as traT, sitA, cvaC or fyuA that are also frequently found in human ExPEC (Johnson et al., 2008b). Several E. coli 17–348F-RifR genes (cvaC, etsA, hlyA, iroN, iss, ompT, and sitA) have also been detected in one of the plasmids of a swine virulent ExPEC (Lemaitre et al., 2013; Liu et al., 2015). The difference in growth performance between IPro and INT pigs may result from different breeding conditions during the suckling period, with, due to facility constraints, non-treated sows and their piglets being placed in slightly more comfortable rooms.
In our experimental conditions, using an inoculum of 6.4 log10 CFU per pig, we observed that the ESCR strain readily colonized the animals and was excreted or detected in the digestive tract in all but one of the non-treated animals up to the end of the trial, four weeks after inoculation. During the experiment, the titres in faeces remained relatively low, most often between 1 log10 and 3 log10/g. Four INT pigs tested negative on three consecutive weeks (day 36, day 42 and day 49), but carried E. coli 17–348 M according to results obtained for the PM samples. This may result from reinfection between animals due to the burrowing behaviour of the pigs, or to differences between the excreted E. coli population and the E. coli populations present or adhering to the different parts of the digestive tract, as discussed below.
Amongst the 264 E. coli isolates obtained from pigs and 28 E. coli isolates obtained from manure samples on MC—CTX, all were rifampicin-resistant, suggesting no transfer of cefotaxime resistance to other E. coli (including to the probiotics), contrary to our previous trials with the M63 strain (Fleury et al., 2015; Mourand et al., 2017), and to the usually very efficient conjugation system of IncI1 plasmids (Carattoli, 2013). Several attempts of in vitro conjugation between the E. coli 17–348 strain and three different receptor commensal E. coli strains, as well as conjugation between E. coli 17–348F-RifR and each probiotic strain, were unsuccessful, possibly due to non-functional plasmid conjugative transfer genes, as suggested by the truncated sequences coding for SogS, PilN, PilJ, PilL and TraQ (data not shown).
All 257 faecal E. coli isolates obtained on MC-Rif were resistant to ESC and carried the blaCTX−M gene, but we detected one E. coli isolate resistant to rifampicin, but not to other antimicrobials, amongst the 26 tested manure isolates. WGS of this isolate confirmed that it belonged to the same ST 767 as the inoculated strain, but had lost the IncI1 replicon and the blaCTX−M-1, sul2 and tetA genes present in this plasmid (data not shown). These data suggest that, overall, the loss of the ESCR gene occured rather rarely, probably due to the in vivo stability of this resistance plasmid in E. coli 17–348F-RifR.
PCR on DNA obtained from faecal and PM samples was used to detect the presence of the two probiotics. The excretion of probiotics appeared higher during the suckling period and decreased thereafter, with large variations between piglets. This difference in excretion is probably due to the administration methods: the individual oral administration of probiotics to suckling piglets allowed each piglet to receive the desired dose, whereas — although we confirmed the stability of the stored suspensions of probiotics over one week — in-feed administration may reduce the actual ingested doses. It is also well known that individual food intake depends on several factors such as temperature, type of housing and feeding systems and social rank (Soraci, Amanto, Tapia, de la Torre & Toutain, 2014). The modification of the intestinal microbiota after weaning may also have a strong impact on the competition between the different bacterial species of the flora and the probiotics. These different factors may partly explain why the numbers of ED1a-positive samples were quite low for the last sampling days. Our in vitro co-culture of the two probiotics showed that they can grow together at rather similar rates, suggesting the absence of competition mechanisms between these two E. coli strains, such as production of bacteriocins (Hrala et al., 2021) or phages, but we cannot exclude competition between the probiotics and other gut bacteria. The sensitivities of the E. coli ED1a PCR and the EcN PCR have previously been shown to be 31 and 8 cells per assay, respectively (Mourand et al., 2017). The better sensitivity of the latter strain may account for the more frequent detection of EcN in faecal and PM samples compared with ED1a. The low sensitivity of the ED1a PCR method performed on DNA extracts from faecal and PM samples was later confirmed in a comparison of results obtained on DNA from a few faecal and PM samples and those obtained on lysates prepared from the pool of E. coli colonies that appeared on non-supplemented MC agar for the same samples. Nevertheless, the different test methods confirmed that both probiotics were still present in several IPro pigs up to day 57, eight days after the end of the administration of probiotics.
Methods to decrease the prevalence of ESCR Enterobacterales in animal production must first include biosecurity measures to prevent the introduction of resistant strains or administration of feed additives (Roth et al., 2017).
In addition, in poultry, competitive exclusion (CE) has been studied for many years (Nurmi, Nuotio & Schneitz, 1992). Because the early environment can have a major influence on the microbiota in chicks, it is tempting to develop intervention strategies that promote microbiota that can better withstand various invaders, such as Salmonella, Campylobacter or resistant bacteria (Dame-Korevaar et al., 2020). Experimental studies have shown that some CE products can reduce colonization, excretion and transmission of ESBL- or AmpC-producing E. coli (Ceccarelli et al., 2017; Dame-Korevaar et al., 2020; Methner, Friese & Rösler, 2019; Nuotio, Schneitz & Nilsson, 2013). Because the use of well-characterized strains is preferable to products of unknown composition, we used the probiotic E. coli ED1a and EcN strains. In addition to the already mentioned characteristics of these two strains, we showed that they did not inhibit each other, and, in our in vitro and in vivo conditions, the resistance plasmid of E. coli 17–348F-RifR was not transferred to them by conjugation. In our pig trial, for faecal samples, a positive impact of these probiotics was observed only on day 49, with significantly lower ESCR E. coli titres in treated pigs. However on day 30, day 31, day 33 and day 42, no beneficial impact of the probiotic was observed, and a negative impact on the excretion was even observed on day 36, perhaps in relation with the diarrhoea symptoms presented by the IPro group on that day. Thus, overall, as confirmed by the manure samples, the administration of probiotics did not reduce ESCR E. coli excretion.
However, the PM samples gave a different picture, because the ESCR E. coli titres were slightly — but significantly — lower for colon, caecum and rectum samples and more INT pigs were positive than IPro pigs. These results may be considered as a confirmation of the results obtained on day 49, and as a surprisingly delayed positive effect of the probiotics. Another explanation may be that the probiotics have a more pronounced effect on the E. coli present or adhering to the different parts of the digestive tract than on the excreted population. For instance, Bednorz et al. (Bednorz et al., 2013) reported that a probiotic may primarily affect mucosa-adherent E. coli, with the influence of the overall population of E. coli being minor. In their experiment, the use of Enterococcus faecium NCIMB 10,415 as a probiotic for pigs led to a reduction in isolates harbouring ExPEC-virulence genes adherent to the mucosa of the colon, suggesting a prophylactic effect. According to their observations, they also concluded that relying on faecal sample results is questionable. Because our PM samples contained a mixture of digesta and scraped mucosa, it is difficult to evaluate whether the beneficial effect of the treatment observed at PM is associated with the difference of sampling (digesta and mucosa vs faeces) or a delayed effect. Further trials with the collection of PM samples at different times and separate analysis of faeces, digesta and mucosa may help to understand the impact of the E. coli probiotics on the carriage and excretion of the inoculated ESCR E. coli and on the strain of E. coli and ExPEC. In case of positive effects, the probiotics may be of interest for preventing ExPEC infections.
Conclusion
Although the two probiotics were given to the sows before farrowing, then to the piglets at high doses from birth to 49 days of age (i.e. four weeks before and three weeks after challenge), they could not prevent or significantly and durably reduce the excretion of E. coli 17–348F-RifR. Analysis of PM samples suggested a slight, but significant decrease in the E. coli population in the colon, caecum and rectum, but other trials with separate analysis of tissues and digesta or faeces collected at different times are needed to further investigate the phenomenon.
Funding information
This work was supported by a grant 2017–389 from the Ecoantibio Plan — a public policy programme set up by the French Ministry of Agriculture, Agro-Food and Forestry to reduce the risks of antibiotic resistance in veterinary medicine —, ANSES (the French Agency for Food, Environmental and Occupational Health & Safety) and the Côtes d'Armor Departmental Council. ED was partially supported by the “Fondation pour la Recherche Médicale” (Equipe FRM 2016, grant number DEQ20161136698). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Ethical statement
The experiments were performed in accordance with French animal welfare regulations and the protocol was approved by the ANSES/ENVA/UPEC ethical committee and the French Ministry for Higher Education, Research and Innovation (ComEth authorization 2019/01/17–5, no. APAFIS: 2,018,121,812,512,422 (# 18,136)).
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to C. Miossec (Da Volterra, Paris) for sharing the EcN PCR protocol and to Yann Bailly (ANSES) for his technical assistance.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.vas.2021.100217.
Appendix. Supplementary materials
References
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednorz C., Guenther S., Oelgeschläger K., Kinnemann B., Pieper R., Hartmann S., et al. Feeding the probiotic Enterococcus faecium strain NCIMB 10415 to piglets specifically reduces the number of Escherichia coli pathotypes that adhere to the gut mucosa. Applied and Environmental Microbiology. 2013;79:7896–7904. doi: 10.1128/AEM.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet O., Bourdelier E., Glodt J., Clermont O., Denamur E. Diversity of the auxotrophic requirements in natural isolates of Escherichia coli. Microbiology (Reading) 2017;163:891–899. doi: 10.1099/mic.0.000482. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. International Journal of Medical Microbiology. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Ceccarelli D., van Essen-Zandbergen A., Smid B., Veldman K.T., Boender G.J., Fischer E.A.J., et al. Competitive exclusion reduces transmission and excretion of extended-spectrum-β-lactamase-producing Escherichia coli in broilers. Applied and Environmental Microbiology. 2017;83:16. doi: 10.1128/AEM.03439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ye W., Zhang Y., Xu Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Lescat M., O'Brien C.L., Gordon D.M., Tenaillon O., Denamur E. Evidence for a human-specific Escherichia coli clone. Environmental Microbiology. 2008;10:1000–1006. doi: 10.1111/j.1462-2920.2007.01520.x. [DOI] [PubMed] [Google Scholar]
- Dame-Korevaar A., Kers J.G., van der Goot J., Velkers F., Ceccarelli D., Mevius D., et al. Competitive exclusion prevents colonization and compartmentalization reduces transmission of ESBL-producing Escherichia coli in broilers. Frontiers in Microbiology. 2020;11:1–12. doi: 10.3389/fmicb.2020.566619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E., Clermont O., Bonacorsi S., Gordon D. The population genetics of pathogenic Escherichia coli. Nature Reviews in Microbiology. 2021;19:37–54. doi: 10.1038/s41579-020-0416-x. [DOI] [PubMed] [Google Scholar]
- EFSA Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA Journal. 2011;9:2322. [Google Scholar]
- EFSA/ECDC The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA Journal. 2020;18(3):166. doi: 10.2903/j.efsa.2020.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA/ECDC The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA Journal. 2021;19(4):67–112. doi: 10.2903/j.efsa.2021.6490. 6490,6179 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury M.A., Mourand G., Jouy E., Touzain F., Le Devendec L., De Boisseson C., et al. Impact of ceftiofur injection on gut microbiota and Escherichia coli resistance in pigs. Antimicrobial Agents and Chemotherapy. 2015;59:5171–5180. doi: 10.1128/AAC.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet J.P., Firmesse O., Gourmelon M., Bridonneau C., Tap J., Mondot S., et al. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiology Ecology. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- Hancock V., Dahl M., Klemm P. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. Journal of Medical Microbiology. 2010;59:392–399. doi: 10.1099/jmm.0.008672-0. [DOI] [PubMed] [Google Scholar]
- Hrala M., Bosak J., Micenkova L., Krenova J., Lexa M., Pirkova V., et al. Escherichia coli Strains Producing Selected Bacteriocins Inhibit Porcine Enterotoxigenic Escherichia coli (ETEC) under both In Vitro and In Vivo Conditions. Applied and Environmental Microbiology. 2021;87 doi: 10.1128/AEM.03121-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.J., Wannemuehler Y., Doetkott C., Johnson S.J., Rosenberger S.C., Nolan L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. Journal of Clinical Microbiology. 2008;46:3987–3996. doi: 10.1128/JCM.00816-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.J., Wannemuehler Y., Johnson S.J., Stell A.L., Doetkott C., Johnson J.R., et al. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Applied and Environmental Microbiology. 2008;74:7043–7050. doi: 10.1128/AEM.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre C., Mahjoub-Messai F., Dupont D., Caro V., Diancourt L., Bingen E., et al. A conserved virulence plasmidic region contributes to the virulence of the multiresistant Escherichia coli meningitis strain S286 belonging to phylogenetic group C. PloS one. 2013;8:e74423. doi: 10.1371/journal.pone.0074423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM ». arXiv preprint arXiv:1303.3997, 2013. https://arxiv.org/abs/1303.3997.
- Li Y., Ma X., Li C., Dai X., Zhang L. Occurrence and genomic characterization of ESBL-producing Escherichia coli ST29 strains from swine with abundant virulence genes. Microbial Pathogenesis. 2020:1–7. doi: 10.1016/j.micpath.2020.104483. [DOI] [PubMed] [Google Scholar]
- Liu C., Zheng H., Yang M., Xu Z., Wang X., Wei L., et al. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC genomics. 2015;16:1–18. doi: 10.1186/s12864-015-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas P., Jouy E., Le Devendec L., de Boisseson C., Perrin-Guyomard A., Jove T., et al. Characterization of plasmids harboring blaCTX-M genes in Escherichia coli from French pigs. Veterinary Microbiology. 2018;224:100–106. doi: 10.1016/j.vetmic.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Methner U., Friese A., Rösler U. Competitive exclusion: A tool to combat extended-spectrum β-lactamase-producing Escherichia coli strains in chickens. Research in Veterinary Science. 2019;123:124–128. doi: 10.1016/j.rvsc.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Mourand G., Paboeuf F., Fleury M.A., Jouy E., Bougeard S., Denamur E., Kempf I., et al. Escherichia coli Probiotic Strain ED1a in Pigs Has a Limited Impact on the Gut Carriage of Extended-Spectrum-beta-Lactamase-Producing E. coli. Antimicrob Agents and Chemotherapy. 2017;61:1–16. doi: 10.1128/AAC.01293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuotio L., Schneitz C., Nilsson O. Effect of competitive exclusion in reducing the occurrence of Escherichia coli producing extended-spectrum beta-lactamases in the ceca of broiler chicks. Poultry Science. 2013;92:250–254. doi: 10.3382/ps.2012-02575. [DOI] [PubMed] [Google Scholar]
- Nurmi E., Nuotio L., Schneitz C. The competitive exclusion concept: Development and future. International Journal of Food Microbiology. 1992;15:237–240. doi: 10.1016/0168-1605(92)90054-7. [DOI] [PubMed] [Google Scholar]
- Roth N., Mayrhofer S., Gierus M., Weingut C., Schwarz C., Doupovec B., et al. Effect of an organic acids based feed additive and enrofloxacin on the prevalence of antibiotic-resistant e. Coli in cecum of broilers. Poultry Science. 2017;96:4053–4060. doi: 10.3382/ps/pex232. [DOI] [PubMed] [Google Scholar]
- Soraci A.L., Amanto F., Tapia M.O., de la Torre E., Toutain P.L. Exposure variability of fosfomycin administered to pigs in food or water: Impact of social rank. Research in Veterinary Science. 2014;96:153–159. doi: 10.1016/j.rvsc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Thomsen M.C., Ahrenfeldt J., Cisneros J.L., Jurtz V., Larsen M.V., Hasman H., et al. A Bacterial Analysis Platform: An Integrated System for Analysing Bacterial Whole Genome Sequencing Data for Clinical Diagnostics and Surveillance. PloS one. 2016;11(6) doi: 10.1371/journal.pone.0157718. e0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verliat F., Hemonic A., Chouet S., Le Coz P., Liber M., Jouy E., et al. An efficient cephalosporin stewardship programme in French swine production. Veterinay Medicine Science. 2021;7:432–439. doi: 10.1002/vms3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2019). WHO advisory group on integrated surveillance of antimicrobial resistance (AGISAR): Critically important antimicrobials for human medicine, 6th revision (https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/).
- Woodford N., Fagan E.J., Ellington M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. Journal of Antimicrobial Chemotherapy. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.