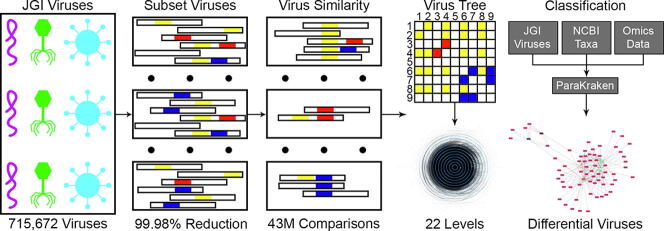
Graphical abstract
Keywords: Metagenomics, Viriome, Metatranscriptomics, Microbiome, Autism spectrum disorder, Populus
Abstract
Viruses are an underrepresented taxa in the study and identification of microbiome constituents; however, they play an essential role in health, microbiome regulation, and transfer of genetic material. Only a few thousand viruses have been isolated, sequenced, and assigned a taxonomy, which limits the ability to identify and quantify viruses in the microbiome. Additionally, the vast diversity of viruses represents a challenge for classification, not only in constructing a viral taxonomy, but also in identifying similarities between a virus’ genotype and its phenotype. However, the diversity of viral sequences can be leveraged to classify their sequences in metagenomic and metatranscriptomic samples, even if they do not have a taxonomy. To identify and quantify viruses in transcriptomic and genomic samples, we developed a dynamic programming algorithm for creating a classification tree out of 715,672 metagenome viruses. To create the classification tree, we clustered proportional similarity scores generated from the k-mer profiles of each of the metagenome viruses to create a database of metagenomic viruses. The resulting Kraken2 database of the metagenomic viruses can be found here: https://www.osti.gov/biblio/1615774 and is compatible with Kraken2. We then integrated the viral classification database with databases created with genomes from NCBI for use with ParaKraken (a parallelized version of Kraken provided in Supplemental Zip 1), a metagenomic/transcriptomic classifier. To illustrate the breadth of our utility for classifying metagenome viruses, we analyzed data from a plant metagenome study identifying genotypic and compartment specific differences between two Populus genotypes in three different compartments. We also identified a significant increase in abundance of eight viral sequences in post mortem brains in a human metatranscriptome study comparing Autism Spectrum Disorder patients and controls. We also show the potential accuracy for classifying viruses by utilizing both the JGI and NCBI viral databases to identify the uniqueness of viral sequences. Finally, we validate the accuracy of viral classification with NCBI databases containing viruses with taxonomy to identify pathogenic viruses in known COVID-19 and cassava brown streak virus infection samples. Our method represents the compulsory first step in better understanding the role of viruses in the microbiome by allowing for a more complete identification of sequences without taxonomy. Better classification of viruses will improve identifying associations between viruses and their hosts as well as viruses and other microbiome members. Despite the lack of taxonomy, this database of metagenomic viruses can be used with any tool that utilizes a taxonomy, such as Kraken, for accurate classification of viruses.
1. Introduction
The number of phages on Earth is estimated to be as high as 4.80 × 1031 [1], implying the total number of viruses that might exist is much greater. Viruses play an essential role in the regulation of the microbiome [2]. Additionally, viruses are also omnipresent within the human microbiome, even in the absence of disease [3], [4]. The roles that viruses play, independent of disease, are also not well understood, making identification and classification of viruses necessary for better understanding interactions between host, virus, and microbiome. Despite the large number of viruses and their importance in the microbiome, only a small proportion have been sequenced or characterized. Metagenomics and metatranscriptomics in general have led to the discovery of more viruses than isolation and sequencing has previously allowed; however, knowledge of their taxonomy is limited, making accurate identification in -omic samples challenging. While updates have been made to classify viruses by the International Committee on Taxonomy of Viruses (ICTV) [5], only 5560 viral species have been assigned a taxonomy as of 2019. In contrast, the Joint Genome Institute’s (JGI) Integrated Microbial Genomes & Microbiomes (IMG) [6] reports over 8000 viral isolates and IMG/VR [7] lists >715,000 metagenomic viruses, the majority of which are devoid of taxonomic classification because they are sampled from a mixture of organisms and have not been isolated. Metagenomic viruses often lack phenotypic characteristics as well as host information, which creates challenges for understanding their basic biology [8] and requires different methodologies for classification [9].
Additionally, methods that rely on sequence homology or NCBI/RefSeq databases for classification do not work where there is either no taxonomy or no homology to anything with a taxonomy, such as the majority of metagenomic viruses. For example, ViromeScan [10] relies on known taxonomy from NCBI to identify viruses from RefSeq in metagenomic samples. Tools such as vConTACT [11], Low et al [12], and Metavir [13] require homology to known viruses to assign taxonomy. As far as we are aware, there are no known methods or databases for classifying metagenomic viruses without taxonomy or homology to a virus with taxonomy in RNASeq or DNASeq samples. Despite taxonomic and biological hurdles, identifying viruses in meta-omic experiments allows for novel insights into host-virus interactions, in addition to other interactions throughout the microbiome and phytobiome.
JGI’s effort to assemble metagenomic viruses [14] has led to an unprecedented number of viral sequences that can be utilized to classify the microbial dark matter that can make up the bulk of metagenomic and metatranscriptomic samples. To the best of our knowledge, our k-mer based approach is the only extant method able to classify and quantify viruses at the scale provided by IMG-VR. One of the major challenges of classifying viruses, especially in the absence of phenotypic information, is their diversity [8] and the poor relationship between sequence and evolution [15]. While the inclusion of highly divergent sequences increases the difficulty of creating a detailed, fine-scale viral taxonomy, the presence of unique sequences can aid in the classification of viruses in -omic samples, i.e. we know they are likely different at the species level or greater. Methods such as natural vector representation [16], [17], pairwise sequence comparisons [18], and pairwise evolutionary distances [19] have been developed to better identify phylogenetic similarity among viruses, but k-mer-based methods can provide the speed and scale that is necessary for highly efficient and accurate classification of millions to billions of sequencing reads against databases of taxonomic sequences [20]. Additionally, these k-mer based methods can be extended to sequences without taxonomy, allowing for both a greater fidelity in classifying reads and a greater understanding of the types of organisms that may be present in the microbiome.
In this paper, we create a methodology for generating a classification tree of 715,672 viruses from IMG to VR [7] for use in identifying viral sequences in metagenomic and metatranscriptomic studies (Fig. 1). Given the infeasibility of comparing all viruses to each other, we first subset the viruses to identify which pairs have k-mer overlaps for calculating similarity scores. Subsetting resulted in the reduction of the comparison space by 99.98%, allowing for quantitative proportional similarity coefficients [21] to be calculated for each virus pair with a non-zero similarity. The algorithm Hip-MCL [22], was then used to cluster similar viruses for use in generating a hierarchical tree based on multiple inflation values (Supplemental File 1). The classification tree was integrated with NCBI’s taxonomy, allowing for taxonomic classification of reads from metagenomic and metatranscriptomic samples. Finally, the pseudo-taxonomy was used to create a Kraken [20] database of all the metagenomic viruses (available here: https://www.osti.gov/biblio/1615774) for use in Kraken2. We utilized ParaKraken [23] to illustrate the broad use of our viral classification method to expand more traditional metagenomic analysis to include a greater diversity of viruses than can be provided by NCBI alone.
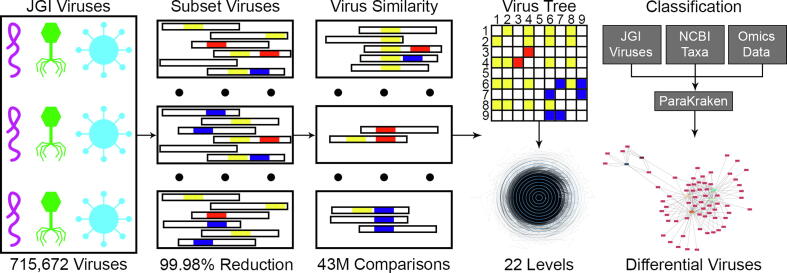
Fig. 1.
Creation and use of a viral classification tree. To first identify which of the 715,672 metagenome viruses have non-zero similarity scores with other viruses, we subset the viruses to identify k-mer overlaps. We identified ∼43 million pairs with nonzero similarity scores, a reduction in the number of calculations by 99.98%. Clusters of viruses were then created by running HipMCL on quantitative proportional similarity coefficients with the following inflation values: 1.4, 2.0, 3.0, 4.0, and 6.0. MCLCM was used to analyze the inflation clusters to generate a hierarchy, and then neighbor joining was performed on clusters with >3 members. The metagenome virus tree was then integrated with NCBI’s taxonomy for use in classifying metatranscriptomic and metagenomic samples. We analyzed Populus and ASD data sets with ParaKraken using the NCBI whole genomes and JGI metagenome viruses databases, allowing us to classify taxa and identify differential abundance across conditions. The network shows differential abundance of viruses (violet-red) within the Populus genotypes (green – hybrid, orange – P. deltoides). Virus abundances across genotypes were similar in the soil (lightest) and the rhizosphere (middle) compartments, but the endosphere (darkest) was dissimilar to the other compartments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To do this, we applied the same metagenomic classification methodology, including our Kraken databases, to a plant dataset containing rhizosphere, endosphere, and soil samples of two different Populus genotypes and a dataset of post mortem brain samples from individuals with Autism Spectrum Disorder (ASD) and controls [24]. Both plant and human samples were chosen to show that the methodology presented here is agnostic to the host and microbiome, with higher fidelity in the viriome. Additionally, both of these hosts have well studied microbiomes in relation to bacteria and are often lacking for organisms such as viruses. Furthermore, viruses both influence and are influenced by the health of these host organisms, making both ideal for exploration of viruses within the microbiome. For example, we specifically chose ASD brain tissue samples because prior literature has shown an association between ASD and viruses in brain tissues. A study found an increased number of polyomaviruses in brain tissue of individuals with ASD [35], and the number of viruses within an individual has been shown to be correlated with decreased neuropsychological development [36], supporting the idea that there is an association between ASD and the viruses present in brain tissue. We also utilize NCBI’s viral sequences as a positive control to estimate the uniqueness of viral sequences in individual isolates and to identify known viruses under infection conditions, such as COVID-19 infection in a bronchoalveolar lavage fluid (BALF) sample [25] and cassava brown streak virus in a cassava sample [26].
2. Materials and methods
2.1. Virus and taxonomy downloads
Metagenomic viruses were downloaded from JGI-IMG/VR [7] (N = 715,672 in January 2018). NCBI [27] whole genome sequences containing 116 k (∼67 k unique species/strains) eukaryotes, prokaryotes, archaea, and viruses were downloaded in November 2017. The resulting classification tree from the metagenome viruses was then integrated with NCBI’s taxonomy, which can be used with Kraken [20], ParaKraken [23] (Supplemental Zip 1), or any other classification method that utilizes NCBI’s taxonomic tree.
2.2. Calculating similarity
K-mers of size 31, using a sliding window of size one, were utilized to generate the classification tree as it allows for direct incorporation with our NCBI databases in Kraken or ParaKraken; however, the methodology presented here is agnostic to the k-mer size. Due to the inability to both store and quickly access all unique k-mers for all viruses (the number of unique k-mers was greater than the number of keys available in a hashtable), the viruses were first broken into 20 subsets. Each subset was compared to all other subsets, resulting in 71,566 viruses in each comparison set. First, all k-mers were generated for all viruses in each subset pair, with only the unique k-mers stored in a hashmap, along with any/all associated viruses containing that k-mer. Two k-mers were treated as the same if there was an exact match for the 31-mer, utilizing both the forward and reverse complement for the determination of an exact match.
The forward and the reverse complement of each sequence was used because viral genomes can be a mixture of DNA/RNA and single/double-stranded viruses, the strandedness of some viruses are different at different life cycle stages, and some viruses have a genome that is partially single and partially double stranded. While the metagenomic viruses are composed of DNA viruses, the methodology was developed to be agnostic to the type of virus. K-mers with ambiguous bases were not stored. K-mers with more than one virus associated with the sequence are indicative of overlap between two or more viruses. Viruses with at least one overlapping k-mer with another virus were stored in pairs to calculate similarity scores. Any virus pair without any k-mer overlaps were assigned a similarity score of zero. The initial subset decreased the number of similarity scores to be calculated from ∼256 billion to only ∼43 million (a decrease of over 99.98%), making the computation more feasible. The decrease was achieved due to the sparsity of virus pairs that had any overlapping k-mers, resulting in the majority of virus pairs having a similarity score of zero.
Quantitative proportional similarity coefficients were calculated between the sets of k-mers for each virus pair with overlapping k-mers. Due to memory limitations of 500G per node and speed limitations of calculating all similarities in a linear fashion, the ∼43 million overlapping virus pairs were broken into 400 subsets. K-mer profiles from each virus in each subset were generated beforehand to eliminate the need for regenerating profiles with every comparison (as a given virus can appear in multiple subsets). However, to decrease the memory overhead (at the cost of a longer run), the k-mer profiles can be generated for each virus and for each comparison. Each of the subsets was then run on the Summit supercomputer in parallel. K-mer overlap was calculated as before with a k-mer and its reverse complement being treated as the same k-mer. The result is a matrix of quantitative proportional similarity coefficients for all virus pairs.
2.3. Virus classification tree
Groups of similar viruses were identified by running HipMCL [22], a parallel Markov clustering algorithm, on the triples of virus k-mer similarity scores. Inflation values of 1.4, 2.0, 3.0, 4.0, and 6.0 were utilized to identify a range of cluster sizes. The resulting clusters were integrated into a hierarchy by MCLCM [28]. Clusters of more than three viruses (excluding the 32,084 viruses that had no similarity to any other virus) were reclustered using neighbor joining to increase granularity, resulting in a hierarchy of viruses consisting of 22 levels (one of the cluster trees shown in Supplemental Fig. 1). Pseudo-taxonomic IDs were then assigned to each level and virus in order to integrate with NCBI’s taxonomy in order to create Kraken databases from the metagenomic viruses. The Kraken2 database of the JGI metagenome viruses can be found here: https://www.osti.gov/biblio/1615774 [29].
2.4. Autism and Populus datasets
Bulk RNA-Seq from 22 ASD and 19 control post mortem brain samples were obtained from Velmeshev et al [24]. FASTQ files were trimmed using Atropos [30] and mapped to the GRCh38 human reference genome using STAR [31]. P. deltoides and P. deltoides × P. trichocarpra endosphere, rhizosphere, and soil samples (five samples of each compartment and genotype combination; one P. deltoides endosphere was removed for quality reasons) were obtained from JGI (https://gold.jgi.doe.gov/biosamples?Study.GOLD+Study+ID=Gs0103573). Endosphere samples underwent differential centrifugation to decrease the concentration of plant host material [32] followed by paired-end sequencing. Reads were trimmed using skewer [33] and filtered against P. deltoides and P. trichocarpa reference genomes (∼0.2% of reads aligned to the host). Unmapped reads from both the Populus and ASD datasets were run through ParaKraken [23]. A median normalization was applied, taxa with <75% coverage across samples were removed, taxa making up <0.01% of the reads at a species level in the ASD data and 0.001% of the reads at a species level in the Populus data were also removed. Differential abundance was then assessed using fcros [34] with p-value <0.05 and f-value >0.9, and networks were visualized using Cytoscape [35].
2.5. Cassava, coronavirus and updated viral genomes datasets
A cassava root RNA-Seq sample with a confirmed brown streak virus was obtained from Amisse et al. [26] and analyzed using ParaKraken [23] to classify taxa. A BALF sample with a confirmed COVID-19 infection was obtained from Wu et al. [25]. Reads were trimmed using Atropos [30] and then run through ParaKraken for taxa classification. We then downloaded the latest RefSeq viral genomes from NCBI in Feb 2020 [27], as the updated version contained the virus responsible for COVID-19. We then reran ParaKraken on the BALF sample to compare the read counts for coronavirus pre- and post-COVID-19 genome inclusion. To assess the uniqueness of viruses in the Feb 2020 version of RefSeq, we created reads from all 67,519 RefSeq viruses using a read length of 200 and a sliding window of one nucleotide. The resulting 1.06 billion reads were then analyzed with ParaKraken. The taxonomic assignment was then compared to the taxonomy of the viruses in which the reads originated.
2.6. JGI virus accuracy assessment
Reads from the JGI viruses included in the database were simulated by selecting 100 reads of 200 nucleotides in length at random for each viral sequence. Any read that started in the same position as another read in the same virus was excluded and another random read was selected. Any reads with ambiguous base pairs were excluded. Viruses in which 100 reads of 200 bp in length could not be created were also excluded (resulting in a removal of two viruses due to high numbers of ambiguous reads). Reads were then mutated at 5, 10, and 25 randomly selected nucleotides. Mutations were conserved for a given read such that reads with 25 mutations included all of the mutations generated in the 10 mutation read, and the 10 mutation read contained all of the mutations generated in the 5 mutation read. Once all reads were generated, they were analyzed with ParaKraken using both the NCBI and JGI databases to assess accuracy. A read was determined to be accurately classified if it matched the exact virus of interest or a parent of that virus (equivalent to other methods that assess accuracy based upon NCBI taxonomy). A read was determined to be inaccurately classified in JGI if it matched another JGI virus or a node that was not a parent to the virus used to generate the read. Any read that was unclassified, classified as an NCBI taxon, or classified as ambiguous (represented by the NCBI root) was classified as “other”.
3. Results
The virus classification tree can be integrated into the NCBI taxonomy for classifying viral sequences from metagenomic and metatranscriptomic samples. To illustrate the power of having a classification tree of 715,652 viruses, we applied ParaKraken [23] to data produced from two distinct studies. The first study is a Populus metagenome sampled from three different compartments (endosphere, rhizosphere, and soil) in two different genotypes (P. deltoides and P. deltoides × P. trichocarpa). Each genotype-compartment pair had five replicates (with one P. deltoides endosphere sample removed for quality control reasons). Without the viral databases, the resulting metagenomic classification was unable to identify taxa for 50% of the reads. The high amount of unknown microbial dark matter allows for a low-end estimation to be made of how many viral sequences may exist in microbiome samples. The second study is an ASD study comparing post mortem brain biopsies between ASD patients and controls. While the vast majority of reads in the ASD study are human, we captured a portion of the brain microbiome, including the viriome.
3.1. Populus metagenome
To identify differences in the phytobiome between compartments and genotypes, we applied ParaKraken to reads produced from endosphere, rhizosphere, and soil samples from P. deltoides and P. deltoides × P. trichocarpa (a hybrid). To mirror commonly used mapping methods in recent publications aimed at characterizing plant viriomes [36], [37], the samples were initially analyzed using only the NCBI databases of all publicly available genomes from prokaryotes, archaeas, eukaryotes and 8000 viral taxa. The NCBI databases resulted in 50% of the 4.8 billion reads being assigned to taxa, with the rest representing unknown microbial dark matter (Fig. 2A). To illustrate the increased mapping ability of the k-mer based viral databases, we then analyzed the samples using our k-mer based viral databases in combination with the NCBI databases. The inclusion of the 715,672 JGI metagenome viruses resulted in an increase in mapping by 347 million reads. Metagenome viruses make up between 6% and 20% (mean 15%) of the total mapped reads for a given sample, greatly increasing the coverage of the viriome (Supplemental Tables 1 and 2). In addition to the increased read coverage, viral sequences also differ between compartments and genotypes leading to potential associations between viruses, the host, and other community members.
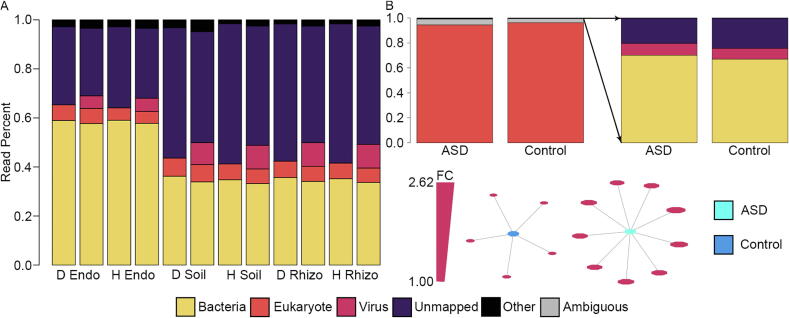
Fig. 2.
Classification of viruses in metagenomic and metatranscriptomic samples. A) Effect of the virus databases on the number of reads mapped in the Populus deltoides (D) and hybrid (H) data. The first bar in each group represents ParaKraken results before the viral databases were included and the second bar represents classification after the inclusion of the viral databases. Metagenome viruses averaged 15% of the mapped reads with a higher percent mapping in the rhizosphere (Rhizo) and soil (Soil) relative to the endosphere (Endo), suggesting that viruses make up a substantial portion of the microbiome. B) Differences in viruses between ASD brains and control brains. Unsurprisingly, eukaryotes make up the vast majority of reads in human samples. However, we were able to identify sequences associated with viruses, eight of which were significantly higher in ASD versus controls (p-value <0.05 and f-value >0.9, >2 fold change). The graph shows all significant differential abundance viruses (irrespective of fold change), with size of the viruses representing fold change (smallest – 1.00 FC, largest 2.62 FC). While there were five viruses with p-value <0.05 and f-value >0.9 in controls, average fold change was 1.09 compared to 2.23 for the nine viruses higher in ASD, suggesting ASD brains may have higher viral counts. Other – NCBI viruses, viroids, ambiguous sequences. FC – fold change.
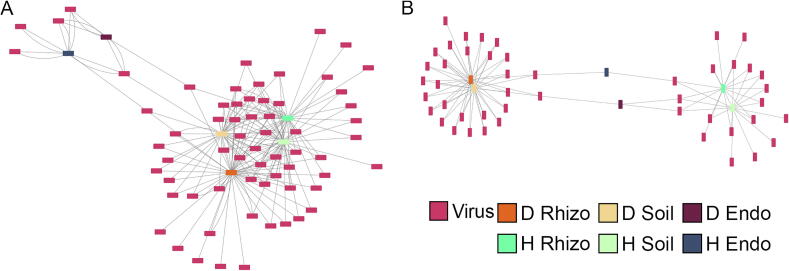
To better explore the differences between compartments and genotypes, we ran differential abundance analyses comparing both within and between genotypes, focusing on viruses. For the within-genotype comparison, we compared endosphere vs. rhizosphere, rhizosphere vs. soil, and soil vs. endosphere for each genotype (Fig. 3A). There were 65 viral sequences that were significantly differentially abundant across the comparisons (p-value <0.05 and f-value >0.9). Rhizosphere (48 and 37 sequences) and soil (36 and 39 sequences) had much higher amounts of differentially abundant viral sequences compared to the endosphere (four and six sequences), likely due to the lower amounts of detected viral reads in the endosphere. Rhizosphere and soil profiles are similar to each other, with only 10 significant sequences unique to one compartment and genotype. Additionally, rhizosphere and soil samples within a genotype are more similar than rhizosphere and soil across genotypes. The two endosphere samples shared three out of the six unique significant endosphere viral sequences.
Fig. 3.
Differential abundance of viral sequences in Populus genotypes and compartments. In the A) within genotype comparison, the rhizosphere and soil samples had similar significant (p-value <0.05 and f-value >0.9) viral sequences across genotypes; however, the significant soil and rhizosphere viral sequences are more similar within genotypes than across genotypes. The two endosphere samples have few significant viral sequences, and they have more in common with each other than other compartments. In the B) between genotype comparison, the soil and rhizosphere samples for a given genotype have similar significant differentially abundant viruses. Additionally, the endosphere samples have many fewer significant differential sequences compared to the rhizosphere and soil, likely due to the overall lower abundance of viral sequences. Both graphs suggest there is a host or microbiome mediated selection of viral sequences that has some genotype and compartment specificity. D – P. deltoides, H – hybrid, Endo – endosphere, Rhizo – rhizosphere.
In addition to the within genotype comparisons, a within compartment comparison was performed as well comparing: P. deltoides soil to hybrid soil, P. deltoides rhizosphere to hybrid rhizosphere, and P. deltoides endosphere to hybrid endosphere (Fig. 3B). There were 48 significantly differential viral sequences across the comparisons. Similar to the within genotype comparison, the rhizosphere (28 and 15 sequences) and soil (27 and 16 sequences) had more significant viruses compared to the endosphere (three and two sequences). Additionally, the soil and rhizosphere samples for a given genotype have nearly identical significant viral sequences with one virus unique to the P. deltoides rhizosphere, three unique to the hybrid rhizosphere, and four unique to the hybrid soil. There were no significant viruses shared across genotypes for the rhizosphere and soil. There were no unique viruses associated only with the endosphere, and each endosphere sample shared at least one significant virus with the hybrid rhizosphere/soil and P. deltoides rhizosphere/soil.
3.2. Autism spectrum disorder metatranscriptome
To better understand the association between viral sequences and human health, we analyzed post mortem brain tissue metatranscriptomic samples [24] from ASD individuals and controls (Fig. 2B). We first aligned reads to the GRCh38 human reference genome, resulting in 67.5% of reads (2.99 of the total 4.43 billion reads) mapping to the reference. The unmapped reads were then processed with our ParaKraken pipeline. Unsurprisingly, 95% of the unmapped reads were assigned to eukaryotes, most likely due to ambiguous mappings and differences between the patient and the human reference genome (Supplemental Table 3). Despite the high percentage of human sequences in the unmapped reads, metagenomic viral reads were readily identified in all samples, ranging from 5 k to 125 k reads (avg 0.06% of reads; whereas, bacteria make up 0.57% of reads). To assess the uniqueness of the JGI viruses, we quantified the percent of reads that mapped at the metagenome virus level. Only 8.9% of reads mapped at a higher taxonomic level than the individual virus, suggesting that the JGI viruses are highly unique and unambiguous in the sample set.
To further understand if there is an association between ASD and viral sequences in the brain biopsies, differential abundance was performed comparing ASD to controls. Eight metagenome viral sequences were significantly more abundant in ASD cases relative to controls at a >2× fold change, compared to zero significantly more abundant in the controls at that fold change. When comparing all of the significant viruses (p-value <0.05 and f-value >0.9; irrespective of fold change), the average fold change of the nine viruses that were significantly higher in ASD was 2.23 (1.99–2.62) compared to 1.09 (1.00–1.16) for the five viruses significantly higher in controls (Fig. 2B), suggesting that brain tissue from those with ASD may have relatively higher abundances of viral sequences (at least for the viruses contained in our databases). The JGI IDs for the nine viruses higher in ASD are: Ga0114997_10000721, Ga0099847_1000845, Ga0180434_10000080, Ga0181583_10003850, C687J26657_10000305, Ga0080013_1000178, Ga0114957_1000600, Ga0007809_10000384, Ga0181563_10001177, and the five higher in controls: Ga0075122_10000531, DelMOWin2010_c10003738, Ga0075125_10000004, Ga0121002_100132, Ga0160422_10001594. These metagenomic viruses represent the closest known organisms by sequence homology for reads within the brain tissue samples.
3.3. Assessment of virus uniqueness and abundance
To assess the uniqueness of viral sequences, we downloaded an updated version of NCBI’s viral taxonomy in Feb. 2020 [27]. We first computationally generated reads from all of NCBI’s viral sequences with a length of 200 bp and a sliding window of one, resulting in 1.06 billion reads. We then analyzed these reads with ParaKraken using both the NCBI and JGI databases. A slight majority of the reads (53.4%) mapped only to the individual viral isolate in which the read was generated. A small percentage of the reads (8.3%) mapped to the NCBI taxonomy root, which means that the viral reads either had homology to another superkingdom or to the metagenomic viruses. The uniqueness of the reads within the NCBI’s database suggests that much of the viral diversity is undiscovered, there are few viruses in which there are multiple similar isolates sequenced, and there are few overlaps between NCBI’s viruses and the viruses from JGI. The JGI viruses in the Autism metagenome had much lower than expected ambiguity based on NCBI’s results. The ratio of reads that mapped to the individual isolate compared to non-NCBI root viral reads was 1.4 (53.4/38.3) for the viruses in NCBI compared to the 10.2 (91.1/8.9) for the JGI metagenome virus in the Autism dataset.
To further understand the uniqueness of NCBI’s viruses, we used two different databases to classify viruses in a COVID-19 BALF sample [25] (Supplemental Table 4). The first database consists of the original NCBI and JGI parakraken databases, while the second database includes the Feb. 2020 version of NCBI’s viruses (which includes the SARS-CoV-2 genome that causes COVID-19). Without the isolate of interest, 26,878 reads (0.2% of the total reads) mapped to different Coronavirinae, with the top hit of 22,238 being SARS coronavirus (the previous coronavirus taxonomy). With the addition of the SARS-CoV-2 isolate, 62,480 reads (0.5% of total reads) mapped to Coronavirinae with the majority 62,461 mapping to the specific virus taxa of interest: severe acute respiratory syndrome coronavirus 2. SARS-CoV-2 has an 89.1% sequence homology with another SARS-like coronavirus [25], which is partly why we were able to identify coronavirus reads without the exact isolate of interest. However, despite having highly closely related viruses already sequenced, the inclusion of the exact isolate of interest increased the mapping by 2.3× (reaffirming the prior results that the majority of viral sequences are unique given the sparsity of the number of viruses that have been sequenced).
In addition to the COVID-19 sample, we also analyzed a cassava sample from Mozambique that was confirmed to be infected with a cassava brown streak virus [26] (Supplemental Table 5). We applied ParaKraken to the original NCBI database and the JGI databases on the sample and confirmed the presence of the virus of interest. ParaKraken identified 1942 reads (0.1% of total reads) associated with the cassava brown streak virus. As expected, neither the coronavirus nor the brown streak virus was identified by ParaKraken in the Autism brain samples or the plant metagenome samples. Both the SARS-CoV-2 and cassava brown streak viruses demonstrate that ParaKraken can identify viruses of interest during an active infection, and that viruses contain highly unique sequences, partially due to the lack of viruses sequenced and viruses with taxonomy.
3.4. Accuracy of the JGI viral databases
To assess the accuracy of the JGI viral genomes we randomly simulated 100 reads from each genome, resulting in 71.6 million reads. We also simulated mutations at a rate of 2.5%, 5%, and 12.5% to determine how minor changes in the nucleic acid sequence influence accuracy (Table 1). ParaKraken with the Kraken2 JGI metagenomic and NCBI databases were able to identify the correct virus (including its parent in the classification tree) 82% of the time. Additionally, 9.5% of reads mapped to other JGI viruses, with 8.5% of reads mapping to simply “other” (other includes NCBI taxa, unmapped reads, and ambiguous reads). Mutations of 2.5% and 5% had minimal influence on accuracy, but when we introduced a mutation rate of 12.5%, there was a decrease in accuracy of 56%. Although the accuracy is 10–15% lower than that produced by Kraken2 (the underlying classifier for ParaKraken) using NCBI databases with prokaryote genera and viruses with a genus classification [40], much of the decrease in accuracy is likely due to the larger numbers of genomes used in our assessment and the much higher diversity of both the viruses and viral sequences in the JGI databases compared to the subset of data that Kraken2 used for their accuracy assessment. As such, any reduction in the kmer/lmer space and subsampling used in creating a more compact database from Kraken2 will have a greater impact on organisms with high sequence diversity, such as the JGI viruses used in this study. However, despite the slight decrease in accuracy, we are still able to identify metagenomic viruses with high accuracy, which leads to improved identification of associations between viruses and the host and viruses and other members of the microbiome.
Table 1.
Accuracy of the JGI Databases. To determine the accuracy of the JGI databases, 100 reads of 200 nucleotides in length were randomly selected from each virus (generating 71.6 million reads). Reads were then mutated to include 5, 10, and 25 mutations. Reads were then analyzed using ParaKraken on the Kraken2 JGI and NCBI Databases. A read was determined to be correctly classified if the read matched the exact virus or a parent of that virus. A read was determined to be incorrectly classified in JGI if it mapped to another JGI virus or a node that was not a parent of the virus in which the read was generated. If a read was unmapped, ambiguous, or mapped to an NCBI read it was classified as “other”. Mutations of 5–10 per read had minimal impact on accuracy.
| Sample Type | Correct JGI Hit | Incorrect JGI Hit | Other |
|---|---|---|---|
| JGI Virus Reads | 82.05% | 9.45% | 8.50% |
| 1.25% Mutation Rate | 81.65% | 9.26% | 9.09% |
| 2.5% Mutation Rate | 80.96% | 8.60% | 10.44% |
| 12.5% Mutation Rate | 26.1% | 3.12% | 70.78% |
4. Discussion
Identifying constituents that comprise the microbiome and their relationship to the host is crucial to understanding human health, plant health, and how microorganisms impact phenotypes of other organisms in general. Current methodologies for microbiome analyses are largely focused on bacteria using 16S rRNA sequencing, fungi with ITS sequencing, and other organisms using taxonomy assessment through metagenomic sequencing; however, very little work has been done to quantify and understand viral reads in metagenome and metatranscriptome samples outside of what can be achieved with the few viruses that have been isolated and assigned to a taxonomy. Additionally, viruses have high sequence diversity, with the vast majority of viruses having no close relative in NCBI as illustrated by the unique metagenomic viruses identified by IMG/VR.
Addressing current limitations in virus identification is critical because viruses play an important but understudied role in many biological systems. For example, bacteriophages can modulate the metabolome of gut microbiomes in mice, including influencing the bacteria that the phage does not directly impact [2]. Additionally, end-stage viral infection in chimpanzees can cause a destabilization of the bacteria in the gut microbiome, likely through alteration of the host immune system [41]. The microbiome can also play a protective role, helping to decrease the risk of viral infections [42]. Much of a virus’ role in microbiome samples is unknown due to the lack of methodology for their detection and the fact that the majority of information gleaned from microbiome samples is from bacteria.
Additionally, prior to this paper, there had been a lack of methodology to quantify viral taxa, study host-virus interactions, or interactions between viruses and other microbiome constituents at large scales (scales orders of magnitudes larger than can be achieved by viruses with taxonomy). Directly measuring known viruses, viral antigens, and viral antibodies in human samples has led to the identification of associations between polyomaviruses and ASD [38], other viruses with neurodevelopment in general [39], different herpes viruses with multiple sclerosis [43] and peripheral neuropathies [44], HIV and peripheral arterial disease [45], and hepatitis C and kidney disease [46], etc. Additionally, autoimmunity, both in the mother and in the child, has been shown to play an important role in both the development of autism and in neurodevelopment [47]. Furthermore, multiple viruses have been associated with congenital autism, such as: rubella [48], influenza [49], [50], cytomegalovirus [51], and polyomavirus [38]. It is highly likely that there are many human-related and microbiome-related viruses that have implications for human health that await discovery, either directly or by interacting with other taxa in the microbiome. In plants undergoing drought stress, pathogenic viruses have been shown to have an amplifying effect with drought, increasing both infection-related stress and drought-related stress [52]; however, different infectious viruses have been shown to decrease drought severity by reducing water loss, ultimately leading to improved tolerance of both infection and drought [52], [53]. Furthermore, the effect of combined viral stress and abiotic stressors (such as drought, heat, salinity, etc.) are affected by the overlap in epigenetic responses to individual stressors, which can produce positive or negative effects depending on plant-specific responses to each stress [54]. More thorough knowledge and improved methods for identifying viruses on large scales is needed to better understand the wide variety of direct and indirect effects on the host and microbiome caused by viruses.
IMG/VR [7], [42] offers a suitable starting point for methodology development and hypothesis generation in identifying unknown viruses and their associations by providing the most extensive known collection of metagenomic viruses. While their work identifies viral assemblies in a single sample, we have utilized their large collection of assemblies to quantify viral sequences in diverse metagenomic and metatranscriptomic samples. To classify viral sequences, we have developed a dynamic programming algorithm that allows one to create a classification tree from metagenomic viruses that can be integrated with NCBI’s taxonomy. While it is impractical to compare all viruses to each other, the method initially identifies which viruses have a non-zero similarity score, reducing the number of similarity calculations by 99.98%. The reduction makes the calculation of similarities and construction of the classification tree feasible, providing the first step in identifying viral sequences in metagenomic and metatranscriptomic samples. For samples that show relationships between a virus and a phenotype of interest, that virus can be isolated for better characterization.
The classification tree’s utility is demonstrated through identification of viral sequences in Populus genotypes and tissue compartments and in ASD and control brain biopsies. We found significant differences in viral sequences associated with the P. deltoides, the hybrid, and the different compartments suggesting host factors and differences in microbiome community composition may select for different viruses. However, whether the lower number of viruses associated with the endosphere samples relative to the rhizosphere/soil is due to differential centrifugation, some host-related factor, or some database bias is unknown. While the individual virus counts identified in the two datasets are orders of magnitude lower than cassava brown streak virus and COVID-19 infection samples, neither the Populus nor ASD samples presented with any known virus-induced pathophysiology. However, the Populus metagenome had a total viral load ∼15% of reads on average, which is higher than the 0.3% of reads seen in the cassava sample with active infection. The higher total viral counts in a non-infected sample could be explained by the fact soil contains almost no host, and the samples underwent differential centrifugation to limit host, allowing more of the microbiome to be represented by the sequencing, in addition to any potential database biases.
5. Conclusion
Viruses are a vastly understudied component of microbiomes. The method we present here for creating a classification tree from metagenomic viruses can be utilized with any taxonomy-based classification tool to better identify viruses and their impacts in the microbiome. Although the 715,672 metagenome viruses that JGI has identified potentially make up only a small fraction of viruses that exist, it is still much greater than the number of viruses with taxonomy. As such, methods that require a taxonomy or homology to viruses with taxonomy will not work until a method is devised to handle creating taxonomies for viruses at large scale. Until such a time, we show that it is possible to identify viral sequences in metagenomic Populus genotype and compartment samples and metatranscriptomic ASD samples without the need of a taxonomy. More specifically, we identified eight significant differential viral sequences that are significantly higher and with a FC > 2.0 in ASD patients than in controls. We also show that our method can accurately identify viruses by utilizing NCBI’s viral genomes to identify known viruses in COVID-19 and cassava brown streak virus infection samples. Through the use of NCBI’s viral databases we also show that viral sequences are highly specific to the individual viral isolate, and that the JGI metagenome viruses have a higher uniqueness than the NCBI viruses. While the uniqueness and diversity of the JGI viruses makes them more difficult to classify in samples with Kraken2, our method is still 82% accurate in identifying the correct JGI viral sequences and over 90% accurate in identifying the sequence as a JGI virus. In addition to classification and quantification, further downstream analyses on viral reads, such as assembly, homology, and functional annotation can be performed to predict features of the potential virus or viral sequence. Ultimately, a better understanding of the effects that viruses have on both the microbiome and the host will lead to a better comprehension of human health and plant biology.
CRediT authorship contribution statement
Benjamin J. Garcia: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Ramanuja Simha: Data curation, Formal analysis, Methodology. Michael Garvin: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Anna Furches: Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Piet Jones: Methodology, Software, Writing - original draft, Writing - review & editing. Joao G.F.M. Gazolla: Methodology, Software. P. Doug Hyatt: Methodology, Software. Christopher W. Schadt: Data curation, Funding acquisition. Dale Pelletier: Data curation, Funding acquisition, supervision. Daniel Jacobson: Conceptualization, Methodology, Funding acquisition, Project administration, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research used resources of the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725. The plant viriome research was supported by the Plant-Microbe Interfaces Scientific Focus Area in the Genomic Science Program, the Office of Biological and Environmental Research (BER) in the U.S. Department of Energy Office of Science, and by the Department of Energy, Laboratory Directed Research and Development (LDRD) funding (ProjectID 8321), at the Oak Ridge National Laboratory. The coronavirus work was supported by an ORNL LDRD (ProjectID 10074). The Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the US DOE under contract DE-AC05-00OR22725. This research used resources of the Compute and Data Environment for Science (CADES). Support for DOI 10.13139/OLCF/1615774 dataset is provided by the U.S. Department of Energy, project syb105 under Contract DE-AC05-00OR22725. Project syb105 used resources of the Oak Ridge Leadership Computing Facility at Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725. This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.029.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
References
- 1.Cobián Güemes A.G., Youle M., Cantú V.A., Felts B., Nulton J., Rohwer F. Viruses as winners in the game of life. Annu Rev Virol. 2016;3(1):197–214. doi: 10.1146/annurev-virology-100114-054952. [DOI] [PubMed] [Google Scholar]
- 2.Hsu B.B., Gibson T.E., Yeliseyev V., Liu Q., Lyon L., Bry L., et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe. 2019;25(6):803–814.e5. doi: 10.1016/j.chom.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghose C., Ly M., Schwanemann L.K., Shin J.H., Atab K., Barr J.J., et al. The virome of cerebrospinal fluid: viruses where we once thought there were none. Front Microbiol. 2019;10:2061. doi: 10.3389/fmicb.2019.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbas A.A., Taylor L.J., Dothard M.I., Leiby J.S., Fitzgerald A.S., Khatib L.A., et al. Redondoviridae, a family of small, circular DNA viruses of the human oro-respiratory tract associated with periodontitis and critical illness. Cell Host Microbe. 2019;26(2):297. doi: 10.1016/j.chom.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Dempsey D.M., Dutilh B.E., et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019) Arch Virol. 2019;164:2417–2429. doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]
- 6.Markowitz V.M., Chen I.-M.A., Palaniappan K., Chu K., Szeto E., Grechkin Y., et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paez-Espino D, Chen I-MA, Palaniappan K, Ratner A, Chu K, Szeto E, et al. IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 2017;45:D457–65. [DOI] [PMC free article] [PubMed]
- 8.Simmonds P., Adams M.J., Benkő M., Breitbart M., Brister J.R., Carstens E.B., et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15(3):161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 9.Paez-Espino D, Pavlopoulos GA, Ivanova NN, Kyrpides NC. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data [Internet]. Nature Protocols. 2017. p. 1673–82. Available from: http://dx.doi.org/10.1038/nprot.2017.063. [DOI] [PubMed]
- 10.Rampelli S., Soverini M., Turroni S., Quercia S., Biagi E., Brigidi P., et al. ViromeScan: a new tool for metagenomic viral community profiling. BMC Genomics. 2016;17 doi: 10.1186/s12864-016-2446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang HB, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, et al. Gene sharing networks to automate genome-based prokaryotic viral taxonomy [Internet]. Available from: http://dx.doi.org/10.1101/533240.
- 12.Low S.J., Džunková M., Chaumeil P.-A., Parks D.H., Hugenholtz P. Evaluation of a concatenated protein phylogeny for classification of tailed double-stranded DNA viruses belonging to the order Caudovirales. Nat Microbiol. 2019;4(8):1306–1315. doi: 10.1038/s41564-019-0448-z. [DOI] [PubMed] [Google Scholar]
- 13.Roux S., Tournayre J., Mahul A., Debroas D., Enault F. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinf. 2014;15:76. doi: 10.1186/1471-2105-15-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., et al. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 15.Simmonds P. Methods for virus classification and the challenge of incorporating metagenomic sequence data. J Gen Virol. 2015;96:1193–1206. doi: 10.1099/jgv.0.000016. [DOI] [PubMed] [Google Scholar]
- 16.Yu C., Hernandez T., Zheng H., Yau S.-C., Huang H.-H., He R.L., et al. Real time classification of viruses in 12 dimensions. PLoS ONE. 2013;8(5):e64328. doi: 10.1371/journal.pone.0064328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng M, Yu C, Liang Q, He RL, Yau SS-T. A novel method of characterizing genetic sequences: genome space with biological distance and applications. PLoS One. 2011;6:e17293. [DOI] [PMC free article] [PubMed]
- 18.Bao Y., Chetvernin V., Tatusova T. Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Arch Virol. 2014;159(12):3293–3304. doi: 10.1007/s00705-014-2197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauber C., Gorbalenya A.E. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J Virol. 2012;86(7):3890–3904. doi: 10.1128/JVI.07173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weighill D.A., Jacobson D. Network metamodeling: effect of correlation metric choice on phylogenomic and transcriptomic network topology. Adv Biochem Eng Biotechnol. 2017;160:143–183. doi: 10.1007/10_2016_46. [DOI] [PubMed] [Google Scholar]
- 22.Azad A, Pavlopoulos GA, Ouzounis CA, Kyrpides NC, Buluç A. HipMCL: a high-performance parallel implementation of the Markov clustering algorithm for large-scale networks. Nucleic Acids Res. 2018;46:e33. [DOI] [PMC free article] [PubMed]
- 23.Garcia BJ, Labbé JL, Jones P, Abraham PE, Hodge I, Climer S, et al. Phytobiome and Transcriptional Adaptation of Populus deltoides to Acute Progressive Drought and Cyclic Drought [Internet]. Phytobiomes Journal. 2018. p. 249–60. Available from: http://dx.doi.org/10.1094/pbiomes-04-18-0021-r.
- 24.Velmeshev D., Schirmer L., Jung D., Haeussler M., Perez Y., Mayer S., et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science. 2019;364(6441):685–689. doi: 10.1126/science.aav8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. Nature Publishing Group. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amisse J.J.G., Ndunguru J., Tairo F., Ateka E., Boykin L.M., Kehoe M.A., et al. Analyses of seven new whole genome sequences of cassava brown streak viruses in Mozambique reveals two distinct clades: evidence for new species. Plant Pathol. 2019;68(5):1007–1018. doi: 10.1111/ppa.13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia BJ, Simha R, Garvin M, Furches A, Jones P, Hyatt PD, et al. Kraken2 Metagenomic Virus Database [Internet]. 2020. Available from: https://www.osti.gov/biblio/1615774.
- 30.Didion JP, Martin M, Collins FS. Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ. 2017;5:e3720. [DOI] [PMC free article] [PubMed]
- 31.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed]
- 32.Utturkar S.M., Cude W.N., Robeson M.S., Yang Z.K., Klingeman D.M., Land M.L., et al. Enrichment of root endophytic bacteria from populus deltoides and single-cell-genomics analysis. Appl Environ Microbiol. 2016;82(18):5698–5708. doi: 10.1128/AEM.01285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang H., Lei R., Ding S.-W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dembélé D., Kastner P. Fold change rank ordering statistics: a new method for detecting differentially expressed genes. BMC Bioinf. 2014;15:14. doi: 10.1186/1471-2105-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo Y., Lian S., Chu H., Cho J.K., Yoo S.-H., Choi H., et al. Peach RNA viromes in six different peach cultivars. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-20256-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Marais A, Lefebvre M, Theil S, Svanella-Dumas L, Faure C, et al. Phytovirome analysis of wild plant populations: comparison of double-stranded RNA and virion-associated nucleic acid metagenomic approaches. J Virol [Internet]. 2019; Available from: http://dx.doi.org/10.1128/JVI.01462-19. [DOI] [PMC free article] [PubMed]
- 38.Lintas C., Altieri L., Lombardi F., Sacco R., Persico A.M. Association of autism with polyomavirus infection in postmortem brains. J Neurovirol. 2010;16(2):141–149. doi: 10.3109/13550281003685839. [DOI] [PubMed] [Google Scholar]
- 39.Karachaliou M., Chatzi L., Roumeliotaki T., Kampouri M., Kyriklaki A., Koutra K., et al. Common infections with polyomaviruses and herpesviruses and neuropsychological development at 4 years of age, the Rhea birth cohort in Crete. Greece J Child Psychol Psychiatry. 2016;57:1268–1276. doi: 10.1111/jcpp.12582. [DOI] [PubMed] [Google Scholar]
- 40.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbian HJ, Li Y, Ramirez M, Klase Z, Lipende I, Mjungu D, et al. Destabilization of the gut microbiome marks the end-stage of simian immunodeficiency virus infection in wild chimpanzees. Am J Primatol [Internet]. 2018;80. Available from: http://dx.doi.org/10.1002/ajp.22515. [DOI] [PMC free article] [PubMed]
- 42.Tsang TK, Lee KH, Foxman B, Balmaseda A, Gresh L, Sanchez N, et al. Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin Infect Dis [Internet]. 2019; Available from: http://dx.doi.org/10.1093/cid/ciz968. [DOI] [PMC free article] [PubMed]
- 43.Engdahl E, Gustafsson R, Huang J, Biström M, Bomfim IL, Stridh P, et al. Increased serological response against human herpesvirus 6A is associated with risk for multiple sclerosis [Internet]. Available from: http://dx.doi.org/10.1101/737932. [DOI] [PMC free article] [PubMed]
- 44.Brizzi KT, Lyons JL. Peripheral Nervous System Manifestations of Infectious Diseases [Internet]. The Neurohospitalist. 2014. p. 230–40. Available from: http://dx.doi.org/10.1177/1941874414535215. [DOI] [PMC free article] [PubMed]
- 45.Beckman J.A., Duncan M.S., Alcorn C.W., So-Armah K., Butt A.A., Goetz M.B., et al. Association of human immunodeficiency virus infection and risk of peripheral artery disease. Circulation. 2018;138(3):255–265. doi: 10.1161/CIRCULATIONAHA.117.032647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabrizi F., Donato F.M., Messa P. Association between hepatitis C virus and chronic kidney disease: a systematic review and meta-analysis. Ann Hepatol. 2018;17(3):364–391. doi: 10.5604/01.3001.0011.7382. [DOI] [PubMed] [Google Scholar]
- 47.Meltzer A, Van de Water J. The Role of the Immune System in Autism Spectrum Disorder [Internet]. Neuropsychopharmacology. 2017. p. 284–98. Available from: 10.1038/npp.2016.158. [DOI] [PMC free article] [PubMed]
- 48.Hutton J. Does rubella cause autism: a 2015 reappraisal? Front Hum Neurosci. 2016;10:25. doi: 10.3389/fnhum.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atladottir H.O., Henriksen T.B., Schendel D.E., Parner E.T. Autism after infection, febrile episodes, and antibiotic use during pregnancy: an exploratory study. Pediatrics. 2012;130(6):e1447–e1454. doi: 10.1542/peds.2012-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Lv C.-C., Tian J., Miao R.-J., Xi W., Hertz-Picciotto I., et al. Prenatal and perinatal risk factors for autism in China. J Autism Dev Disord. 2010;40(11):1311–1321. doi: 10.1007/s10803-010-0992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libbey J., Sweeten T., McMahon W., Fujinami R. Autistic disorder and viral infections. J Neurovirol. 2005;11(1):1–10. doi: 10.1080/13550280590900553. [DOI] [PubMed] [Google Scholar]
- 52.Cui Z.-H., Bi W.-L., Hao X.-Y., Xu Y., Li P.-M., Walker M.A., et al. Responses of in vitro-grown plantlets (Vitis vinifera) to Grapevine leafroll-associated virus-3 and PEG-induced drought stress. Front Physiol. 2016;7:203. doi: 10.3389/fphys.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramegowda V., Senthil-Kumar M. The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J Plant Physiol. 2015;176:47–54. doi: 10.1016/j.jplph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Pandey P., Ramegowda V., Senthil-Kumar M. Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms. Front Plant Sci. 2015;6:723. doi: 10.3389/fpls.2015.00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.