Abstract
Filamentous soil bacteria are known to produce diverse specialized metabolites. Despite having enormous potential as a source of pharmaceuticals, they often produce bioactive metabolites at low titers. Here we show that inactivation of the pactamycin, NFAT-133, and conglobatin biosynthetic pathways in Streptomyces pactum ATCC 27456 significantly increases the production of the mitochondrial electron transport inhibitors piericidins. Similarly, inactivation of the pactamycin, NFAT-133, and piericidin pathways significantly increases the production of the heat shock protein (Hsp) 90 inhibitor conglobatin. In addition, four new conglobatin analogues (B2, B3, F1, and F2) with altered polyketide backbones, together with the known analogue conglobatin B1, were identified in this mutant, indicating that the conglobatin biosynthetic machinery is promiscuous toward different substrates. Among the new conglobatin analogues, conglobatin F2 showed enhanced antitumor activity against HeLa and NCI-H460 cancer cell lines compared to conglobatin. Conglobatin F2 also inhibits colony formation of HeLa cells in a dose-dependent manner. Molecular modeling studies suggest that the new conglobatins bind to human Hsp90 and disrupt Hsp90/Cdc37 chaperone/co-chaperone interactions in the same manner as conglobatin. The study also showed that genes that are involved in piericidin biosynthesis are clustered in two different loci located distantly in the S. pactum genome.
Keywords: conglobatin, Streptomyces pactum, polyketide synthase, genetic engineering, anticancer, molecular docking
Graphical Abstract
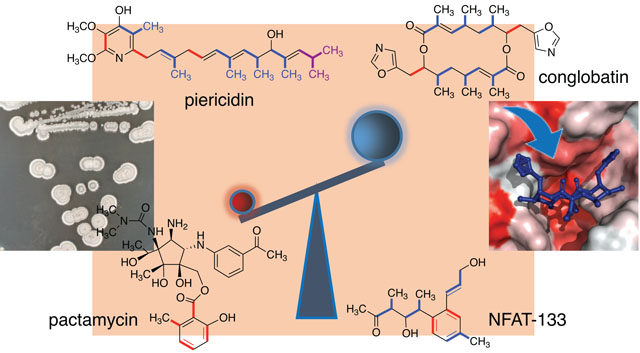
INTRODUCTION
The strains of Streptomyces, a genus of filamentous bacteria found in soil and water, are known to be a rich source of bioactive natural products.1 They produce a wide variety of specialized metabolites, e.g., polyketides, non-ribosomal peptides, terpenes, and aminocyclitols, with excellent biological activities.2–4 However, those metabolites are often produced at low concentrations, diminishing their prospects as commercially viable drug candidates. In bacteria, the production of specialized metabolites is normally controlled by certain regulatory systems. In addition to global and pathway specific regulators,5–7 which may be influenced by pH, temperature, osmotic pressure, nutrients and cell density,8 the production of specialized metabolites may also be affected by the availability of the biosynthetic precursors and co-factors in the cells.9,10 The latter element is thought to be more significant in the situations where several major biosynthetic machineries compete for the same source of building blocks.
Recently, we reported a draft genome sequence of Streptomyces pactum ATCC 27456,11 the producer of the antitumor antibiotic pactamycin (1) (Figure 1).12 In silico analysis of the genome sequence uncovered 31 putative BGCs, among which nine are polyketide synthase (PKS) gene clusters, including ptmQ, which encodes an iterative type-I PKS responsible for the production of 6-methylsalicylate in pactamycin biosynthesis.10,13–15 In addition to pactamycin, the ATCC 27456 strain also produces the polyketides NFAT-133 (2), piericidins (3–6), and conglobatin (7) (Figure 1).11,16,17 The aromatic polyketide NFAT-133, which is the major product of this strain, has immunosuppressive, antidiabetic, and antitrypanosomal activities. It inhibits the nuclear factor of activated T cells (NFAT), which is responsible for its immunosuppressive activity, and activates the AMPK pathway, which causes the increase of insulin-stimulated glucose uptake in L6 myotubes. In S. pactum, NFAT-133 is synthesized by modular type-I PKSs whose genes are highly disorganized and inconsistent with collinearity of regular modular type-I polyketide assembly lines.11
Figure 1.
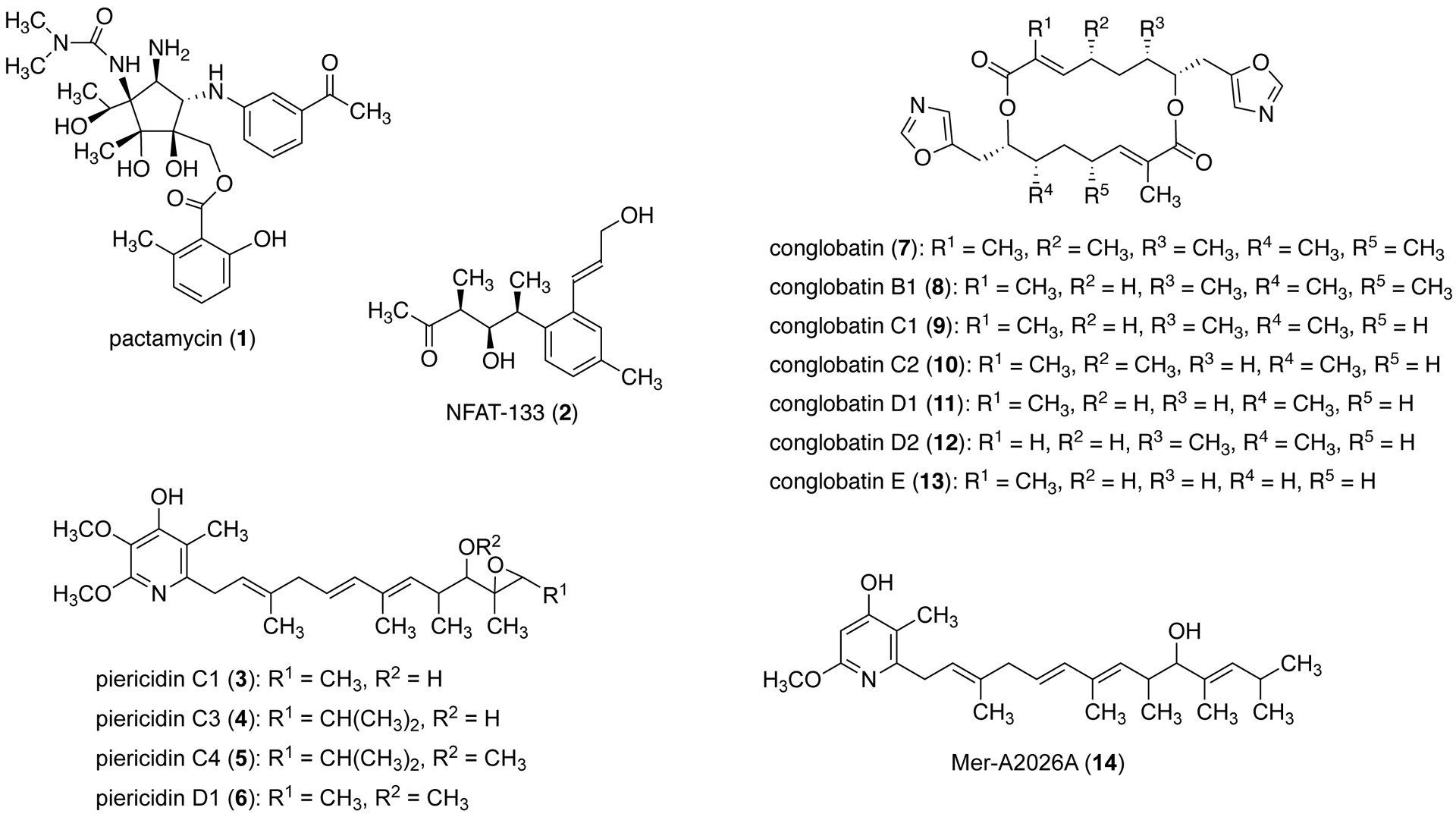
Chemical structures of pactamycin, NFAT-133, conglobatins, and piericidins.
Piericidins are also polyketide natural products which are synthesized by modular type-I PKSs.18,19 Compounds from this family of natural products have been shown to have various biological activities, such as inhibition of mitochondrial electron transport, inhibition of phosphatidylinositol turnover, inhibition of cell division of fertilized starfish eggs, and vasodilation.20–22 Piericidins A1 and B1 were found to be major components of the antibiotic cocktail produced by the symbiotic Streptomyces bacteria of beewolves from across geographical areas.23
The oxazole-containing C2-symmetrical 16-membered macrodiolide conglobatin is also synthesized by modular type-I PKSs.16,24 This antitumor compound was first isolated from a soil actinomycete, Streptomyces conglobatus ATCC 31005,25 but later was also found in other strains of Streptomyces.16,26,27 Conglobatin can cause G2/M cell-cycle arrest, induce apoptosis, and downregulate the expression of heat-shock protein Hsp90.26 Recently, six new conglobatin analogues (8–13) (Figure 1) were reported from an Australian Streptomyces strain MST-91080.28 Many of them have enhanced cytotoxic activity against the NS-1 myeloma cell line, raising hope that structural modifications of conglobatin may lead to better compounds.
To modulate the production of specialized metabolites in S. pactum ATCC 27456 and to discover new bioactive compounds, we genetically engineered mutant strains of S. pactum ATCC 27456, in which the BCGs of pactamycin, NFAT-133, conglobatin, and/or piericidins are individually or in combination inactivated. Here, we report multi-fold enhancement in the production of piericidins or conglobatin in some of these mutants, as well as the isolation, structure characterization, biosynthesis, and bioactivity profiles of four new conglobatin analogues.
RESULTS AND DISCUSSION
Inactivation of Competing Pathways Increases Piericidin Production.
Earlier studies have shown that pactamycin, NFAT-133, and/or conglobatin are synthesized by either modular or iterative type I PKSs. The 6-methylsalicylyl moiety of pactamycin is formed by the iterative type I PKS PtmQ,13 whereas NFAT-133 and conglobatin are synthesized by modular type I PKSs encoded in S. pactum ATCC 27456 by BGC-1.2 and BGC-1.23, respectively.11,16 Previously, we generated a mutant strain of S. pactum in which several pactamycin biosynthetic genes including ptmQ were inactivated (S. pactum ΔptmTDQ).11 While the production of pactamycin was diminished, the gene inactivation did not seem to significantly affect the production level of other metabolites (Figure 2). Subsequently, we inactivated the NFAT-133 BGC in the pactamycin mutant – to produce S. pactum ΔptmTDQ/ΔBGC-1.2 – and analyzed the metabolites of the mutant.11 The results showed that the inactivation of the NFAT-133 BGC substantively enhanced the production of piericidins and to a lesser extent also enhanced the production of conglobatin (Figure 2), indicating that there is interconnectivity between the NFAT-133 and the other two PKS pathways. The enhanced production of piericidins and conglobatin in this mutant may be due to the increased availability of the polyketide building blocks and/or some other factors, e.g., cross regulations between secondary metabolites. Isolation and structure characterization of the major peaks revealed the identity of the compounds to be conglobatin (7), piericidins C1 (3), C3 (4), C4 (5), D1 (6), and Mer-A2026A (14) (Figure 1). Quantitative analysis, based on the area-under-curve calculations of their HPLC peaks, showed that the production of piericidins C1, C3, C4, D1, and Mer-A2026A increased by 2- to 2.5-fold (Figure 2a–b).
Figure 2.
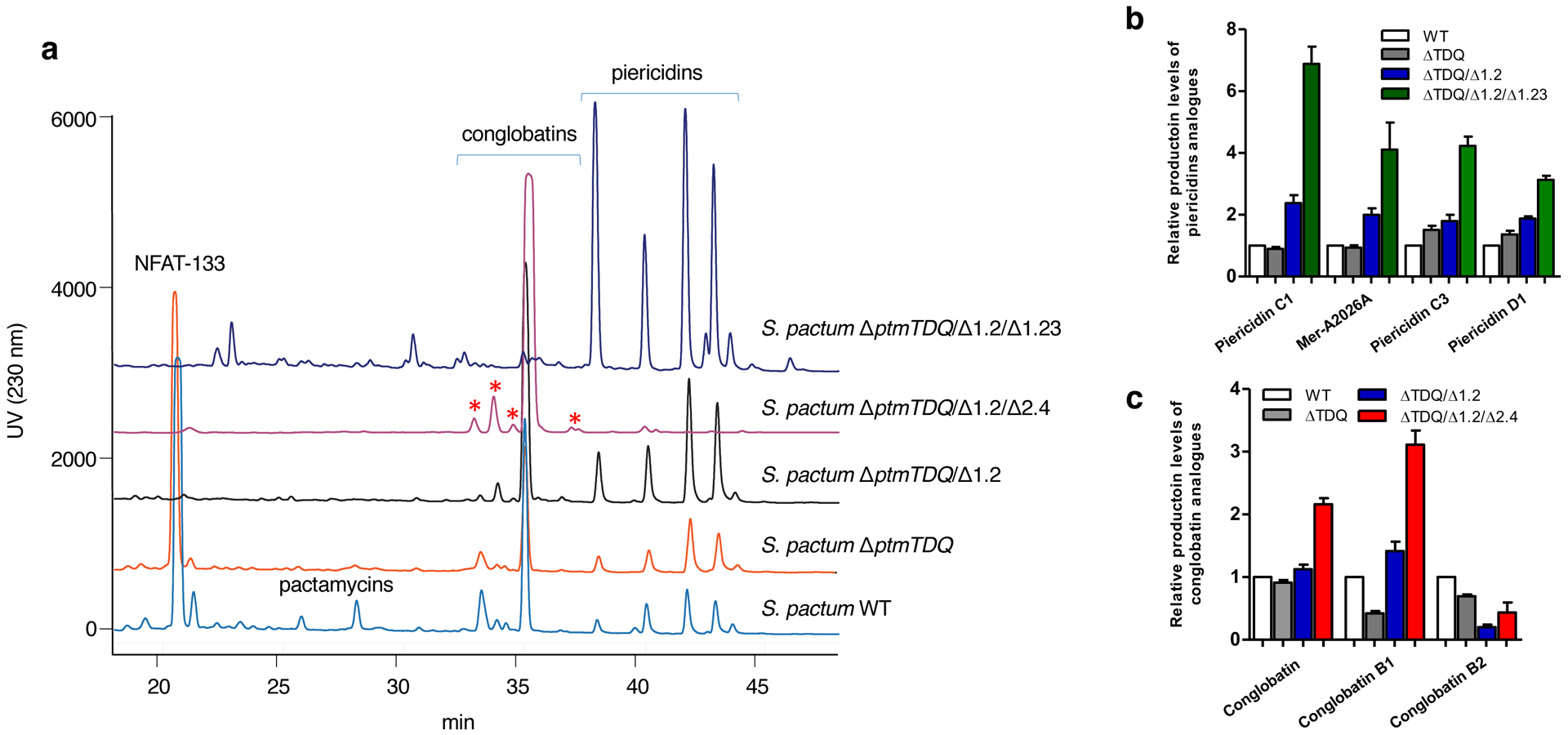
Phenotypic analysis of Streptomyces pactum mutants. (a) HPLC traces of the mutants; (b) comparisons of the production yields of the piericidins by various mutants; (c) comparisons of the production yields of conglobatins by various mutants. Error bars indicate s.d. (n=3 analytical replicates).
To further investigate the effects of polyketide biosynthetic pathways on the production of other polyketide compounds, we inactivated the conglobatin pathway (BGC-1.23) in the ΔptmTDQ/ΔBGC-1.2 strain to produce a triple-BGC knockout mutant (ΔptmTDQ/ΔBGC-1.2/BGC-1.23). This triple-BGC knockout mutant was generated by in-frame deletion of a 2909-bp DNA fragment part of the congC1 and congC2 genes in BGC-1.23 according to the procedure reported previously.16 The mutant was confirmed genetically by comparing the PCR fragments of the mutant and the wild-type (Supplementary Figure 1). Subsequently, the mutant was cultured in a liquid BTT medium for 7 days at 30 °C and the culture broth was extracted with EtOAc. The extract was then analyzed by HPLC and mass spectrometry. Compared to the other two mutants, this strain produced piericidins in significantly increased yields, where piericidins C1, C3, D1, and Mer-A2026A were produced 7, 4, 3, 4-fold, respectively, higher than the wild-type (Figure 2a–b). The results suggest that the production yields of the piericidins directly correlate with the availability of their biosynthetic precursors, i.e., malonyl-CoA and methylmalonyl-CoA.
Identification of the Piericidin BGC in S. pactum.
The biosynthetic gene cluster of piericidin A1 has been reported in Streptomyces sp. SCSIO 03032.18,19 It consists of six modular type I PKS genes (pieA1 – pieA6) flanked upstream by a regulatory gene (pieR) and downstream by five tailoring genes (pieB1, pieC, pieD, pieB2, and pieE). An inspection of the draft genome sequence of S. pactum (consists of two contigs) using AntiSMASH resulted in two loci that contain PKS genes similar to those of piericidin A1, but they appeared to be incomplete or separated.11 The first locus, BGC-1.26, only contains a modular type I PKS gene similar to pieA6 and five tailoring genes similar to pieB1, pieC, pieD, pieB2, and pieE (Figure 3). The second locus (BGC-2.4) consists of a gene homologous to pieR and five PKS genes similar to pieA1 – pieA5. There is an approximately 537-kb gap between these two loci in the chromosome, suggesting that the piericidin BGC in S. pactum has been split into two loci. We manually confirmed the separation of these two loci by Sanger sequencing of the chromosomal DNA fragments around the 3´-end of pieA5 and its downstream region (total size sequenced 446 bp) and the 5´-end of pieA6 and its upstream region (total size sequenced 859 bp). Overall, the similarity between the piericidin BGC in Streptomyces sp. SCSIO 03032 and BGC-1.26 and BGC-2.4 are 41% and 58%, respectively. The isolation of piericidin C1, piericidin C3, piericidin C4, Mer-A2026A, and piericidin D1 from S. pactum ATCC 27456 indicates that the loading module in PieA1 can use either acetyl or isopropyl as a starter unit. It is postulated that the production of various piericidin analogues is due to the relaxed substrate specificity of the loading domain, in combination with the post PKS tailoring processes.
Figure 3.
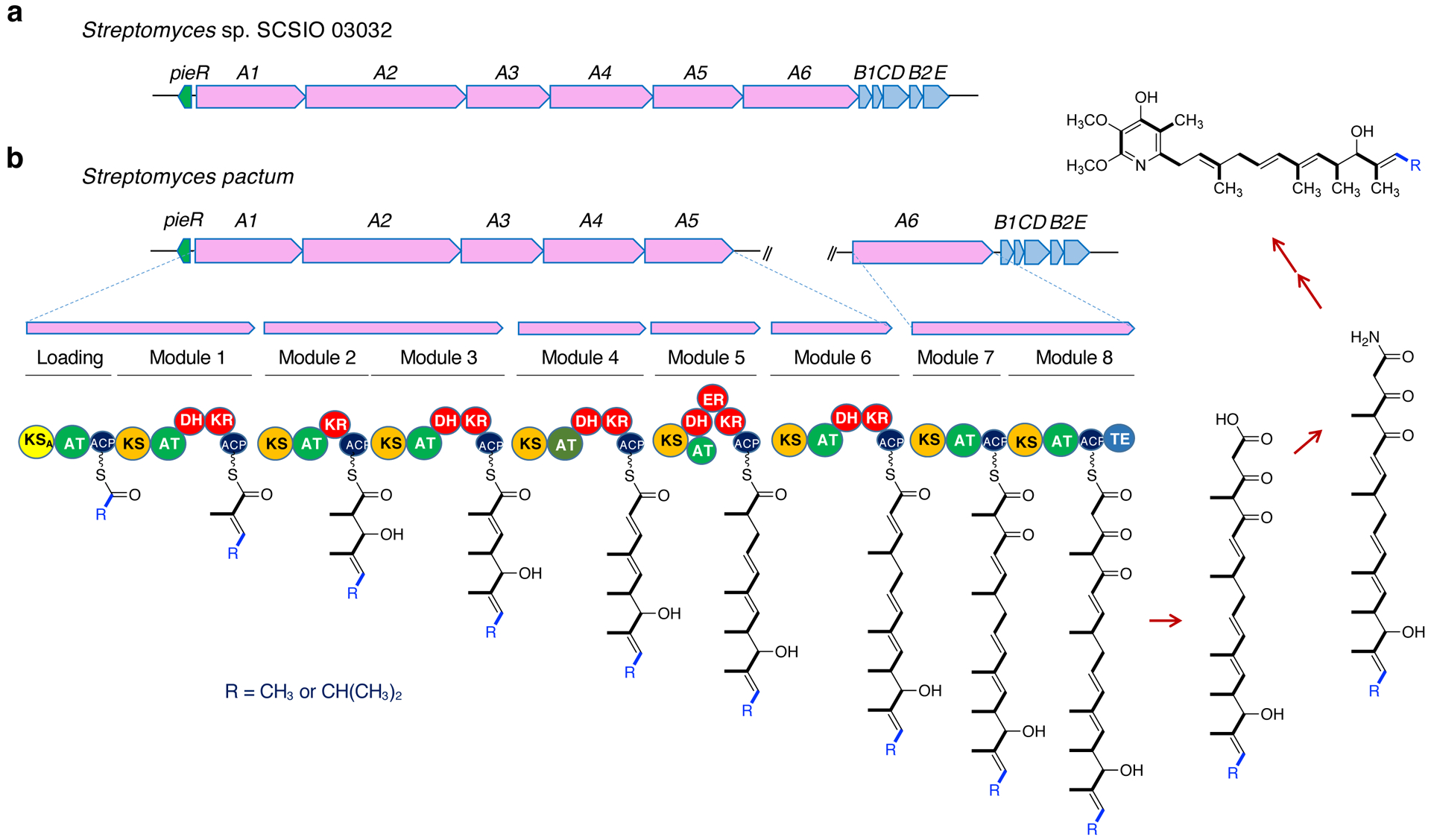
Genetic organization of the piericidin BGC in S. pactum. (a) reported piericidin A1 biosynthetic gene cluster from Streptomyces sp. SCSIO 03032; (b) biosynthetic gene cluster and proposed polyketide assembly line of piericidins in S. pactum ATCC 27456. The piericidin cluster in S. pactum splits into two loci. The loading module can use acetyl or isopropyl as a starter unit (blue). AT, acyltransferase; ACP, acyl carrier protein; KS, ketosynthase; DH, dehydratase; KR, ketoreductase; TE, thioesterase; KSA, KS domain in the loading module that does not contain the conserved Cys-His-His residues as a catalytic triad.
Inactivation of Other BGCs Increased Conglobatin Production.
To confirm that the BGC-2.4 and BGC-1.26 loci are indeed part of the piericidin BGC and to further determine the interconnectivity between the polyketide pathways, BGC-2.4 and BGC-1.26 were inactivated in S. pactum ΔptmTDQ/ΔBGC-1.2 to give two triple-BGC knockout mutants, S. pactum ΔptmTDQ/ΔBGC-1.2/ΔBGC-2.4 and ΔptmTDQ/ΔBGC-1.2/ΔBGC-1.26. A 2160-bp DNA fragment part of the pieA5 gene in the BGC-2.4 locus and a 1161-bp DNA fragment part of the pieA6 gene in the BGC-1.26 locus were individually deleted in-frame from the chromosome of S. pactum ΔptmTDQ/ΔBGC-1.2. The mutants were cultured in a liquid BTT medium for 7 days at 30 °C and the broths were extracted with EtOAc. LC-MS analysis of the EtOAc extracts from the culture broths of these mutants showed complete abolishment of piericidin production, indicating that BGC-2.4 and BGC-1.26 are indeed part of the piericidin BGC. In addition, the production of conglobatin in these two mutants increased by 2-fold (Figure 2c, Supplementary Figure 1), and a number of previously unknown conglobatin analogues were produced in higher yields (Figure 2a). Similar to that observed in the piericidin overproducing mutant, the increase in production yields of conglobatins in S. pactum ΔptmTDQ/ΔBGC-1.2/ΔBGC-2.4 or ΔptmTDQ/ΔBGC-1.2/ΔBGC-1.26 is most likely due to the increased availability of their biosynthetic precursors, malonyl-CoA and methylmalonyl-CoA, in the cell. However, it is also possible that NFAT-133 and/or piericidins somehow downregulate the conglobatin pathway, where in the absence of the former compounds conglobatins are produced in higher yields. While the ratio of conglobatins produced in the mutant has seemingly shifted compared to the wild-type, the reason for that is unclear.
Isolation and Structure Characterization of the New Conglobatins.
To identify the chemical structures of the new conglobatin analogues, the EtOAc extract of the culture broth was subjected to successive isolation procedures using reverse-phase HPLC to give four new conglobatins, namely conglobatin B2 (15), B3 (16), F1 (17), and F2 (18) (Figure 4) – on the basis of their chemical structures – along with a known conglobatin analogue, conglobatin B1 (8).28
Figure 4.
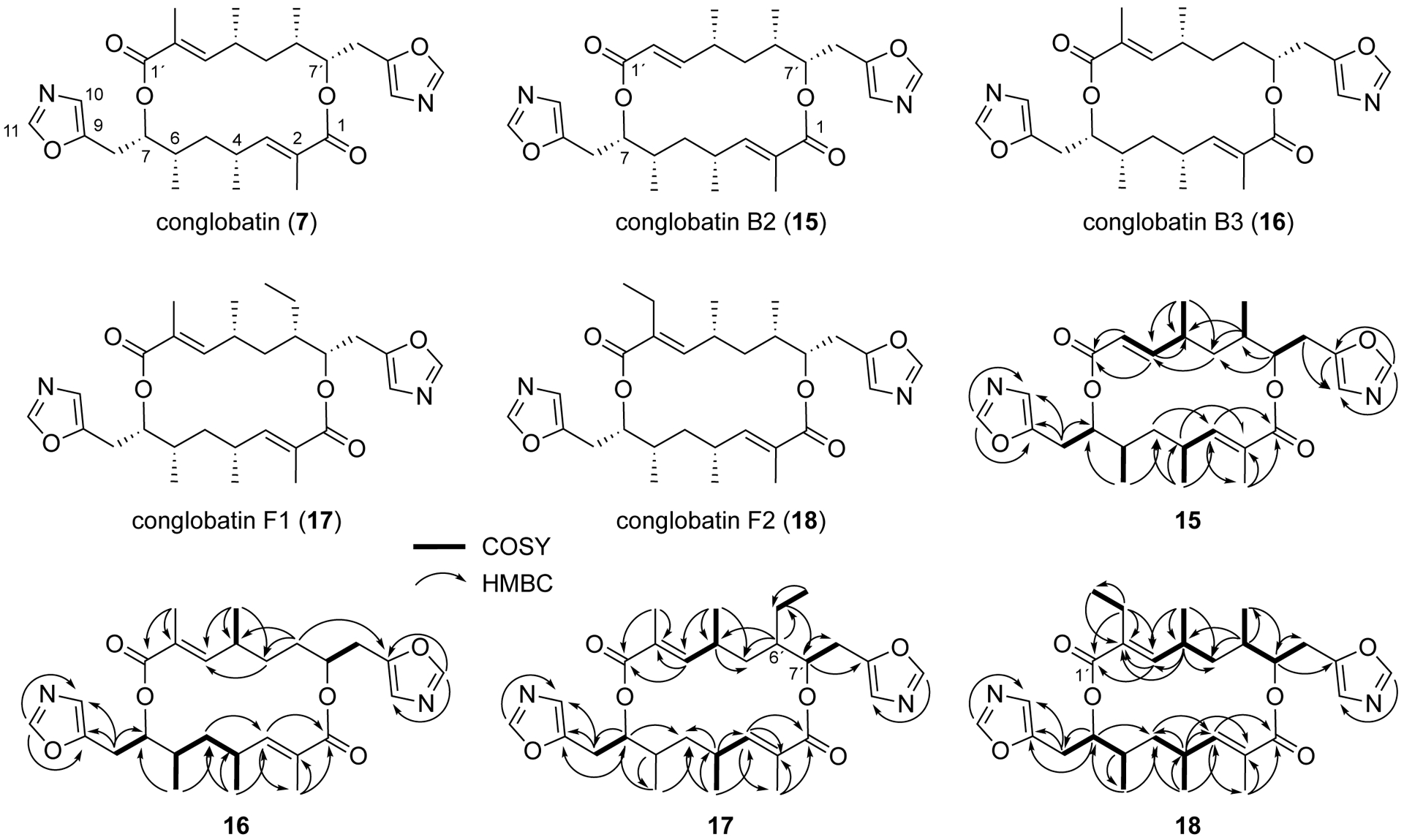
Chemical structures of new conglobatins isolated from Streptomyces pactum ΔptmTDQ/ΔBGC1.2/ΔBGC2 and selected COSY and HMBC correlations of conglobatins B2, B3, F1 and F2.
Conglobatin B2 (15) was isolated as a yellow oil with a postulated molecular formula of C27H36N2O6 (HRESIMS m/z 485.2646 [M+H]+), which is one carbon and two hydrogen atoms fewer than conglobatin. The 1H and 13C NMR spectra of conglobatin B2 showed the presence of a set of resonances similar to those of conglobatin and another set of resonances that closely resemble the first one. Among them are resonances of four pendant methyl groups [δH 0.92 (d, J = 6.9 Hz, 6-Me), 0.96 (d, J = 6.0 Hz, 6´-Me), 1.05 (d, J = 6.4 Hz, 4-Me) and 1.08 (d, J = 6.5 Hz, 4´-Me)], and an olefinic methyl group [δH 1.64 (s, 2-Me)] (Table 1). In contrast to conglobatin which is a C2-symmetric macrodiolide, conglobatin B2 appears to be an asymmetric analogue that lacks an olefinic methyl group at C-2´. Consequently, the carbonyl carbons (C-1 and C-1´) of conglobatin B2 were observed as two separate resonances (δC 166.1 and 164.2) in the 13C NMR spectrum. The planar structure of conglobatin B2 was further confirmed by HSQC and HMBC experiments, where the α,β-unsaturated esters were determined based on HMBC correlations between H-2/2´, H-3/3´ and C-1/1´, whereas the presence of oxazole rings was confirmed by HMBC correlations between H-11/11´ and C-9/9´, C-10/10´, and between H8/8´ and C-10/10´, C-7/7´. Other HMBC correlations are consistent with those expected for conglobatin-type macrodiolides (Figure 4). The absolute configuration of conglobatin B2 was proposed to be 4R, 6S, 7S, 4´R, 6´S, 7´S on the basis of biosynthetic consideration and its similar specific optical rotation (–47°) to that of conglobatin (–44°).
Table 1.
1H and 13C NMR data for conglobatins B2 (15), B3 (16), F1 (17), and F2 (18) in DMSO-d6.
| No | 15 | 16 | 17 | 18 | ||||
|---|---|---|---|---|---|---|---|---|
| δc | δH, mult. (J in Hz) |
δc | δH, mult. (J in Hz) |
δc | δH, mult. (J in Hz) |
δc | δH, mult. (J in Hz) |
|
| 1 | 166.1 | 166.2 | 166.3 | 166.0 | ||||
| 2 | 126.8 | 127.0 | 126.8 | 126.8 | ||||
| 2-Me | 12.8 | 1.64, s | 12.7 | 1.70, s | 12.8 | 1.63a, s | 12.7 | 1.66, s |
| 3 | 147.2 | 6.28, d (10.4) | 147.6 | 6.36, dd (10.4, 1.3) | 147.6 | 6.32a, m | 147.4 | 6.35, dd (10.8, 1.3) |
| 4 | 30.9 | 2.57, m | 30.7 | 2.60, m | 31.0 | 2.57a, m | 30.7 | 2.58, m |
| 4-Me | 21.2 | 1.05, d (6.4) | 21.0 | 1.06, d (5.9) | 21.2 | 1.05a, m | 21.6 | 1.07a, d (5.6) |
| 5 | 37.4 | 1.20a, m | 37.6 | 1.69, m 1.27, m |
37.6 | 1.64a, m 1.25a, m |
37.5 | 1.67a, m 1.25a, m |
| 6 | 35.0 | 1.23a, m | 32.8 | 2.38, m | 35.1 | 1.22a, m | 35.3 | 1.26a, m |
| 6-Me | 16.3 | 0.92, d (6.9) | 16.1 | 0.95, d (6.0) | 16.3 | 0.95, d (6.1) | 16.2 | 0.96, d (6.0) |
| 7 | 74.3 | 4.99, m | 74.5 | 5.18, m | 74.7 | 5.23, m | 74.5 | 5.02a, m |
| 8 | 24.1 | 2.98a, dd (15.9, 3.0) 2.79a, dd (15.8, 10.5) |
24.0 | 2.98a, dd (15.9, 3.0) 2.79a, dd (15.8, 10.5) |
24.1 | 2.96a, m 2.79a, m |
24.0 | 2.98a, m 2.78a, m |
| 9 | 149.5 | 149.4 | 149.6 | 149.4 | ||||
| 10 | 122.9 | 6.82, s | 123.1 | 6.94, s | 123.0 | 6.82, s | 122.9 | 6.83, s |
| 11 | 151.4 | 8.21, s | 151.5 | 8.25, s | 151.5 | 8.20, s | 151.4 | 8.22, s |
| 1´ | 164.2 | 166.0 | 166.1 | 165.4 | ||||
| 2´ | 121.2 | 5.66, d (15.6) | 126.8 | 126.7 | 133.3 | |||
| 2´-Me | 12.6 | 1.64, s | 12.8 | 1.63a, s | 14.1 | |||
| 2´-Et | 19.9 | 2.14, m; 2.09, m; 0.80, t (7.0) | ||||||
| 3´ | 154.3 | 6.54, dd (15.6, 10.0) | 147.4 | 6.33, dd (10.4, 1.3) | 147.4 | 6.32a, m | 146.6 | 6.24, dd (10.1, 1.3) |
| 4´ | 30.9 | 2.57, m | 29.8 | 1.60, m | 30.9 | 2.57a, m | 30.5 | 2.58, m |
| 4´-Me | 21.1 | 1.08, d (6.5) | 20.8 | 1.05, d (6.1) | 21.2 | 1.05a, m | 21.1 | 1.07a, d (5.6) |
| 5´ | 37.0 | 1.71a, m | 35.3 | 1.27, m | 36.8 | 1.64a, m 1.25a, m |
37.5 | 1.67a, m 1.25a, m |
| 6´ | 34.7 | 1.38, m | 29.5 | 1.68, m 1.14, m |
42.1 | 1.0, m | 35.1 | 1.26a, m |
| 6´-Me | 16.2 | 0.96, d (6.0) | 12.4 | 16.1 | 0.93, d (6.0) | |||
| 6´-Et | 23.3 | 1.53, m; 1.18, m; 0.89, t (7.0) | ||||||
| 7´ | 74.7 | 5.02, m | 70.0 | 5.03, m | 71.4 | 5.00, m | 74.2 | 5.02a, m |
| 8´ | 23.8 | 2.98a, dd (15.9, 3.0) 2.79a, dd (15.8, 10.5) |
27.1 | 2.94, m | 24.0 | 2.96a, m 2.79a, m |
24.0 | 2.98a, m 2.78a, m |
| 9´ | 149.5 | 148.7 | 149.6 | 149.4 | ||||
| 10´ | 122.9 | 6.84, s | 122.9 | 6.83, s | 122.9 | 6.82, s | 122.9 | 6.80, s |
| 11´ | 151.4 | 8.21, s | 151.4 | 8.22, s | 151.5 | 8.20, s | 151.3 | 8.20, s |
overlapped resonances
Similar to conglobatin B2, conglobatin B3 (16) was also isolated as a yellow oil. The HRESIMS data for conglobatin B3 suggested a molecular formula of C27H36N2O6 (m/z 485.2648 [M+H]+), indicating that conglobatin B3 also contains one carbon and two hydrogen atoms fewer than conglobatin. The 1H and 13C NMR spectra of conglobatin B3 are very similar to those of conglobatin B2, except that one of the pendant methyl group resonances is missing. On the other hand, two methylene proton resonances (δH 1.14 and 1.68) were observed (Table 1). Analysis of the 1H-1H COSY, HSQC and HMBC spectra of conglobatin B3 confirmed the lack of a pendant methyl group at C-6 (Fig. 4), indicating that conglobatin B3 is another desmethyl analogue of conglobatin.
The third compound, conglobatin F1 (17), was also isolated as a yellow oil with a suggested molecular formula of C29H40N2O6 (m/z 513.2961 [M+H]+). In contrast to conglobatin B2 and conglobatin B3, conglobatin F1 contains one carbon and two hydrogen atoms more than 1. The 1H and 13C NMR spectra of conglobatin F1 are similar to those of conglobatin, except for an up-field shift of a methyl proton resonance (δH 0.89, t, J = 7.0 Hz) and the presence of two additional methylene proton resonances [δH 1.18 (m) and 1.53 (m)] in conglobatin F1 (Table 1). Analysis of the 1H-1H COSY, HSQC and HMBC spectra of conglobatin F1 indicated that it contains an ethyl group at C-6´. The specific position of the ethyl group was assigned based on HMBC correlations between H-6´ and the methylene carbon and between the methylene protons and C-7. Thus, the chemical structure of conglobatin F1 was deduced as shown in Figure 4.
The fourth compound, conglobatin F2 (18), also a yellow oil, was determined to have a molecular formula of C29H40N2O6 (m/z 513.2966 [M+H]+), which is one carbon and two hydrogens more than conglobatin. The 1H and 13C NMR spectra of conglobatin F2 are also similar to those of conglobatin F1 and conglobatin (Table 1). Similar to conglobatin F1, the presence of a shielded methyl proton resonance (δH 0.80, t, J = 7.0 Hz) and two additional methylene proton resonances [δH 2.09 (m) and 2.14, (m)] suggested that conglobatin F2 also contains an ethyl group. The ethyl group was assigned to be on C-2´ based on HMBC correlations between the methylene protons and C-1´, C-2´, C-3´ and between the shielded methyl protons and C-2´. Therefore, the chemical structure of conglobatin F2 was deduced as shown in Figure 4.
Relaxed Substrate Specificity of Conglobatin PKSs.
The biosynthesis of conglobatin is mediated by a nonribosomal peptide synthetase (NRPS)-like loading module (encoded by congA) coupled with five polyketide synthase modules within the CongB–CongD proteins, and a cyclodehydratase (encoded by congE) (Figure 5a).16,24 The formation of the macrodiolide structure of conglobatin involves a cyclase/thioesterase (TE) domain that acts iteratively, couples two polyketide chains head-to-tail, rebinds the product, and then cyclizes it.24 The formation of asymmetrical macrodiolides conglobatins B1, B2, B3, F1 and F2 indicates that the conglobatin PKSs, particularly modules 4, 5, and 6, are somewhat tolerant to different extender units. However, genetic analysis of the AT domains of modules 4,5, and 6 showed that only the AT domain of module 5 (Cong-AT3) appears to be promiscuous toward various extender units, as it does not contain the typical conserved motifs for malonyl-, methylmalonyl-, or ethylmalonyl-CoA.29 On the other hand, the AT domains of modules 4 and 6 (Cong-AT2 and Cong-AT4) contain conserved motifs for methylmalonyl-CoA (e.g., RVDVVQ, GHSQG and YASH) (Figure 5b). This finding suggests that the commonly known conserved motifs for substrate recognition within the AT domains may not be strictly selective. Alternatively, other parts of the proteins may play a role in their substrate specificity or promiscuity. Interestingly, while Cong-AT1 (which is part of module 3) does not contain conserved motifs for malonyl-CoA, it appears to be very specific toward malonyl-CoA.
Figure 5. The biosynthetic gene cluster of conglobatin.
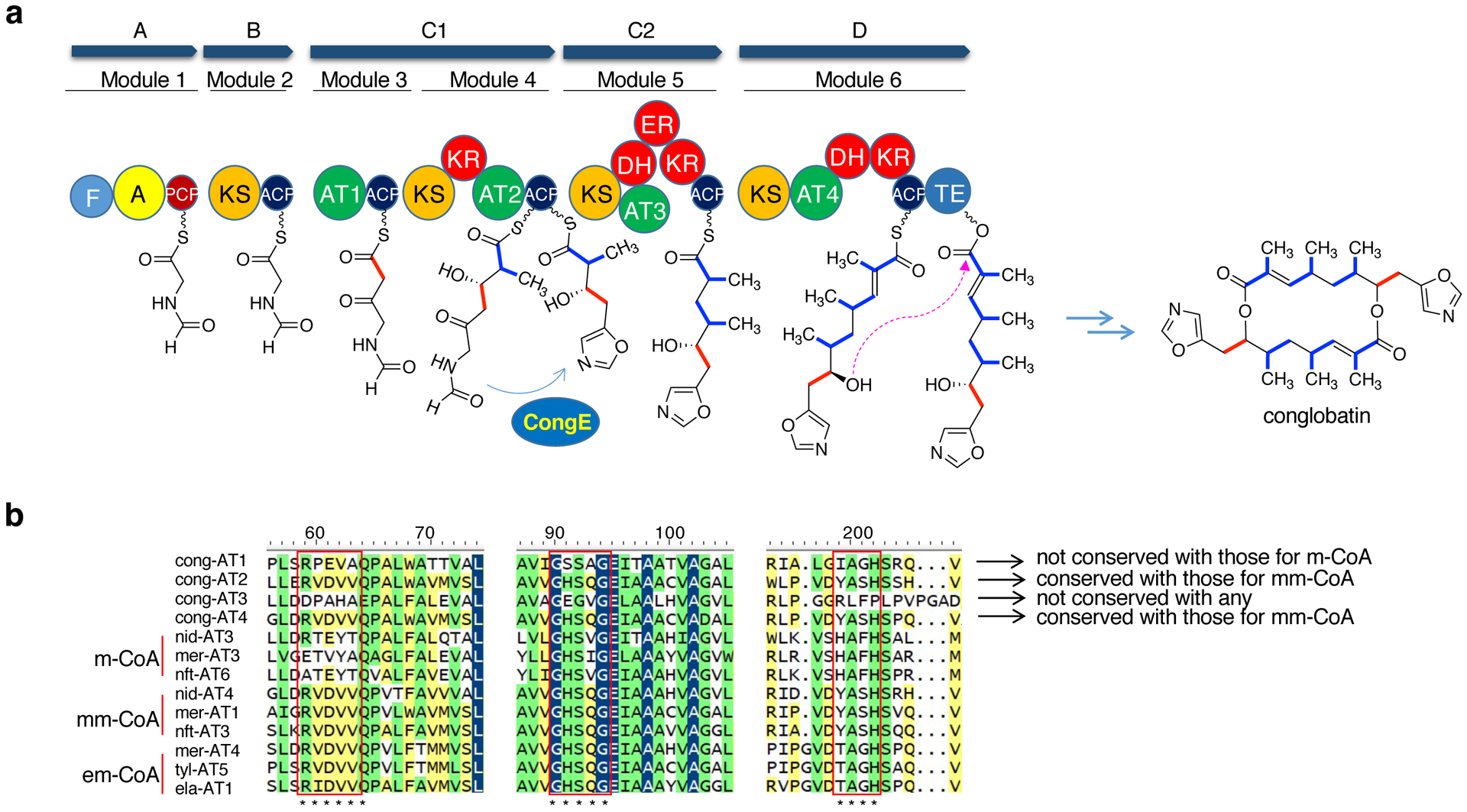
(a) genetic organization of conglobatin BGC; (b) Partial multiple amino acid alignment of AT domains of PKS enzymes. In the 60th-70th aa region, the [RQSED]V[DE]VVQ motif is commonly found in methylmalonyl-AT and the ZTX$[AT][QE] motif is commonly found in malonyl-AT, in which Z is a hydrophilic residue and $ is an aromatic residue. In the 100th aa region, the GHSXG motif, when the ‘X’ residue is Q, the AT domain prefers methylmalonyl-CoA, and either leucine or valine exhibits a preference for malonyl-CoA over methylmalonyl-CoA. In the 200th aa region, [YVW]ASH for methylmalonyl-AT, [HTVY]AFH for malonyl-AT, TAGH for ethylmalonyl-CoA. cong, conglobatin; nid, niddamycin; tyl, tylactone; ela, elaiophylin; nft, NFAT-133; mer, meridamycin. m-CoA, malonyl-CoA; mm-CoA, methylmalonyl-CoA; em-CoA, ethylmalonyl-CoA.
Biological Activity of the New Conglobatins.
All of the new conglobatin analogues were screened for their cytotoxicity against three human cancer cell lines and mice macrophages. As shown in Figure 6a–e, most of the new conglobatins showed different levels of cytotoxic activity against human cancer cell lines (HeLa, NCI-H460, and MCF-7) in a dose-dependent manner. Particularly, conglobatin F2 showed relatively higher toxicity against the cervical cancer cells (HeLa) and the lung cancer cells (NCI-H460) with IC50 values of 3.72 μM and 7.43 μM, respectively (Supplementary Table 1). Those were approximately two times more active than conglobatin. However, most of the conglobatin analogues only have moderate toxicity against breast cancer cell line (MCF-7), whereas conglobatin F1 is not cytotoxic to this cell line (> 100 μM). Interestingly, all of the conglobatin analogues were virtually non-toxic to Abelson murine leukemia virus-induced tumor cell line RAW264.7, whereas conglobatin only showed weak activity (IC50, 83.29 μM) (Supplementary Table 1), suggesting that the conglobatins are more active against human tumor cell lines than murine tumor cell lines.
Figure 6.
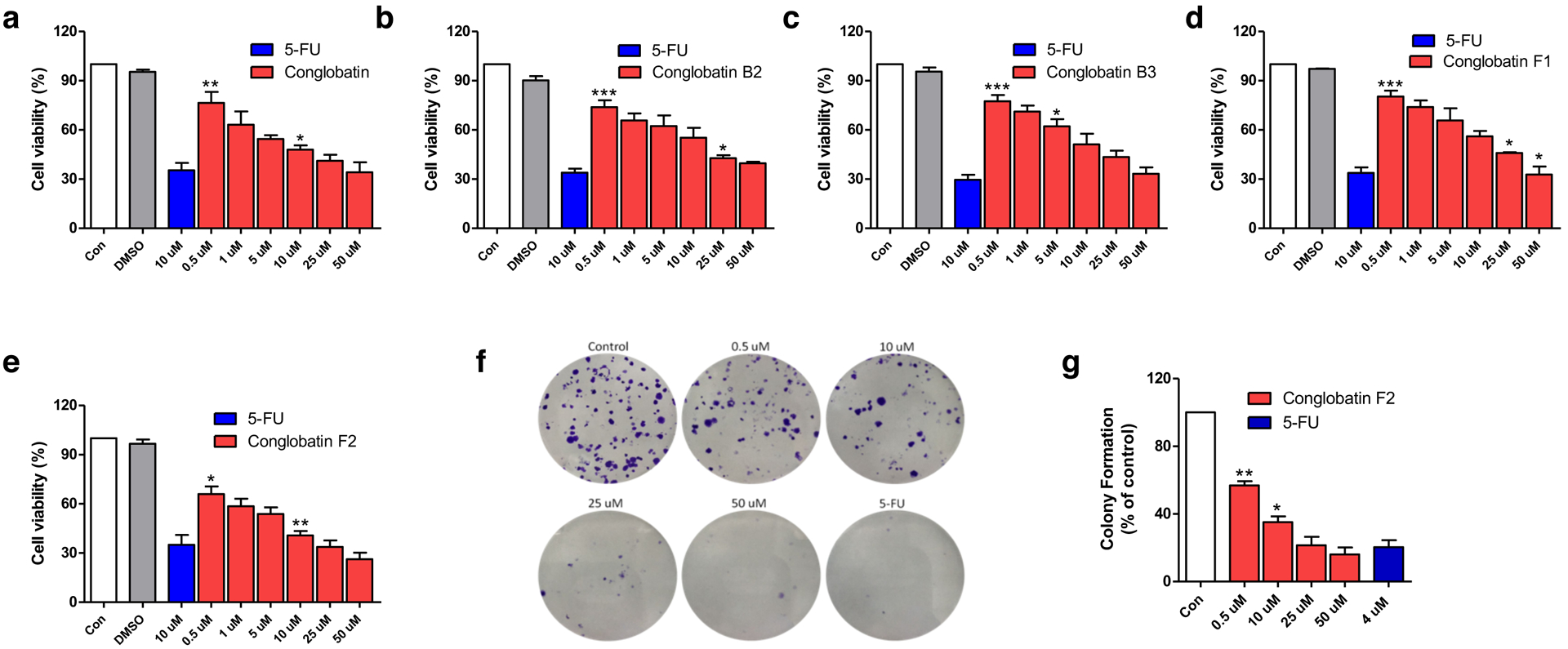
Cytotoxicity of conglobatins on HeLa cells. (a–e) Dose-dependent growth inhibition of the conglobatins. Cells were treated with different concentrations of conglobatins for 48 h. Con: cell was treated with the same amount of vehicle. DMSO was used as a negative control. 5-FU: Fluorouracil 10 μM was used as a positive control; (f) and (g) Colony formation assay of conglobatin F2. Cells were treated with conglobatin F2 for 48 h at varied concentrations. Con: cell was treated with the same amount of vehicle. 5-FU: Fluorouracil 4 μM was used as a positive control. Cell viability ratios are presented as mean ± SD (n=3). * p < 0.05, ** p < 0.01, *** p < 0.001 vs control.
As conglobatin F2 showed better cytotoxic activity than the other analogues, we subsequently investigated its anti-proliferative activity against HeLa cells using the colony formation assay (Figure 6f–g). The result showed that conglobatin F2 moderately attenuated colony formation of HeLa cells in a concentration-dependent manner, where the activity of conglobatin F2 at the concentration of 25 μM was comparable to that of 4 μM fluorouracil.
It is noteworthy that similar to conglobatin none of the new conglobatin analogues have any anti-bacterial activity against S. aureus, B. subtilis, P. aeruginosa and E. coli (MIC > 100 μg/mL).
Binding Model of the Conglobatin Analogues to Heat Shock Protein 90.
Heat shock protein (Hsp) 90 is a highly abundant and conserved chaperone protein which plays an essential role in many cellular processes including cell cycle control, cell survival, hormone and other signaling pathways. Consequently, Hsp90 has been recognized as an important therapeutic target of cancer.30 Normally, Hsp90 requires a couple of co-chaperones to form a protein complex in order to function, and the most reported co-chaperone is cell division cycle protein (Cdc37). In addition, Hsp90 was reported to contain three functional domains, the N-terminal nucleotide binding domain (NBD) for ATP/ADP binding, the middle domain (MD) for client protein binding, and the C-terminal dimerizing domain, each of which playing an essential role in the function of the protein.31 The N-terminal ATP/ADP domain is the target of the anticancer natural product geldanamycin.32
Conglobatin binds to Hsp90 via a novel action that is distinct from that of geldanamycin.26 It binds to the N-terminal domain of Hsp90 and disrupts Hsp90/Cdc37 interaction, resulting in disassociation of Hsp90/Cdc37/client complexes and degradation of the Hsp90 client protein (Supplementary Figure 2).26 These surprising findings illuminated the pharmacological properties of conglobatin and refreshed interest in its development as a potential candidate for a new anticancer drug.
Our molecular modelling study showed that the new conglobatin analogues B2, B3, F1, and F2 also bind Hsp90 at the same binding site as conglobatin (Figure 7 and Supplementary Figure 3). They also showed similar binding affinity values (around −8.0 Kcal/mol, Supplementary Table 2), suggesting that they can bind to Hsp90 as effectively as conglobatin. Therefore, the somewhat different anti-proliferative activity between conglobatin and the new conglobatin analogues against different cancer cell lines is intriguing and merits further investigation.
Figure 7.
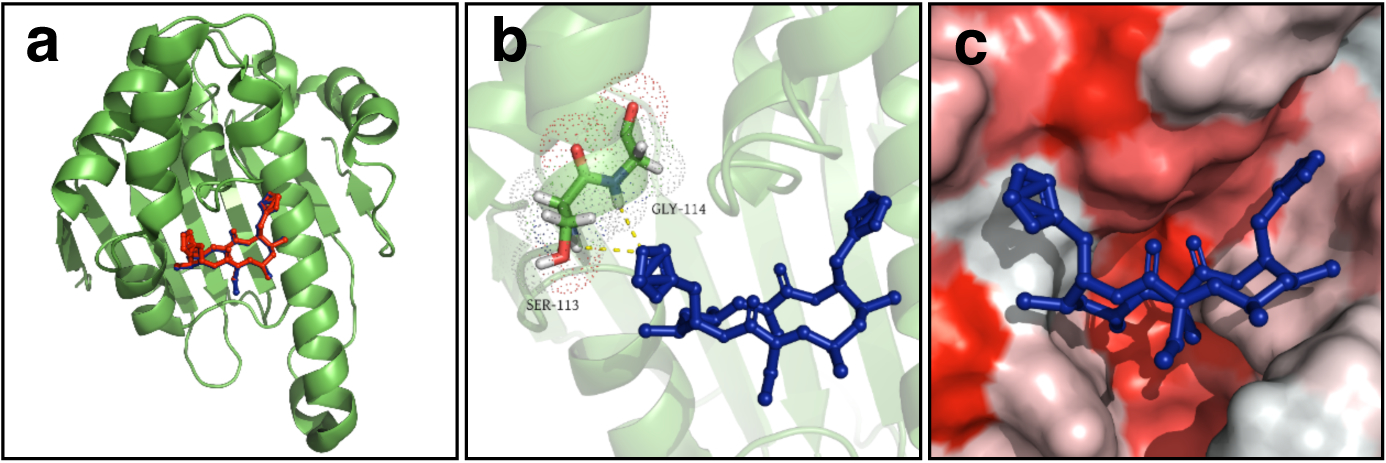
Docking structures of conglobatins/Hsp90 binding (Protein Data Bank [PDB] ID: 2K5B). (a) Conglobatin and conglobatin F2 bind to the N-terminal domain of Hsp90 in the ATP binding pocket. Hsp90 is shown in the green ribbon view; conglobatin and conglobatin F2 are shown in red and blue stick, respectively; (b) Zoomed view of the hydrogen bonding interactions between conglobatin F2 and Hsp90; (c) The hydrophobic and hydrophilic regions of Hsp90 LBD are shown in red and white, respectively.
In summary, we have engineered a number of mutant strains of S. pactum and shown that the production of the antitumor polyketides piericidins and conglobatins significantly increased when other polyketide biosynthetic pathways were inactivated. We found that the piericidin biosynthetic genes are located in two different genetic loci in the chromosome of S. pactum. Our study also showed that both piericidin and conglobatin biosynthetic pathways contain PKS enzymes with relaxed substrate specificity. Consequently, we were able to identify a number of new conglobatin analogues with altered polyketide backbones. Some of the new conglobatin analogues showed enhanced antitumor activity against HeLa and NCI-H460 cancer cell lines compared to conglobatin. Molecular modeling studies suggest that the new conglobatins bind to human heat shock protein (Hsp90) and disrupt Hsp90/Cdc37 chaperone/co-chaperone interaction in the same manner as conglobatin. This report provides insights into the interconnectivity between polyketide biosynthetic pathways in Streptomyces, which may be useful for modulating the production of specialized metabolites in other bacteria.
EXPERIMENTAL SECTION
General Experimental Procedures.
For details of the general experimental procedures and the instruments used, see the Supporting Information.
Bacterial Strains and Plasmids.
Streptomyces pactum ATCC 27456 was purchased from American Type Culture Collection (ATCC). E. coli DH5α was used for DNA manipulation. E. coli ET12567/pUZ8002 was used as a donor strain for intergeneric conjugation. pTMN002, a pIJ101 derivative containing an oriT transfer element, was used as a knockout plasmid.33
General DNA Manipulations.
Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. Chemicals were purchased from Sigma-Aldrich. Plasmid DNA was isolated using the Plasmid Mini Kit I (Omega BioTek) according to the manufacturer’s protocol. PCR amplification was carried out using a BioRad T-100™ thermal cycler and Platinum Taq DNA polymerase (Invitrogen). The nucleotide sequences of the gene fragments were determined at the Center for Genome Research and Biocomputing (CGRB) Core Laboratories, Oregon State University.
Construction of S pactum Mutants.
Gene disruptions were performed according to the general protocols reported in our previous papers.15,33 For details, see the Supporting Information.
Strain Cultivation and Compound Isolation.
S. pactum ATCC 27456 mutants were grown on BTT agar plates [glucose (10 g), yeast extract (1 g), beef extract (1 g), casein hydrolysate (2 g), agar (15 g) in ddH2O (1 L)] at 30 °C for 3 days. The colonies were used to inoculate BTT liquid medium [glucose (10 g L−1), yeast extract (1 g L−1), beef extract (1g L−1), soytone (2 g L−1), trace element (0.5 mL), distilled water 100 mL] in 500 mL flasks (n = 10) and the cultures were agitated at 30 °C, 200 rpm, for 2 days. The seed cultures were then used to inoculate liquid BTT medium (500 mL) in 2 L flasks (n = 10). The cultures were agitated at 30 °C, 200 rpm, for 7 days. The cultures were centrifuged, and the supernatants were collected and extracted three times with EtOAc. The EtOAc extracts were fractionated by reverse-phase HPLC (C18-YMC-Pack ODS-A, S-5 μm, flow rate 2.0 mL min−1, 230/254 nm detection) using a stepped solvent gradient (10%, 30%, 60%, 80%, 90%, and 100% CH3CN in H2O). Fractions obtained from the 60% and the 80% CH3CN elution were further purified by reverse-phase HPLC (C18-YMC-Pack ODS-A, S-5 μm, CH3CN–H2O 70:30, flow rate 2.0 mL min−1, 230/254 nm detection) to yield conglobatin (55 mg), conglobatin B2 (3.5 mg), conglobatin B1 (7.6 mg), conglobatin B3 (0.5 mg), conglobatin F1 (1.5 mg), and conglobatin F2 (1.5 mg). Fractions obtained from the 90% and the 100% CH3CN elution were further purified by reverse-phase HPLC (C18-YMC-Pack ODS-A, S-5 μm, CH3CN–H2O 80:20, flow rate 2.0 mL min−1, 230/254 nm detection) to yield piericidin C1 (6.3 mg), piericidin C3 (6.1 mg), Mer-A2026A (2.0 mg), and piericidin D1 (2.3 mg).
Conglobatin B2: a yellow oil, = −47° (c = 0.03, MeOH); UV (MeOH) λmax (log ε) 216 (4.38) nm. 1H NMR data and 13C NMR data: Table 1. HRESIMS at m/z 485.2646 ([M + H] +, C27H36N2O6; calc. 485.2652).
Conglobatin B3: a yellow oil, = −31° (c = 0.01, MeOH); UV (MeOH) λmax (log ε) 216 (4.33) nm. 1H-NMR data and 13C-NMR data: Table 1. HRESIMS at m/z 485.2648 ([M + H] +, C27H36N2O6; calc. 485.2652).
Conglobatin F1: a yellow oil, = −29° (c = 0.01, MeOH); UV (MeOH) λmax (log ε) 218 (4.37) nm. 1H NMR data and 13C NMR data: Table 1. HRESIMS at m/z 513.2961 ([M + H] +, C29H40N2O6; calc. 513.2965).
Conglobatin F2: a yellow oil, = −29° (c = 0.01, MeOH); UV (MeOH) λmax (log ε) 218 (4.42) nm. 1H NMR data and 13C NMR data: Table 1. HRESIMS at m/z 513.2966 ([M + H] +, C29H40N2O6; calc. 513.2965).
In Vitro Anti-bacterial Test of Conglobatins.
For details of the in vitro antibacterial test, see the Supporting Information.
In Vitro Cytotoxicity Test of Conglobatins.
For details of the in vitro cytotoxicity test, see the Supporting Information.
Colony Formation Inhibitory Activity Test of Conglobatins.
HeLa cells were seeded in a 6-well plate with a density of calc. 0.5 × 103 cells/well for 12 h and then treated with conglobatin F2 (0.5 – 50 μM) or 5-FU (4 μM). After 10 days, the colonies were stained with crystal violet and quantified by glacial acetic acid (30%) using an SpectraMax 190 Reader (Molecular Devices) at a wavelength of 520 nm.
Statistical Analysis.
The significance of intergroup differences was determined by ANOVA. Results are expressed as the mean ± SD of the indicated numbers of independent experiments. Values of p < 0.05 were considered statistically significant.
Molecular Modeling.
For details of the docking calculations, see the Supporting Information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grant AI129957 (to T.M.) from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not represent the official views of the National Institute of Allergy and Infectious Diseases, or the National Institutes of Health (NIH). We acknowledge the support of the Oregon State University NMR Facility funded in part by the National Institutes of Health, HEI Grant 1S10OD018518, and by the M. J. Murdock Charitable Trust grant #2014162.
Footnotes
Supporting Information
Additional experimental details, materials, and methods, cytotoxicity data, calculated binding affinity data, gel electrophoresis, HPLC chromatograms, 1D and 2D NMR spectra for all new compounds, UV spectra, and high-resolution mass spectra.
This material is available free of charge via the internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Takahashi Y, and Nakashima T (2018) Actinomycetes, an inexhaustible source of naturally occurring antibiotics, Antibiotics 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Baltz RH (2019) Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities, J. Ind. Microbiol. Biotechnol 46, 281–299. [DOI] [PubMed] [Google Scholar]
- (3).Mahmud T, Flatt PM, and Wu X (2007) Biosynthesis of unusual aminocyclitol-containing natural products, J. Nat. Prod 70, 1384–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fotso S, Mahmud T, Zabriskie TM, Santosa DA, Sulastri, and Proteau PJ (2008) Angucyclinones from an Indonesian Streptomyces sp., J Nat Prod 71, 61–65. [DOI] [PubMed] [Google Scholar]
- (5).Xie P, Sheng Y, Ito T, and Mahmud T (2012) Transcriptional regulation and increased production of asukamycin in engineered Streptomyces nodosus subsp. asukaensis strains, Appl. Microbiol. Biotechnol 96, 451–460. [DOI] [PubMed] [Google Scholar]
- (6).Lu W, Alanzi AR, Abugrain ME, Ito T, and Mahmud T (2018) Global and pathway-specific transcriptional regulations of pactamycin biosynthesis in Streptomyces pactum, Appl. Microbiol. Biotechnol 102, 10589–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Millan-Oropeza A, Henry C, Lejeune C, David M, and Virolle MJ (2020) Expression of genes of the Pho regulon is altered in Streptomyces coelicolor, Sci. Rep 10, 8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Alanzi AR, Demessie AA, and Mahmud T (2018) Biosynthesis and metabolic engineering of pseudo-oligosaccharides, Emerg. Top Life Sci 2, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Li S, Li Z, Pang S, Xiang W, and Wang W (2020) Coordinating precursor supply for pharmaceutical polyketide production in Streptomyces, Curr. Opin. Biotechnol 69, 26–34. [DOI] [PubMed] [Google Scholar]
- (10).Almabruk KH, Lu W, Li Y, Abugreen M, Kelly JX, and Mahmud T (2013) Mutasynthesis of fluorinated pactamycin analogues and their antimalarial activity, Org. Lett 15, 1678–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhou W, Posri P, Abugrain ME, Weisberg AJ, Chang JH, and Mahmud T (2020) Biosynthesis of the nuclear factor of activated T cells inhibitor NFAT-133 in Streptomyces pactum, ACS Chem. Biol 15, 3217–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bhuyan BK (1962) Pactamycin production by Streptomyces pactum, Appl. Microbiol 10, 302–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ito T, Roongsawang N, Shirasaka N, Lu W, Flatt PM, Kasanah N, Miranda C, and Mahmud T (2009) Deciphering pactamycin biosynthesis and engineered production of new pactamycin analogues, ChemBioChem 10, 2253–2265. [DOI] [PubMed] [Google Scholar]
- (14).Abugrain ME, Brumsted CJ, Osborn AR, Philmus B, and Mahmud T (2017) A highly promiscuous β-ketoacyl-ACP synthase (KAS) III-like protein is involved in pactamycin biosynthesis, ACS Chem. Biol 12, 362–366. [DOI] [PubMed] [Google Scholar]
- (15).Abugrain ME, Lu W, Li Y, Serrill JD, Brumsted CJ, Osborn AR, Alani A, Ishmael JE, Kelly JX, and Mahmud T (2016) Interrogating the tailoring steps of pactamycin biosynthesis and accessing new pactamycin analogues, ChemBioChem 17, 1585–1588. [DOI] [PubMed] [Google Scholar]
- (16).Zhou W, Posri P, and Mahmud T (2021) Natural occurrence of hybrid polyketides from two distinct biosynthetic pathways in Streptomyces pactum, ACS Chem. Biol 16, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhou W, Posri P, Liu X-J, Ju Z, Lan W-J, and Mahmud T (2021) Identification and biological activity of NFAT-133 congeners from Streptomyces pactum, J. Nat. Prod 84, 2411–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu Q, Yao F, Chooi YH, Kang Q, Xu W, Li Y, Shao Y, Shi Y, Deng Z, Tang Y, et al. (2012) Elucidation of piericidin A1 biosynthetic locus revealed a thioesterase-dependent mechanism of alpha-pyridone ring formation, Chem. Biol 19, 243–253. [DOI] [PubMed] [Google Scholar]
- (19).Chen Y, Zhang W, Zhu Y, Zhang Q, Tian X, Zhang S, and Zhang C (2014) Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis, Org. Lett 16, 736–739. [DOI] [PubMed] [Google Scholar]
- (20).Nishioka H, Imoto M, Imaoka T, Sawa T, Takeuchi T, and Umezawa K (1994) Antitumor effect of piericidin B1 N-oxide, J. Antibiot 47, 447–452. [DOI] [PubMed] [Google Scholar]
- (21).Kominato K, Watanabe Y, Hirano S, Kioka T, Terasawa T, Yoshioka T, Okamura K, and Tone H (1995) Mer-A2026A and B, novel piericidins with vasodilating effect. I. Producing organism, fermentation, isolation and biological properties, J. Antibiot 48, 99–102. [DOI] [PubMed] [Google Scholar]
- (22).Kubota NK, Ohta E, Ohta S, Koizumi F, Suzuki M, Ichimura M, and Ikegami S (2003) Piericidins C5 and C6: new 4-pyridinol compounds produced by Streptomyces sp. and Nocardioides sp, Bioorg. Med. Chem 11, 4569–4575. [DOI] [PubMed] [Google Scholar]
- (23).Engl T, Kroiss J, Kai M, Nechitaylo TY, Svatos A, and Kaltenpoth M (2018) Evolutionary stability of antibiotic protection in a defensive symbiosis, Proc. Natl. Acad. Sci. U.S.A 115, E2020–E2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhou Y, Murphy AC, Samborskyy M, Prediger P, Dias LC, and Leadlay PF (2015) Iterative mechanism of macrodiolide formation in the anticancer compound conglobatin, Chem. Biol 22, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Westley JW, Liu CM, Evans RH, and Blount JF (1979) Conglobatin, a novel macrolide dilactone from Streptomyces conglobatus ATCC 31005, J. Antibiot 32, 874–877. [DOI] [PubMed] [Google Scholar]
- (26).Huang W, Ye M, Zhang LR, Wu QD, Zhang M, Xu JH, and Zheng W (2014) FW-04–806 inhibits proliferation and induces apoptosis in human breast cancer cells by binding to N-terminus of Hsp90 and disrupting Hsp90-Cdc37 complex formation, Mol. Cancer 13, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hashida J, Niitsuma M, Iwatsuki M, Mori M, Ishiyama A, Namatame M, Nishihara-Tsukashima A, Matsumoto A, Ara I, Takahashi Y, et al. (2012) Panowamycins A and B, new antitrypanosomal isochromans produced by Streptomyces sp. K07–0010, J. Antibiot 65, 197–202. [DOI] [PubMed] [Google Scholar]
- (28).Lacey HJ, Booth TJ, Vuong D, Rutledge PJ, Lacey E, Chooi YH, and Piggott AM (2020) Conglobatins B-E: cytotoxic analogues of the C2-symmetric macrodiolide conglobatin, J. Antibiot 73, 756–765. [DOI] [PubMed] [Google Scholar]
- (29).Sheng Y, Lam PW, Shahab S, Santosa DA, Proteau PJ, Zabriskie TM, and Mahmud T (2015) Identification of elaiophylin skeletal variants from the Indonesian Streptomyces sp. ICBB 9297, J. Nat. Prod 78, 2768–2775. [DOI] [PubMed] [Google Scholar]
- (30).Maloney A, and Workman P (2002) HSP90 as a new therapeutic target for cancer therapy: the story unfolds, Expert Opin. Biol. Ther 2, 3–24. [DOI] [PubMed] [Google Scholar]
- (31).Biebl MM, and Buchner J (2019) Structure, function, and regulation of the Hsp90 machinery, Cold Spring Harb. Perspect. Biol 11, a034017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, and Pearl LH (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin, J. Med. Chem 42, 260–266. [DOI] [PubMed] [Google Scholar]
- (33).Lu W, Roongsawang N, and Mahmud T (2011) Biosynthetic studies and genetic engineering of pactamycin analogs with improved selectivity toward malarial parasites, Chem. Biol 18, 425–431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


