Abstract
Background
More than half of Neuroblastoma (NB) patients presented with distant metastases and the relapse of metastatic patients was up to 90%. It is urgent to explore a biomarker that could facilitate the prediction of metastasis in NB patients.
Methods and results
In the present study, we systematically analyzed Gene Expression Omnibus datasets and focused on identifying the critical molecular networks and novel key hub genes implicated in NB metastasis. In total, 176 up-regulated and 19 down-regulated differentially expressed genes (DEGs) were identified. Based on these DEGs, a PPI network composed of 150 nodes and 452 interactions was established. Through PPI network identification combined with qRT-PCR, ELISA and IHC, S100A9 was screened as an outstanding gene. Furthermore, in vitro tumorigenesis assays demonstrated that S100A9 overexpression enhanced the proliferation, migration and invasion of NB cells.
Conclusions
Taken together, our findings suggested that S100A9 could participate in NB tumorigenesis and progression. In addition, S100A9 has the potential to be used as a promising clinical biomarker in the prediction of NB metastasis.
Keywords: Neuroblastoma, Bioinformatics analysis, S100A9, Metastasis, Biomarker
Introduction
Neuroblastoma (NB), the most aggressive form of solid tumor of infants, accounting for 15% of cancer deaths in children [1, 2]. Until now, the main preferred treatment choices still remain surgery, chemotherapy and radiotherapy, which inevitably lead to tremendous toxicity and drug resistance. For high-risk NB patients, the recurrence and progression ratio are about 50% [3]. Thus, it is urgent for us to identify new effective biomarker for early diagnosis, metastasis prediction and ideal therapeutic target for NB patients.
S100A9, which is also called calgranulin B or migration inhibitory factor-related protein 14 (MRP-14), belongs to the low-molecular-weight calcium-binding S100 protein family [4]. Evidence has shown that S100A9 protein is elevated in metastatic melanoma and prostate cancer [5, 6]. In these tumors, increased expression of S100A9 was correlated with tumorigenesis and poor differentiation. Although S100A9 have been studied in many types of cancers, the biological function in malignancies was still remains contradictory and poorly understood. For example, elevated S100A9 in malignant tissues were associated with significantly shorter cancer survival, while downregulated S100A9 is correlated with tumor proliferation, inflammation invasion and angiogenesis [7–13]. To our knowledge, this is the first research to investigate the effects of S100A9 in NB patients.
In the present study, bioinformatic analysis was performed based on the GEO database [14]. After DEGs screening and functional analysis, PPI net-work of DEGs were further analyzed. And we found that the expression of S100A9 is pretty high in metastatic NB patients. In addition, we investigated the biological functions of S100A9 in NB cell line. These results may help us identify new effective biomarker for early diagnosis, metastasis prediction and ideal therapeutic target for NB patients, and provide valuable biological information for further investigation of NB.
Materials and methods
Bioinformatic analysis
Data processing and analysis
The public microarray dataset GSE90121, which was obtained based on the Affymetrix GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), was downloaded from the Gene expression omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) [14]. This dataset was deposited by David Kaplan et al., containing information from human NB SK-N-AS metastatic subpopulations isolated after in vivo selection, aimed to identify genomic signatures that regulate metastasis and candidate therapeutics for NB patients. A total of 16 samples, including 12 metastatic samples and four primary samples, were enrolled in the current dataset. Robust multi-array average (RMA) affy package of Bioconductor was used to adjust the raw data. The processed gene expression data was then filtered to include those probe sets with annotations which reference the new version annotation files. To identify DEGs, we used the Linear Models (Microarray Data package in Bioconductor) to compare the expression levels of genes between the metastatic group and the localized tumor group [15]. An adjusted p-value of < 0.05 and a |log2FC (fold change) | of ≥ 2 was used as the threshold.
Functional enrichment analysis of DEGs
Database for annotation, visualization and integrated discovery (DAVID) integrates a set of functional enrichment tools to distinguish functional genes underlying diseases processing (http://david.abcc.ncifcrf.gov/) [16]. Gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs were performed based on DAVID. P value < 0.05 and count ≥ 2 was regarded as statistically significant differences.
Protein–protein interaction network construction by STRING
We used the Search Tool for the Retrieval of Interacting Genes (STRING), an online tool and biological database for prediction of interactions between proteins, to construct the PPI network [17]. According to our analysis based on STRING, score (median confidence) > 0.15 was the standard of PPIs for DEGs selections. The Cytoscape software was used to visualize the PPI network [18]. The proteins that have many interaction partners, were defined as the hub proteins, constituting the extremely important nodes in the PPI network. To identify such hub proteins in the PPI network, we utilized six bioinformatic tools, namely Closeness, Degree, EPC, MNC, Radiality and Stress centrality. Sub-network analysis was then conducted to help us discover the outstanding genes.
NB patients, tissue samples and cell lines
Primary tumor tissues were obtained from 9 NB patients with bone marrow metastasis and 10 NB patients without bone marrow metastasis who had undergone tumor resection surgery at the Affiliated Hospital of Qingdao University. None of the included patients was treated with chemotherapy, hormonal therapy or radiotherapy before the tumor resection surgery. Written informed consent was obtained from all the participants. The current research was conducted with the permission of the Medical Ethical Committee of Affiliated Hospital of Qingdao University (Qingdao, China).
Human NB cell line SH-SY5Y was kindly provided by Professor Xiao from the Guizhou Medical University. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) contained 10% fetal bovine serum (FBS, Hyclone, USA) under the conditions of 37 °C, 5% CO2.
Quantitative real-time PCR
The expression of TAC1, PTGS2 and FGF1was examined by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The collected tissues were immediately frozen at − 80 °C after surgery. Total RNA was extracted from cancer tissues by TRIzol reagent (Invitrogen Life Technologies). Then the extracted RNA was transcribed into cDNA using random primers and analyzed with an ABI 7000 Real-Time PCR System (Applied Biosystems). PCR primers were as following: TAC1 primers: (forward) 5′-TGA TCT GAA TTA CTG GTC CGA CT-3′ and (reverse) 5′-TCC GGC AGT TCC TCC TTG A-3′; PTGS2 primers: (forward) 5′-TAA GTG CGA TTG TAC CCG GAC-3′ and (reverse) 5′-TTT GTA GCC ATA GTC AGC ATT GT-3′; FGF1 primers: (forward) 5′-CTC CCG AAG GAT TAA ACG ACG-3′ and (reverse) 5′-GTC AGT GCT GCC TGA ATG CT-3′; GAPDH primers (forward) 5′-CAG CGA CAC CCA CTC CTC-3′ and (reverse) 5′-TGA GGT CCA CCA CCC TGT-3′.Reactions were performed in triplicate using SYBR Green master mix (TaKaRa, Japan) and normalized to GAPDH mRNA level using the ΔΔCt method.
NB serum samples and ELISA
After blood samples centrifuged at 3000 × g for 10 min at 4 °C, the serum samples were aliquoted and stored at− 80 °C until further processing. To quantify levels of S100A9 in serum, ELISA was performed as previously described [19]. By using human S100A9 (JYM0539Hu, JYM, China) ELISA kits, S100A9 in serum of the NB patients were detected according to the manufacturer’s recommended procedure.
IHC staining
In brief, the formalin fixed, paraffin-embedded tissues sections were deparaffinized, rehydrated and boiled in 0.01 M citrate buffer for 10 min, then incubated with 0.3% H2O2 in methanol to block endogenous peroxidase activity. Then the sections were incubated with the anti-S100A9 antibody (Cell Signaling Technology, USA), followed by incubation with secondary antibody tagged with the peroxidase enzyme for 30 min at room temperature, finally visualized with 0.05% DAB (3,3′-diaminobenzidine) until the desired brown reaction product was obtained. All slides were observed under an OLYMPUS BX41 Microscope, and representative photographs were taken.
Construction of plasmids and establishment of stably transfected cells
SBI-piggyBac vector, GST-S100A9, were kindly provided by Professor T.C. He from the University of Chicago. To construct an S100A9 overexpression plasmid, the complete coding sequence of human S100A9 gene was subcloned into the SBI-piggyBac vector. For S100A9 silencing, siRNAs targeting human S100A9 with the sequences of 5′- GCA AGA CGA TGA CTT GCA A -3′ and 5′- TTG CAA GTC ATC GTC TTG C -3′ were synthesized and assembled into the SBI-piggyBac vector, resulting in SBI-siS100A9. After transfecting SH-SY-5Y NB cells with the constructed plasmids, the stably transfected cells were selected by incubation with puromycin for one week. The stable transfected cell lines, namely Control (SH-SY5Y transfected with SBI empty vector), S100A9 (SH-SY5Y transfected with SBI-S100A9) and siS100A9 (SH-SY5Y transfected with SBI-siS100A9) briefly.
Cell viability assay
The viability of SH-SY5Y cells was assessed by 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphe-nyltrazolium bromide (MTT) assay. Briefly, stably transfected SH-SY5Y cells were seeded in 96-well plates (1000 cells/well). The cells were incubated in DMEM supplemented with 1% FBS for 24, 48, 72, 96 and 120 h, then incubated with MTT reagent (Progema, Madison, WI, USA, 20 µL/well) for another 4 h at 37 °C to allow the formation of formazan. After that, 100 µL of dimethyl sulfoxide was added into the cell culture medium for another 10-min incubation at room temperature. At last, in every day of the next five days, a microplate reader (Bio-rad, iMark) was used to measure the absorbance at 492 nm of each well. Each sample included three independent replicates.
Colony formation assay
Exponentially growing stably transfected SH-SY5Y cells were seeded at a low density (100 cells/well) in cell culture medium containing 1% FBS in 6-well plates. The cells were allowed to grow for about 10 days to form colonies. The culture medium was refreshed every 3–4 days. Crystal violet was used to stained the colonies. Colony numbers from 3 wells were used to calculated the average colony number.
Scratch wound healing assay
The scratch wound healing assay was performed as described previously [20, 21]. Briefly, stably transfected SH-SY5Y cells were seeded in 6-well plates and grown to ~ 90% confluency. Then, sterile micro-pipette tips were used to scratch the monolayer formed by SH-SY5Y cells to create a wound. After that, the medium (DMEM containing 1% FBS) was refreshed every day to remove the floating cells. Bright field microscopy was used to monitor the wound healing status at 24 h, 48 h, and 72 h after the wound was created. Each assay was repeated three times. ImageJ software was used to calculate the wound healing ratio.
Transwell invasion assay
A chamber coated with non-type I-collagen (Millipore, USA) was used for the transwell assay. The upper chamber coated with ECM gel (Sigma, USA) was filled with 400µLof serum-free DMEM and seeded with exponentially growing stably transfected SH-SY5Y cells (1 × 104 cells). The lower chamber was filled with 500µL of DMEM supplemented with 20% FBS, which served as a chemoattractant. After 24 h of incubation, the cells migrated across the transwell membrane were dried, fixed with methanol, and then stained with hematoxylin–eosin (H and E). Cotton swabs were used to remove the cells on the upper surface of the transwell membrane. At last, an inverted microscope (× 100 magnification) was used to count the number of cells migrated across the transwell membrane. Five randomly-selected fields were examined to obtain the mean value of the number of cells migrated across the transwell membrane. The experiment was repeated three times.
Statistical analysis
All data are presented as means ± standard deviations. T test was performed in the GraphPad Prism software to determine the statistical significance of differences between groups. A P value of less than 0.05 was considered statistically significant.
Results
Enrichment analyses of DEGs
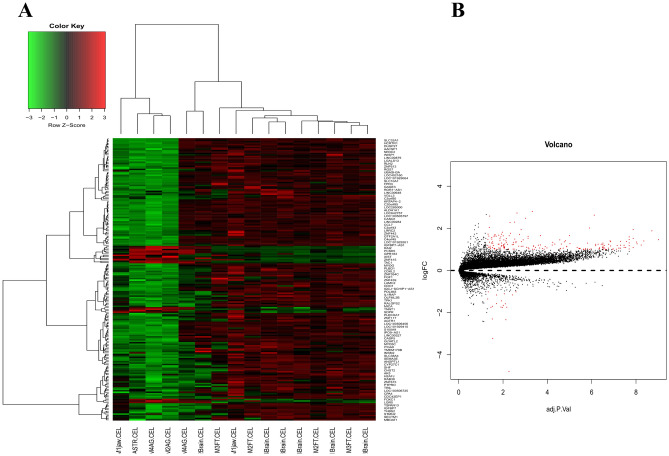
In general, 176 up-regulated DEGs and 19 down-regulated DEGs were identified based on the cut-off criteria (p-value < 0.05 and count ≥ 2), which were used to generate a heatmap for the NB patients with and without metastasis (Fig. 1A). Functional enrichment analysis revealed that the top 18 mostly enriched GO terms and top two mostly enriched KEGG pathways were associated with regulation of nucleosome assembly, innate immune response in mucosa, extracellular matrix organization, angiogenesis and innate immune response, etc. (Table 1). In addition, volcano plot was able to quick identify the expression changes within the gene sets by combination of statistical tests (adjusted p-value) and magnitude of change (Fig. 1B).
Fig. 1.
A Heat map for the differentially expressed genes (DEGs). B Volcano plots reflecting number, significance and reliability of differentially expressed mRNA in NB. The red dots indicate dysregulated mRNAs
Table 1.
The tops enriched GO terms of the differentially expressed genes
| Category | ID | Term | p-value | Gene count | Genes |
|---|---|---|---|---|---|
| BP | GO:0001558 | Regulation of cell growth | 3.13E-04 | 6 | NANOS1, NRG3, IGFBP2, AGTR1, IGFBP7, WISP1 |
| GO:0006954 | Inflammatory response | 5.07E-04 | 11 | CCL7, TRIL, C3AR1, FPR3, IL1RAP, CDO1, PTGS2, TAC1, HDAC9, S100A9, CHST2 | |
| GO:0007204 | Positive regulation of cytosolic calcium ion concentration | 0.00316801 | 6 | AGTR1, C3AR1, ADM, FPR3, LPAR3, TAC1 | |
| GO:0030335 | Positive regulation of cell migration | 0.011814659 | 6 | CCL7, GPNMB, CDH13, SEMA3E, LAMC2, FGF1 | |
| GO:0051384 | Response to glucocorticoid | 0.012309797 | 4 | IGFBP2, ADM, CDO1, PTGS2 | |
| GO:0,007,200 | Phospholipase C-activating G-protein coupled receptor signaling pathway | 0.012827699 | 4 | HTR1D, AGTR1, C3AR1, FPR3 | |
| GO:0051482 | Positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G- | 0.018038462 | 3 | GPR65, AGTR1, LPAR3 | |
| GO:0008285 | Negative regulation of cell proliferation | 0.027623627 | 8 | MYOG, KAT2B, GPNMB, CDH13, IGFBP7,ADM,PTGS2,MYO16 | |
| GO:0030819 | Positive regulation of cAMP biosynthetic process | 0.035152385 | 3 | GPR65, RLN2, ADM | |
| GO:0007267 | Cell–cell Signaling | 0.040239268 | 6 | CCL7, ADM, TAC1, PCSK5, S100A9, WISP1 | |
| CC | GO:0043005 | Neuron projection | 0.007369421 | 7 | SLC32A1, CASP5, PTPRO, STMN2,CDH13,PTGS2,POU4F1 |
| GO:0005615 | Extracellular space | 0.007817435 | 19 | GDF10, IGFBP2, SECTM1, ADM, LAMC2, SEMA3E, FGF1, PCSK5, WISP1, CCL7, NRG3, PLBD1, CDH13, IGFBP7, TAC1, S100A9, ANGPTL1, MICA, CPA4 | |
| GO:0005576 | Extracellular region | 0.011007958 | 21 | OLFML2B, IGFBP2, PSG1, ADM, LAMC2, SEMA3E, IL1RAP, FGF1, THBS2, FBLN2, APOLD1, CCL7, NRG3, RLN2, FDCSP, IGFBP7, CCDC3, VIP, TAC1, S100A9, CHGB | |
| GO:0005887 | Integral component of plasma membrane | 0.012602605 | 19 | GABRB1, VIPR1, SLC10A1, GPR65, PTPRO, HTR1D, LPAR3, FPR3, IL1RAP, HCRTR1, TSPAN13, GPNMB, NRG3, GPR183, C3AR1, AGTR1, LGR5, MICA, SLC38A4 | |
| GO:0005901 | Caveola | 0.079588673 | 3 | SDPR, CDH13, PTGS2 | |
| MF | GO:0008201 | Heparin binding | 0.007960505 | 6 | CCL7, GPNMB, LAMC2, THBS2, FGF1, WISP1 |
| GO:0003705 | Transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding | 0.014352101 | 4 | MYOG, FOXC1, POU4F1, VGLL3 | |
| GO:0005179 | Hormone activity | 0.03603451 | 4 | RLN2, ADM, VIP, CHGB |
| Category | ID | Term | Gene symbol | p-value |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa04080 | Neuroactive ligand-receptor interaction | GABRB1, VIPR1, HTR1D, AGTR1, C3AR1, FPR3, LPAR3, HCRTR1 | 9.46E-04 |
| hsa04512 | ECM-receptor interaction | LAMC2, THBS2, ITGA9 | 0.090573261 |
PPI network analysis
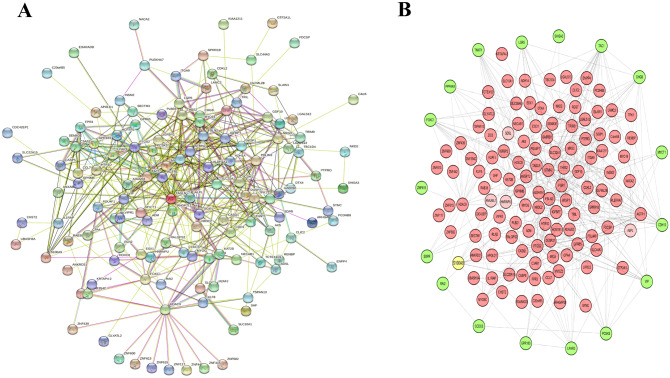
To identify the potential biomarkers that predict metastasis in NB patients, the DEGs were delineated to construct a PPI network. A DEG with a combined score (median confidence) > 0.15 was regarded as a significantly differentially expressed gene. The Cystoscope software was used to visualize the PPI network. As shown in Fig. 2A, the PPI network constructed by STRING is composed of 150 nodes and 452 interactions. The top 10 hub proteins in the PPI network determined by Closeness, Degree, EPC, MNC, Radiality and Stress centrality are listed in Table 2. The results showed that TAC1, PTGS2 and FGF1were the most outstanding genes and maybe play an important role in the NB metastasis. Sub-network analysis suggested that S100A9 was the outstanding hub protein (Fig. 2B).
Fig. 2.
A Protein-protein interaction network constructed with the dysregulated differentially expressed genes. B PPI network was visualized by Cystoscope software
Table 2.
Top 10 genes that evaluated by betweenness, BottleNeck, Closeness, Clustering coefficient, Degree, DMNC, EcCentricity, EPC, MCC, MNC, Radiality and Stress centrality respectively in the protein–protein interaction network
| Gene name | Clinenms centrality | MNC, Radiality and stress centrality respectively in the protein–protein interaction network | Gene name | Stress centrality | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Degree centrality | Gene name | EPC centrality | Gene name | MNC centrality | Gene name | Radiality centrality | ||||
| HDAC9 | 70.66667 | 5LC22A15 | 25 | TRIM9 | 22.245 | TAC1 | 25 | TAC1 | 4.87121 | PTGS2 | 7892 |
| PTGS2 | 70.08333 | ZNF43P | 25 | GPR183 | 22.121 | STMN2 | 23 | FGF1 | 4.86344 | HDAC9 | 7010 |
| TAC1 | 69.75 | URASH3A | 23 | AK5 | 22.046 | SLC32A1 | 23 | PTGS2 | 4.84091 | TAC1 | 6490 |
| KAT2B | 64.41667 | CHST2 | 20 | CASQ1 | 19.554 | RGS7 | 18 | TAGLN | 4.79545 | FGF1 | 6136 |
| FOXC1 | 66.25 | SEMA3E | 19 | WISP1 | 19.156 | PTGS2 | 17 | CDH13 | 4.7803 | FOXC1 | 4312 |
| FGF1 | 65.83333 | HCRTR 1 | 18 | RGS7 | 19.142 | FGF1 | 17 | AGTR1 | 4.72727 | STMN2 | 4228 |
| STMN2 | 65.75 | GPR183 | 17 | NRG3 | 18.906 | CHGB | 16 | STMN2 | 4.7197 | KAT2B | 4132 |
| MYOG | 64.41667 | FPR3 | 17 | ANGPTL1 | 18.77 | IGFBP7 | 14 | MYOG | 4.69697 | CDH13 | 4034 |
| CDH13 | 63.45 | CCL7 | 17 | SEMA3E | 18.362 | THBS2 | 14 | KAT2B | 4.68939 | TAGLN | 3990 |
| TAGLN | 62.83333 | NRG3 |
16 15 |
GPR65 | 17.728 | NRG3 | 13 | SLC32A1 | 4.68182 | AGTR1 | 3838 |
Validation the outstanding genes in NB patients
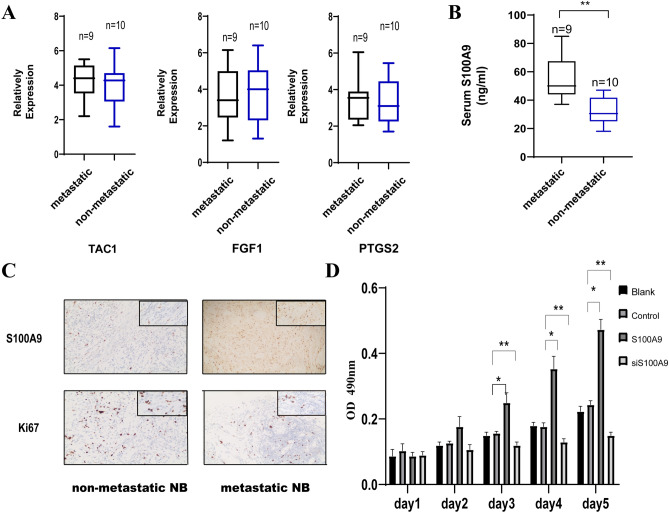
To verify the expression of TAC1, PTGS2 and FGF1 in microarray data, qPCR was performed to detect the expression of those three genes. The qRT-PCR results showed that the expression of three genes were no significant differences between the 9 NB patients with bone marrow metastasis and 10 NB patients without bone marrow metastasis (Fig. 3A). Next, ELISA and IHC were performed to detect the protein levels of S100A9 in NB patients (9 NB patients with bone marrow metastasis and 10 NB patients without bone marrow metastasis).The serum levels of S100A9 were higher from NB patients with bone marrow metastasis than without bone marrow metastasis (Fig. 3B). Further, IHC staining was also used to examined the expression of S100A9 from NB patients’ tissues with and without bone marrow metastasis (Fig. 3C). The results showed that the expression levels of S100A9 were significantly higher in NB patients with bone marrow metastasis than NB patients without bone marrow metastasis.
Fig. 3.
A qRT-PCR analysis for the expression of hub genes. B ELISA analysis for serum levels of S100A9 in metastatic (n=9) and non-metastatic NB (n=10) patients. C Representative IHC staining for S100A9 in tissue sections from metastatic and non-metastatic NB patients. **p < 0.01. D MTT analysis for Blank/Control/S100A9/siS100A9 SH-SY-5Y cells for sequential 5 days. *p<0.05, **p < 0.01.
S100A9 overexpression promoted the proliferation, migration and invasion of NB cells
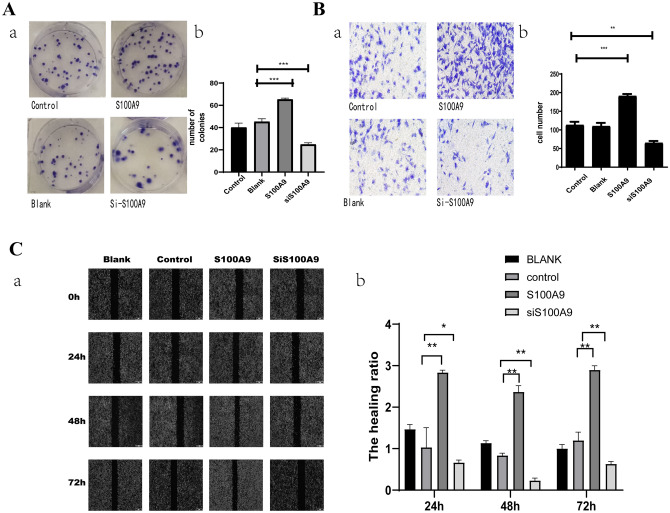
To investigate the effects of S100A9 on the proliferation of NB cells, the coding sequence of human S100A9 gene expressed in GST-S100A9 vector was subcloned into the SBI-piggyBac plasmid to overexpress S100A9 in SH-SY5Y cells. According to the MTT assay results, the SH-SY5Y cells overexpressing S100A9 (defined as the S100A9 group) exhibited higher proliferation ability than the SH-SY5Y cells transfected with empty vector (defined as the control group) at days 3, 4, and 5 (p < 0.05). The S100A9-knockdown SH-SY5Y cells (defined as the siS100A9 group) exhibited significantly slower proliferation when compared with the control group at days 3, 4, and 5 (p < 0.001) (Fig. 4A). Colony formation assay showed that the S100A9 group formed more colonies compared with the control group. Quantitatively, the number of colonies formed in the S100A9 group was approximately double than that of the control group (Fig. 4B). These results indicated that S100A9 overexpression accelerated the proliferation of SH-SY5Y cells. In addition, as revealed by wound healing and transwell assays, the results exhibited that the migration and invasion of SH-SY5Y cells were significantly active by S100A9 (Fig. 4A and B).
Fig. 4.
A a. Colony formation assay for Blank/Control/S100A9/siS100A9 SH-SY-5Y cells. b. The representative images of transmembrane cells are shown in the right panel. The mean numbers of transmembrane cells ± SD per microscopic field of three independent experiments are quantified in the right panel. **p < 0.01, ***p < 0.001. B a.Transwell invasion assay for Blank/Control/S100A9/siS100A9 SH-SY-5Y cells for 24 h. b.The representative images of transmembrane cells are shown in the right panel. The mean numbers of transmembrane cells ± SD per microscopic field of three independent experiments are quantified in the right panel. Magnification, ×100, **p < 0.01, ***p < 0.001. C a.Wound healing assay for Blank/Control/S100A9/siS100A9 SH-SY-5Y cells for 72 h. b.The incision width of different sites was measured and average healing rate was calculated and shown in the right panel. *p < 0.05, **p < 0.01
Discussion
High-throughput genome sequencing technologies have been widely used nowadays. Using these technologies, several research groups have identified genetic variations in human NB patients [22]. Among the identified genetic variations, MYCN amplification (32.1%), 11q loss (47.5%) and 17q gain (80.4%) were the most frequently observed ones (around 90% in total) in individuals with a high risk of developing NB [23]. Furthermore, two research groups found that recurrent genomic rearrangements affecting genomic regions close to the telomerase reverse transcriptase (TERT) gene locus led to significant transcriptional upregulation of TERT [24, 25]. However, even though great progresses have been made in understanding the genetic basis of NB tumor occurrence and development, effective biomarkers for prediction of metastasis in NB patients are still lacking.
In the current study, the GSE90121 dataset which was deposited by David Kaplan were downloaded and analyzed by bioinformatics method to identify potential crucial genes associated with NB metastasis. A total of 195 genes including 19 down-regulated and 176 up-regulated genes were obtained. Besides, the significantly enriched GO terms were mainly focused in regulation of cell growth, inflammatory response, positive regulation of cell migration. Hub genes of the regulatory network were then selected and conducted with PPI network module. Dysregulated TAC1, PTGS2 and FGF1 were the top three outstanding genes based on both six methods (Closeness, Degree, EPC, MNC, Radiality, and Stress centrality) evaluation. The expression levels of TAC1, PTGS2 and FGF1 in resected specimen of NB patients with or without metastasis were then validated by qRT-PCR. Although the expression of TAC1, PTGS2 and FGF1 were related with many kinds of tumorigenesis, such as non-small cell lung cancer, pancreatic ductal cancer, colorectal cancer, squamous cell carcinoma, gastric cancer and clear cell renal cell carcinoma[26–33], but these genes expression did not exhibit significant change as expected as the microarray results, indicating that these three genes may not the pivotal gene that participate in the metastasis of NB.
After go through the differentially expressed genes list and reviewing the relevant literatures, we found that S100A9 exhibits a broad range of biological functions involving in various cancer progression [34–36]. Characterized by calcium-binding EF hand motifs, S100 family comprise of more than 20 homologous proteins. Studies have revealed that S100A9 induced activation of NF-kB which participate in a broad range of intracellular and extracellular functions by regulating angiogenesis, tumor migration, wound healing, cell apoptosis, proliferation, differentiation, and inflammation [9, 37–42]. In addition, it has become increasingly evident that S100A9 acts as a potent amplifier of inflammation in tumor. S100A9 have been reported to be as Damage-associated molecular patterns (DAMPs) and involved in almost all aspects of cancer biology, such as proliferation, tumorigenesis, apoptosis, invasion, metastasis and angiogenesis. To our knowledge, fewer researches investigated the expression and biological function of S100A9 in NB. In the present study, the promoted expression levels of S100A9 were verified to be consistent with the microarray results. To further validate these results, our results suggested that elevated S100A9 promoted the proliferation and migration of NB cancer cells.
Conclusions
In conclusion, the current observations indicate that S100A9 may be an important carcinogenic factor in the occurrence and progression of NB and may serve as a promising biomarker for metastasis prediction of NB patients. Nevertheless, well designed and multi-ethnics clinical researches with large sample will be necessary to verify and strengthen the metastatic role of S100A9 in NB patients.
Acknowledgements
We thank Professor Yan Xiao who kindly provided SH-SY-5Y cells.
Author contributions
YYZ conceived the study. XC, YKX, JF, QWT and QW perform the study. Statistical analysis was undertaken by XC. All authors read and approved the final version of the manuscript. QW and YYZ wrote the manuscript.
Funding
This study is fully supported by the National Natural Science Foundation of China (No. 81601821 to YYZ), the Natural Science Foundation of Shandong Province (No. ZR201910230488 to XC and No. ZR202102180776 to YYZ), China Postdoctoral Science Foundation (No. 2016M592144 to XC and No. 2016M590621 to YYZ), Postdoctoral innovation project of Shandong Province (No. 201603069 to YYZ), Shangdong Province Key Research and Development Program (2019GSF108108 to QW).
Data availability
The following information was supplied regarding data availability: The data is available at NCBI GEO: GSE90121.
Declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Qingdao University.
Consent to participate
Informed consent were obtained from the participants or their parents.
Consent for publication
All authors had gone over the manuscript and consent to submit.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yunyuan Zhang and Qing Wang have contributed equally to this work.
Contributor Information
Yunyuan Zhang, Email: hellozyycool@163.com.
Qing Wang, Email: wangqing0533@aliyun.com.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brignole C, Bensa V, Fonseca NA, Del Zotto G, Bruno S, Cruz AF, Malaguti F, Carlini B, Morandi F, Calarco E, Perri P, Moura V, Emionite L, Cilli M, De Leonardis F, Tondo A, Amoroso L, Conte M, Garaventa A, Sementa AR, Corrias MV, Ponzoni M, Moreira JN, Pastorino F. Cell surface nucleolin represents a novel cellular target for neuroblastoma therapy. J Exp Clin Cancer Res. 2021;40:180. doi: 10.1186/s13046-021-01993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, Weiss WA. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 4.Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, Angel P, Mayer D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11:5146–5152. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 5.Wagner NB, Weide B, Gries M, Reith M, Tarnanidis K, Schuermans V, Kemper C, Kehrel C, Funder A, Lichtenberger R, Sucker A, Herpel E, Holland-Letz T, Schadendorf D, Garbe C, Umansky V, Utikal J, Gebhardt C (2019) Tumor microenvironment-derived S100A8/A9 is a novel prognostic biomarker for advanced melanoma patients and during immunotherapy with anti-PD-1 antibodies. J Immunother Cancer 7:343. 10.1186/s40425-019-0828-1 [DOI] [PMC free article] [PubMed]
- 6.Aberg AM, Bergstrom SH, Thysell E, Tjon-Kon-Fat LA, Nilsson JA, Widmark A, Thellenberg-Karlsson C, Bergh A, Wikstrom P, Lundholm M. High monocyte count and expression of S100A9 and S100A12 in peripheral blood mononuclear cells are associated with poor outcome in patients with metastatic prostate cancer. Cancers (Basel) 2021 doi: 10.3390/cancers13102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidehag V, Hammarsten P, Egevad L, Granfors T, Stattin P, Leanderson T, Wikstrom P, Josefsson A, Hagglof C, Bergh A. High density of S100A9 positive inflammatory cells in prostate cancer stroma is associated with poor outcome. Eur J Cancer. 2014;50:1829–1835. doi: 10.1016/j.ejca.2014.03.278. [DOI] [PubMed] [Google Scholar]
- 8.De Veirman K, De Beule N, Maes K, Menu E, De Bruyne E, De Raeve H, Fostier K, Moreaux J, Kassambara A, Hose D, Heusschen R, Eriksson H, Vanderkerken K, Van Valckenborgh E. Extracellular S100A9 protein in bone marrow supports multiple myeloma survival by stimulating angiogenesis and cytokine secretion. Cancer Immunol Res. 2017;5:839–846. doi: 10.1158/2326-6066.CIR-17-0192. [DOI] [PubMed] [Google Scholar]
- 9.Laouedj M, Tardif MR, Gil L, Raquil MA, Lachhab A, Pelletier M, Tessier PA, Barabe F. S100A9 induces differentiation of acute myeloid leukemia cells through TLR4. Blood. 2017;129:1980–1990. doi: 10.1182/blood-2016-09-738005. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Du G, Wang D. The S100 protein family in lung cancer. Clin Chim Acta. 2021 doi: 10.1016/j.cca.2021.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Zheng S, Liu L, Xue T, Jing C, Xu X, Wu Y, Wang M, Xie X, Zhang B. Comprehensive analysis of the prognosis and correlations with immune infiltration of S100 protein family members in hepatocellular carcinoma. Front Genet. 2021;12:648156. doi: 10.3389/fgene.2021.648156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondet J, Chevalier S, Mossuz P. Pathogenic roles of S100A8 and S100A9 proteins in acute myeloid and lymphoid leukemia: clinical and therapeutic impacts. Molecules. 2021 doi: 10.3390/molecules26051323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Liu S, Deng P, Liang Y, Xiao R, Tang LQ, Chen J, Chen QY, Guan P, Yan SM, Huang X, Hong JH, Chen J, Sun Y, Teh BT, Yu Q, Mai HQ, Tan J. Targeting the IRAK1-S100A9 axis overcomes resistance to paclitaxel in nasopharyngeal carcinoma. Cancer Res. 2021;81:1413–1425. doi: 10.1158/0008-5472.CAN-20-2125. [DOI] [PubMed] [Google Scholar]
- 14.Seong BK, Fathers KE, Hallett R, Yung CK, Stein LD, Mouaaz S, Kee L, Hawkins CE, Irwin MS, Kaplan DR. A metastatic mouse model identifies genes that regulate neuroblastoma metastasis. Cancer Res. 2017;77:696–706. doi: 10.1158/0008-5472.CAN-16-1502. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cesaro A, Anceriz N, Plante A, Page N, Tardif MR, Tessier PA. An inflammation loop orchestrated by S100A9 and calprotectin is critical for development of arthritis. PLoS ONE. 2012;7:e45478. doi: 10.1371/journal.pone.0045478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luu HH, Zhou L, Haydon RC, Deyrup AT, Montag AG, Huo D, Heck R, Heizmann CW, Peabody TD, Simon MA, He TC. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Lett. 2005;229:135–148. doi: 10.1016/j.canlet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Duan L, Wu R, Zhang X, Wang D, You Y, Zhang Y, Zhou L, Chen W. HBx-induced S100A9 in NF-kappaB dependent manner promotes growth and metastasis of hepatocellular carcinoma cells. Cell Death Dis. 2018;9:629. doi: 10.1038/s41419-018-0512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsubota S, Kadomatsu K. Origin and initiation mechanisms of neuroblastoma. Cell Tissue Res. 2018 doi: 10.1007/s00441-018-2796-z. [DOI] [PubMed] [Google Scholar]
- 23.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Kramer A, Roncaioli JL, Sand F, Heuckmann JM, Ikram F, Schmidt R, Ackermann S, Engesser A, Kahlert Y, Vogel W, Altmuller J, Nurnberg P, Thierry-Mieg J, Thierry-Mieg D, Mariappan A, Heynck S, Mariotti E, Henrich KO, Gloeckner C, Bosco G, Leuschner I, Schweiger MR, Savelyeva L, Watkins SC, Shao C, Bell E, Hofer T, Achter V, Lang U, Theissen J, Volland R, Saadati M, Eggert A, de Wilde B, Berthold F, Peng Z, Zhao C, Shi L, Ortmann M, Buttner R, Perner S, Hero B, Schramm A, Schulte JH, Herrmann C, O'Sullivan RJ, Westermann F, Thomas RK, Fischer M. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ, Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 26.Liu B, Ricarte Filho J, Mallisetty A, Villani C, Kottorou A, Rodgers K, Chen C, Ito T, Holmes K, Gastala N, Valyi-Nagy K, David O, Gaba RC, Ascoli C, Pasquinelli M, Feldman LE, Massad MG, Wang TH, Jusue-Torres I, Benedetti E, Winn RA, Brock MV, Herman JG, Hulbert A. Detection of promoter DNA methylation in urine and plasma aids the detection of non-small cell lung cancer. Clin Cancer Res. 2020;26:4339–4348. doi: 10.1158/1078-0432.CCR-19-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maekawa H, Ito T, Orita H, Kushida T, Sakurada M, Sato K, Hulbert A, Brock MV. Analysis of the methylation of CpG islands in the CDO1, TAC1 and CHFR genes in pancreatic ductal cancer. Oncol Lett. 2020;19:2197–2204. doi: 10.3892/ol.2020.11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Z, Williams M, Cheng YY, Leung WK. Roles of methylated DNA biomarkers in patients with colorectal cancer. Dis Markers. 2019;2019:2673543. doi: 10.1155/2019/2673543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunzmann AT, Murray LJ, Cardwell CR, McShane CM, McMenamin UC, Cantwell MM. PTGS2 (Cyclooxygenase-2) expression and survival among colorectal cancer patients: a systematic review. Cancer Epidemiol Biomarkers Prev. 2013;22:1490–1497. doi: 10.1158/1055-9965.EPI-13-0263. [DOI] [PubMed] [Google Scholar]
- 30.Vene R, Costa D, Augugliaro R, Carlone S, Scabini S, Casoni Pattacini G, Boggio M, Zupo S, Grillo F, Mastracci L, Pitto F, Minghelli S, Ferrari N, Tosetti F, Romairone E, Mingari MC, Poggi A, Benelli R. Evaluation of glycosylated PTGS2 in colorectal cancer for NSAIDS-based adjuvant therapy. Cells. 2020 doi: 10.3390/cells9030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shintani T, Higaki M, Okamoto T. Heparin-binding protein 17/Fibroblast growth factor-binding protein-1 knockout inhibits proliferation and induces differentiation of squamous cell carcinoma cells. Cancers (Basel) 2021 doi: 10.3390/cancers13112684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Zhang J, Pang X, Chen Z, Zhang Z, Lei L, Xu H, Wen L, Zhu J, Jiang Y, Cui Y, Chen G, Wang X. MiR-205-5p suppresses angiogenesis in gastric cancer by down regulating the expression of VEGFA and FGF1. Exp Cell Res. 2021;404:112579. doi: 10.1016/j.yexcr.2021.112579. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Wang Z, Zeng Z, Shen N, Wang B, Zhang Y, Shen H, Lu W, Wei R, Ma W, Wang C. Bioinformatic analysis identifying FGF1 gene as a new prognostic indicator in clear cell renal cell carcinoma. Cancer Cell Int. 2021;21:222. doi: 10.1186/s12935-021-01917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang Y, Zhou L. S100A9 promotes human hepatocellular carcinoma cell growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK pathways. Exp Cell Res. 2015;334:228–238. doi: 10.1016/j.yexcr.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Duan L, Wu R, Ye L, Wang H, Yang X, Zhang Y, Chen X, Zuo G, Zhang Y, Weng Y, Luo J, Tang M, Shi Q, He T, Zhou L. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/beta-catenin pathway. PLoS ONE. 2013;8:e62092. doi: 10.1371/journal.pone.0062092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen KT, Kim PD, Jones KA, Devarajan K, Patel BB, Hoffman JP, Ehya H, Huang M, Watson JC, Tokar JL, Yeung AT. Potential prognostic biomarkers of pancreatic cancer. Pancreas. 2014;43:22–27. doi: 10.1097/MPA.0b013e3182a6867e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JS, Lee NR, Kashif A, Yang SJ, Nam AR, Song IC, Gong SJ, Hong MH, Kim G, Seok PR, Lee MS, Sung KH, Kim IS. S100A8 and S100A9 promote apoptosis of chronic eosinophilic leukemia cells. Front Immunol. 2020;11:1258. doi: 10.3389/fimmu.2020.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zha H, Li X, Sun H, Duan L, Yuan S, Li H, Li A, Gu Y, Zhao J, Xie J, Zhou L. S100A9 promotes the proliferation and migration of cervical cancer cells by inducing epithelialmesenchymal transition and activating the Wnt/betacatenin pathway. Int J Oncol. 2019;55:35–44. doi: 10.3892/ijo.2019.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Kong F, Jin C, Hu E, Shao Q, Liu J, He D, Xiao X. The expression of S100A8/S100A9 is inducible and regulated by the Hippo/YAP pathway in squamous cell carcinomas. BMC Cancer. 2019;19:597. doi: 10.1186/s12885-019-5784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider RK, Schenone M, Ferreira MV, Kramann R, Joyce CE, Hartigan C, Beier F, Brummendorf TH, Germing U, Platzbecker U, Busche G, Knuchel R, Chen MC, Waters CS, Chen E, Chu LP, Novina CD, Lindsley RC, Carr SA, Ebert BL. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22:288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Y, Cao R, Che B, Sun D, Tang Y, Jiang L, Bai Q, Liu Y, Morozova-Roche LA, Zhang C. Proinflammatory S100A9 regulates differentiation and aggregation of neural stem cells. ACS Chem Neurosci. 2020;11:3549–3556. doi: 10.1021/acschemneuro.0c00365. [DOI] [PubMed] [Google Scholar]
- 42.Marinkovic G, Koenis DS, de Camp L, Jablonowski R, Graber N, de Waard V, de Vries CJ, Goncalves I, Nilsson J, Jovinge S, Schiopu A. S100A9 links inflammation and repair in myocardial infarction. Circ Res. 2020;127:664–676. doi: 10.1161/CIRCRESAHA.120.315865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability: The data is available at NCBI GEO: GSE90121.