Abstract
The Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C) is essential for EBV-dependent immortalization of human primary B lymphocytes. Genetic analysis indicated that amino acids 365 to 992 are important for EBV-mediated immortalization of B lymphocytes. We demonstrate that this region of EBNA3C critical for immortalization interacts with prothymosin alpha (ProTα), a cellular protein previously identified to be important for cell division and proliferation. This interaction maps to a region downstream of amino acid 365 known to be involved in transcription regulation and critical for EBV-mediated transformation of primary B lymphocytes. Additionally, we show that EBNA3C also interacts with p300, a cellular acetyltransferase. This interaction suggests a possible role in regulation of histone acetylation and chromatin remodeling. An increase in histone acetylation was observed in EBV-transformed lymphoblastoid cell lines, which is consistent with increased cellular gene expression. These cells express the entire repertoire of latent nuclear antigens, including EBNA3C. Expression of EBNA3C in cells with increased acetyltransferase activity mediated by the EBV transactivator EBNA2 results in down-modulation of this activity in a dose-responsive manner. The interactions of EBNA3C with ProTα and p300 provide new evidence implicating this essential EBV protein EBNA3C in modulating the acetylation of cellular factors, including histones. Hence, EBNA3C plays a critical role in balancing cellular transcriptional events by linking the biological property of mediating inhibition of EBNA2 transcription activation and the observed histone acetyltransferase activity, thereby orchestrating immortalization of EBV-infected cells.
Epstein-Barr Virus (EBV) is a human gammaherpesvirus predominantly infecting epithelial cells of the oropharynx and human primary B lymphocytes (41, 63). EBV is the etiological agent of infectious mononucleosis and is also associated with various human malignancies, including Burkitt's lymphoma, nasopharyngeal carcinoma, non-Hodgkin's disease, AIDS immunoblastic lymphomas, and lymphoproliferative disease (3, 63). Infection of the oropharyngeal epithelium is predominantly a lytic type of infection with the production of progeny virus (33, 61, 63, 73). Infection of human primary B lymphocytes by EBV transforms them into continuously proliferating lymphoblastoid cell lines (LCLs) in vitro (11, 29). Recent studies have demonstrated that EBV utilizes two major cellular signaling pathways for transforming B cells, the NOTCH1 signaling pathway and the TNF signaling pathway (6, 34, 57).
After initial infection of B lymphocytes, EBV typically establishes a latent infection with the expression of 11 viral transcripts (41, 63). These genes are the six EBV nuclear antigens (EBNAs), three latent membrane proteins (LMPs), and the EBV early RNAs (41). Only a selected number of these genes are necessary for EBV-mediated immortalization of B lymphocytes (65). EBNA2, EBNA3A, EBNA3C, and LMP1 are essential for EBV-induced immortalization of B lymphocytes; however, EBNA3B, EBV early RNAs, and LMP2 are dispensable for B lymphocyte immortalization (11, 40, 47, 49, 76–78). EBNA1 is important for the persistence of the EBV episome in infected cells (1, 89).
Previous genetic analysis of EBNA3C demonstrated that introduction of an amber stop codon at amino acid (aa) 365 in EBNA3C renders the recombinant EBV incapable of immortalizing human primary B lymphocytes (78). This suggests that interactions with cellular or viral factors that occur downstream of aa 365 of the EBNA3C protein are critical for EBV immortalization of B lymphocytes. EBNA3C is an essential viral transcription factor with motifs similar to those of the cJun/cFos family of transcription factors (41, 69, 78). The basic structure of the protein sequence (see Fig. 1B) shows a large polypeptide of 992aa with a putative nuclear localization signal, leucine zipper motif, acidic domains, and proline- and glutamine-rich domains (41, 69, 78). EBNA3C demonstrates an ability to act as both a repressor and an activator of transcription in transient-reporter assays (7, 53, 64, 66, 67, 90). In transient-reporter assays the two acidic domains have been reported to function as a negative regulator of transcription and the glutamine-rich region has been reported to function as an activator when fused to the GAL4-DNA binding domain (GAL4DBD) (7, 44, 53, 66). The amino-terminal portion of EBNA3C can interact with a ubiquitous, sequence-specific cellular transcription factor, RBP-Jκ (67, 90). This interaction results in disruption of RBP-Jκ with its cognate sequence (67, 90). EBNA3C also competes with EBNA2, the EBV transactivator for binding to RBP-Jκ (66). Therefore EBNA3C acts as a modulator of transcription through interaction with and inhibition of RBP-Jκ from binding to DNA or other transcriptional regulators such as the EBV transactivator EBNA2 (53, 66, 67, 90). These functions resemble that of the Drosophila melanogaster protein Hairless in regulating Suppressor of Hairless (SuH), the Drosophila homolog of RBP-Jκ (8, 64).
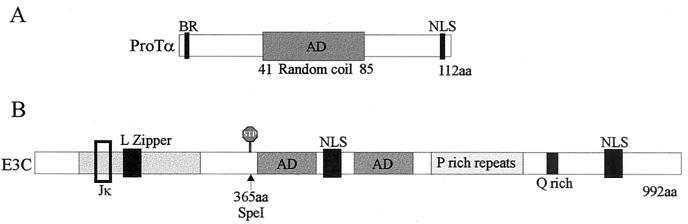
FIG. 1.
ProTα was isolated from a yeast two-hybrid cDNA library screen as a cellular molecule interacting with EBNA3C. (A) The sequence of the cDNA obtained from screen was matched against the previously known ProTα sequence found with a BLAST search in GenBank. Shown is a schematic representation of the ProTα protein, showing the locations of the central acidic domain (AD) that may have a random coil conformation, the putative nuclear localization signal (NLS), and the amino-terminal basic region (Br) (16, 19, 26). (B) Diagram showing a schematic representation of the EBNA3C protein. The RBP-Jκ binding site is indicated by the open boxed region. The putative leucine zipper motif (L), the ADs, the NLS, the proline-rich repeats (P), and glutamine-rich regions (Q) are indicated on the diagram. The stop signal indicates position aa 365 where the amber codon was introduced in the genetic analysis of the EBNA3C gene (41, 69).
To identify cellular proteins interacting with the region of EBNA3C downstream of the RBP-Jκ binding site, we used a yeast two-hybrid screen with an EBV-positive lymphoblastoid cell line-derived cDNA library (31). A truncated form of EBNA3C (aa 365 to 992) was cloned into a yeast expression vector fused to the GAL4DBD and used to screen for interacting cellular proteins crucial for EBV-induced B-lymphocyte immortalization. A cellular protein isolated multiple times from the screen was identified as prothymosin alpha (ProTα), known to be important for cell division and proliferation (26, 68, 71, 86).
ProTα is a 112-aa, highly acidic nuclear protein ubiquitously expressed in most eukaryotic cells (21, 24, 70). Although the specific functions of this highly conserved protein remain unknown, ProTα has been implicated as a crucial factor in cell division and proliferation (26, 68, 71, 86). Previous studies to support this notion have shown that expression of ProTα is elevated in proliferating lymphocytes and is activated by the proto-oncogene c-myc, a cellular oncogene known to be involved in driving cell proliferation (15, 25). ProTα has also been shown to be present during all stages of the cell cycle, increasing at the S/G2 transition point (81). In addition, antisense oligonucleotides directed against ProTα mRNA inhibited cell division and proliferation in myeloma cells (71). ProTα has also been associated with a number of human tumors (56). The mRNA levels of ProTα correlate with that of the c-myc transcripts in human colon cancers, and elevated levels of the protein were seen in malignant tissues of the intestine and breast (13, 79).
ProTα also interacts with histones in vitro and may be involved in chromatin remodeling (12, 23, 39). Moreover, cellular gene expression is regulated at the level of chromatin structure through histone acetylation by the recruitment of histone acetyltransferases (HATs) like p300 and CREB-binding protein (CBP) (32, 35, 43). The addition of acetyl groups to core histones results in disassociation of the chromatin and nucleosomal structure, thereby rendering the transcriptional regulatory sites accessible to the transcription machinery (22, 72, 74, 75). These findings prompted us to investigate the potential role that the interaction of EBNA3C and ProTα may have in regulation of HAT activity. These studies demonstrate the first association of EBNA3C with acetyltransferases and show that EBNA3C may be recruited to the chromatin through a specific nuclear factor, ProTα, thereby modulating histone acetylation balancing transcriptional events. These results suggest a finely tuned mechanism for immortalization of human B lymphocytes.
MATERIALS AND METHODS
Cell lines and antibodies.
BJAB is an EBV-negative Burkitt's lymphoma cell line obtained from Elliott Kieff (41, 52). BJAB EBNA3C and BJAB neo control are BJAB cells transfected with pZipneo eukaryotic expression vector with EBNA3C or vector alone (66). LCL1 and LCL2 are recently immortalized LCLs transformed by EBV. All cells were cultured in RPMI 1640 supplemented with 2 mM glutamine, 25 U of penicillin-streptomycin per ml, 200 μg of neomycin (Gibco-BRL) per ml when necessary, 10% fetal bovine serum, and 20 μg of gentamicin (Gemini-Bioproducts Inc.) per ml. 293 embryonic kidney epithelial cells were cultured in Dulbecco's modified Eagle medium supplemented with 2 mM glutamine, 25 U of penicillin-streptomycin (Gibco-BRL) per ml, 10% fetal bovine serum, and 20 μg of gentamicin (Gemini-Bioproducts Inc.) per ml.
A10 is a mouse monoclonal hybridoma antibody against EBNA3C obtained from Martin Rowe (54). The anti-EBNA3C serum is a rabbit polyclonal that was made from a glutathione S-transferase (GST) fusion protein of EBNA3C by Cocalico Inc. 9E10 is a mouse monoclonal antibody directed against the myc tag. The antibodies used for detection of p300 were purchased from Santa Cruz. Histone H1 monoclonal antibody was purchased from Upstate Biotechnology. The anti-ProTα rabbit polyclonal serum was initially obtained from Fernando Dominquez and was also made from a GST fusion of the entire ProTα gene by Cocalico Inc.
Plasmids and constructs.
pAS1ΔEBNA3C encodes a truncated form of EBNA3C (aa 365 to 992) created by removing the amino-terminal 364 aa by SpeI digestion and ligating the carboxy fragment (aa 365 to 992) into the pAS1 vector at the NdeI site. All yeast vectors and the cDNA library were previously described and were obtained from Stephen Elledge (30). A GST-ProTα fusion protein was obtained from Fernando Dominguez as clone pAV1 (46). pA3MProTα is a ProTα-myc fusion protein constructed by ligating the entire coding sequence of ProTα that was amplified out of pAV1 using Vent Polymerase (NEB) in frame with the myc epitope in pA3M (6). Primers flanking the coding sequence of ProTα contained EcoRI and EcoRV restriction sites in the forward and reverse primers, respectively (forward primer, 5′GGAATTCCATGTCAGACGCAGCCGTAGACA3′, and reverse primer, 5′GGATATCGGGTCATCCTCGTCGGTCTTCTG3′). pSG5EBNA3C and 3′ and 5′ P300 clones have been previously described (59, 66).
For construction of GST fusions of specific regions of EBNA3C, we digested EBNA3C with AluI and RsaI restriction enzymes. Fragments from these digestions were isolated and then fused to GST in the pGEX2T vector. All fusions obtained from RsaI digestion were denoted as R1 to R10, and all fusions obtained from AluI digestions were denoted as A1 to A10 (numbered from largest to smallest fragment). AluI fragments were cloned into the EcoRI site of pGEX2T, and the RsaI fragments were cloned into the SmaI or EcoRI site.
Yeast two-hybrid cDNA screen.
An EBV-positive B-lymphoblastoid cell line-derived cDNA library was screened using a yeast two-hybrid system as previously described (30, 57). Briefly, Y190 yeast cells were first stably transformed with pASIΔEBNA3C. The Y190 expressing the truncated EBNA3C molecule was then transformed with the pACTcDNA library, and resulting transformants growing on media lacking histidine, tryptophan, and leucine were tested for β-galactosidase (β-Gal) reactivity. To confirm positive reactivity, positive clones were retransformed into yeast with pASIΔEBNA3C and screened again for positive β-Gal reactivity. Yeast cells that produced blue color within 15 min to 1 h were considered positive clones. After isolation of plasmids and retesting of β-Gal activity, the positive clones were then sequenced using an automated sequencing core facility or by manual sequencing using the Thermo-Sequenase kit (Amersham). Sequences obtained were compared to other known sequences in GenBank by using the BLAST program.
Northern blot analysis.
[32P]dCTP-labeled cDNA probes were used in Northern blot analysis of poly(A) RNA from multiple human tissues (CLONTECH Laboratories Inc.) and EBV-positive and -negative B-lymphocyte cell lines. A 32P-labeled ProTα probe was made using a random primer kit and purified using a Nuc-Trap probe purification column (Stratagene). A [32P]dCTP-labeled cDNA probe was created using β-actin control cDNA that was provided with the multiple human blots (CLONTECH Laboratories Inc.). DNA used for the GAPDH probe was from a cDNA clone obtained from Fred Wang. Hybridization conditions were as suggested by the manufacturer (CLONTECH Laboratories Inc.).
In vitro protein binding assays.
EBNA3C, 3′P300, and 5′P300 were translated in vitro (TNT system; Promega) from T7 expression plasmids using 35S-met/cys Express label (NEN-Dupont). In competition binding experiments, nonlabeled EBNA3C was in vitro translated using the same method without 35S-met/cys translabel. The GST-ProTα fusion protein was expressed from pAV1 in Escherichia coli DH5α and purified using glutathione-Sepharose beads. All binding experiments were done as previously described (6, 66). Dried gels were analyzed using a PhosphorImager (Molecular Dynamics), and signals were quantified using ImageQuant software.
In pull-down assays, 108 cells were lysed as described below (“Immunoprecipitations and immunoblotting”). Lysates were incubated with glutathione-bound GST beads for 30 min at 4°C. After brief centrifugation to remove beads, cell lysates were incubated with glutathione-bound GST-ProTα overnight at 4°C. Beads were collected by brief centrifugation, washed four times in radioimmunoprecipitation assay (RIPA) buffer, and denatured in sodium dodecyl sulfate (SDS)–β-mercaptoethanol lysis buffer. Proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to 0.45-μm-pore-size nitrocellulose membranes.
For mapping the region of EBNA3C interacting with ProTα, the plasmid containing the ProTα gene was used in an in vitro translation reaction using the Promega TNT system. In vitro-translated ProTα was used in binding experiments with GSTΔEBNA3C fusion proteins. Bound proteins were fractionated by SDS-PAGE, dried, and exposed to a PhosphorImager as described above.
Immunoprecipitations and immunoblotting.
A total of 108 cells were lysed in RIPA buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin [1 μg/ml], and pepstatin [1 μg/ml]) for 1 h on ice with brief vortexing every 15 min. A portion of the lysate was removed for use as the control. Lysates were precleared by incubation with 2 μl of normal rabbit serum for 30 min at 4°C, and bound precipitates were collected with protein A-Sepharose beads (Sigma). In some cases, lysates were precleared using the same incubation with protein A-Sepharose beads alone. Anti-EBNA3C monoclonal (A10), anti-ProTα, or anti-5′ p300 (Santa Cruz) antibodies were incubated with lysates overnight at 4°C. Immunoprecipitates were collected with protein A-Sepharose beads and washed four times in RIPA buffer. For p300 analysis, cells were subjected to Dounce homogenization with 10 strokes to remove cell membrane and cytoplasm. The nuclear fraction was then collected and lysed in RIPA buffer for 1 h on ice with brief vortexing every 15 min. Proteins were heated in SDS–β-mercaptoethanol lysis buffer and then analyzed by SDS-PAGE. Western blot analyses were done using specific antibodies for detection of EBNA3C, ProTα, and p300.
Transfections and colocalizations.
Five million 293 cells were transfected with 20 μg of pA3M, pA3M-ProTα, pSG5/EBNA3C, and pA3M/ProTα, or with no DNA as a control, using a Bio-Rad electroporation apparatus at 210 V and 975 μF. Cells were resuspended in 10 ml of Dulbecco's modified Eagle medium prepared as described above. Cells were collected and used for immunoprecipitations 24 h posttransfection. Cells were washed once in 1× phosphate-buffered saline (PBS). Immunoprecipitation was performed as described above. Anti-myc ascites antibody was used to obtain immunoprecipitates. Anti-ProTα and anti-EBNA3C antibodies were used for Western blot analysis.
For colocalizations of ProTα and EBNA3C in EBV-immortalized human primary B lymphocytes, 1 million cells were transferred to a 1.5-ml Eppendorf tube and washed in sterile PBS. A sample of the cells was fixed onto slides with methanol-acetone (1:1) for 10 min at −20°C. Cells were blocked with 20% goat serum. Primary antibodies used were anti-rabbit ProTα (1:100) and anti-mouse EBNA3C A10 ascites (1:1,000). Secondary antibodies used were goat anti-rabbit Texas red and goat anti-mouse fluorescein isothiocyanate (FITC) (1:1,000). Cells were subjected to four 5-min washes with 1× PBS after each antibody. Cells were visualized using standard fluorescence microscopy on an Olympus BX60 microscope. Digital pictures were obtained using an Olympus digital camera and Esprit computer software program (version 1.2).
In vivo HAT assay.
Immunoprecipitates were collected as described above (“Immunoprecipitations and immunoblotting”) using ProTα or 5′ p300 antibodies from lysates of EBV-infected LCLs (LCL1 and LCL2) and B cells expressing EBNA3C or no EBNA as a control. For transient transfections BJAB cells were transfected at 210 V and 960 μF using a Bio-Rad electroporator. Expression constructs containing EBNA2 or EBNA2 and EBNA3C were mixed with BJAB cells, electroporated, and then incubated for 24 h. Cells were collected by centrifugation and lysed in RIPA buffer. Protein complexes containing acetylases were obtained by immunoprecipitation with polyclonal antibodies against the 5′ region of p300. Immunoprecipitates were washed twice in RIPA buffer (same as previously described except with 0.5% Nonidet P-40) followed by two washes in HAT assay buffer (50 mM Tris-HCl [pH 8.0], 10 mM sodium butyrate, 10% glycerol, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride). Immunoprecipitates were incubated in HAT buffer with 25 μg of crude histones (Sigma) and 500 nCi of [3H]coenzyme A for 1 h at 30°C. After brief centrifugation to pellet protein A-Sepharose beads, the supernatant was spotted onto p-81 filters (Whatman) and washed in 0.2 M sodium carbonate three times using vacuum filtration. Filters were allowed to dry under a heat lamp before being placed in ScintiVerse scintillation cocktail and analyzed in a Beckman LS3801 scintillation counter.
RESULTS
A yeast two-hybrid screen indicates that a cellular protein interacting with the critical carboxy terminus of the EBNA3C protein is ProTα.
A truncated form of EBNA3C (aa 365 to 992) was fused in frame to the GAL4DBD. This fusion protein was used in a two-hybrid screen for identifying interacting cDNAs in an EBV-positive LCL-derived cDNA library (31). The GAL4 activating domain was fused to the cDNA clones in the library made from LCLs (30). As a control, the RBP-Jκ molecule cloned into the vector containing the activating domain did not interact with our GAL4DBDΔEBNA3C (aa 365 to 992) and resulted in a negative result for β-Gal activity. As expected, the full-length EBNA3C fused to the GAL4DBD in pAS1 scored positive for β-Gal activity, with RBP-Jκ demonstrating the specificity for EBNA3C-interacting proteins (E3CIP) to aa 365 to 992 of EBNA3C (Table 1). A number of β-Gal-positive transformants growing on medium lacking His, Leu, and Trp were detected in the presence of 50 mM 3-aminotriazole. These clones were analyzed further by replating on medium lacking His, Leu, and Trp and tested for β-Gal activity. Two of the interacting clones were consistently positive for β-Gal activity within 30 min. The two E3CIP interacted with the truncated EBNA3C (aa 365 to 992) fused to the GAL4DBD on replating as determined by β-Gal activity (Table 1). These clones were sequenced and found to be identical to a molecule identified through a BLAST sequence search in GenBank as ProTα. Our submitted sequence was 99% identical to the known GenBank sequences (16, 25, 26). All other deposited sequences were identical with a single deleted GAG codon except for one sequence, which contained this additional codon 3′ to aa 39 (16). Analysis of the amino acid sequence revealed a central acidic domain from aa 41 to 85, rich in glutamic acid and asparagine, with potential for random coil formation (21). A small basic region is seen in the amino-terminal region of ProTα, and a nuclear localization signal is at the carboxy end (shown in the schematic diagram in Fig. 1A) (24, 51, 81). The insert isolated from the yeast cDNA clone was approximately 1.2 kb in size and contained the entire open reading frame of the ProTα gene, including a 138-bp region of leader sequence.
TABLE 1.
ProTα interacts with ΔEBNA3C (aa 365 to 992) in the yeast two-hybrid assay as determined by β-galactosidase activitya
| GAL4DBD | GAL4-AD | β-Gal activity |
|---|---|---|
| pAS1ΔEBNA3C (aa 365 to 992) | − | |
| pAS1ΔEBNA3C (aa 365 to 992) | pACTcDNA (E3CIP) | ++++ |
| pAS1ΔEBNA3C (aa 365 to 992) | pACTRBP-Jκ | − |
| pAS1EBNA3C | pACTRBP-Jκ | ++++ |
The yeast strain Y190 was transformed with the pAS1 constructs, and the transformed colonies were isolated and checked for expression of the fusion proteins. The transformants were then transformed with the individual pACT plasmid constructs as indicated. As expected, the control experiments show that EBNA3C interacts with RBP-Jκ in the yeast system (53, 66). The truncated EBNA3C with the deletion of the 5′ amino-terminal portion encoding the RBP-Jκ binding site does not interact with RBP-Jκ in this assay. The truncated EBNAC fusion molecule pAS1ΔEBNA3C (aa 365 to 992) does not activate in 50 mM aminotriazole. However, E3CIP demonstrated strong interaction with the truncated EBNA3C (aa 365 to 992) molecule by β-Gal activity. The intensity of the β-Gal signal is denoted as follows: ++++, strongly positive; −−−, negative.
A stop codon inserted at aa 365 of EBNA3C (see Fig. 1B) results in the inability of EBV to transform B lymphocytes, suggesting that cellular molecules interacting at the carboxy terminus are potentially an important requirement for EBV to transform primary B lymphocytes. Therefore, this newly identified interaction of ProTα and ΔEBNA3C (aa 366 to 992) may also be critical for immortalization of B lymphocytes.
Expression of ProTα in human tissues and cell lines.
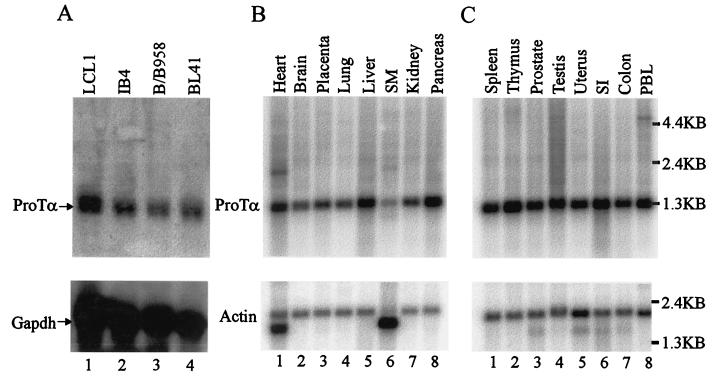
Northern blot analyses of poly(A) RNA from human tissues and EBV-positive and -negative cell lines were performed to determine the levels of ProTα expression in tissues and in previously transformed cell lines infected with EBV. We also wanted to determine if virus infection leads to upregulation of the ProTα transcript. An α-32P-labeled ProTα DNA probe was hybridized to membranes containing poly(A) RNA isolated from various cell lines and tissues (top panels in Fig. 2). The ProTα transcripts are ubiquitously expressed in all tissues and cell lines examined (Fig. 2). In BL41, a Burkitt's lymphoma, EBV-negative cell line, ProTα is readily detected. No dramatic upregulation was seen in the BL41 EBV-infected (isolate B95-8) cell line BL41/B95-8 or the primary B-cell lines transformed by EBV, IB4, and LCL1 (see Fig. 2A, lanes 1 to 4), suggesting that similar levels of ProTα are expressed in EBV-negative and -positive Burkitt's lymphoma-derived cell lines. However, ProTα may be induced as a consequence of the initiation of the transformation event in primary B cells after EBV infection or may be a consequence of other changes in cellular events leading to cell proliferation (26, 68, 71, 86).
FIG. 2.
ProTα is expressed in various tissue types and in EBV-infected cells. (A) Northern blot analysis of poly(A) RNA in EBV-infected LCLs and Burkitt's lymphoma cell lines shows ProTα. LCL1, IB4, and B/B958 are EBV-positive cell lines, while BL41 is an EBV-negative Burkitt's lymphoma cell line. The top panel shows Northern blot analysis using a 32P-labeled ProTα DNA probe. The lower panel shows Northern blot analysis using a [32P]GAPDH DNA probe. (B and C). Northern blot analysis of poly(A) RNA in Multiple Tissue Northern Blots (CLONTECH Laboratories Inc.). The top panels show Northern blot analysis using a 32P-labeled ProTα DNA probe. The lower panels show Northern blot analysis using a 32P-labeled human β-actin cDNA probe.
We then wanted to determine the comparative levels of ProTα expression in tissue. An increase in the level of ProTα transcripts was seen in the heart, liver, pancreas, thymus, small intestine, and peripheral blood leukocytes (Fig. 2B and C). In skeletal muscle the levels of ProTα transcripts were lower (at least a fourfold decrease) than those in the other tissues (Fig. 2B, lane 6). The two transcripts seen in the control β-actin lanes (Fig. 2B and C) for heart and skeletal muscle are due to alternative splicing of β-actin in these tissues (CLONTECH Laboratories Inc.). Another alternative transcript for ProTα of approximately 1.8 to 2.0 kb was predominantly seen in heart (Fig. 2B, lane 1), and an approximately 4.4-kb transcript was seen in primary blood leukocytes (Fig. 2C, lane 8), indicating that in some tissues there are possible alternatively spliced transcripts or forms of ProTα not previously documented in other studies (50, 80).
ProTα interacts with EBNA3C in vitro.
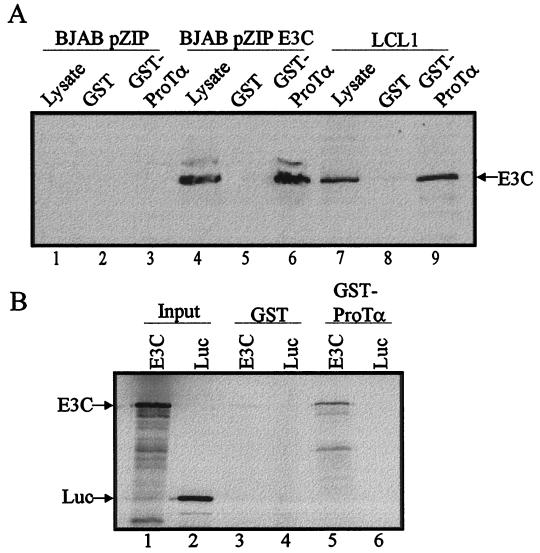
To verify EBNA3C and ProTα interaction, GST-ProTα coupled to glutathione-Sepharose beads was incubated with cell lysates from an EBV-positive LCL and BJAB cells converted with the pZipneo eukaryotic expression vector containing either full-length EBNA3C or no insert as a vector control. Bound proteins were collected, fractionated by SDS-PAGE, and analyzed by Western blotting. EBNA3C was found to interact with GST-ProTα in BJAB cells expressing EBNA3C and in a recently transformed EBV-positive LCL, LCL1, expressing appropriate levels of EBNA3C required to maintain growth transformation of human primary B cells that is mediated by EBV latent gene expression (Fig. 3A, lanes 6 and 9). To further corroborate this finding, an in vitro binding experiment was conducted using in vitro-translated 35S-labeled EBNA3C and a GST fusion protein containing the full-length ProTα used in the above-mentioned binding experiment. Luciferase protein was also in vitro translated with 35S-translabel and incubated with GST-ProTα coupled to glutathione-Sepharose beads as a binding control. 35S-labeled EBNA3C bound to GST-ProTα protein in this experiment at levels approximately 20-fold over binding with GST beads alone as determined by arbitrary counts using the Molecular Dynamics software ImageQuant supplied with the PhosphorImager unit (Fig. 3B, compare lane 3 with lane 5). No detectable signal was seen in the negative control lane using luciferase incubated with GST-ProTα, indicating that the interaction of EBNA3C with GST-ProTα is a specific interaction in vitro.
FIG. 3.
EBNA3C and ProTα associate in cell lysates and interact in vitro. (A) BJAB cells converted with a eukaryotic expression vector containing EBNA3C (BJAB pZIP E3C) or no insert (BJAB pZIP) and an EBV-positive LCL (LCL1) were used in GST pull-down analysis. Cell lysates from 100 million cells were incubated with GST fusion protein coupled to glutathione-Sepharose beads followed by GST-ProTα fusion protein coupled to glutathione-Sepharose beads. Lysates (1%) were used as a control. Proteins were fractionated by SDS–7% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for EBNA3C using the monoclonal antibody A10 (55). (B) GST and GST-ProTα fusion protein coupled to glutathione-Sepharose beads were incubated with 35S-labeled in vitro-translated EBNA3C or luciferase (Luc). A 10% input of in vitro-translated proteins was run as a control. Bound proteins were fractionated by SDS–7% PAGE, dried, and analyzed on a PhosphorImager (Molecular Dynamics) using ImageQuant software.
ProTα associates with EBNA3C in cotransfected cells.
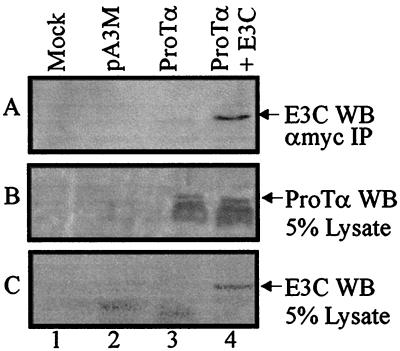
Cotransfection experiments were conducted to further support the association of ProTα and EBNA3C in human cells. 293 cells were transfected with eukaryotic expression vectors containing either no insert, ProTα-myc alone, or EBNA3C and ProTα-myc. Transfections were balanced using pSG5 vector to provide equivalent amounts of DNA per transfection. Additionally, no DNA was used as a mock control. At 24 h posttransfection, cell lysates were incubated with anti-myc ascites antibodies and immunoprecipitates were fractionated by SDS-PAGE followed by Western blot analysis. EBNA3C was detected as an immunoprecipitate of anti-myc antibodies where ProTα and EBNA3C were transiently transfected into 293 cells (Fig. 4A, lane 4). Cell lysates were analyzed by SDS-PAGE to demonstrate protein expression for ProTα and EBNA3C (Fig. 4B and C, respectively). To detect ProTα by Western blotting we used a 0.2-μm-thick transfer membrane to minimize the loss of the small proteins that occurred frequently with the 0.45-μm-pore-size nitrocellulose membranes (39, 51). These results demonstrate that EBNA3C and ProTα can associate with each other in transiently transfected 293 cells.
FIG. 4.
EBNA3C coimmunoprecipitates with ProTα in transiently transfected cells. 293 cells were transfected with pA3M, pA3M/ProTα, pA3M/ProTα and pSG5/EBNA3C, or no DNA (Mock). Anti-myc ascites antibodies were used to collect immunoprecipitates. (A) Proteins were fractionated by SDS–7% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for EBNA3C (A10). Cell lysates (5%) were also analyzed by SDS-PAGE to serve as a control. (B) ProTα was analyzed on a 15% gel, transferred to 0.2-μm-pore-size nitrocellulose membranes, and Western blotted with rabbit anti-ProTα antibody (1:100). (C) EBNA3C was detected by Western analysis as described for panel A.
ProTα interacts with EBNA3C in a region downstream from aa 365.
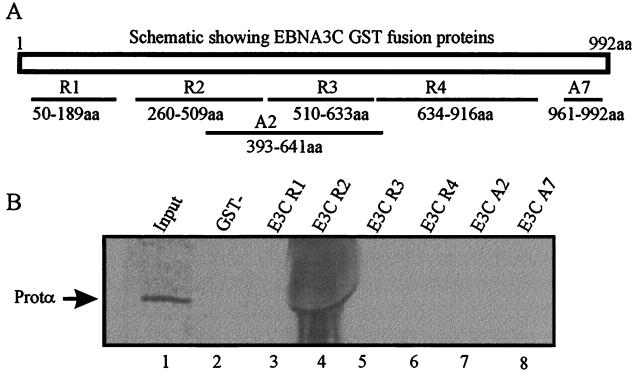
To determine the region of EBNA3C interacting with ProTα we constructed overlapping clones of regions of EBNA3C fused to GST protein (Fig. 5A). These fusion proteins were then used in binding experiments to determine the approximate position of interaction. In vitro-translated ProTα was made and incubated with equivalent amounts of fusion proteins bound to glutathione beads. The resulting bound ProTα was fractionated and exposed to a PhosphorImager screen. The results indicate that a fusion protein (R2) containing a region of EBNA3C from aa 260 to 509 bound to ProTα (Fig. 5B). Additionally, another fusion protein (A2) which overlaps with R2 from aa 393 to 509 did not bind to ProTα, suggesting that the interaction domain lies between aa 260 and 393 (Fig. 5).
FIG. 5.
ProTα interacts with EBNA3C 3′ to aa 365. GSTΔEBNA3C fusion proteins were incubated with in vitro-translated ProTα. Bound proteins were fractionated by SDS–12% PAGE, dried, and then exposed to a PhosphorImager. (A) EBNA3C fragments fused to GST in pGEX vector. The appropriate amino acids for each fragment are indicated, and the names of each fragment begin with R for RsaI fragments and with A for AluI fragments. (B) Results of binding experiment indicating that the EBNA3C fragment from aa 260 to 509 interacts with ProTα. Another fragment, aa 393 to 641, does not interact, indicating that the region of interaction lies before aa 393. Lane 1, input in vitro-translated ProTα; lane 4, ProTα bound to R2 fragment.
The data from the two-hybrid screen where GAL4DBDΔEBNA3C365-992 amino acids interacted with ProTα (Table 1) strongly indicated that the domain of interaction lies within the carboxy terminus of EBNA3C. This information along with the GST binding data above puts the interaction domain within a 28-aa stretch which lies between aa 365 and 393 of the EBNA3C sequence.
ProTα coimmunoprecipitates with EBNA3C and p300 in LCLs.
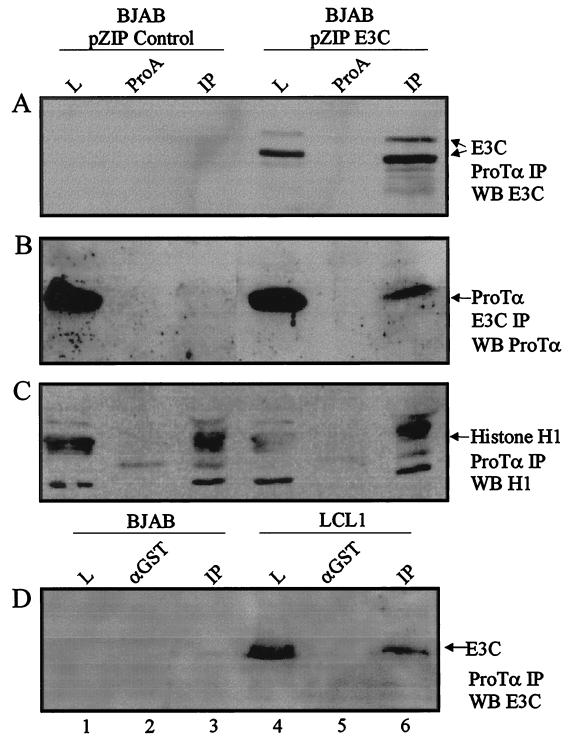
Recent studies on ProTα have shown that it may be involved in chromatin remodeling and interacts with histones in vitro (12, 23). We therefore decided to address the possibility that ProTα is involved in recruiting transcription factors involved in modifying the histones for activating or repressing transcriptional activities. It was possible that EBNA3C is recruited to the chromatin structure in association with p300 and other cellular factors regulating transcription. To determine if EBNA3C, p300, and histone H1 can associate with ProTα in B-lymphoblastoid cells we immunoprecipitated cellular factors associated with ProTα using a rabbit polyclonal antiserum against ProTα. The complexes were analyzed by Western blotting for the presence of EBNA3C, p300, and histone H1. Immunoprecipitates were collected from BJAB cells converted with the pZipneo resistant eukaryotic expression vector containing full-length EBNA3C compared to no insert as a control. Western blot analysis with mouse anti-EBNA3C identified specific bands migrating at the correct size for EBNA3C in the cell lysate as well as in the immunoprecipitate lane (Fig. 6A, lanes 4 and 6). The multiple alternative bands seen in the position where EBNA3C migrates is due to posttranslational modification of the EBNA3C protein as seen in previous studies (36, 67). Moreover, Western blot analysis with mouse anti-histone H1 indicated that ProTα can associate with histone H1 in B-lymphoblastoid cells, as shown in the immunoprecipitate of ProTα in the control EBNA3C-negative BJAB cells as well as in BJAB cells expressing EBNA3C (Fig. 6C, compare lanes 1 and 3 with 4 and 6). Note that histone H1 migrates as a tight doublet on SDS-PAGE gels (27).
FIG. 6.
ProTα coimmunoprecipitates with EBNA3C and histone H1 in B-lymphoblastoid cells. BJAB cells converted with the pZIP eukaryotic expression vector containing EBNA3C (BJAB pZIP E3C) or no insert (BJAB pZIP control) were lysed in RIPA buffer and precleared with either protein A-Sepharose beads (ProA) or GST rabbit polyclonal serum (GST). Immunoprecipitates were collected with anti-ProTα rabbit polyclonal antibody. Cell lysates (1%) were fractionated as a control. (A) Immunoprecipitates fractionated by SDS–7% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for EBNA3C using the monoclonal antibody A10. EBNA3C coimmunoprecipitated with ProTα in EBNA3C-expressing BJAB cells (lane 6) but not in control B cell lines (lane 3). (B) Immunoprecipitates were fractionated by SDS–15% PAGE, transferred to 0.2-μm-pore-size nitrocellulose membranes, and Western blotted for ProTα using the rabbit polyclonal antiserum. ProTα coimmunoprecipitated with EBNA3C seen in the immunoprecipitate lane 6, similar to lysate in lane 4, but not in the EBNA3C-negative BJAB cell lines. (C) Immunoprecipitates fractionated SDS–12% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for histone H1 using the monoclonal antibody from Upstate Biotechnology. Histone H1 was detected as a doublet in the immunoprecipitate lanes 3 and 6, similar to that seen in the lysate lanes 1 and 4. (D) Immunoprecipitation performed as described above except 40 million BJAB (EBV-negative) and LCL1 (EBV-positive) cells were used and 5% cell lysates were used as a control. Cell lysates were precleared with anti-GST rabbit serum. Immunoprecipitates were fractionated by SDS–8% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for EBNA3C using monoclonal antibody A10. EBNA3C immunoprecipitated with ProTα in the EBV-infected LCL (lanes 4 and 6), but not in the control EBV-negative cells (lanes 1 and 3). Quantitative analysis of the signals for changes in immunoprecipitation was done using SCION, NIH Image software.
To determine if EBNA3C associates with p300 and ProTα in LCLs, we immunoprecipitated cellular complexes associating with EBNA3C using anti-EBNA3C polyclonal antibodies. Immunoprecipitates were fractionated, transferred to membranes, and analyzed for detection of ProTα and p300 with EBNA3C antibodies. Both ProTα and p300 coimmunoprecipitated with EBNA3C in B-lymphoblastoid cells. In BJAB cells expressing EBNA3C, ProTα was detected in the immunoprecipitate lane (Fig. 6B, lane 6) but not in the immunoprecipitate of BJAB cells with vector alone (control) (Fig. 6B, lane 3).
We also wanted to determine if the association of ProTα with EBNA3C occurred in recently transformed LCLs in the context of EBV infection and immortalization of human primary B lymphocytes. A similar immunoprecipitation was performed using cell lysates from an EBV-negative cell line (BJAB) and a recently transformed EBV-infected LCL (LCL1) expressing the entire repertoire of latent antigens. Western blot analysis for EBNA3C showed that EBNA3C associates with ProTα in EBV-infected cells in vivo (Fig. 6D, lanes 4 and 6). Taken together, these results suggest that EBNA3C exists in a complex with ProTα, p300, and histone H1 in EBV-transformed human B lymphocytes.
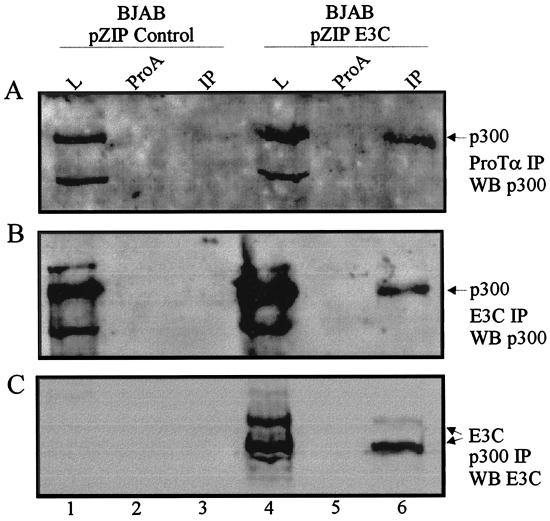
Coimmunoprecipitation of p300 using anti-ProTα antibody demonstrated that p300 levels associated with the complex were consistently lower in BJAB cells with vector alone than in the BJAB-expressing EBNA3C line in multiple experiments (Fig. 7A, compare lanes 1 and 3 with lanes 4 and 6). Thus, p300 was immunoprecipitated with anti-ProTα polyclonal antibody from BJAB cells expressing EBNA3C (Fig. 7A, lanes 6). Moreover, p300 was shown to coimmunoprecipitate with EBNA3C in BJAB cells expressing EBNA3C (Fig. 7B, lane 6). In the reciprocal experiment EBNA3C coimmunoprecipitated with p300, suggesting a tight association between these two molecules in EBNA3C-expressing B-LCLs (Fig. 7C, lanes 4 and 6). These results suggest that EBNA3C is directly associating in a complex with ProTα and p300 in B-lymphoblastoid cells. It is possible that ProTα and/or other transcription factors, including p300, recruit EBNA3C to the chromatin structure. These data do not exclude the possibility that other cellular molecules are also be in the complex and that the interaction is regulated by other associated molecules that are as yet unidentified.
FIG. 7.
The acetyltransferase p300 coimmunoprecipitates with ProTα and EBNA3C in B-lymphoblastoid cells. BJAB cells converted with the pZIP eukaryotic expression vector containing EBNA3C (BJAB pZIP E3C) or no insert (BJAB pZIP control) were lysed in RIPA buffer and precleared with either protein A-Sepharose beads (ProA) or GST rabbit polyclonal serum (GST). Immunoprecipitates were collected with anti-ProTα rabbit polyclonal antibody. Cell lysates (1%) were fractionated as a control. (A) Immunoprecipitates were fractionated by SDS–6% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for p300. The p300 signal seen in the immunoprecipitate lane 6 is barely detectable in the EBNA3C-negative cell line in lane 3. (B) Immunoprecipitates were fractionated by SDS–6% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for p300. Similarly, the p300 signal was seen in the EBNA3C-expressing cell line (lane 6) but not in the EBNA3C-negative cell line (lane 3). (C) Immunoprecipitates were fractionated by SDS–6% PAGE, transferred to 0.45-μm-pore-size nitrocellulose membranes, and Western blotted for EBNA3C. EBNA3C signal was clearly observed in lane 6 where immunoprecipitates from EBNA3C-expressing cells were fractionated.
EBNA3C competes with ProTα for interaction with the 3′ terminus of p300.
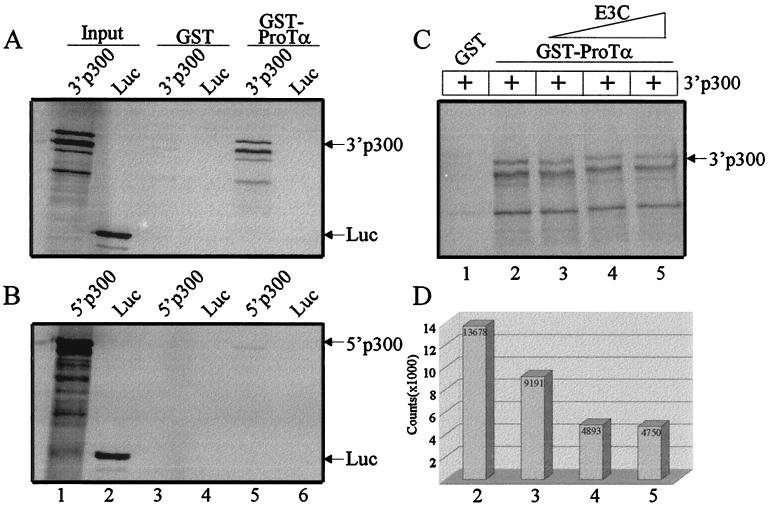
To demonstrate the region of p300 associating with EBNA3C and ProTα, two polypeptides of p300 were utilized. Two p300 polypeptides, corresponding to the 3′ and 5′ regions of p300, were in vitro translated and labeled with [35S]methionine/cysteine. In vitro GST binding experiments demonstrated that GST-ProTα interacts with the 3′ portion of p300 at a level approximately 68-fold over binding with GST alone (Fig. 8, compare lanes 3 and 5). Little or no binding was seen with the 5′ portion of the in vitro-translated p300 (Fig. 8, lanes 3 and 5). Counts of binding to the 5′ p300 were about 1.2-fold over that of the control protein with GST alone. To investigate a potential association or competition of EBNA3C with the p300 and ProTα complex, we added cold in vitro-translated EBNA3C in increasing amounts to the GST-ProTα-bound 3′ p300 complex. EBNA3C competed with 3′ p300 for binding with GST-ProTα, resulting in decreased binding of the truncated 3′ p300 molecule in a dose-dependent manner (Fig. 8C and D). The association of p300 with ProTα as determined by the coimmunoprecipitation was greater in EBNA3C-expressing cell lines in multiple experiments, suggesting a possible stabilization of the ProTα and p300 complex when EBNA3C is present. However, the nature of these associations may be transitory and may depend on whether or not p300 and ProTα exist as a strong complex within a larger complex in the absence of EBNA3C. Additionally, it is also possible that separate complexes of ProTα and EBNA3C as well as ProTα and p300 also exist in vivo and cannot be clearly distinguished in these experiments. Nonetheless, these are new and novel interactions that are indicative of functional changes related to chromatin modifications.
FIG. 8.
The cellular acetylase-coactivator p300 interacts with ProTα. GST and GST-ProTα fusion protein coupled to glutathione-Sepharose beads were incubated with 35S-labeled in vitro-translated 3′ p300, 5′ p300, or luciferase (Luc). Bound proteins as well as 10% input protein were fractionated by SDS–7% PAGE, dried, and analyzed on a PhosphorImager (Molecular Dynamics). (A) GST-ProTα binds the 3′ p300 polypeptide at a level approximately 20-fold (lane 5) over GST (lane 3). (B) The 5′ polypeptide of p300 bound at a level only about 1.2-fold over GST alone. No signal was seen with luciferase nonspecific control polypeptide. (C) GST (lane 1) and GST-ProTα fusion protein coupled to glutathione-Sepharose beads (lanes 2 to 5) were incubated with 35S-labeled 3′ p300 and 2, 5, and 10 μl, respectively, of nonlabeled in vitro-translated EBNA3C. Bound proteins were fractionated by SDS–7% PAGE, dried, and analyzed on a PhosphorImager (Molecular Dynamics). (D) Histogram representing arbitrary counts corresponding to the in vitro binding analysis in panel C analyzed by ImageQuant software (Molecular Dynamics). Column numbers refer to lane numbers in panel C. Numbers indicate actual values obtained. Addition of increasing amounts of EBNA3C to the reaction results in reduction of 3′ p300 signal.
ProTα colocalizes with EBNA3C and p300 in the nucleus of B cells.
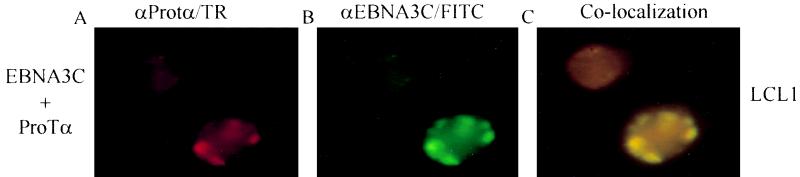
To determine if EBNA3C colocalizes with ProTα in LCLs we performed immunofluorescence assays using antibodies specific to EBNA3C and ProTα. Polyclonal rabbit antiserum against ProTα was detected using anti-rabbit Texas red secondary antibodies (Fig. 9A). Mouse monoclonal antibody against EBNA3C was detected using anti-mouse FITC secondary antibodies (Fig. 9B). Interestingly, in nascently derived LCLs, ProTα and EBNA3C localized in specific nuclear structures in the perinuclear regions (Fig. 9). However, p300 also localized to these regions but could also be seen as diffuse staining throughout the nucleus (data not shown). Additionally, the signals for p300 and ProTα in EBNA3C-expressing cell lines, and particularly in LCLs, were not limited to specific nuclear structures, suggesting that, as expected, they may be involved in numerous other interactions in addition to that seen with EBNA3C. Taken together, these findings support our in vivo data showing association between these proteins and hint at the possibility that a complex exists in the nucleus in infected B lymphocytes.
FIG. 9.
Immunofluorescence analysis demonstrates nuclear colocalization of EBNA3C and ProTα. Cells from LCL1, a nascently derived EBV-immortalized cell line, were fixed in methanol-acetone (1:1) and incubated with antibodies against ProTα (rabbit polyclonal at 1:500 dilution) and against EBNA3C (mouse monoclonal at 1:1,000). Secondary antibodies to detect primary rabbit polyclonal antibodies against ProTα were goat anti-rabbit Texas red (A), and secondary antibodies to detect primary monoclonal antibodies against EBNA3 were goat anti-mouse FITC (B). (C) Overlay of panels A and B demonstrating colocalization in perinuclear regions. These data indicate that EBNA3C, ProTα, and p300 colocalize in the nucleus.
EBV increases histone acetylation in nascently transformed human LCLs.
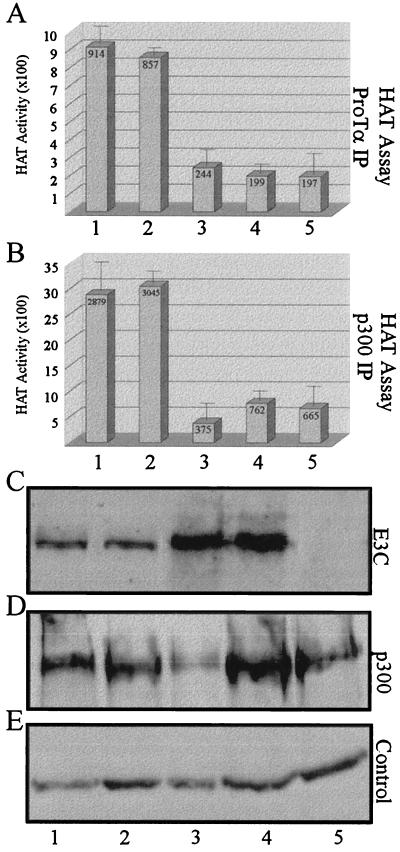
The association of ProTα and EBNA3C with p300 prompted us to look at the acetylation of core histones in EBV-infected and EBNA3C-overexpressing cells through the interaction of ProTα with the cotransactivator-acetylase p300. HAT assays were conducted using immunoprecipitates from two recently transformed EBV-positive B-LCLs expressing the entire set of latent nuclear antigens (EBNAs), two cell lines overexpressing EBNA3C, and a control B-cell line that does not express any of the EBNAs. Protein complexes coimmunoprecipitating with ProTα were collected and used in the HAT assay. In EBV-transformed B-cell lines expressing all the essential latent viral genes, including the major transactivators (EBNA1 and EBNA2), an approximately fourfold increase in acetylation was observed compared to the uninfected B-cell line (Fig. 10A, compare lanes 1 and 2 with lane 5). When HAT activity was determined for B-cell lines overexpressing the EBNA3C protein, a significantly lower HAT activity (approximately fourfold), similar to that in the uninfected control B-cell line (compare lanes 1 and 2 with lanes 3 and 4 in Fig. 10A), was also observed.
FIG. 10.
EBV increases histone acetylation in nascently transformed human LCLs. (A) Results of HAT assay in cell lines expressing the EBV nuclear antigens in recently transformed LCLs, in B-cell lines overexpressing EBNA3C, and in a B-cell line that does not express any of the EBNA proteins. Immunoprecipitates from an anti-ProTα immunoprecipitation demonstrated that a significant HAT activity was seen in the two isogenic B-cell lines expressing all the EBNA proteins compared to the control (compare lanes 1 and 2 with lane 5), whereas this activity was much lower (approximately fourfold) in isogenic B-cell lines overexpressing EBNA3C (lanes 3 and 4), similar to that of the EBNA-negative isogenic B-cell line control (lane 5). A histogram of the results of counts obtained from HAT assays performed using ProTα immunoprecipitates is shown in panel A. (B) Histogram of the radioactive counts obtained from the HAT assay performed using p300 immunoprecipitates. (C) Western blot analysis showing levels of EBNA3C expression in cells used in HAT assays. Lanes correspond to the numbered columns in panels A and B. Lanes 1 and 2 represent assays done with two isogenic B-cell lines infected with EBV, lanes 3 and 4 also contain isogenic B-cell lines overexpressing EBNA3C, and lane 5 contains cells from the same isogenic B cell line used as a control which does not express any of the EBV EBNA proteins. (D) Western blot analysis to determine p300 levels as a control for each cell line used in HAT assays. (E) Internal protein loading controls on the SDS-PAGE gel, transferred to nitrocellulose and stained with Ponceau S.
Another HAT assay using the same cell lines was also performed to determine if protein complexes immunoprecipitated using antibodies to the known acetylase p300 would result in similar acetylation activities. Again the EBV-infected cell lines expressing all the essential latent proteins showed a high level of histone acetylation, approximately four- to sevenfold (Fig. 10B, compare lanes 1 and 2 with lanes 3 to 5) greater than that observed in the EBNA3C-overexpressing cell lines (Fig. 10B, lane 3 and 4) and the control B-cell line control expressing no EBV proteins (Fig. 10B, lane 5). The level of EBNA3C expressed in the B-cell lines was determined by Western blotting. The results show EBNA3C expression which is typical of that seen in EBV-infected B-cell lines expressing all the essential viral proteins compared to relatively high levels being expressed in the B-cell lines overexpressing EBNA3C from a heterologous promoter (Fig. 10C, compare lanes 1 and 2 with lanes 3 and 4). No EBNA3C signal was detected in the control EBV-negative B-cell line (Fig. 10C, lane 5). The B-cell lines overexpressing EBNA3C and containing the control vector alone are isogenic from the same parental Burkitt's cell line. Additionally, Western blotting indicated that the levels of p300 present in these cell lines (Fig. 10D) were comparable to those protein loading control (Fig. 10E), suggesting that the changes in the HAT activity are not due to varying levels of the acetyltransferase p300. Therefore, the increase in HAT activity seen in Fig. 10, lanes 1 and 2, is possibly due to the presence of other EBV or EBV-induced cellular molecules capable of associating with acetyltransferases and their transcription factors, forming large transcription complexes. Recent data demonstrated that expression of the EBV transactivator EBNA2 in cooperation with p300 and pCAF histone acetylases results in increased transcription mediated by EBNA2 (84). Additionally, it has also been demonstrated that EBNA3C can associate with the deacetylase HDAC1 at its amino terminus, suggesting that the ability of EBNA3C to recruit HDAC1 to promoters may have a significant role in repression of transcription mediated by EBNA3C (62).
EBNA3C down-modulates EBNA2-mediated HAT activity.
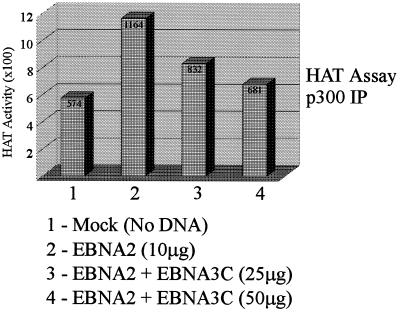
We wanted to investigate the effects of expression of EBNA3C on the HAT activity when EBNA2 is expressed in cells. Previous studies demonstrated that EBNA3C down-modulates EBNA2-mediated transactivation of the major LMP1 promoter (66). We therefore wanted to determine if EBNA3C could down-modulate the HAT activity mediated by EBNA2 and acetylases. Transfection of 293T cells with specific expression constructs followed by immunoprecipitation with anti-p300 antibody and then measurement of the HAT activity indicated that EBNA3C down-modulates the HAT activity in a dose-responsive manner (Fig. 11). This provides one explanation for the ability of EBNA3C to down-modulate transcription activation. It should be noted that EBNA3C reduces the HAT activity to levels similar to those of the mock-transfected cells (compare Fig. 11, lanes 1 and 4).
FIG. 11.
EBNA3C down-modulates HAT activity mediated by EBNA2 and acetyltransferases. In a representative experiment, 293T cells were transfected with expression constructs containing EBNA2 or EBNA2 and EBNA3C. Transfected cells were incubated for 24 h and then harvested and lysed in RIPA buffer. Complexes were collected by immunoprecipitation using the 5′ p300 polyclonal antibody from Santa Cruz. HAT activity was tested as described in Materials and Methods. Addition of EBNA3C results in depression of the HAT activity in a dose-responsive manner.
In previous studies EBNA3C has been shown to down-modulate the transactivation activity of EBNA2 on the major EBV latent promoters (44, 53, 64, 66). Further experiments are currently under way to specifically determine the nature of these observed interactions and the ability of the other essential viral and cellular molecules to mediate the induction of HAT activities in EBV-infected cells. Since EBNA1 and EBNA2 are known activators of transcription, they may have a role in activating or recruiting acetylase complexes, including other cellular transcription factors, as shown by recent work (20, 42, 82, 83).
DISCUSSION
Studies investigating the role of EBNA3C in EBV-mediated B-cell growth transformation have been limited to the association of EBNA3C with the cellular transcription factor RBP-Jκ, which results in down-modulation of transcription from the major EBV latent promoters (53, 64, 66). EBNA3C competes with EBNA2 for binding to RBP-Jκ and also disrupts RBP-Jκ from binding to its cognate sequence (66, 67). These activities are important for driving B-cell transformation after infection by EBV, since the interaction of RBP-Jκ and EBNA2 is essential for B-cell immortalization (87). EBNA3C is an essential protein in this process as shown by the genetic studies where a stop codon inserted at aa 365 renders the recombinant virus incompetent for B-cell immortalization (78). This study focuses on identifying cellular molecules that interact with the portion of EBNA3C, aa 365 to 992, essential for primary B-cell growth transformation as seen by genetic analysis (Fig. 1B) (76). We have identified a cellular molecule, ProTα, through a yeast two-hybrid screen and have confirmed its association with EBNA3C in established B-LCLs and newly transformed LCLs. ProTα is a small protein with a large acidic domain in the central portion of the molecule, a basic region at the amino terminus, and a putative nuclear localization signal at the carboxy terminus (21, 24, 26). ProTα has no real homolog to other known proteins, but it may support a random coil conformation in the central portion of the protein (21) and associates with core histone proteins as shown by in vitro binding studies (12) and linker histone H1 in NIH 3T3 cells (39). Although a large body of data suggests that histone H1 may be acting as a global repressor, other studies show that it is also associated with active genes (85). This provides a role for histone H1 in contributing to gene activation. The ProTα transcript is also upregulated in proliferating lymphoblastoid cells and in cells where the expression of c-myc is induced (15, 25). Inhibition of ProTα expression by antisense oligonucleotides also results in arrest of cell division, suggesting a role in cell cycle regulation (71, 81).
We have shown that ProTα associates with the carboxy terminus of EBNA3C (aa 365 to 992), the region of the protein demonstrated to be critical for EBV-mediated cell growth transformation. Further mapping studies have indicated that the binding region lies between aa 366 and 393, which are conserved among EBNA3C in type I and type II EBV (69). In a number of different assays we have demonstrated that EBNA3C can strongly interact with ProTα in vitro and in vivo under physiological conditions. We have also shown that ProTα association with p300 at the 3′ terminus is about 70-fold greater than that seen with the 5′ terminus of p300. Moreover, EBNA3C can compete with p300 for interaction with ProTα and disrupts this interaction in a dose-dependent manner in vitro. In addition to its association with EBNA3C, ProTα also associates with p300 as well as histone H1 in EBV-infected B cells. These findings provide a molecular basis for elucidating the effects of EBNA3C and ProTα on regulating cell transcription activities through modification of chromatin structure via acetylation of the histones. The identification of the other viral and cellular molecules potentially involved in regulation of these activities is currently under investigation.
The fact that ProTα and EBNA3C may compete for similar regions on the 3′ terminus of p300 is important since other viral oncoproteins, including E1A and large T antigen, bind to the 3′ region of p300 adjacent to the domain that contains the HAT activity and the bromo domain of p300 (14, 58). Our studies have not thoroughly investigated the association of the pCAF molecule, which is also known to associate within the same region of p300 as E1A and large T antigen (9, 10, 37, 48, 55, 58; Z. Arany, W. R. Sellers, D. M. Livingston, and R. Eckner, Letter, Cell 77:799–800, 1994). However, we are currently investigating other acetylases that may be regulated by ProTα and other EBNAs. It is possible that ProTα and EBNA3C also compete with pCAF for p300 association as a critical step in regulation of histone acetylation mediated by EBV essential viral molecules.
The molecular mechanism by which ProTα and EBNA3C modulate HAT activity in EBV-transformed B cells is not fully understood. However, the fact that E1A as well as the transcription factor Twist inhibits HAT activities of p300 and pCAF suggests similarities in their mechanisms of inhibition (10, 28). ProTα and EBNA3C may bind similarly to the CH3 domain of p300, regulating the activity of the HAT domain. Alternatively, they may also directly interact with the HAT domain, based on the conformation of p300 in EBV-infected cells. If these activities are cell cycle regulated then we can envision a scenario in which p300 conformation is temporally regulated, thus providing access to factors like ProTα and viral proteins available to interact and regulate its HAT activity.
The identification of ProTα is a result of studies investigating the role of EBNA3C in EBV-mediated B-cell growth transformation. The biochemical and genetic experiments in the context of EBV-transformed cells establish a connection between an essential EBNA and regulation of histone acetylation. As shown in this study, EBNA3C is shown to be a modulator of this activity in EBV-infected cells. Moreover, previous work indicates that EBNA3C modulates the transactivation activity of another essential EBV latent antigen, EBNA2, through its interaction with the cellular repressor RBP-Jκ (53, 66). The EBNA3C molecule is highly hydrophilic, containing two acidic domains, a proline-rich domain and a glutamine-rich region at the carboxy terminus potentially involved in transcription activation, a leucine zipper that may be involved in homo- or heterodimerization with other similar proteins, and nuclear localization signals responsible for nuclear translocation of EBNA3C (41, 69). EBNA3C is essential for EBV transformation of B lymphocytes; interacts with the transcription repressor RBP-Jκ, which is ubiquitously expressed; and is potentially involved in the NOTCH1 signaling pathway as a downstream target for transcription regulation by intracellular activated NOTCH1 (6, 8, 64). The interaction with RBP-Jκ results in disruption of its binding to the cognate RBP-Jκ sequence. RBP-Jκ binds to a corepressor complex containing the histone deacetylase, and on activation of NOTCH1 signaling the intracellular form of NOTCH1 is cleaved and then translocates to the nucleus, where it interacts with RBP-Jκ (4, 5). This interaction results in the displacement of the deacetylase-corepressor complex, which leads to activation as the promoters become derepressed (38). Since both EBNA1 and EBNA2 are known viral transactivators (20, 42, 45, 82, 83, 91) we suggest that EBNAs (e.g., EBNA1 and/or EBNA2) may act as functional homologs of intracellular activated NOTCH1, potentially displacing corepressors and deacetylases as they recruit the coactivator p300 in cooperation with ProTα, resulting in transcription activation (38). This transcriptional activation may be a consequence of acetylation of the histones by the activation complex and acetylases that may include other EBNA proteins, ProTα, and p300. This increased acetylation does not go unchecked but is stringently regulated by the essential EBV modulator EBNA3C competing with p300 for binding to ProTα, resulting in down-modulation of the acetylation activity.
This line of investigation reveals a novel function of the essential EBV latent nuclear antigen EBNA3C, i.e., associating with ProTα and the coactivator-acetylase p300 to modulate histone acetylation in EBV-infected cells. The mechanism of this modulation is not clearly understood, but preliminary data suggest that EBNA3C disrupts the interaction of ProTα with p300. ProTα may be an important molecule in recruiting p300 and possibly other acetylases to the nucleosomes in proliferating cells through associations with histones (12, 23, 39). It is known that ProTα is highly upregulated in proliferating lymphocytes and may have a major role in increasing acetylation of core histones through recruitment of acetylases (68).
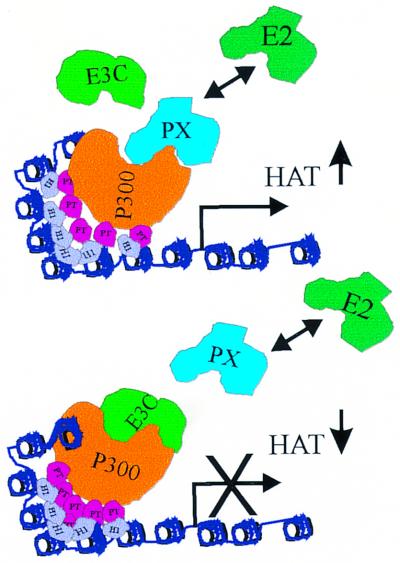
We therefore propose a model whereby EBV infection of primary human B lymphocytes results in induction of viral and cellular gene expression through the cooperation of a number of factors, including ProTα, histone H1, and p300 as well as other viral (e.g., EBNA2) and/or cellular transactivators (denoted as PX). This destabilizes the corepressor complex, resulting in increased histone acetylation and transcription (38). The major EBV latent promoter Cp is then activated and EBNA3C is expressed (2, 41). EBNA3C, once expressed negatively, regulates major EBV latent promoters as well as other cellular promoters by modulating the transactivation activity of the activator complexes through displacement and sequestration of these megacoactivator complexes. The corepressor complex that includes EBNA3C, possibly in cooperation with other corepressors, can then be positioned to modulate HAT activity (Fig. 12).
FIG. 12.
Schematic diagram illustrating a possible model for EBNA3C association with ProTα and p300. ProTα may recruit the acetylase p300 and other viral or cellular transactivators denoted as protein X (PX). It is possible that corepressor complexes are also displaced by ProTα as the coactivator-acetylase p300 is recruited to promoters (38). This leads to acetylation of histone and upregulation of transcription. The expression of EBNA3C then displaces the megacoactivator complex, which may include p300, ProTα, and other coactivators like PX, leading to down-modulation of the histone acetylation activity. The net result of this activity leads to modulation of transcriptional activity.
The identification of p300 and ProTα as cellular targets associated with one of the essential EBNAs involved in mediating B-cell growth transformation places this viral antigen, EBNA3C, in the context of chromatin regulation and suggests a mechanism through which it influences B-cell proliferation. This is a novel report of an EBV antigen that specifically modulates the acetylation activity of cellular proteins involved in histone acetylation. Other viral proteins have been shown to cooperate with p300 in regulating transcriptional activities. These include the human immunodeficiency virus accessory protein, Vpr, involved in regulation of NF-κB and the basal transcription machinery resulting in human immunodeficiency virus gene expression (18); the oncogenic E1A adenoviral protein; and the simian virus 40 large T antigen (17, 60, 72, 88). We are currently investigating the domains of EBNA3C as well as p300 that interact with ProTα in an effort to determine the mechanism whereby EBV regulates histone acetylation and thereby mediates B-lymphocyte proliferation.
ACKNOWLEDGMENTS
We are grateful to S. Elledge for the GAL4 cDNA activation library and the reagents necessary for use in the yeast two-hybrid assay. We thank Elliott Kieff for the EBNA3C reagents and Andrew Cooper, Kenneth Izumi, Jeffrey Lin, and George Mosialos for helpful advice. We also thank Gary Nabel for critical comments during the preparation of the manuscript and the p300 reagents. Jennifer Callahan provided technical assistance on this project. Fernando Dominguez provided the ProTα construct and polyclonal antibody, for which we are grateful. We also extend special thanks to Vojo Deretic and Eric Fearon for use of their fluorescence microscope and digital imaging camera system.
This work was supported by grant AHA9650467N and by a grant from the National Cancer Institute (CA072150-01) (to E.S.R.). E.S.R. is a Scholar of the Leukemia and Lymphoma Society of America. M.A.C. is supported by funds from the Medical Scientist Training Program of the National Institute of General Medical Sciences (NIH5 T32 GM07863) and is a fellow of the Lady Tata Memorial Trust.
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. . (Erratum, 185:946.) [DOI] [PubMed] [Google Scholar]
- 3.Ambinder R F. Human lymphotropic viruses associated with lymphoid malignancy: Epstein-Barr and HTLV-1. Hematol Oncol Clin N Am. 1990;4:821–833. [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 5.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 6.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 7.Bain M, Watson R J, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila suppressor of hairless and of its human homolog, KBF2/RBP-J kappa, by direct protein-protein interaction with Drosophila hairless. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Jullien C, Perez-Estevez A, Covelo G, Freire M. Prothymosin alpha binds histones in vitro and shows activity in nucleosome assembly assay. Biochim Biophys Acta. 1996;1296:219–227. doi: 10.1016/0167-4838(96)00072-6. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez F, Magdalena C, Cancio E, Roson E, Paredes J, Loidi L, Zalvide J, Fraga M, Forteza J, Regueiro B J, et al. Tissue concentrations of prothymosin alpha: a novel proliferation index of primary breast cancer. Eur J Cancer. 1993;6:893–897. doi: 10.1016/s0959-8049(05)80433-2. [DOI] [PubMed] [Google Scholar]
- 14.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 15.Eilers M, Schirm S, Bishop J M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eschenfeldt W H, Berger S L. The human prothymosin alpha gene is polymorphic and induced upon growth stimulation: evidence using a cloned cDNA. Proc Natl Acad Sci USA. 1986;83:9403–9407. doi: 10.1073/pnas.83.24.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felzien L K, Farrell S, Betts J C, Mosavin R, Nabel G J. Specificity of cyclin E-Cdk2, TFIIB, and E1A interactions with a common domain of the p300 coactivator. Mol Cell Biol. 1999;19:4241–4246. doi: 10.1128/mcb.19.6.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frangou-Lazaridis M, Clinton M, Goodall G J, Horecker B L. Prothymosin alpha and parathymosin: amino acid sequences deduced from the cloned rat spleen cDNAs. Arch Biochem Biophys. 1988;263:305–310. doi: 10.1016/0003-9861(88)90640-6. [DOI] [PubMed] [Google Scholar]
- 20.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gast K, Damaschun H, Eckert K, Schulze-Forster K, Maurer H R, Muller-Frohne M, Zirwer D, Czarnecki J, Damaschun G. Prothymosin alpha: a biologically active protein with random coil conformation. Biochemistry. 1995;34:13211–13218. doi: 10.1021/bi00040a037. [DOI] [PubMed] [Google Scholar]
- 22.Giles R H, Peters D J M, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Marquez J, Rodriguez P. Prothymosin alpha is a chromatin-remodelling protein in mammalian cells. Biochem J. 1998;333:1–3. doi: 10.1042/bj3330001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Marquez J, Segade F. Prothymosin alpha is a nuclear protein. FEBS Lett. 1988;226:217–219. doi: 10.1016/0014-5793(88)81425-x. [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Marquez J, Segade F, Dosil M, Pichel J G, Bustelo X R, Freire M. The expression of prothymosin alpha gene in T lymphocytes and leukemic lymphoid cells is tied to lymphocyte proliferation. J Biol Chem. 1989;264:8451–8454. [PubMed] [Google Scholar]
- 26.Goodall G J, Dominguez F, Horecker B L. Molecular cloning of cDNA for human prothymosin alpha. Proc Natl Acad Sci USA. 1986;83:8926–8928. doi: 10.1073/pnas.83.23.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graziano V, Gerchman S E, Schneider D K, Ramakrishnan V. Histone H1 is located in the interior of the chromatin 30-nm filament. Nature. 1994;368:351–354. doi: 10.1038/368351a0. [DOI] [PubMed] [Google Scholar]
- 28.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 29.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 30.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 31.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartzog G A, Winston F. Nucleosomes and transcription: recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- 33.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 36.Johannsen E, Miller C L, Grossman S R, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 38.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karetsou Z, Sandaltzopoulos R, Frangou-Lazaridis M, Lai C Y, Tsolas O, Becker P B, Papamarcaki T. Prothymosin alpha modulates the interaction of histone H1 with chromatin. Nucleic Acids Res. 1998;26:3111–3118. doi: 10.1093/nar/26.13.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieff E. Epstein-Barr virus and its replication. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 42.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 44.Le Roux A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 45.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loidi L, Vidal A, Zalvide J B, Puente J L, Reyes F, Dominguez F. Development of ELISA to estimate thymosin alpha1, the N terminus of prothymosin alpha, in human tumors. Clin Chem. 1997;43:59–63. [PubMed] [Google Scholar]
- 47.Longnecker R. Biochemical and genetic studies of Epstein-Barr virus latent membrane protein 2. Leukemia. 1994;8(Suppl. 1):S46–S50. [PubMed] [Google Scholar]
- 48.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 49.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manrow R E, Berger S L. GAG triplets as splice acceptors of last resort. An unusual form of alternative splicing in prothymosin alpha pre-mRNA. J Mol Biol. 1993;234:281–288. doi: 10.1006/jmbi.1993.1583. [DOI] [PubMed] [Google Scholar]
- 51.Manrow R E, Sburlati A R, Hanover J A, Berger S L. Nuclear targeting of prothymosin alpha. J Biol Chem. 1991;266:3916–3924. [PubMed] [Google Scholar]
- 52.Marchini A, Longnecker R, Kieff E. Epstein-Barr virus (EBV)-negative B-lymphoma cell lines for clonal isolation and replication of EBV recombinants. J Virol. 1992;66:4972–4981. doi: 10.1128/jvi.66.8.4972-4981.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maunders M J, Petti L, Rowe M. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J Gen Virol. 1994;75:769–787. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 55.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 56.Mori M, Barnard G F, Staniunas R J, Jessup J M, Steele G D, Jr, Chen L B. Prothymosin-alpha mRNA expression correlates with that of c-myc in human colon cancer. Oncogene. 1993;8:2821–2826. [PubMed] [Google Scholar]
- 57.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 58.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 59.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 60.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 61.Raab-Traub N, Webster-Cyriaque J. Epstein-Barr virus infection and expression in oral lesions. Oral Dis. 1997;3(Suppl. 1):S164–S170. doi: 10.1111/j.1601-0825.1997.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 62.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rickinson, A., and E. K. 1996. Epstein-Barr virus, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 64.Robertson E S. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Rep. 1997;4:143–150. [Google Scholar]
- 65.Robertson E S, Kieff E. Genetic analysis of Epstein-Barr virus in B lymphocytes. Epstein-Barr Virus Rep. 1995;2:73–80. [Google Scholar]
- 66.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez P, Vinuela J E, Alvarez-Fernandez L, Buceta M, Vidal A, Dominguez F, Gomez-Marquez J. Overexpression of prothymosin alpha accelerates proliferation and retards differentiation in HL-60 cells. Biochem J. 1998;331:753–761. doi: 10.1042/bj3310753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sburlati A R, Manrow R E, Berger S L. Human prothymosin alpha: purification of a highly acidic nuclear protein by means of a phenol extraction. Protein Expr Purif. 1990;1:184–190. doi: 10.1016/1046-5928(90)90014-p. [DOI] [PubMed] [Google Scholar]
- 71.Sburlati A R, Manrow R E, Berger S L. Prothymosin alpha antisense oligomers inhibit myeloma cell division. Proc Natl Acad Sci USA. 1991;88:253–257. doi: 10.1073/pnas.88.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shikama N, Lyon J, LaThangue N. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–235. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 73.Sixbey J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 74.Sternglanz R. Histone acetylation: a gateway to transcriptional activation. Trends Biochem Sci. 1996;21:357–358. [PubMed] [Google Scholar]
- 75.Struhl K. Histone acetylation and transcription regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 76.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomkinson B, Kieff E. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsitsiloni O E, Stiakakis J, Koutselinis A, Gogas J, Markopoulos C, Yialouris P, Bekris S, Panoussopoulos D, Kiortsis V, Voelter W, et al. Expression of alpha-thymosins in human tissues in normal and abnormal growth. Proc Natl Acad Sci USA. 1993;90:9504–9507. doi: 10.1073/pnas.90.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vareli K, Frangou-Lazaridis M, Tsolas O. Prothymosin alpha mRNA levels vary with c-myc expression during tissue proliferation, viral infection and heat shock. FEBS Lett. 1995;371:337–340. doi: 10.1016/0014-5793(95)00938-6. [DOI] [PubMed] [Google Scholar]
- 81.Vareli K, Tsolas O, Frangou-Lazaridis M. Regulation of prothymosin alpha during the cell cycle. Eur J Biochem. 1996;238:799–806. doi: 10.1111/j.1432-1033.1996.0799w.x. [DOI] [PubMed] [Google Scholar]
- 82.Wang F, Kikutani H, Tsang S F, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984;38:17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- 86.Wu C L, Shiau A L, Lin C S. Prothymosin alpha promotes cell proliferation in NIH3T3 cells. Life Sci. 1997;61:2091–2101. doi: 10.1016/s0024-3205(97)00882-5. [DOI] [PubMed] [Google Scholar]
- 87.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]
- 88.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 89.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]