Abstract
Purpose
This current study investigated the effect of metformin treatment on hepatic oxidative stress and inflammation associated with nonalcoholic fatty liver disease (NADLD) in high fat diet (HFD) fed rats.
Method
Wistar rats were fed with a HFD or laboratory chow diet for 8 weeks. Metformin was administered orally at a dose of 200 mg/kg. Body weight, food and water intake were recorded on daily basis. Oral glucose tolerance test (OGTT), biochemical analysis and histological examinations were conducted on plasma and tissue samples. Antioxidant and anti-inflammatory mRNA expression was analyzed using reverse transcription polymeric chain reaction (RT-PCR).
Results
Metformin treatment for 8 weeks prevented HFD-induced weight gain and decreased fat deposition in HFD fed rats. Biochemical analysis revealed that metformin treatment significantly attenuated nitro-oxidative stress markers malondialdehyde (MDA), advanced protein oxidation product (APOP), and excessive nitric oxide (NO) levels in the liver of HFD fed rats. Gene expression analysis demonestrated that metformin treatment was associated with an enhanced expression of antioxidant genes such as Nrf-2, HO-1, SOD and catalase in liver of HFD fed rats. Metformin treatment also found to modulate the expression of fat metabolizing and anti-inflammatory genes including PPAR--γ, C/EBP-α, SREBP1c, FAS, AMPK and GLUT-4. Consistent with the biochemical and gene expression data, the histopathological examination unveiled that metformin treatment attenuated inflammatory cells infiltration, steatosis, hepatocyte necrosis, collagen deposition, and fibrosis in the liver of HFD fed rats.
Conclusion
In conclusion, this study suggests that metformin might be effective in the prevention and treatment of HFD-induced steatosis by reducing hepatic oxidative stress and inflammation in the liver.
Keywords: Inflammation, Lipid peroxidation, Metformin, Non-alcoholic fatty liver disease, Obesity
Abbreviations: ALT, alanine aminotransferase; ALP, alkaline phosphatase; AMPK, AMP-activated protein kinase; APOP, advanced protein oxidation product; AST, aspartate aminotransferase; ATP, Adinosine triphosphate; AUC, area under the curve; CAT, catalase; FAS, Fatty acid synthase; HDL, high density lipoprotein; HF, High fat; HSCs, Hepatic stellate cells; IACUC, Institutional Animal Care and Use Committee; IL-6, interleukin-6; LDL, low density lipoprotein; MDA, Malondialdehyde; Met, Metformin; MPO, Myeloperoxidase; NAFLD, nonalcoholic fatty liver disease; NO, nitric oxide; OGTT, Oral glucose tolerance test; PBS, Phosphate buffer saline; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1; PPAR-γ, peroxisome proliferator-activated receptor γ; ROS, reactive oxygen species; SOD, Superoxide dismutase; SREBP1c, sterol regulatory element-binding protein 1c; TBA, Thiobarbituric acid; TBARS, Thiobarbituric acid reactive substances
Highlights
-
•
High fat diet in rats developed glucose intolerance and oxidative stress in liver.
-
•
Metformin restored antioxidant genes expression in liver of HFD fed rats.
-
•
Metformin also inhibited the inflammatory genes expression and fibrosis in liver.
-
•
Moreover, metformin treatment may ameliorate fatty liver in HFD fed rats.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease, affecting about 25% of the population worldwide [1]. While the exact cause of NAFLD is not fully understood, The development of NAFLD may linked to the consumption of high sugar and/or high fat containing foods, lack of physical activity and genetic factors [2,3], which may eventually lead to more aggressive form of non-alcoholic steatohepatitis (NASH), to end-stage liver disease, liver cirrhosis and death [4,5]. NAFLD development is also linked to the progression of hyperlipidaemia and insulin resistance [6,7]. Oxidative stress and inflammation are the two most frequently observed pathological events associated with the development of NAFLD [6,8]. An earlier study showed that high fat diet (HFD)-induced fatty liver has increased oxidative stress markers and decreased endogenous antioxidant enzymes, elevated inflammatory cells infiltration and elevated myeloperoxidase activity [9], which are consistent with the symptoms of NAFLD. Despite some advances in our understanding about the pathogenesis of NAFLD, there is currently no FDA-approved treatment for NAFLD. Lifestyle modifications are often recommended for the management of this disease, which failed to produce the desired outcomes. A prospective study showed that weight loss of ≥10%, caused a significant reduction of fibrosis and NASH in affected individuals [10]. However, only a small percentage of patients were able to achieve such weight loss goal, indicating a critical need for the development of novel pharmacological treatments beyond just lifestyle modifications [11].
Given the clinical presentation of NAFLD, a few recent studies investigated the efficacy of metformin for the treatment of NAFLD, but produced contrasting findings [2,12]. A recent sytemetic review of several randomized controlled trials on hypoglycemic agents including metformin showed small benefit in the treatment of NAFLD [13]. However, mentformin treatment did not produce clinically significant out come to prevent or treat liver fibrosis [13]. As one of the most widely prescribed biguanide for the management of type 2 diabetes mellitus (T2DM), metformin works by decreasing glucose absorption from intestine, improving cellular glucose utilization, lowering blood glucose concentrations, reducing fasting plasma insulin levels and decreasing insulin resistance. Metformin is also known to improve lipid metabolism by activating AMP-activated protein kinase (AMPK), an important regulator of energy metabolism. Metformin in general has been shown to alleviate oxidative stress [14,15] and inflammation [16], which are believed to be responsible for the anti-aging, cardiovascular and neuroprotective effects. In diabetic patients, metformin was reported to prevent oxidative stress partially by restoring antioxidant enzymes [17]. Recent investigations suggest that metformin is effective in reversing in HFD-induced metabolic dysfunction in animal models. Metformin treatment reduces weight gain in HFD fed mice [18] the reduction of body weight gain by metformin treatment is explained by the reduction of visceral fat deposition and improved energy expanditure in human and animals [19]. Metformin treatment has been shown to improve hepatic steatosis and inflammation in HFD fed C57BL/6J mice [20]. Another report suggests that metformin may prevent ischemia reperfusion-induced oxidative stress in fatty liver of HFD fed rats [21]. Along with a variety of beneficial effects, metformin treatment in HFD fed rats has been shown to improve insulin resistance [22].
However, the ameliorative effect of metformin in the prevention of hepatic damage due to lipid accumulation and oxidative stress in HFD induced steatosis remained unclear. Moreover, the prevention of steatosis and fibrosis development in the liver of HFD fed animals by metformin needs to be addressed properly. Considering the above facts, the present study was designed and conducted to observe the effectiveness of metformin treatment in the prevention of hepatic oxidative stress, fibrosis and steatosis development in HFD fed rats.
2. Materials and methods
2.1. Chemicals
Beef tallow for preparing high fat diet, was purchased from Dhaka New Market, Bangladesh. The kits for assaying alanine aminotransferase (ALT), alkaline phosphatase (ALP), triglyceride, total cholesterol, low density lipoprotein (LDL) and high density lipoprotein (HDL) were purchased from DCI diagnostics (Budapest, Hungary). Thiobarbituric acid (TBA) was purchased from Sigma Aldrich (St. Louis, MO, USA) and metformin was a gift sample from Eskayef Pharmaceuticals Limited, Bangladesh. All other reagents used in this study were of analytical grade, collected from reagents store of Department of Pharmaceutical Sciences, North South University.
3. Animals and treatment protocols
All experiments involving animals were conducted following protocols that were previously examined and approved by the Institutional Animal Care and Use Committee (IACUC) of the North South University, Dhaka, Bangladesh (AEC-004-2017). Animals were treated in accordance with the Guide for the Care and Use of Laboratory Animals (8th edition, National Academies Press). Twenty four Wistar male rats, ranging from 10 to 12 weeks of age and weighing between 185 and 220 g, were obtained from the Animal House of the Department of Pharmaceutical Sciences, North South University, Dhaka, Bangladesh for this study. Animals were caged (one animal in each cage) in a controlled room environment, with a temperature 22 ± 2 °C; 55%, humidity and maintained a 12-h light/dark cycles.
For the experiment, rats were randomly categorized into four experimental groups consisting of six rats in each group. The composition of standard chow diet and high fat diet used in this study was obtained from our previously published work (Supplement Table 1). Group I animals received standard chow diet containing 14% proteins, 57% carbohydrates and 13.5% fat, and served as the control (Control). Group III rats received standard chow diet and metformin at 200 mg/kg dose daily by oral gavage for 8 weeks (Control + MET). Group II animals received high fat diet (HFD) containing 14% proteins, 37% carbohydrates and 48% fat, whereas, rats of group IV were given both HFD and metformin at 200 mg/kg every day by oral gavage for 8 weeks (HF + MET). All animals were allowed to drink water ad libitum. Oral glucose tolerance test (OGTT) was done on all groups of animals before the start of the study and at the end of the study to assess the glucose clearance from the blood due to the feeding of HFD in rats. The body weight, food and water intake data were noted on daily basis.
Table 1.
Effect of metformin on body weight, food and water intake in HFD fed rats.
| Parameters | Control | HF | Control + Met | HF + Met |
|---|---|---|---|---|
| Initial body weight (g) | 192.77 ± 7.30ns | 181.85 ± 2.42 | 180.07 ± 3.01 | 170.55 ± 2.17 |
| Final Body Weight (g) | 243.72 ± 6.67 | 307.90 ± 3.58** | 245.52 ± 2.57 | 237.33 ± 6.84 |
| Food intake (g) | 17.60 ± 0.19ns | 14.95 ± 0.33 | 20.74 ± 0.10 | 15.08 ± 0.16 |
| Water intake (mL) | 23.96 ± 0.37ns | 21.11 ± 0.19 | 21.00 ± 0.73 | 20.09 ± 0.20 |
Data are presented as mean ± SEM, N = 6. Statistical analysis was done by One way ANOVA followed by Tukey post hoc test. Statistical significance was considered at p < 0.05 in all cases. ** is significantly different from control and other groups.
3.1. Euthanasia and tissue harvesting
At the end of the treatment protocol, animals were euthanized by intraperitoneal injection of pentobarbital (65 mg/kg) injection. Blood was collected into heparinized tubes. Blood samples were centrifuged at 7000 g for 15 min at 4 °C within 30 min of collection to separate plasma. Plasma samples which were then stored at −20 °C for further analysis. Heart, kidney, spleen, and liver were surgically removed and weighed immediately, stored and preserved for biochemical and microscopic analyses. Adipose tissues in various regions such as peritoneal, epididymal and mesenteric fat pads were also surgically removed and weighed immediately. For histological analysis, the organs were preserved in neutral buffered formalin (NBF) (pH 7.4) after collection. All other parts of organ samples were also stored in −20 °C freezer for further analysis.
3.2. Oral glucose tolerance test
Oral glucose tolerance test (OGTT) was performed before and after starting of HFD feeding following procedures described earlier [9]. Rats were placed on overnight fasting (∼12 h) before the test. However, all animals had access to pure drinking water during the food deprivation period. Blood was drawn from tail vein to measure basal blood glucose concentrations using glucometer (Bionim Corporation, Bedford, MA, USA). The rats were given 2 g/kg body weight glucose as a 40% aqueous solution via oral gavage. Tail vein blood samples were withdrawn at 30, 60, 90 and 120 min following glucose administration and glucose concentrations recorded using a glucometer.
3.3. Assessment of liver function marker
Liver function marker enzymes such as alanine aminotransferase (ALT) and alkaline phosphatase (ALP) activities were assessed in plasma by using commercially available kits, following the manufacturer's protocol.
3.4. Measurement of plasma cholesterol and triglyceride levels
The plasma levels of cholesterol, LDL and triglyceride were estimated by using the cholesterol, LDL and triglyceride assay kits. Assays were performed following the instructions given by the manufacturer.
3.5. Determination of oxidative stress markers
Liver was homogenized by adding phosphate buffer (pH 7.4) and the homogenate mixture was centrifuged at 7000 g for 15 min at 4 °C to isolate the supernatant, which was stored at −20 °C for further analysis. Supernatants were also used to measure oxidative stress markers in liver tissue.
3.6. Estimation of lipid peroxidation
One of the indicators of lipid peroxidation and oxidative stress is the presence of thiobarbituric acid reactive substances (TBARS) in plasma. Lipid peroxidation in plasma and liver tissue was measured colorimetrically by determining thiobarbituric acid reactive substances (TBARS) as described earlier [23]. Briefly, two mL of (1:1:1 ratio) TBA-concentrated acetic acid-HCl reagent was added in 0.1 mL of liver tissue homogenate or plasma and kept in water bath for 15 min and allowed to cool down. The absorbance of clear supernatant was read in ELISA plate reader against reference blank at 535 nm.
3.7. Assay of nitric oxide (NO)
NO levels in plasma and liver tissue homogenates were measured using modified Griess-Illosvoy reagents as described earlier [24,25]. A pink colour chromophore was formed when the reaction mixture containing plasma or tissue homogenates, PBS and the reagent was incubated for 150 min at 25 °C. The absorbance was taken against a corresponding blank solution at 540 nm. NO levels were measured and stated as nmol/mL or nmol/g of tissue using a standard curve.
3.8. Advanced protein oxidation products (APOP) assay
A previously described method [26,27] was modified and used to measure the level of advanced protein oxidation products (APOP). In brief, 2 mL of plasma was diluted in PBS at 1:5 ratio and then 0.1 mL of 1.16 M potassium iodide was added in each tube. After 2 min, 0.2 mL acetic acid was added. Absorbance reading of the reaction mixture was immediately taken at 340 nm after reading a blank containing 2 mL of PBS, 0.1 mL of KI, and 0.2 mL of acetic acid. The chloramines -T absorbance at 340 nm is linear within the range of 0–100 mmol/L. APOP concentrations were expressed as μmol·L−1 chloramine -T equivalents.
3.9. Catalase (CAT) activity assay
CAT activities in plasma and liver tissue homogenates were analyzed following previously described protocols [25,28]. Absorbance changes were measured at 240 nm for the reaction mixture containing 50 mmoL phosphate buffer (pH 5.0), 5.9 mmol hydrogen peroxide, and 0.1 mL enzyme extract. An absorbance change of 0.01 units/min was interpreted as one unit of CAT activity.
3.10. Estimation of superoxide dismutase (SOD) activity
SOD activity in the plasma and liver tissue homogenates was determined following previously described methods [25,29]. Reaction mixture containing enzymes were prepared and absorbance read at 480 nm for 1 min at 15 s intervals. A blank solution without tissue homogenates was run in parallel. Auto-oxidation of epinephrine present in the assay system was calculated and 50% inhibition is defined as the one unit of SOD enzyme activity.
3.11. Gene expression analysis by quantitative real-time PCR
mRNA was isolated and purified from the liver of all four groups of rats by GeneJET RNA Purification Kit (Thermo-Fisher Scientific, Waltham, MA, USA). After measurement of concentration by NanoDrop 2000 spectrophotometer (Bio-Rad, California, USA), mRNA was used for the synthesis of cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo-Fisher Scientific, USA) by T100 Thermal Cycler (Bio-Rad, USA). This cDNA and enzymes using Maxima SYBR Green qPCR master mix (Thermo Scientific, USA) were used for the quantification of mRNA levels of oxidative stress and inflammation-related factors. The process was carried out in a CFX96C1000 Touch Real-Time PCR Detection System (Bio-Rad, USA) and data were analyzed by CFX Manager TM Software (CFX Manager TM Software) according to the manufacturer's protocol. Quantitative real-time PCR was conducted and according to a program followed by Khan et al. [30], using forward and reverse primers designed by Primer 3 online software (Supplement Table 2). The mRNA levels for each of the target genes were measured in relative to the mRNA level of β-actin of the corresponding sample.
3.12. Histopathological examination
Neutral buffered formalin (NBF) was used to fix the liver and intestinal tissues and then embedded in paraffin block. Paraffin-embedded tissues were sectioned at 5 μm thickness using a rotary microtome and slices were stained with hematoxylin-eosin (H&E) to examine the structural features of hepatic tissue and inflammatory cells infiltration. Grading of steatosis was also performed in each group of animals according to a previously published protocol [31]. The grading score was considered as steatosis (<5% = 0; 5–33% = 1; 33–66% = 2; >66% = 3); lobular inflammation (none = 0; 2 foci = 1; 2–4 foci = 2; >4 foci = 3); and hepatocellular ballooning (none = 0; few = 1; prominent = 2). All features were scored in a blinded manner in at least three rats from each group and five fields of view in each sample.
The number of rats selected for histological staining as n = 3, is based on the power analysis (using GPower software, version 3.1) for minimum samples required for statistical significance. Picrosirius red was used to stain the liver tissue sections to analyse the presence and extent of fibrosis. Moreover, Periodic Acid Schiff (PAS) staining was performed on intestinal sections to evaluate the mucus producing goblet cells. Percentage of fibrosis was estimated using Image-J free software (downloaded from NIH website, USA) [32]. Tissues were also stained with Prussian blue staining to examine free iron deposition in them [25,32]. The stained sections were visualized and photographed at either 10× or 40× magnifications using a light microscope (Zeiss Axioscope), and images were processed for further analyses.
3.13. Statistical analysis
For the statistical analysis of data, Graph Pad Prism software (version 6.0) was used. Mean ± standard error of mean (SEM) was used to express all data. For the comparison of data, One way ANOVA along with Tukey post hoc test was used. Comparisons were made between Control vs HF groups and HF vs HF + MET and Control + MET groups. In all cases, p < 0.05 was considered statistically significant.
4. Results
4.1. Metformin inhibits weight gain in rats induced by HFD
Body weight, food and water intake of the animals were noted daily during the course of the experiment (Table 1). Differences between the initial and final body weights are presented in Table- 1. Data showed that HFD fed rats exhibited a striking ∼41% body weight gain compared to control which had ∼20% weight gain. Importantly, metformin treatment reduced such weight gain in HFD fed rats to ∼28%, which is almost similar to the weight gain for control rats (∼26%) receiving metformin. This result indicates that metformin at 200 mg/kg dose suppresses HFD-associated weight gain in rats (Table 2).
Food and water intake behavior was also observed during the 56-day period of treatment. We found that food and water intake were similar across the four different groups of animals throughout the experimental period (Table 2). This data suggest that the diet composition or metformin treatment did not alter food and water consumption in the experimental animals that could potentially account for body weight gain and other pathophysiological changes.
4.2. Metformin inhibits HFD-induced hepatic, retroperitoneal, mesenteric and epididymal fat deposition
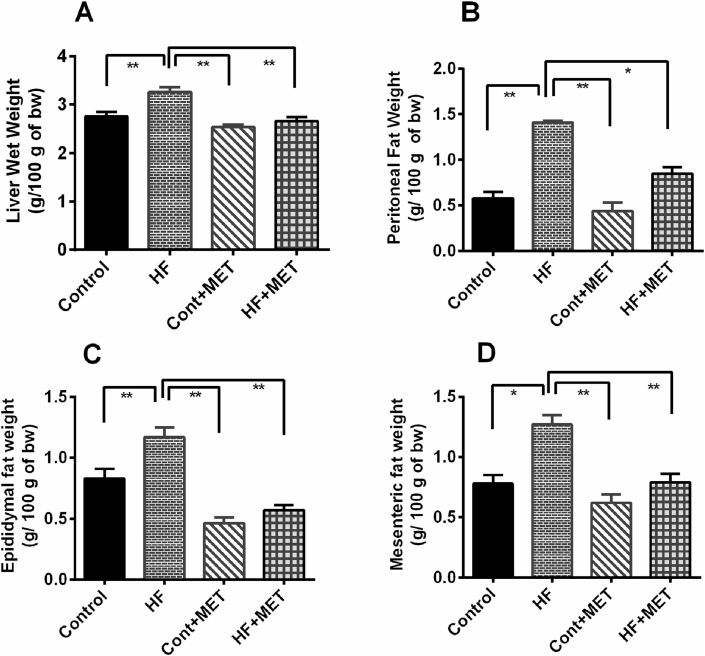
The wet weights of liver are presented in Fig. 1A. Our data showed that there is a significant increase (p < 0.05) in liver weight in HFD fed rats compared to control rats and metformin treatment decreased such inreases in liver weight in HFD fed rats. Metformin administration in control animals caused a slight reduction in liver weight (Contol + MET) (Fig. 1A).
Fig. 1.
Effect of metformin on liver wet weights in HFD fed rats (A) and accumulations of fat in (B) peritoneal, (C) epididymal and (D) mesenteric area. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05. * is significant at p < 0.05 and ** is significant at p < 0.001.
Next, we measured the weight of retroperitoneal, mesenteric and epididymal fat deposition in all groups of experimental animals. Consistent with our previous findings, weight of retroperitoneal, mesenteric and epididymal fat pads was markedly increased in HFD fed rats compared to the control rats (p < 0.05) (Fig. 1B–1D). In addition, metformin treatment markedly decreased the wet weight of peritoneal and epididymal fat in HF + MET group compared to HFD fed rats (Fig. 1B and C). Moreover, there was significant difference in mesenteric fat deposition between the HF + MET rats and the HFD fed rats (Fig. 1D). The wet weight of these three different fat pads was significantly (p < 0.05) reduced in control + MET rats compared to the HFD fed rats (Fig. 1B–1D). On the contrary, there was no significant difference in fat deposition between the Control + MET and Control rats (Fig. 1B–1D). Collectively, our data suggest that metformin reduces fat deposition in HFD fed rats.
4.3. Metformin ameliorates impaired glucose metabolism in HFD fed rats
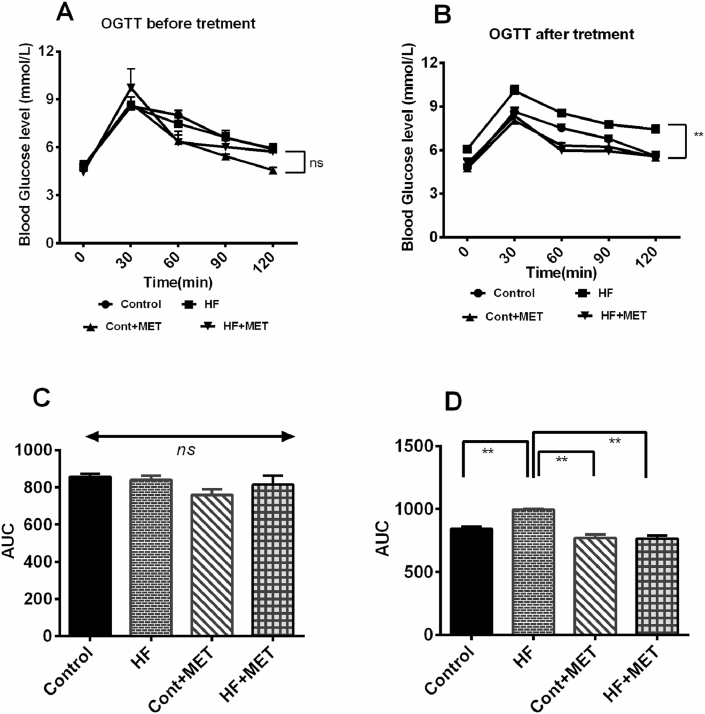
Since NAFLD is related with reduced blood glucose removal and insulin resistance [6,7], we performed oral glucose tolerance test (OGTT) to evaluate glucose metabolism of the rats under different experimental conditions as shown in Fig. 2. At the beginning of the experiment, blood glucose concentrations were similar among the four groups of rats (Fig. 2A). However, at the end of the experimental period, control rats showed blood glucose level of ∼8 mmol/L at 60 min and which reduced to a basal level of ∼5 mmol/L at 120 min, indicating normal glucose utilization. In contrast, in HFD fed obese rats, the blood glucose level reached a peak of∼10 mmol/L after 60 min and remained at a higher level ∼8 mmol/L at 120 min (Fig. 2B), suggesting an impaired glucose metabolism. As expected, a significant drop in blood glucose level, which is 5 mmol/L at 120 min, was observed in HFD fed rats treated with metformin compared to the blood glucose level in control rats at 120 min (Fig. 2B).
Fig. 2.
Effect of metformin on oral glucose tolerance test (OGTT) before (A) and after (B) the treatment in HFD fed rats. C and D are AUC data for OGTT test respectively. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05. * is significant at p < 0.05 and ** is significant at p < 0.001.
Moreover, the area under the curve (AUC) for the OGTT before the initiation of the study exhibited no significant changes among the groups (Fig. 2C). However, the AUC for OGTT at the end of the treatment period, showed that HFD feeding increased AUC in rats compared to control rats significantly (p < 0.05) (Fig. 2C). Metformin treatment normalized the AUC value in HFD fed rats significantly (p < 0.05) (Fig. 2C). Taken together, our data indicated that metformin treatment in HFD fed rats maintained normal physiological glucose metabolism that was compromised in HFD fed rats.
4.4. Metformin suppresses HFD-induced elevation of liver function markers in the plasma
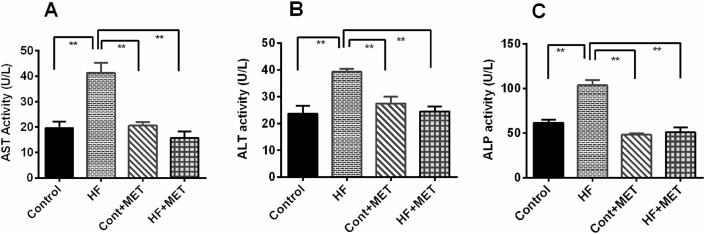
Next, we examined the efficacy of metformin in reducing obesity-induced oxidative damage to the liver by measuring ALT, AST and ALP enzymes and found that their levels were elevated in the plasma, presumably because of the liver injury caused by oxidative stress [9,25]. Feeding of HFD in rats markedly elevated the levels of ALT, AST and ALP enzymes in plasma (Fig. 3) compared to controls (Fig. 3A–C). As anticipated, treatment with metformin significantly (p < 0.05) attenuated HFD associated elevation of plasma AST, ALT and ALP activities. These data revealed that oxidative damage in liver was caused by HFD, and metformin is effective in reversing such HFD induced oxidative damage and may provide protection to the liver.
Fig. 3.
Effect of metformin on liver function markers such as AST (A), ALT (B) and ALP (C) activities in plasma of HFD fed rats. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark.
4.5. Metformin reduces triglyceride, total cholesterol and LDL concentrations in HFD fed rats
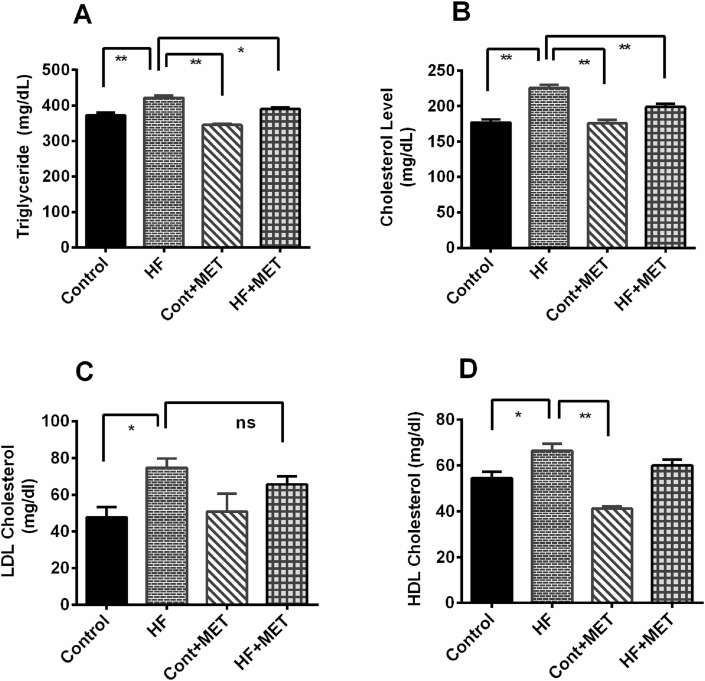
After establishing beneficial effects of metformin in preventing HFD induced fat deposition and obesity, we set out to examine the effect of metformin on the levels of circulating lipids in the blood such as triglyceride, total cholesterol, LDL and HDL. Our results revealed that HFD caused a significant elevation of plasma levels of harmful lipids such as triglyceride, total cholesterol and LDL in HFD fed rats compared to the control rats (p < 0.05) (Fig. 4). Metformin treatment prevented the rise of triglyceride and total cholesterol concentrations (p < 0.05) in HFD fed rats. However, Metformin treatment did not alter the LDL and HDL concentration significantly in HFD fed rats compared to HFD rats (Fig. 4D). Collectively, these results suggest that metformin not only reduced the retroperitoneal, mesenteric and epididymal fats deposition but also decreased the levels of harmful circulating lipids.
Fig. 4.
Effect of metformin on triglyceride (A), total cholesterol (B), LDL (C) and HDL (D) level in plasma of HFD fed rats. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark.
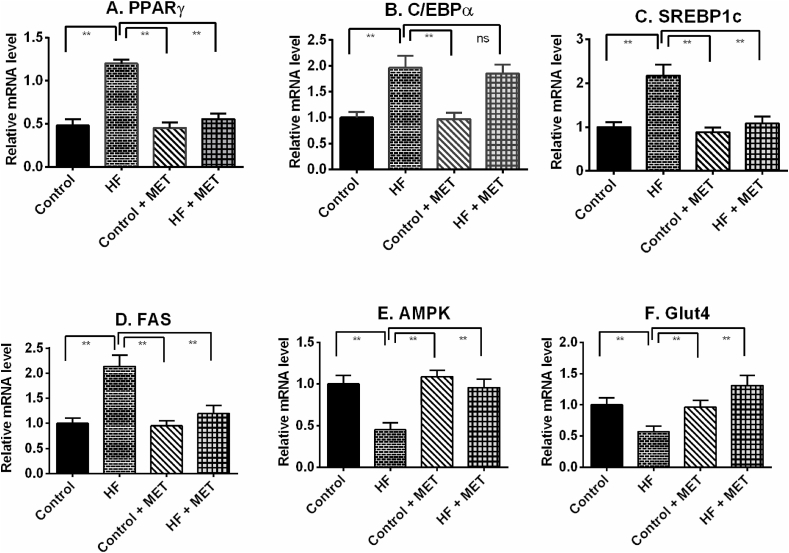
4.6. Metformin modulates the expression of adipogenesis and inflammation related genes in liver of HFD fed rats
Increasing fat metabolism is a way of clearing excess fat deposits in the liver. Our RT-PCR data showed that HFD feeding in rats up-regulated peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT-enhancer-binding protein alpha (C/EBPα) expression in liver tissues significantly (P > 0.05) compared to control rats (Fig. 5). Metformin treatment in HFD fed rats prevented the augmentation of PPARγ mRNA level, but the mRNA level of C/EBPα was not normalized by metformin treatment in HFD fed rats (Fig. 5). Moreover, HFD fed rats showed increased SREBP1c and FAS mRNA expression in the liver (Fig. 5). Metformin treatment in HFD fed rats normalized the SREBP1c and FAS mRNA expression in liver (Fig. 5).
Fig. 5.
Effect of metformin on fat metabolizing genes such as PPAR-γ (A), C/EBP-α (B), SREBP1c (C), FAS (D), AMPK (E) and GLUT-4 (F) expression in liver of HFD fed rats. Data are presented as mean ± SD, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark.
HFD fed rats also showed decreased mRNA level of AMPK and GLUT-4 in liver compared to control rats (Fig. 5). Metformin treatment elevated the AMPK and GLUT-4 mRNA expression to near normal in HFD fed rats (Fig. 5); which may explain the cholesterol lowering and improvement of glucose tolerance effects in HFD fed rats.
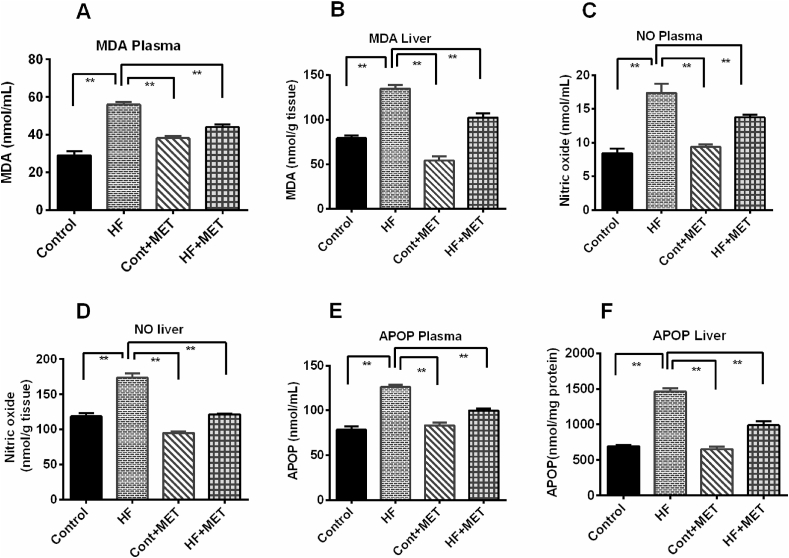
4.7. Metformin treatment improves oxidative stress markers in HFD fed rats
Since, metformin treatment reduced the high fat-related oxidative damage to the liver and prevented mobilization of liver function enzymes into blood; we next measured oxidative stress markers such as MDA and NO in the plasma and liver. Our data demonstrated that MDA and NO levels markedly increased in HFD fed rats compared to control rats (Fig. 6). Plasma levels of these oxidative stress markers are correlating well with their levels in the liver as well (Fig. 6). Metformin treatment reduced the MDA and NO levels in the plasma as well as in the liver tissue homogenates in HFD fed rats (p < 0.05) (Fig. 6). However, there was no significant difference in MDA levels between the control + MET and control rats (Fig. 6).
Fig. 6.
Effect of metformin on oxidative stress markers such as MDA (A, B), NO (C, D) and APOP (E, F) in plasma and liver of HFD fed rats. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark. * is significant at p < 0.05 and ** is significant at p < 0.001.
Next, we measured the advanced protein oxidation products (APOP) in the plasma and liver. As expected, APOP level was significantly (p < 0.05) elevated in HFD fed rats compared to control rats (Fig. 6). Treatment with metformin significantly (p < 0.05) decreased the APOP concentrations in HFD fed rats compared to the HFD fed rats (Fig. 6). However, Control + MET rats did not show any significant changes in APOP concentrations compared to control rats (Fig. 6). In summary, our data suggest that metformin possesses in-vivo antioxidant action that may prevent harmful effects of free radicals and reactive oxygen species in liver.
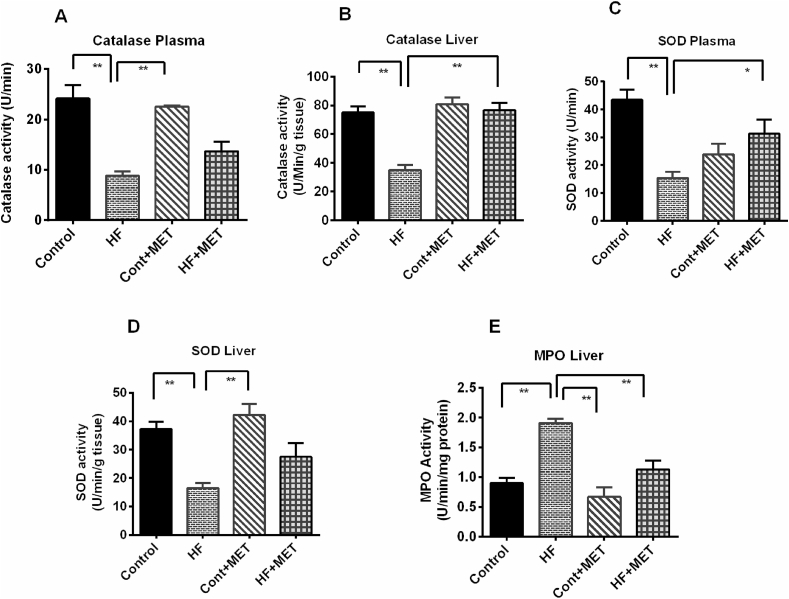
4.8. Metformin restores catalase and superoxide dismutase (SOD) levels in HFD fed rats
Oxidative stress and tissue injury are often linked to elevated production of reactive oxygen species and/or reduced expression of endogenous antioxidant enzymes such as catalase and SOD [33]. Metformin reduced the levels of oxidative stress markers and tissue injuries. Therefore, we next examined, whether metformin could restore endogenous antoxidant enzymes, namely catalase and SOD activities. This study showed that catalase activities in the plasma and liver were significantly (p < 0.05) lowered in HFD fed rats compared to the control rats (Fig. 7A and B). Metformin treatment restored catalase activity in liver of HFD fed rats significantly (p < 0.05) (Fig. 7B). However, metformin treatment caused a slight rise in plasma catalase activity compared to the HFD rats which was not restored the catalase activity significantly (Fig. 7A).
Fig. 7.
Effect of metformin on antioxidant enzyme such as catalase (A, B) and SOD (C, D) activities in plasma and liver as well as MPO (E) activities in liver of HFD fed rats. Data are presented as mean ± SEM, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark. * is significant at p < 0.05 and ** is significant at p < 0.001.
Moreover, SOD activity in both plasma and liver of HFD fed rats was also found to be markedly decreased (p < 0.05) compared to the control rats (Fig. 7C and D). Consistent with previous findings, SOD activity was also restored in plasma of HFD fed rats treated with metformin significantly (p < 0.05) (Fig. 7C).
HFD fed rats also showed increased myeloperoxidase (MPO) activity in liver compared to control rats; which is a sign of neutrophil infiltration (Fig. 7E). Metformin treatment ameliorated the MPO activity in liver of HFD fed rats (Fig. 7E). However, metformin treatment did not alter the MPO activity in Control + Met rats compared to control rats (Fig. 7E).
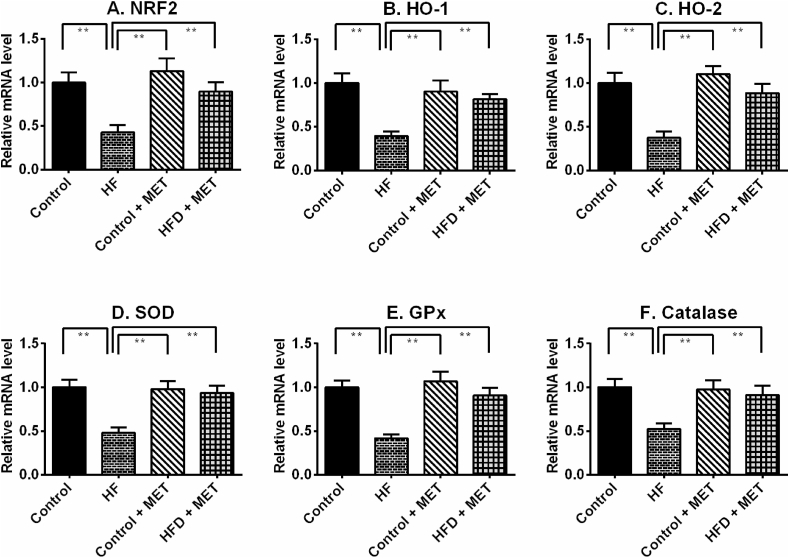
4.9. Metformin augmented antioxidant gene expression in liver of HFD fed rats
HFD fed rats showed decreased antioxidant enzymes activities which may have contributed to the increased oxidative stress in the liver. Such reduction of antioxidant defence might be due to a reduction of genes and proteins required for the neutralization of free radicles in tissues. Nrf-2 is a master regulator of oxidative stress in tissues. Nrf-2 mRNA expression was lowered in the liver of HFD fed rats which was restored by metformin treatment (Fig. 8). In line with this finding, mRNA levels of two downstream proteins directly linked to Nrf-2, is HO-1 and HO-2, were also reduced in HFD fed rats (Fig. 8). Metformin treatment restored the mRNA levels of both isoforms of HO enzymes in the liver of HFD fed rats (Fig. 8). Additionally, this study also revealed that metformin treatment rescued the transcript levels of SOD, catalase and GPx enzymes significantly (p < 0.05), which were also lowered in liver of HFD fed rats (Fig. 8).
Fig. 8.
Effect of metformin on oxidative stress regulator genes such as Nrf-2 (A), HO-1 (B), HO-2 (C), SOD (D), GPx (E) and catalase (F) expression in liver of HFD fed rats. Data are presented as mean ± SD, n = 6. One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark.
4.10. Metformin ameliorates fat deposition and inflammation in the liver of HFD fed rats
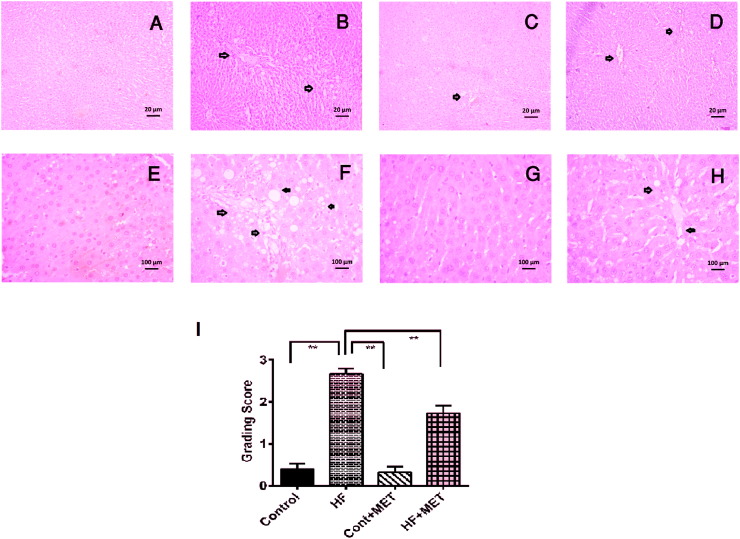
To further confirm the role of metformin in lessening tissue inflammation, we next assessed the effects of metformin treatment on the penetration of inflammatory cells and necrosis in liver by Hematoxylin and Eosin staining. We found that the liver sections of control rats showed normal histoarchitecture (Fig. 9A). However, liver sections from HFD fed rats exhibited marked inflammatory cells infiltration and necrotic changes as well as fat deposits (Fig. 9B, E). These features appear to recapitulate the pathological alterations seen in the liver during the development of NAFLD.
Fig. 9.
Effect of metformin on hepatic inflammation and fat deposition in the liver of HF diet fed rats. (A, E), Control; liver section showed normal histoarchitecture and a clean blood vessel. (B, F), HF; high amount of fat droplets are deposited in liver section. (C, G), Control + Met, showed similar structure of control rats. (D, H), HF + Met, Metformin treatment decreased the inflammatory cells infiltration and fat droplets in the liver section (Upper panel have low magnification, ×10; lower panel have high magnification, ×40). (I), grading score for steatosis is shown in different groups. Liver sections were taken from three rats per group and pictures were taken from five random areas from each section (15 random areas from each group) (please see materials and methods). One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark. * is significant at p < 0.05 and ** is significant at p < 0.001.
As anticipated, metformin treatment attenuated liver damage in HF + MET rats as evidenced by a reduction in necrosis and inflammatory cell infiltration in the liver compared to HFD fed rats (Fig. 9D). Moreover, steatosis score also increased significantly (p < 0.05) in HFD fed rats compared to control rats (Fig. 9F), and metformin treatment reversed the increased steatosis score in HF + MET rats (Fig. 9F).
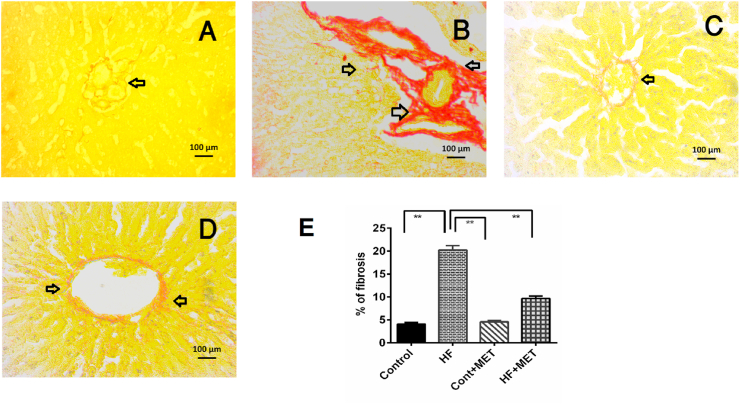
4.11. Metformin ameliorates collagen deposition and fibrosis in the liver of HFD fed rats
Collagen deposition and alignments in liver sections from our experimental rats were done using Picrosirius red staining. HFD fed rats showed a dramatic increase in collagen and fibrosis in liver (Fig. 10), an effect that was reversed by metformin treatment in HF + MET group (Fig. 10D). Altogether, our data indicated that metformin treatment decreased the infiltration of inflammatory cells, excessive collagen deposition and fibrosis in the liver, that are hallmark symptoms of NAFLD.
Fig. 10.
Effect of metformin on hepatic fibrosis in the liver of HF diet fed rats. A, Control; B HF; C, Control + Met; D, HF + Met; E, % of fibrosis is shown in different groups. Liver sections were taken from three rats per group and pictures were taken from five random areas from each section (15 random areas from each group) (please see materials and methods). One-way ANOVA followed by Tukey post hoc test was done for statistical comparison. Values are considered significant at p < 0.05 and marked as asterisk mark. * is significant at p < 0.05 and ** is significant at p < 0.001.
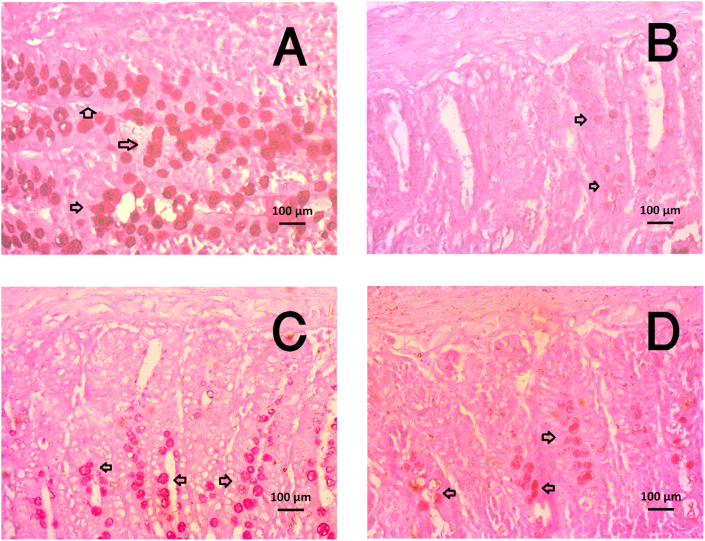
4.12. Metformin treatment restored the goblet cell population in the HFD fed rats
This investigation revealed that HFD fed rats showed fewer number of mucus-secreting goblet cells in the small intestine compared to the control rats (Fig. 11). Metformin treatment in HFD fed rats restored the goblet cells population in the small intestine, which are the mucus secreting cells in the intestine (Fig. 11). Metformin treatment did not alter the goblet cells population in control + Met rats (Fig. 11).
Fig. 11.
Effect of metformin on mucus secreting goblet cells population in the small intestine of HF diet fed rats. A, Control; B HF; C, Control + Met; D, HF + Met. Intestinal sections were taken from three rats per group and pictures were taken from five random areas from each section (15 random areas from each group) (please see materials and methods) magnification, ×40.
4.13. Metformin improves NAFLD-like ferritin deposition in the liver of HFD fed rats
Prussian blue staining showed free iron deposition in the liver. The deposition of ferric ion was not visible in the liver of control rats and Control + MET rats (Supplemental Figs. 1A–B). In contrast, excessive ferric ion deposition was observed in the liver of HFD fed rats compared to the controls (Supplemental Fig. 1C). As expected, ferric ion deposition was significantly diminished in the HFD fed rats treated with metformin compared to those on HFD fed rats (Supplemental Figs. 1C–D).
5. Discussion
The liver is the most enzyme-rich and metabolically active organ of our body and hence performs a wide range of physiological and biochemical functions which are vital for metabolic homeostasis in the body. Consumption of HFD has been reported to cause weight gain and obesity as well as induce oxidative stress, abnormalities of lipid metabolism, chronic inflammation and fatty liver disease [32]. HFD-induced obesity in animal models has been extensively characterized and popular among the researchers, because, the biochemical and pathological changes as well as the clinical manifestations resemble human obesity and associated complications [9]. In the current study, we showed that consumption of HFD leads to many biochemical and histological changes such as oxidative stress, impaired glucose metabolism, the progression of inflammation in hepatic tissue, non-alcohoic fatty liver disease (NAFLD), and caused significant weight gain in rats. Metformin treatment significantly ameliorated such pathological changes through its antioxidant and anti-inflammatory actions. Moreover, metformin also prevented the onset and exacerbation of NAFLD which would otherwise progress to liver failure.
According to the previous studies, HFD enriched by saturated fatty acids, are responsible for insulin resistance and impaired glucose utilization [34,35]. In this study, the HFD contains beef tallow as a source of saturated fatty acid. Previous report suggests that saturated fatty acids, found in butter, cheese, and meat promote the accumulation of lipid in skeletal muscles and may induce insulin resistance [36]. Palmitate, a saturated fatty acid, also promotes the secretion of cytokines such as TNFα and IL-6, which may cause insulin resistance and glucose intolerance [37,38]. In addition, de novo fatty acid synthesis from glucose, may also induce the deposition of free fatty acid in the liver and lead to insulin resistance [36].
Our data demonestrated that, HFD fed rats developed glucose intolerance and showed a relatively high blood glucose levels. Our study also confirmed that, metformin treatment improved glucose intolerance and reduced oxidative stress in the liver. Metformin is a hypoglycaemic drug, primarily used for reducing blood glucose levels in diabetic patients. Thus, our data is also in agreement with previous studies that reported that metformin treatment increased the insulin receptor sensitivity and glucose utilization in skeletal muscles and adipose tissues [18,22]. Additionally, the administration of metformin in HFD fed rats alleviated insulin resistance with simultaneous augmentation of the insulin signaling cascade in the hepatocytes [22]. Previous report also suggests that, metformin is a regulator of AMPK, a key kinase essential for ATP synthesis in mitochondria [39]. The improved glucose utilization could be attributed to the increased gene expression of AMPK and GLUT-4 in the liver of metformin-treated rats.
According to a previous study, increased cholesterol and triglyceride levels in plasma were observed in HFD fed rats which may cause the accumulation of fat in the liver [20]. During NAFLD development, fat accumulation in the liver due to HFD consumption and oxidative stress are considered as the most important risk factors [40,41]. In this study, metformin treatment reduced the cholesterol and triglyceride levels in HFD fed rats. Recent study also demonstrated that metformin treatment reduced high cholesterol levels and prevents NAFLD in experimental animals supplied with a HFD [42]. Metformin mediated upregulation of fat oxidizing genes with simultaneous prevention of de novo lipogenesis has been attributed to the cholesterol lowering actions [19]. A recent meta-analysis also supported the idea that metformin treatment is effective in lowering the high circulating lipid levels in blood and decreased the body weight in adult population [43]. This investigation also showed that HFD diet diminished the mucus producing goblet cells population in the intestine. This mucus layer is the first defense barrier against bacterial invasion which also prevents inflammation. Metformin treatment in HFD fed rats showed increased goblet cells in the intestine which may contribute to the lowering of the fatty acids from intestinal parts. This data was supported by previous literature showed that intact goblet cells population is vital for the lowring of total lipids in the HFD fed rats [32].
To get a deeper insight into the HFD-mediated NAFLD development and the molecular pharmacology of metformin, we considered to explore the gene expression of adipogenesis and insulin resistance-related factors and proteins. In the pathophysiology of adipogenesis, and related complications like NAFLD, the transcription factors, peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT-enhancer-binding protein alpha (C/EBPα) plays critical roles [44]. During preadipocyte differentiation and maturation these two transcription factors cross-regulate the gene expression of each other and up-regulate numerous downstream proadipogneic factors and down-regulate the antiadipogenic factors [45]. For this reason, these two proteins are known as the master regulators of adipogenesis. In patients [46] and experimental mammalian animal models [47] of NAFLD, hepatic PPARγ gene expression has been found to be significantly higher than normal. Moreover, the deletion of PPARγ activity in the hepatocytes of the mouse has been found to be beneficial for steatosis [48]. Therefore, with the aim of conceptualizing the molecular mechanism of HFD-mediated NAFLD and the beneficial effects of metformin, we also investigated the gene expression of PPARγ, C/EBPα, and their downstream proteins, factors, and enzymes which are involved in the regulation of adipogenesis. C/EBP transcription factors may induce SREBP1c in adipogenesis and control several gene expression involved in lipogenesis and insulin sensitivity [49]. The gene expression of PPARγ is also enhanced by the gene expression of SREBP1c [50]. Moreover, previous studies have also confirmed that SREBP1c, up-regulates the gene expression of HMG-CoA reductase (HMGCR) and fatty acid synthase (FAS), two most vital among the enzymes involved in the biosynthesis of steroids and fatty acids [50]. Thus augmetation of SREBP gene expression promotes the biosynthesis of cholesterol and fatty acids.
Our data demonestrated that HFD-mediated upregulation of PPARγ, C/EBPα, and SREBP1c and leptin gene expression was significantly suppressed after metformin treatment. Consistent with our observation, a previous study showed that in partial leptin-deficient animals, leptin deficiency is beneficial for obesity prevention and fatty liver development [51]. On the other hand, HFD-mediated downregulation of AMPK, adiponectin, and GLUT-4 transcripts were significantly restored in the metformin treated rats. Thus, the ameliorative effect of metformin on HFD-induced NAFLD could be, at least partially, attributed to its ability to suppress the gene expression of proadipogenic factors with a parallel augmentation of the antiadipogenic gene expression such as AMPK and GLUT-4 [52,53].
In this study, the oxidative stress marker and lipid peroxidation product, malondialdehyde (MDA) level elevated significantly in HFD-fed rats. Treatment with metformin reduced the MDA level in HFD fed rats to near normal as shown in control rats. Likewise, metformin treatment also lowered the HFD-mediated increased levels of NO and APOP. This ameliorating effect might be attributed to the ability of metformin to increase the activities of antioxidant enzymes such as SOD and catalase. The increased activities of these key enzymes were also supported by the increased gene expression of oxidative stress-related factors such as Nrf-2 and HO enzymes by metformin treatment in HFD fed rats. Previous studies also reported that metformin can reduce oxidative stress in rats by increasing the activities of antioxidant enzymes [21].
NAFLD disease is associated with deposition of free iron in the form of ferrous ion (Fe++) which causes iron induced cytotoxicity by triggering redox reaction. The redox reaction, in turn, causes the generation of deleterious oxygen radicals that damages cellular organelles [54]. Metformin treatment resulted in a reduction of free iron deposition in the liver section of HFD fed rats. Moreover, oxidative stress-induced obesity and NAFLD, not only damages local tissues but can quickly spread to global level causing widespread damage to various body organs [55]. When liver tissue starts to damage, leakage of ALT and ALP occurs into the bloodstream and increased their activities in the plasma [56]. In agreement with a previous observation, our data also supported that HFD-mediated oxidative stress caused hepatocyte damage in rats which was revealed as increased ALT and ALP activities in HFD-fed rats [32]. Metformin treatment diminished the elevated ALT and ALP activities in plasma of HFD fed rats. Thus, our findings are in agreement with a previous report showeing that treatment of NAFLD with metformin reduces ALT activities, with a concurrent reduction of body weight [57,58].
Our data also showed that, metformin treatment reduced the inflammatory cell infiltration and decreased MPO activity in the liver of HFD fed rats. In a previous study, it was reported that metformin prevented proinflammatory cells infiltration in the liver [21]. Our findings were also supported by other studies which showed that metformin has anti-inflammatory activity and prevents hepatic steatosis in HFD fed animals [20] and in streptozotocin-induced type 2 diabetic rats [59]. HFD induced inflammation and oxidative stress may also initiate the development of fibrosis and deposition of extracellular matrix proteins in the liver of experimental animals [9,60]. Local fibroblast and hepatic stellate cells (HSCs) are stimulated by oxidative stress to release extracellular matrix proteins in the liver [60]. This investigation also showed that the HFD associated fibrogenic responses in liver and collagen deposition were attenuated by metformin treatment. Metformin treatment also ameliorated inflammation and oxidative stress conditions, which may contribute to reduce the hepatic fibrosis in HFD-fed rats. Similar effects were also observed in a previous study in which metformin prevented CCl4-induced hepatic fibrosis in rats [61]. Additionally, it was also found that metformin prevented ROS production and suppressed hepatic stellate cells proliferation and fibrosis in liver [62,63].
In summary, our study suggests that consumption of HFD may cause generalized hyperlipidemia, glucose intolerance, oxidative stress, and inflammatory changes in the liver which mimicks the pathological features commonly observed in NAFLD. Metformin, through its antioxidant, and anti-inflammatory actions, significantly prevented HFD-induced dyslipidemia, glucose intolerance, and the development of NAFLD-like symptoms in rats. Further studies should be conducted to understand the underlying molecular mechanisms by which metformin-produced multifaceted benefits in obesity and related complications.
Declarations
Funding
This research did not receive any specific grant from any funding agency in the public, commercial, or not-for-profit sectors. Ms Tahmina Yasmin received a monthly stipend from the Ministry of Science and Technology of Bangladesh in the year 2016–2017.
Conflicts of interest/Competing interests
The authors would like to declare that there are no conflict of interests regarding the publication of this paper.
Ethics approval
All experiments involving animals were conducted following protocols that were previously examined and approved by the Institutional Animal Care and Use Committee (IACUC) at the North South University, Dhaka, Bangladesh (AEC-004-2017).
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: There is no conflict of interest.
Availability of data and material
All data were generated in our lab and will be available on proper request.
Authors' contributions
MAA, SR and TY designed the research. MAA and SR supervise and trained TY, MMR, FK, KN and SL who were also involved in animal care, oral glucose tolerance test, and euthanasia and sample collection from animals. FK, TY, MDI and MMR were involved in biochemical analyses. FK, SL, MDI and KN performed histological staining. MAA, RH and TY were involved in data analysis and statistics. TY, FK, MMR performed the mRNA isolation and RT-PCR. TY, MMA, FK and RH contributed to the first draft of the manuscript. RH revised and prepared the final version of the manuscript in consultation with MAA. All authors approved the final version of the manuscript.
Acknowledgment
The research was conducted in Department of Pharmaceutical Sciences, North South University, Bangladesh. The authors gratefully acknowledge the logistic support provided by the Department of Pharmaceutical Sciences, North South University Bangladesh.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101168.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016 Jul;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Brandt A., Hernandez-Arriaga A., Kehm R., et al. Metformin attenuates the onset of non-alcoholic fatty liver disease and affects intestinal microbiota and barrier in small intestine. Sci. Rep. 2019 Apr 30;9(1):6668. doi: 10.1038/s41598-019-43228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirpich I.A., Marsano L.S., McClain C.J. Gut-liver axis, nutrition, and non-alcoholic fatty liver disease. Clin. Biochem. 2015 Sep;48(13–14):923–930. doi: 10.1016/j.clinbiochem.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkhouri N., Feldstein A.E. Noninvasive diagnosis of nonalcoholic fatty liver disease: are we there yet? Metab. Clin. Exp. 2016 Aug;65(8):1087–1095. doi: 10.1016/j.metabol.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotman Y., Sanyal A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017 Jan;66(1):180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 6.Meex R.C.R., Watt M.J. Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat. Rev. Endocrinol. 2017 Sep;13(9):509–520. doi: 10.1038/nrendo.2017.56. [DOI] [PubMed] [Google Scholar]
- 7.Mota M., Banini B.A., Cazanave S.C., et al. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2016 Aug;65(8):1049–1061. doi: 10.1016/j.metabol.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z., Tian R., She Z., et al. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2020;152:116–141. doi: 10.1016/j.freeradbiomed.2020.02.025. 2020/05/20/ [DOI] [PubMed] [Google Scholar]
- 9.Rahman MM, Alam MN, Ulla A, et al. Cardamom powder supplementation prevents obesity, improves glucose intolerance, inflammation and oxidative stress in liver of high carbohydrate high fat diet induced obese rats. Lipids in Health and Disease. 2017 08/14 02/01/received 08/02/accepted;16:151. [DOI] [PMC free article] [PubMed]
- 10.Vilar-Gomez E., Martinez-Perez Y., Calzadilla-Bertot L., et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology. 2015 Aug;149(2):367–378. doi: 10.1053/j.gastro.2015.04.005. e5; quiz e14-5. [DOI] [PubMed] [Google Scholar]
- 11.Alkhouri N., Scott A. An update on the pharmacological treatment of nonalcoholic fatty liver disease: beyond lifestyle modifications. Clinical Liver Disease. 2018;11(4):82–86. doi: 10.1002/cld.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farah S., Nguyen T., Kelsberg G., et al. Metformin for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Am. Fam. Physician. 2019 Feb 15;99(4):262–263. [PubMed] [Google Scholar]
- 13.Mantovani A., Byrne C.D., Scorletti E., et al. Efficacy and safety of anti-hyperglycaemic drugs in patients with non-alcoholic fatty liver disease with or without diabetes: an updated systematic review of randomized controlled trials. Diabetes Metab. 2020 Nov;46(6):427–441. doi: 10.1016/j.diabet.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Bridges H.R., Jones A.J., Pollak M.N., et al. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 2014 Sep 15;462(3):475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Z., Chen H., Li J., et al. Sirtuin 1-mediated cellular metabolic memory of high glucose via the LKB1/AMPK/ROS pathway and therapeutic effects of metformin. Diabetes. 2012 Jan;61(1):217–228. doi: 10.2337/db11-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saisho Y. Metformin and inflammation: its potential beyond glucose-lowering effect. Endocr. Metab. Immune Disord. - Drug Targets. 2015;15(3):196–205. doi: 10.2174/1871530315666150316124019. [DOI] [PubMed] [Google Scholar]
- 17.Diaz-Morales N., Rovira-Llopis S., Bañuls C., et al. Does metformin protect diabetic patients from oxidative stress and leukocyte-endothelium interactions? Antioxidants Redox Signal. 2017 2017/12/10;27(17):1439–1445. doi: 10.1089/ars.2017.7122. [DOI] [PubMed] [Google Scholar]
- 18.Matsui Y., Hirasawa Y., Sugiura T., et al. Metformin reduces body weight gain and improves glucose intolerance in high-fat diet-fed C57BL/6J mice. Biol. Pharm. Bull. 2010;33(6):963–970. doi: 10.1248/bpb.33.963. [DOI] [PubMed] [Google Scholar]
- 19.Tokubuchi I., Tajiri Y., Iwata S., et al. Beneficial effects of metformin on energy metabolism and visceral fat volume through a possible mechanism of fatty acid oxidation in human subjects and rats. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo S.-L., Xu H., Li H., et al. Metformin ameliorates hepatic steatosis and inflammation without altering adipose phenotype in diet-induced obesity. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahova M., Palenickova E., Dankova H., et al. Metformin prevents ischemia reperfusion-induced oxidative stress in the fatty liver by attenuation of reactive oxygen species formation. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309(2):G100–G111. doi: 10.1152/ajpgi.00329.2014. [DOI] [PubMed] [Google Scholar]
- 22.Zabielski P., Hady H.R., Chacinska M., et al. The effect of high fat diet and metformin treatment on liver lipids accumulation and their impact on insulin action. Sci. Rep. 2018 2018/05/08;8(1):7249. doi: 10.1038/s41598-018-25397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niehaus W., Jr., Samuelsson B. Formation of malonaldehyde from phospholipid arachidonate during microsomal lipid peroxidation. Eur. J. Biochem. 1968;6(1):126–130. doi: 10.1111/j.1432-1033.1968.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 24.Tracey W.R., Tse J., Carter G. Lipopolysaccharide-induced changes in plasma nitrite and nitrate concentrations in rats and mice: pharmacological evaluation of nitric oxide synthase inhibitors. J. Pharmacol. Exp. Therapeut. 1995;272(3):1011–1015. [PubMed] [Google Scholar]
- 25.Ulla A., Alam M.A., Sikder B., et al. Supplementation of Syzygium cumini seed powder prevented obesity, glucose intolerance, hyperlipidemia and oxidative stress in high carbohydrate high fat diet induced obese rats [journal article] BMC Compl. Alternative Med. 2017 June 02;17(1):289. doi: 10.1186/s12906-017-1799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiwari B.K., Kumar D., Abidi A., et al. Efficacy of composite extract from leaves and fruits of medicinal plants used in traditional diabetic therapy against oxidative stress in alloxan-induced diabetic rats. ISRN Pharmacol. 2014;2014 doi: 10.1155/2014/608590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witko-Sarsat V., Friedlander M., Capeillère-Blandin C., et al. Advanced oxidation protein products as a novel marker of oxidative stress in uremia. Kidney Int. 1996;49:1304–1313. doi: 10.1038/ki.1996.186. [DOI] [PubMed] [Google Scholar]
- 28.Khan R.A. Protective effects of Sonchus asper (L.) Hill, (Asteraceae) against CCl4-induced oxidative stress in the thyroid tissue of rats [journal article] BMC Compl. Alternative Med. 2012;12(1):181. doi: 10.1186/1472-6882-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misra H.P., Fridovich I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J. Biol. Chem. 1972;247(10):3170–3175. [PubMed] [Google Scholar]
- 30.Khan F., Syeda P.K., Nartey M.N., et al. Pretreatment of cultured preadipocytes with arachidonic acid during the differentiation phase without a cAMP-elevating agent enhances fat storage after the maturation phase. Prostag. Other Lipid Mediat. 2016 Mar;123:16–27. doi: 10.1016/j.prostaglandins.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Kleiner D.E., Brunt E.M., Van Natta M., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005 Jun;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 32.Lasker S., Rahman M.M., Parvez F., et al. High-fat diet-induced metabolic syndrome and oxidative stress in obese rats are ameliorated by yogurt supplementation. Sci. Rep. 2019 Dec 27;9(1):20026. doi: 10.1038/s41598-019-56538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serino A., Salazar G. Protective role of polyphenols against vascular inflammation, aging and cardiovascular disease. Nutrients. 2018 Dec 28;11(1) doi: 10.3390/nu11010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy A., Martinez K., Chuang C.C., et al. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J. Nutr. 2009 Jan;139(1):1–4. doi: 10.3945/jn.108.098269. [DOI] [PubMed] [Google Scholar]
- 35.McArdle M.A., Finucane O.M., Connaughton R.M., et al. Mechanisms of obesity-induced inflammation and insulin resistance: insights into the emerging role of nutritional strategies. Front. Endocrinol. 2013;4:52. doi: 10.3389/fendo.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears B., Perry M. The role of fatty acids in insulin resistance. Lipids Health Dis. 2015;14 doi: 10.1186/s12944-015-0123-1. (]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korbecki J., Bajdak-Rusinek K. The effect of palmitic acid on inflammatory response in macrophages: an overview of molecular mechanisms. Inflamm. Res. 2019 2019/11/01;68(11):915–932. doi: 10.1007/s00011-019-01273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kewalramani G., Fink L.N., Asadi F., et al. Palmitate-activated macrophages confer insulin resistance to muscle cells by a mechanism involving protein kinase C θ and ε. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Wan Q, Guan Q, et al. High-fat diet feeding impairs both the expression and activity of AMPKa in rats' skeletal muscle. Biochem. Biophys. Res. Commun.. 2006 2006/01/13/;339(2):701-707. [DOI] [PubMed]
- 40.Masarone M., Rosato V., Dallio M., et al. Vol. 2018. 2018. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. (Oxidative Medicine and Cellular Longevity). 9547613-9547613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams K.H., McLennan S.V., Twigg S.M., et al. Diabetes and nonalcoholic fatty liver disease: a pathogenic duo. Endocr. Rev. 2013;34(1):84–129. doi: 10.1210/er.2012-1009. [DOI] [PubMed] [Google Scholar]
- 42.Tikoo K., Sharma E., Amara V.R., et al. Metformin improves metabolic memory in high fat diet (HFD)-induced renal dysfunction. J. Biol. Chem. 2016 Oct 14;291(42):21848–21856. doi: 10.1074/jbc.C116.732990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solymár M., Ivic I., Pótó L., et al. Metformin induces significant reduction of body weight, total cholesterol and LDL levels in the elderly – a meta-analysis. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0207947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moseti D., Regassa A., Kim W.-K. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int. J. Mol. Sci. 2016;17(1):124. doi: 10.3390/ijms17010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madsen M.S., Siersbæk R., Boergesen M., et al. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell Biol. 2014;34(6):939–954. doi: 10.1128/MCB.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettinelli P., Videla L.A. Up-regulation of PPAR-γ mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J. Clin. Endocrinol. Metabol. 2011;96(5):1424–1430. doi: 10.1210/jc.2010-2129. [DOI] [PubMed] [Google Scholar]
- 47.Gavrilova O., Haluzik M., Matsusue K., et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 2003 Sep 5;278(36):34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 48.Morán-Salvador E., López-Parra M., García-Alonso V., et al. Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. Faseb. J. 2011;25(8):2538–2550. doi: 10.1096/fj.10-173716. [DOI] [PubMed] [Google Scholar]
- 49.Payne Victoria A., Au W.-S., Lowe Christopher E., et al. C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem. J. 2009;425(1):215–224. doi: 10.1042/BJ20091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi M., Fujii N., Narita T., et al. SREBP-1c-Dependent metabolic remodeling of white adipose tissue by caloric restriction. Int. J. Mol. Sci. 2018;19(11):3335. doi: 10.3390/ijms19113335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S, Li N, Zhu Y, et al. Partial leptin deficiency confers resistance to diet-induced obesity in mice. Molecular Metabolism. 2020 2020/07/01/;37:100995. [DOI] [PMC free article] [PubMed]
- 52.Zheng J., Woo S.-L., Hu X., et al. Metformin and metabolic diseases: a focus on hepatic aspects. Front. Med. 2015;9(2):173–186. doi: 10.1007/s11684-015-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruckbauer A., Banerjee J., Fu L., et al. A combination of leucine, metformin, and sildenafil treats nonalcoholic fatty liver disease and steatohepatitis in mice. Bangladesh Liver J. 2016:9185987. doi: 10.1155/2016/9185987. 2016 2016/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meli R., Mattace Raso G., Irace C., et al. High fat diet induces liver steatosis and early dysregulation of iron metabolism in rats. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manna P., Jain S.K. Obesity, oxidative stress, adipose tissue dysfunction, and the associated health risks: causes and therapeutic strategies. Metab. Syndr. Relat. Disord. 2015;13(10):423–444. doi: 10.1089/met.2015.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Contreras-Zentella M.L., Hernández-Muñoz R. Is liver enzyme release really associated with cell necrosis induced by oxidant stress? Oxidative medicine and cellular longevity. 2016;2016 doi: 10.1155/2016/3529149. 3529149-3529149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loomba R., Lutchman G., Kleiner D.E., et al. Clinical trial: pilot study of metformin for the treatment of non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2009 Jan;29(2):172–182. doi: 10.1111/j.1365-2036.2008.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iogna Prat L., Tsochatzis E.A. The effect of antidiabetic medications on non-alcoholic fatty liver disease (NAFLD) Hormones (Basel) 2018 2018/06/01;17(2):219–229. doi: 10.1007/s42000-018-0021-9. [DOI] [PubMed] [Google Scholar]
- 59.Salman Z.K., Refaat R., Selima E., et al. The combined effect of metformin and L-cysteine on inflammation, oxidative stress and insulin resistance in streptozotocin-induced type 2 diabetes in rats. Eur. J. Pharmacol. 2013 Aug 15;714(1–3):448–455. doi: 10.1016/j.ejphar.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 60.Gandhi C.R. Oxidative stress and hepatic stellate cells: a paradoxical relationship. Trends Cell Mol. Biol. 2012;7:1–10. [PMC free article] [PubMed] [Google Scholar]
- 61.Tripathi D.M., Erice E., Lafoz E., et al. Metformin reduces hepatic resistance and portal pressure in cirrhotic rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2015 Sep 1;309(5):G301–G309. doi: 10.1152/ajpgi.00010.2015. [DOI] [PubMed] [Google Scholar]
- 62.Ko M-T, Huang H-C, Lee W-S, et al. Metformin reduces intrahepatic fibrosis and intrapulmonary shunts in biliary cirrhotic rats. J. Chin. Med. Assoc.. 2017 2017/08/01/;80(8):467-475. [DOI] [PubMed]
- 63.Nguyen G, Park SY, Le CT, et al. Metformin ameliorates activation of hepatic stellate cells and hepatic fibrosis by succinate and GPR91 inhibition. Biochem. Biophys. Res. Commun.. 2018 2018/01/22/;495(4):2649-2656. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data were generated in our lab and will be available on proper request.
Data will be made available on request.