Summary
Maintaining proteostasis is important for animal development. How proteostasis influences spermatogenesis that generates male gametes, spermatozoa, is not clear. We show that testis-specific paralog of ribosomal large subunit protein RPL39, RPL39L, is required for mouse spermatogenesis. Deletion of Rpl39l in mouse caused reduced proliferation of spermatogonial stem cells, malformed sperm mitochondria and flagella, leading to sub-fertility in males. Biochemical analyses revealed that lack of RPL39L deteriorated protein synthesis and protein quality control in spermatogenic cells, partly due to reduced biogenesis of ribosomal subunits and ribosome homeostasis. RPL39/RPL39L is likely assembled into ribosomes via H/ACA domain containing NOP10 complex early in ribosome biogenesis pathway. Furthermore, Rpl39l null mice exhibited compromised regenerative spermatogenesis after chemical insult and early degenerative spermatogenesis in aging mice. These data demonstrate that maintaining proteostasis is important for spermatogenesis, of which ribosome homeostasis maintained by ribosomal proteins coordinates translation machinery to the regulation of cellular growth.
Subject areas: Molecular biology, Male reproductive endocrinology, Developmental biology
Graphical abstract
Highlights
-
•
Rpl39l deletion causes reduced spermatogenesis and subfertility in male mice
-
•
SSC proliferation, mitochondria and sperm flagella compromised in Rpl39l–/– mice
-
•
Rpl39l deletion affects ribosomal LSU formation and protein quality control
-
•
Aberrant proteostasis affects spermatogenesis and regeneration
Molecular biology; Male reproductive endocrinology; Developmental biology
Introduction
Maintaining cellular proteostasis is important for animal development, tissue regeneration, and aging (Dissmeyer et al., 2019). Complex protein quality control (PQC) systems in cells facilitate the rheostat of protein synthesis and degradation (Jayaraj et al., 2020; Sherman and Qian, 2013; Wolff et al., 2014), including molecular chaperone networks, ER associated degradation (ERAD), mitochondrial protein quality control (mtPQC), proteasomes, and autophagy (Galluzzi et al., 2017; Hwang and Qi, 2018; Morimoto, 2008; Ravanelli et al., 2020; Rousseau and Bertolotti, 2016). Since protein synthesis consumes more than 70% of cellular energy, changes of proteostasis often reflect upon mitochondrial functions and cellular metabolic states (Galluzzi et al., 2018; Lu and Guo, 2020). How cells systemically regulate proteostasis that integrates protein synthesis, PQC, and cellular metabolism remains to be fully understood.
Continuously changing cell fate during animal development requires expression of cell type-specific proteomes that accompany cellular functions. Spermatogenesis, the developmental process that generates sperm, encompasses three consecutive stages–mitosis, meiosis, and spermiogenesis (the cellular morphogenesis of haploid spermatids)—during which spermatogenic cells alter cell fate sequentially (Hermann et al., 2018; Hermo et al., 2010; Oakberg, 1956). Disruption of spermatogenesis at any stage due to either genetic or environmental interventions may render male sterility and, in humans, accounts for 50% of infertility (Krausz and Riera-Escamilla, 2018; Visser and Repping, 2010). Although it is conceivable that synthesis and quality of sperm proteome are relevant to sperm quality and male fertility, little is known about how dynamic proteostasis is achieved and maintained during spermatogenesis (D'Amours et al., 2019; Netherton et al., 2018; Schafer et al., 1995).
Proteostasis can be regulated by the synthesis of ribosomal proteins (RPs) and assembly of ribosomes, the core protein synthesis machinery made of 79–80 RPs and 4 RNA species in eukaryotes (Albert et al., 2019; Emmott et al., 2019). Biogenesis of ribosomes occurs in multiple steps, ranging from co-transcriptional assembly of pre-ribosomal complexes in the nucleolus to the maturation of small and large subunits (SSUs and LSUs) in cytoplasm (Klinge and Woolford, 2019). In response to changes of growth conditions and metabolism, cells regulate ribosome homeostasis through intracellular signaling and hundreds of regulatory factors (Badertscher et al., 2015; de la Cruz et al., 2018; Farley-Barnes et al., 2018; Pena et al., 2017). However, RP deficiencies caused by genetic mutations often generate cell or tissue-specific phenotypes, such as Diamond-Blackfan Anemia (DBA), suggesting more diversified RP functions than translation (Bhavsar et al., 2010; Kondrashov et al., 2011; Lipton and Ellis, 2010). Specific roles of RPs may render ribosomes selective to mRNAs’ translation in cell type-specific manner (the ribosome filter hypothesis) (Mauro and Edelman, 2002; Mills and Green, 2017). Varied RP components, ribosomal-associated proteins (RAPs) and post-translational modifications of RPs and rRNAs also generate heterogeneous ribosomes with varied functionality (Genuth and Barna, 2018a). In addition, accumulating evidence shows that ribosomes regulate the quality of nascent polypeptides during protein synthesis via ribosomal quality control (RQC) system so that cells would minimize the resources cost on generating misfolded proteins at the earliest stages (Joazeiro, 2019; Sitron and Brandman, 2020). How different RPs participate in ribosome homeostasis and coordinate PQC requires further investigation.
RPs are expressed heterogeneously in different cell types (Slavov et al., 2015; van de Waterbeemd et al., 2017). In this regard, several mammalian RP paralogs are expressed in testis-specific fashion, including Rps4y2 (paralog to Y-linked Rps4y1), Rps4l, Rpl10l and Rpl39l (paralogs to X-linked Rps4x, Rpl10 and Rpl39, respectively) (Lopes et al., 2010; Sugihara et al., 2010, 2013; Uechi et al., 2002). These retrogenes are thought to originate from sex chromosomes due to the pressure of meiotic sex chromosome inactivation (MSCI) (Wang, 2004). However, we previously found that the expression of Rpl39l is highly enriched in spermatogonial stem cells (SSCs) (Yang et al., 2013), in line with recent single cell RNA sequencing analyses showing the expression of Rpl39l in undifferentiated SSCs (Green et al., 2018; Tan et al., 2020). Although expression of Rpl39l was also found in embryonic stem cells and several cancerous cells (Nadano et al., 2002; Wong et al., 2014), its roles during spermatogenesis remains to be determined. Several studies in yeast and cancerous cells have suggested that RPL39 potentially interacts with nascent polypeptide chain, Ubc, and RNA editor (ADAR) (Dave et al., 2014; Zhang et al., 2013). Deletion of Rpl39 in yeast caused increased translational error, decreased nitric oxide synthase (NOS) expression and impeded cell proliferation (Dave et al., 2014; Dresios et al., 2000), suggesting that RPL39/RPL39L may regulate cellular growth via influences on protein synthesis.
In the present study, we first validated the dominant expression of Rpl39l over Rpl39 in male germ cells. Using a gene knockout mouse model, it was found that RPL39L plays essential roles during mouse spermatogenesis. Lack of RPL39L caused reduction of protein synthesis and aberrant PQC, partly due to the deficiency in biogenesis of large ribosomal subunits. Comparing to the wild type, Rpl39l null mice exhibited impeded spermatogenesis recovery following busulfan treatment. These results demonstrate the importance of a single RP in maintaining ribosome and protein homeostasis during mouse spermatogenesis and may provide novel targets to ameliorate degenerative and aging spermatogenesis.
Results
Deletion of Rpl39l causes sub-fertility in mouse
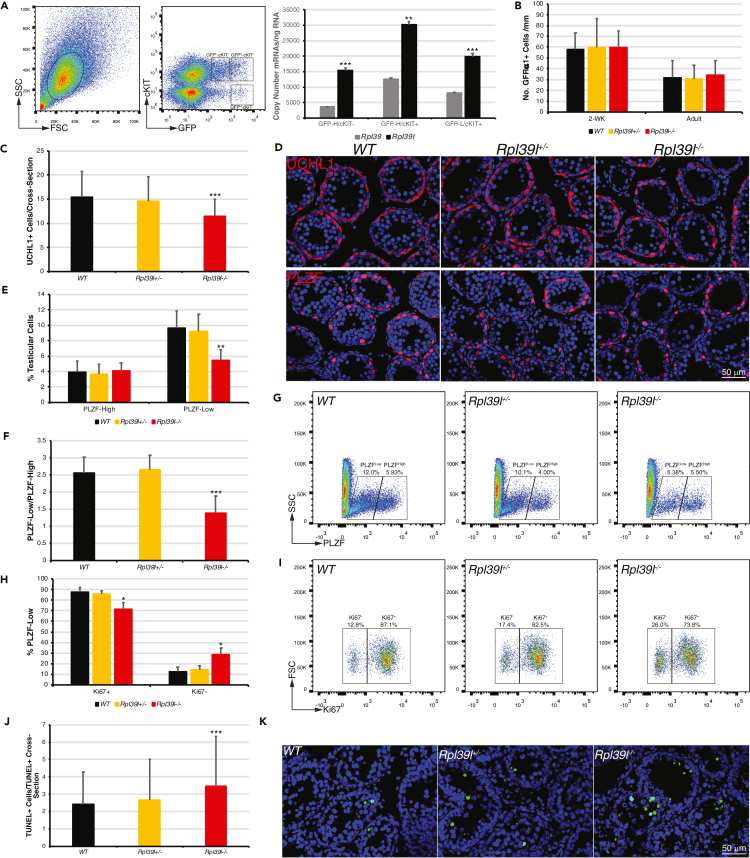
It was suggested that Rpl39l is exclusively expressed in testis, whereas Rpl39 is expressed ubiquitously (Uechi et al., 2002) (Figure S1A). Using quantitative RT-PCR, we found that in mouse testes the expression of Rpl39l was already detectable around 3-dpp (days postpartum) and exhibited increasing expression during post-natal mouse development, reciprocal to that of Rpl39 (Figure S1B). To identify whether Rpl39l is expressed in SSCs, self-renewing (OCT4+) and differentiating (cKIT+) SSCs were isolated by fluorescence-activated cell sorting (FACS) from OG2 mice (transgenic mouse line expressing GFP under the control of the Pou5f1 promoter) (Yoshimizu et al., 1999). Quantitative RT-PCR indicated that sub-populations of OCT4+ SSCs contain more Rpl39l mRNAs than that of Rpl39 (Figure 2A). These data suggest that expression of Rpl39l starts in SSCs, earlier than MSCI commences, and may gradually replace Rpl39 as the main component of ribosomes in differentiating spermatogenic cells.
Figure 2.
Reduced proliferation of spermatogonial stem cells (SSCs) caused by Rpl39l deletion
(A) SSCs from 2-week-old OG2 mice were sorted from intact cells (left panel, circled) using GFP expression and cKIT immunostaining (middle panel). Copy numbers of Rpl39l and Rpl39 transcripts were compared using quantitative RT-PCR (right panel). N = 3 experimental repeats, paired-sample Student’s t-test, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) GFRα1+ SSCs per unit length of seminiferous tubules in 2-week-old and adult mice. N ≥ 40 1-mm long seminiferous tubules.
(C) UCHL1+ spermatogonia per cross-section of 2-week-old mouse testes. N ≥ 183.
(D) Confocal images of testis sections of 2-week-old mice immunostained for UCHL1 (upper panels) or PLZF (lower panels). Scale bar: 50 μm.
(E) Percent of PLZFHigh and PLZFLow SSCs in total testicular cells from 2-week-old mice. N ≥ 7 testes.
(F) Ratio of PLZFLow to PLZFHigh SSCs in 2-week-old mice. N ≥ 7 testes.
(G) FACS for PLZF+ SSCs from 2-week-old mice.
(H) Percent of Ki67-labeled PLZFLow SSCs from 2-week-old mice. N ≥ 3 testes.
(I) FACS for Ki67-labeled PLZFLow SSCs from 2-week-old mice.
(J) TUNEL+ cells per cross-section containing TUNEL+ cells. N ≥ 203.
(K) Confocal images of TUNEL assays on mouse testis sections. Scale bar: 50 μm.
One-way ANOVA Tukey’s test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In order to investigate functional roles of RPL39L during mouse spermatogenesis, an Rpl39l gene null mouse line was established using CRISPR/Cas9 method (Figure S1C) (see STAR Methods). The 19-bp insertion mutation causes frameshift near the starting codon and would eliminate RPL39L protein (Figure S1D). RT-PCR of total RNAs extracted from mouse testes suggested that Rpl39l–/– testis expressed only the mutant mRNA (Figure S1E). Using a pan-RPL39/RPL39L antibody generated from full-length RPL39 (RPL39 and RPL39L are 94% identical with differences in only three amino acids) (Figure S1C), both Western blotting and immunofluorescent staining showed dramatic reduction of protein signals in Rpl39l–/– testis (Figures S1F–S1H). In adult mice, the reduction of protein signals was most evident in elongating spermatids and residue bodies, in which the expression of RPL39 would be turned off by MSCI (Figure S1H). Examination of off-targets found no off-targeting mutations in the established Rpl39l null mouse line (Table S2).
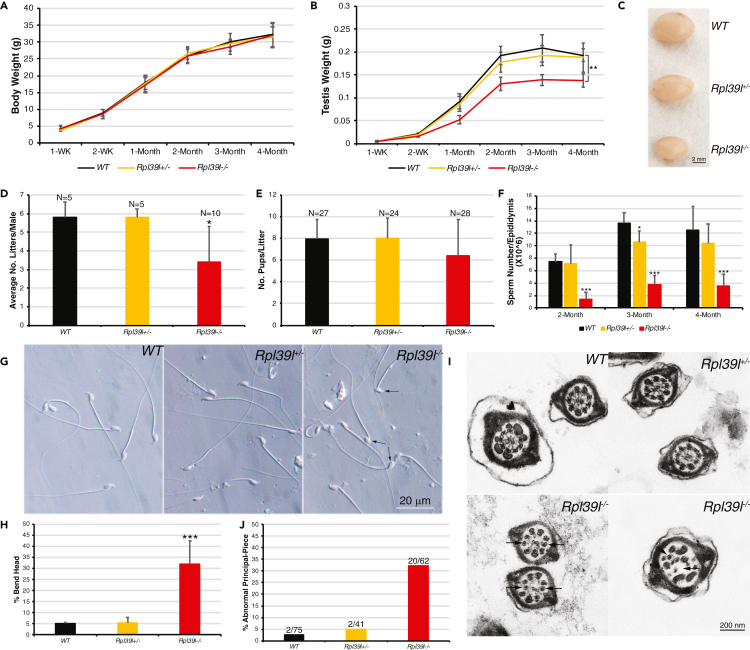
Phenotypically, the overall growth of Rpl39l–/– mice appeared comparable to those of wild type and Rpl39l+/– mice (Figure 1A); however, male gonads were reduced by about 1/3 in weight after a month following birth (Figures 1B and 1C). Male Rpl39l–/– mice sired about 50% less frequently in a recorded time period of three months (Figure 1D), although the average number of pups in each litter from Rpl39l–/– males was only slightly reduced (6.4 ± 3.3, comparing to 7.9 ± 1.8 from wild type and 8 ± 1.9 from Rpl39l+/– males) (Figure 1E). Consistently, numbers of sperm that could be retrieved from cauda epididymis, the storage site for mature sperm, were significantly less in adult Rpl39l–/– mice, compared with those of wild type and Rpl39l+/– counterparts (N ≥ 5, p < 0.001, One-way ANOVA) (Figure 1F). Computer-assisted sperm analysis (CASA) further showed that Rpl39l–/– mice produced significantly less motile sperm than wild type and Rpl39l+/– counterparts (Figure S2). Supporting this, Rpl39l null sperm were more morphologically abnormal, including frequently bend heads (31.86% ± 10.7% vs. 4.99% ± 0.64% in wild type and 5.42% ± 2.34% in Rpl39l+/–, N = 4–5 experimental repeats, p < 0.001, One-way ANOVA) (Figures 1G and 1H). Ultrastructure of sperm revealed by transmission electron microscopy (TEM) also showed that Rpl39l null sperm contained derailed outer dense fiber (ODF) and axoneme in the flagella of sperm tail (Figures 1I and 1J). Taken together, these data indicate that deletion of Rpl39l caused reduced sperm production, defected sperm morphology and motility, leading to sub-fertility in male mice.
Figure 1.
Deletion of Rpl39l causes sub-fertility in male mice
(A) Body weights of post-natal male mice. N ≥ 5 mice at each time point.
(B) Testis weights of post-natal male mice. N ≥ 5 mice at each time point.
(C) Images of testes from mice with different genotypes. Scale bar: 2 mm.
(D) Average number of litters produced by male mice with different genotypes.
(E) Average number of pups per litter.
(F) Average numbers of sperm retrieved from cauda epididymis. N ≥ 5 mice.
(G) Images of epididymal sperm. Arrows indicate bend heads of Rpl39l null sperm. Scale bar: 20 μm.
(H) Percent of sperm with bend heads in total sperm counted from images as in (G). N = 3 experimental repeats.
(I) TEM images of sperm tail cross-sections. Arrows indicate missing or dis-oriented ODF and axoneme of Rpl39l null sperm. Scale bar: 200 nm.
(J) Percent of abnormal principal-piece cross-sections. Numbers of cross-sections containing malformed ODF and axoneme were counted from TEM images as shown in (I).
One-way ANOVA Tukey’s test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Reduced proliferation of spermatogonial stem cells caused by Rpl39l deletion
To find out when the reduced spermatogenesis occurred, we first examined populations of spermatogenic cells using testes from mice at various ages. The precise timing of mouse spermatogenesis provides that spermatogenic cells at different stages appear gradually from basal lamina to the lumen of seminiferous tubules following the birth of an animal. For example, it takes approximately 11 days for SSCs to enter meiosis and 10 more days to complete it. Further 14 days are required for haploid spermatids to undergo cellular morphogenesis in order to generate mature sperm. Histological examination of testis sections revealed that compared with wild type and Rpl39l+/– mice, Rpl39l–/– testes contained more empty tubules at 2- to 4-week postpartum, when meiotic spermatocytes and post-meiotic spermatids should appear respectively (Figures S3A–S3C) and adult Rpl39l–/– mice contained smaller seminiferous tubules and less sperm in cauda epididymis (Figures S3D and S3E). Using a flow cytometry method, it was found that while no gross differences in spermatogenic cells were seen at 1 week, post-meiotic spermatids (1N) were significantly reduced in Rpl39l–/– mice at 4 weeks during the first wave of spermatogenesis (Figures S3A–S3C, right panels, Figures S3F and S3G). The reduced spermatogenesis could cause higher percentages of diploid cells at 2–4 weeks in Rpl39l–/– mice. Supporting this notion, the number of SYCP3+ spermatocytes was less at 2 weeks (Figure S4A) and reduced in adult Rpl39l–/– mice (Figures S4B and S4C). However, no apparent changes in the proportion of spermatocytes at meiosis I and II stages (Figures S4D–S4F) or during the prophase of meiosis I were found (Figures S4G and S4H), suggesting that meiotic procession was not affected. Collectively, these data suggest that Rpl39l deletion caused reduced spermatogenesis before meiosis starts, however, Rpl39l–/– spermatocytes could still complete meiosis.
The expression of Rpl39l in proliferating SSCs suggests potential effects of Rpl39l deletion on SSCs. Sub-populations of SSCs were therefore analyzed using flow cytometry and confocal microscopy following immunostaining of SSCs with various markers. When compared to the wild type, GDNF receptor α1-expressing (GFRα1+) SSCs (the earliest self-renewing SSCs known) were not changed (Figures 2B and S5A), whereas total spermatogonia (Ubiquitin carboxy-terminal hydrolase L1-expressing, UCHL1+) were reduced in Rpl39l–/– mice (Figures 2C and 2D, upper panel). SSCs express PLZF when they enter proliferative stage and gradually reduce its expression when SSCs develop towards cKIT+ differentiation stage (Hobbs et al., 2010; Yoshinaga et al., 1991) (Figure S5E). Analyses of SSCs expressing PLZF showed that PLZFHigh SSCs appeared similar, whereas PLZFLow SSCs were reduced in Rpl39l–/– mice comparing to those of wild type (Figure 2D lower panel, 2E-G and S5B), suggesting that proliferative SSCs were decreased when entering differentiation stage. In the same vein, PLZFHigh SSCs were not changed, regardless of cKIT expression (Figures S5C and S5D). Using Ki67 immunolabeling, it was found that differentiating PLZFLow SSCs contained decreased Ki67+ and increased Ki67– cells in Rpl39l–/– mice, suggesting decreased proliferation of PLZFLow SSCs (Figures 2H and 2I). Furthermore, TUNEL assay suggested that apoptotic cells were slightly more in Rpl39l–/– testis than those of wild type and Rpl39+/− counterparts (Figures 2J, 2K, and S5F). No apparent changes were observed in Sertoli cells, the main somatic cells within seminiferous tubules, suggesting that Rpl39l deletion affected mainly germ cells (Figures S5G and S5H). Collectively, these data suggest that deletion of Rpl39l caused decrease of PLZFLowcKIT+ differentiating SSCs, leading to reduced spermatogenesis.
Rpl39l deletion compromised structure and functions of sperm mitochondria
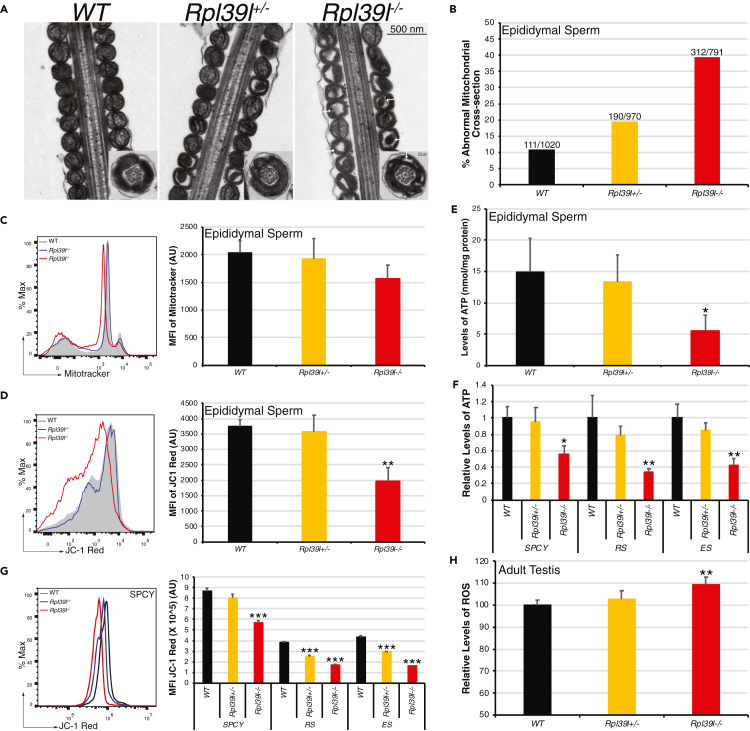
Sperm motility is powered by mitochondria located in the mid-piece of sperm tail and cytoskeletal networks of flagella, of which malformation often causes defected sperm motility. Closer examination of TEM images of sperm showed that other than disorganized ODF and axoneme, mitochondria of Rpl39l null sperm contained hollow matrix and electron dense cristae aggregates, indicating malformed mitochondria sheath (Figures 3A and 3B). To examine whether mitochondrial functions were affected, cauda epididymal sperm and spermatogenic cells were isolated and analyzed using mitochondrial indicators. The results showed that although intensities of MitoTracker Red staining were not significantly altered (Figure 3C), marked reductions were found in mitochondrial membrane potential (JC-1 staining) (Figure 3D) and cellular ATP content (5.56 ± 2.42 nmol/mg protein in Rpl39l–/– sperm, compared with 14.88 ± 5.39 nmol/mg protein in wild type and 13.34 ± 4.23 nmol/mg protein in Rpl39l+/– sperm, respectively, N = 5 experimental repeats, one-way ANOVA, p < 0.05) (Figure 3E), suggesting reduced mitochondrial functions in epididymal sperm. It was further found that mitochondrial functions has already been disrupted during earlier stages of spermatogenesis, as cellular ATP in Rpl39l null spermatocytes, round and elongating spermatids were decreased to 55.33% ± 11.24%, 33.67% ± 4.93% and 42% ± 8.89% of those of wild type cells, respectively (N = 3 experimental repeats, p < 0.05, one-way ANOVA) (Figure 3F), along with reduced mitochondrial membrane potential as revealed by JC-1 staining (Figure 3G). Consistently, testicular cells from Rpl39l–/– mice contained about 10% higher levels of ROS (reactive oxygen species) than those from Rpl39l+/– and wild type mice (N = 4 experimental repeats, p < 0.01, one-way ANOVA) (Figure 3H). Taken together, the above data indicate that the absence of RPL39L has detrimental effect on sperm mitochondrial structure and functions, which contributes to the defective sperm motility.
Figure 3.
Rpl39l deletion compromises structure and functions of sperm mitochondria
(A) TEM images of mid-piece longitudinal sections of sperm tail. Arrows indicate mitochondria with hollowed matrix and cristae condensates. Insets: cross-sections of sperm mitochondria. Scale bar: 500 nm.
(B) Percent of abnormal mitochondrial cross-sections in the mid-piece of sperm tail. Numbers indicate cross-sections containing disorganized matrices in total mitochondrial cross-sections counted from TEM images as shown in (A).
(C) Flow cytometry (left) and the average intensity of MitoTracker Red signals (right) from stained epididymal sperm. N ≥ 3 testes.
(D) Flow cytometry (left) and the average intensity of JC-1 Red signals (right) in epididymal sperm stained with JC-1. N ≥ 3 testes.
(E) Cellular ATP of epididymal sperm. N = 5 cauda epididymides.
(F) Relative contents of cellular ATP in spermatogenic cells separated by BSA gravity sedimentation, using wild type cells as the control. N = 3 mice.
(G) Flow cytometry (left, only spermatocytes shown) and the average intensity of JC-1 (right) in spermatogenic cells. N = 3 testes. In (F) and (G), SPCY: spermatocytes, RS: round spermatids, ES: elongating spermatids. In (C), (D) and (G), MFI: mean fluorescence intensity, AU: arbitrary unit.
(H) Relative levels of ROS in adult testes. N ≥ 4 mice.
One-way ANOVA Tukey's test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Decreased protein synthesis in Rpl39l null testis
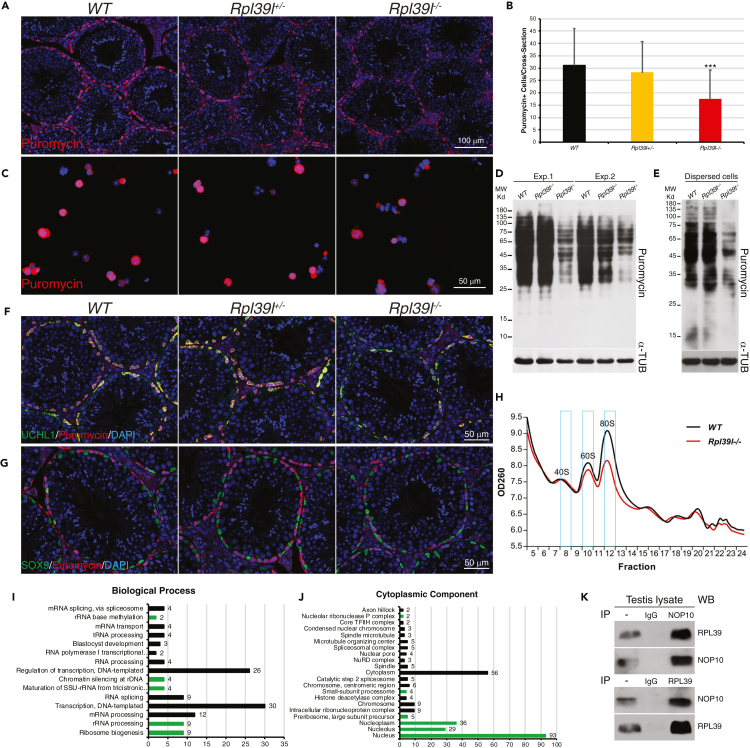
RPL39L is one of the 46 ribosomal LSU proteins whose primary function is to form ribosomes and translate mRNAs. The absence of RPL39L could affect protein synthesis in spermatogenic cells, leading to aberrant production of proteins that are important for mitochondrial functions and spermatogenesis. To test this, mouse testes were intraperitoneally injected with puromycin, an amino acid analog that occupies the A-sites of ribosomes and prevents translation elongation, which can be used to label nascent polypeptides. After 1.5 h, testes were harvested and sectioned for immunostaining with anti-puromycin antibody. As shown in Figure 4A, immunofluorescent signals of puromycin representing nascent proteins were mainly distributed in cells located at the outer rim of seminiferous tubules where SSCs reside. The number of puromycin-labeled cells was significantly lower in Rpl39l–/– testes than those of wild type and Rpl39+/− testes (Figure 4B). It was noted that cells in the inner layers of seminiferous tubules were largely devoid of puromycin labeling under this condition. One possibility is that the blood-testis barrier formed via cell-cell junctions among Sertoli cells segregates meiotic spermatocytes and post-meiotic spermatids from outside environment. This could prevent puromycin from entering and labeling spermatocytes and spermatids. To test this, dispersed spermatogenic cells were cultured in vitro transiently and labeled with puromycin for 1 h, a condition that allowed incorporation of puromycin into spermatocytes and post-meiotic spermatids. Similar to in vivo labeling, cells from Rpl39l–/– mice contained less puromycin signals than those from wild type and Rpl39l+/– mice, suggesting reduced protein synthesis in Rpl39l null cells (Figure 4C). The reduction of protein synthesis in testes and cultured spermatogenic cells was further confirmed by Western blotting of the same samples (Figures 4D and 4E). Additionally, immunostaining showed that puromycin signals were mainly localized in UCHL1+ spermatogonia (Figure 4F) but not SOX9+ Sertoli cells (Figure 4G) following in vivo labeling, suggesting that nascent protein synthesis was mostly detected in germ cells during the short period of labeling time. Together, these data indicate a global decrease of protein synthesis in germ cells in the absence of RPL39L.
Figure 4.
Decreased protein synthesis and ribosome large subunits in the absence of RPL39L
(A) Confocal images of testis cross-sections immunostained with anti-puromycin following in vivo labeling. Scale bar: 100 μm.
(B) Puromycin+ cells per cross-section as shown in (A) N ≥ 41, One-way ANOVA Tukey’s test, ∗∗∗p < 0.001.
(C) Confocal images of dispersed testicular cells immunostained with anti-puromycin following in vitro labeling. Scale bar: 50 μm.
(D) Western blotting of testis lysates following in vivo puromycin labeling. Shown are two experimental repeats.
(E) Western blotting of testicular cell lysates following in vitro puromycin labeling. α-TUBULIN was used as loading control in (D) and (E).
(F and G) Confocal images of testis sections immunostained with anti-puromycin and anti-UCHL1 (F) or anti-SOX9 (G) following in vivo puromycin labeling. Cell nuclei were stained with DAPI. Scale bars: 50 μm.
(H) Sucrose gradient sedimentation of testis lysates. Peaks of 40S SSU, 60S LSU and 80S monosomes are indicated with blue boxes.
(I and J) GO analyses of proteins pulled-down by both GST-RPL39 and GST-RPL39L. GO groups relevant to ribosome biogenesis are indicated in green.
(K) Co-immunoprecipitation of testis lysates with pan-anti-RPL39 or anti-NOP10. NOP10 is common to both GST-RPL39 and GST-RPL39L pull-downs.
Biogenesis of 60S ribosome LSUs affected by Rpl39l deletion
Protein translation efficiency is regulated by multiple factors, including the abundance of translation regulators, mRNA metabolism and ribosome biogenesis, which coordinate with cellular growth and proliferation (Tahmasebi et al., 2019). To find whether the lack of RPL39L caused ribosome deficiency, testis lysates were subjected to sucrose gradient sedimentation. Ultracentrifugation of testis lysates on a 10–50% sucrose gradient resolved 40S SSUs, 60S LSUs, 80S monosomes and polysomes. Compared with the wild type, Rpl39l–/– testes contained similar level of 40S SSUs; however, levels of 60S LSUs and 80S monosomes were reduced (Figure 4H), suggesting that deletion of RPL39L could affect biogenesis or the stability of 60S LSUs and 80S monosomes. Ribosomal subunits are first assembled co-transcriptionally in cell nucleoli before being exported into cytoplasm (Klinge and Woolford, 2019). In line with this, immunostaining showed that RPL39/RPL39L were localized in cell nuclei of early spermatocytes and cytoplasm of SSCs, pachytene spermatocytes (containing sex bodies labeled with γH2AX) and post-meiotic spermatids (Figures S1G, S1H, and S6A). It is not clear how RPL39/RPL39L participate in the ribosome biogenesis. To gain insights into this question, GST-RPL39 and GST-RPL39L expressed in E. coli were used to pull down testis lysates, of which components of protein complexes were then identified by protein mass spectrometry (Figures S6B and S6C). A total of 190 and 164 proteins were identified from RPL39 and RPL39L complexes, respectively (Table S3, see Star Methods). Of these, 141 (66.2%) proteins overlapped between the two groups, suggesting highly conserved functions or pathways involving RPL39 and RPL39L (Figure S6D). GO analysis suggested that overlapped proteins preferentially function in RNA binding, ribosome biogenesis, and rRNA processing (Figures 4I, 4J and S6E). Among them, 9 proteins were ribosome assembly factors (AFs) that have been known to regulate ribosome biogenesis at various steps. Using co-immunoprecipitation of testis lysate, we further verified the potential complexes that formed endogenously between RPL39/RPL39L and NOP10, a known H/ACA domain-containing AF that is also involved in rRNA pseudouridylation and telomere maintenance (Egan and Collins, 2012; Rashid et al., 2006) (Figure 4K). Taken together, these results suggested that RPL39/RPL39L is assembled into ribosomes early in nucleus, partly via NOP10-containing complex.
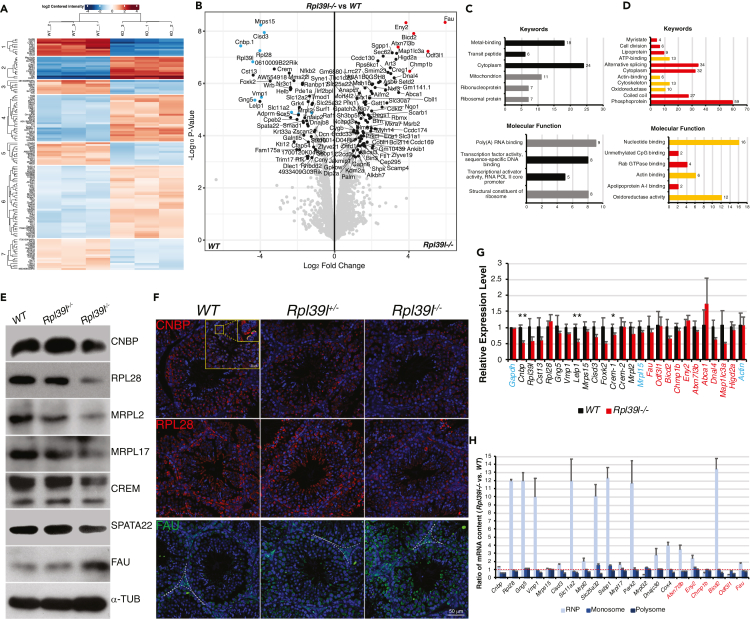
Differentially expressed spermatogenic proteome caused by Rpl39l deletion
Ribosomal proteins facilitate translation of all cellular proteins. However, accumulating evidence suggests that cells respond to translational control differently via either varied mRNA translatability or heterogeneous ribosomes (Genuth and Barna, 2018b; Mauro and Edelman, 2002; Mills and Green, 2017). The malformed mitochondria and flagella in Rpl39l null sperm suggest that Rpl39l deletion preferentially affected certain groups of proteins. To test this, proteins extracted from adult mouse testes were applied to quantitative mass spectrometry. Among ∼8,640 proteins identified by LC-MS/MS, 6,352 of them were quantifiable and 194 differentially expressed proteins (DEPs) were significantly changed (cut-off of FC > 1.5, p < 0.05), of which 81 and 113 proteins were decreased or increased, respectively, in Rpl39l–/– testes compared with those of wild type (Table S4). Heatmaps of DEPs showed high consistency among three experimental repeats (Figure 5A). The small amounts of DEPs revealed by MS indicates that the absence of RPL39L indeed preferentially affects particular protein species (Figure 5B). GO analysis of DEPs revealed that significantly decreased proteins include both cytoplasmic and mitochondrial ribosomal proteins (i.e., RPL28, RPL27A, mRPL2, mRPL17, mRPL32, and mRPS15), RNA binding proteins (e.g., CNBP, cellular nucleic acid binding protein; CPEB2, cytoplasmic polyadenylation elements binding protein 2) and mitochondrial proteins (e.g., CISD3, CDGSH Iron-Sulfur Domain-Containing Protein 3; PARK2, Parkin RBR E3 Ubiquitin Protein Ligase; SURF1, SURF1 Cytochrome C Oxidase Assembly Factor) (Figures 5C and Table S4), consistent with the phenotypes of reduced protein synthesis and compromised mitochondrial functions in Rpl39l–/– germ cells.
Figure 5.
Differentially expressed spermatogenic proteome caused by Rpl39l deletion
(A) Heatmap of differentially expressed proteins (DEPs) between wild type and Rpl39l–/– testes. Shown are three experimental repeats. DEPs are clustered into seven clusters according to their similarities in changes. Red: upregulated DEPs, Blue: downregulated DEPs.
(B) Volcano plot of DEPs between wild type and Rpl39l–/– testes. Blue and red indicate top DEPs down- or up-regulated in Rpl39l–/– testes, respectively.
(C and D) GO analyses of DEPs. Down-regulated GO groups relevant to ribosomes and mitochondria are shown in grey (C). Up-regulated GO groups relevant to metabolism and cytoskeleton are shown in yellow (D).
(E) Western blotting of testis lysates for selected top DEPs. α-TUBULIN was used as loading control.
(F) Immunostaining of testis sections. Insets show ER and acrosomal localizations of CNBP in spermatocytes and elongating spermatids from a different cross-section. Green: PNA. Blue: DAPI. Scale bar: 50 μm. Scale bar in inset: 20 μm.
(G) Quantitative RT-PCR of genes encoding DEPs. Gapdh, Actin and Mrpl15 were used as controls. N = 3 testes, paired-sample Student’s t-test, ∗p < 0.05, ∗∗p < 0.01.
(H) Changes of mRNA distributions. RNAs were extracted from sucrose gradient fractions representing RNPs, monosomes and polysomes and relative mRNAs in each fraction were compared using quantitative RT-PCR. Most mRNAs were found to accumulate in RNPs in Rpl39l–/– testes as indicated by the ratio above 1 (red dotted line). Genes encoding decreased (black) or increased (red) DEPs are indicated in G and (H)
Interestingly, a set of proteins were also increased in Rpl39l–/– testes, including oxidoreductase, ATP binding proteins, and cytoskeletal binding proteins (Figure 5D). Significantly increased proteins include FAU (Finkel-Biskis-Reilly murine sarcoma virus, ubiquitously expressed), ENY2 (Enhancer of Yellow 2 Transcription Factor Homolog), ATXN7L3B (Ataxin 7 Like 3B), ODF3L1 (Outer Dense Fiber of Sperm Tails 3 Like 1) and BICD2 (Drosophila bicaudal-D homolog) (Figure 5B). FAU belongs to the UBA family, of which ubiquitin/ubiquitin-like domains are fused with ribosome proteins at C-termini (RPS30 in the case of FAU) (Casteels et al., 1995; Kas et al., 1992); ENY2 and ATXB7L3B are components of SAGA (Spt-Ada-Gcn5 acetyltransferase) complexes facilitating nuclear acetylation and de-ubiquitination of histones H2A/B during ER stress responses (Komander, 2009; Li et al., 2016; Schram et al., 2013); and ODF3L1 and BICD2 are homologs of ODF3 and DrosophilaBicaudal D, respectively, which adapt cargo complexes to microtubules and maintain structural integrity of cytoskeleton (Petersen et al., 2002; Splinter et al., 2010). Although their functional roles during spermatogenesis remain to be determined, elevation of these proteins hinted at PQC responses induced by the lack of RPL39L.
The expression of representative DEPs was further verified using Western blotting and immunostaining (Figures 5E and 5F). Interestingly, immunostaining showed that these proteins presented heterogeneous sub-cellular localizations in spermatogenic cells at various developmental stages. For example, CNBP, a cytoplasmic RNA binding protein known for its role in regulating G-quadruplex of nucleotides and translation of 5′-TOP (5′-terminal oligopyrimidine) mRNAs (Bezzi et al., 2021; Calcaterra et al., 2010) was mainly localized in cytoplasm of SSCs and spermatocytes, probably associated with the ER, however, became concentrated in acrosomal vesicles of elongating spermatids (Figure 5F, insets). RPL28 and FAU were mainly down- or up-regulated in the cytoplasm of spermatogenic cells, respectively. The multitude of changes of protein translation regulators suggest that RPL39L is required for translation that occurs in various sub-cellular compartments in spermatogenic cells at different developmental stages. Using quantitative RT-PCR, mRNAs encoding the top DEPs were found comparable in Rpl39l–/– and wild type testes, suggesting that changes of DEP expression mainly occurred at post-transcriptional or translational levels (Figure 5G). Supporting this, it was found that the majority of mRNAs of these DEPs was accumulated in RNP fractions of sucrose gradient in Rpl39l–/– mice, suggesting their impeded translation activities (Figure 5H).
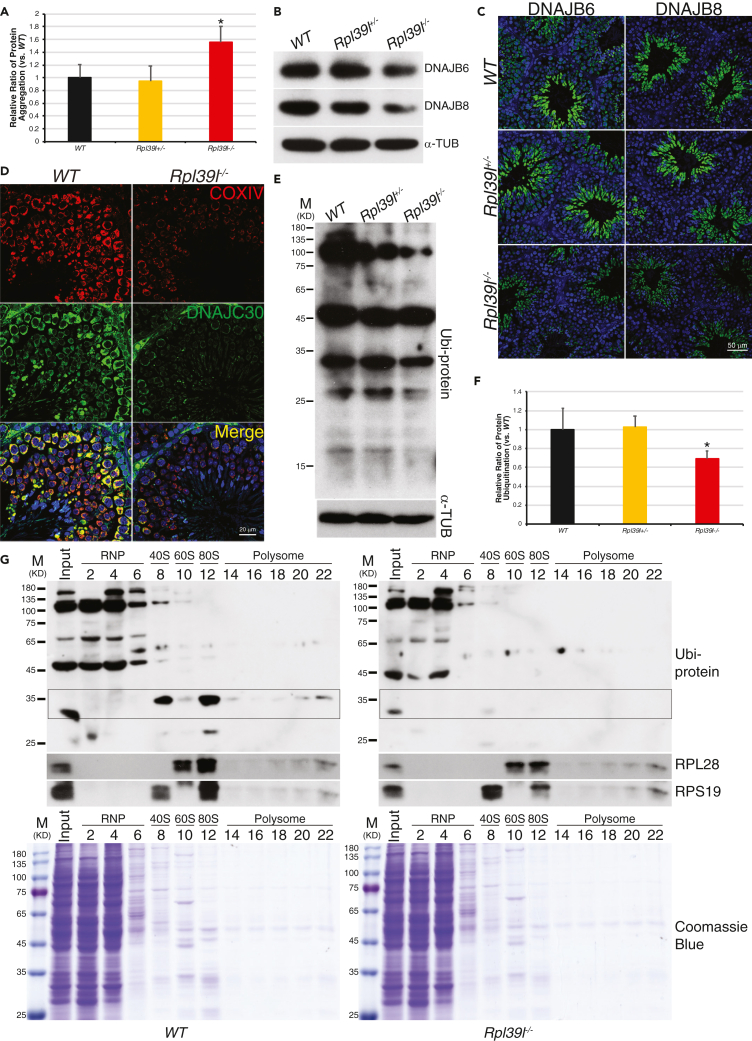
Aberrant protein quality control in spermatogenic cells lacking RPL39L
Aberrant protein translation induces PQCs in order to maintain cellular proteostasis (Wolff et al., 2014). Whether RPL39L participates in the control of protein quality during mouse spermatogenesis is not clear. To address this question, we first examined whether PQC responses would be induced in spermatogenic cells in the absence of RPL39L. To this end, it was found that insoluble protein aggregates increased by 60% in Rpl39l–/– testis, comparing to that of wild type (N = 3, p < 0.05, One-way ANOVA) (Figure 6A). In the same vein, two of the DNAJ molecular chaperons DNAJB6 and DNAJB8 were decreased as revealed by MS (Table S4). DNAJ family proteins have been shown to interact with HSP70s and facilitate the prevention of unfolded protein aggregates (Jayaraj et al., 2020). Western blotting and immunofluorescent staining of testes samples showed that both DNAJB6 and DNAJB8 were indeed decreased in Rpl39l–/– testis comparing to those of wild type (Figures 6B and 6C), in which DNAJB6 was not only expressed in post-meiotic spermatids like DNAJB8 but also presented in pre-meiotic cells (Figure 6C), suggesting that spermatogenic cells contain and utilize differential unfolded protein response machineries. Mitochondria contain over 1,500 proteins, the majority of which are encoded by nuclear genome and synthesized in cytoplasm before being transported into mitochondria (Fang and Lee, 2009). The defected mitochondrial structure and functions, including numerous reduced mitochondrial ribosomal proteins in Rpl39l–/– testis, prompted us to exam whether mitochondrial proteostasis was affected by Rpl39l deletion. For this, we applied crude mitochondria fractions from testicular lysates to protein mass spectrometry (Figure S6F). Quantitative mass spectrometry revealed 134 DEPs (72 decreased and 62 increased) in mitochondria fractions from Rpl39l–/– mice comparing to that of wild type (FC > 2, p < 0.05) (Figures S6G, S6H and Table S5). Among them, mitochondria-specific chaperon DNAJC30 was markedly reduced (Figure 6D). These results suggested that both cytoplasmic and mitochondrial PQCs were altered in the absence of RPL39L.
Figure 6.
Aberrant protein quality control in spermatogenic cells lacking RPL39L
(A) Increased protein aggregations in Rpl39l–/– testis. N = 3 testes, One-way ANOVA Tukey’s test, ∗p < 0.05.
(B) Western blotting of testis lysates. α-TUBULIN was used as loading control.
(C) Immunostaining of testis sections. Scale bar: 50 μm.
(D) Immunostaining of testis sections. DNAJC30 co-localizes with mitochondrial COXIV and both were decreased in Rpl39l–/– testes. Scale bar: 50 μm.
(E) Western blotting of testis lysates. α-TUBULIN was used as loading control.
(F) Quantitation of protein ubiquitination measured from Western blotting as shown in (E). N = 5 experimental repeats, One-way ANOVA Tukey’s test, ∗p < 0.05.
(G) Western blotting of sucrose gradient fractions. Decreased protein ubiquitination was found in ribosome subunits, monosomes and polysomes, especially in boxed area. Coomassie blue staining of the same samples are shown below.
Deleterious un-folded or mis-folded proteins are often ubiquitinated and degraded through proteasome systems in order to maintain proteostasis. In addition, protein ubiquitination also facilitates the maintenance of nascent protein quality and ribosomal proteins under normal physiological conditions (Balchin et al., 2016; Komander and Rape, 2012). In order to distinguish these possibilities, we examined levels of protein ubiquitination in testes. Western blotting of testis lysates with anti-ubiquitin antibody showed an overall reduction of protein ubiquitination in Rpl39l–/– testis comparing to that of wild type and Rpl39l+/– counterparts (Figures 6E and 6F), suggesting that protein ubiquitination was not induced by elevated protein aggregation in Rpl39l–/– testis. This could be caused by the reduced levels of protein synthesis or ribosomal biogenesis in the absence of RPL39L, prompting the reduction of co-translational protein ubiquitination. In support of this notion, Western blotting of sucrose gradient fractionation of testes lysates showed that protein ubiquitination was indeed decreased in fractions containing ribosomal subunits, as well as 80S monosomes and polysomes (Figure 6G). Collectively, the increased protein aggregation and decreased protein ubiquitination indicate aberrant PQCs in the absence of RPL39L.
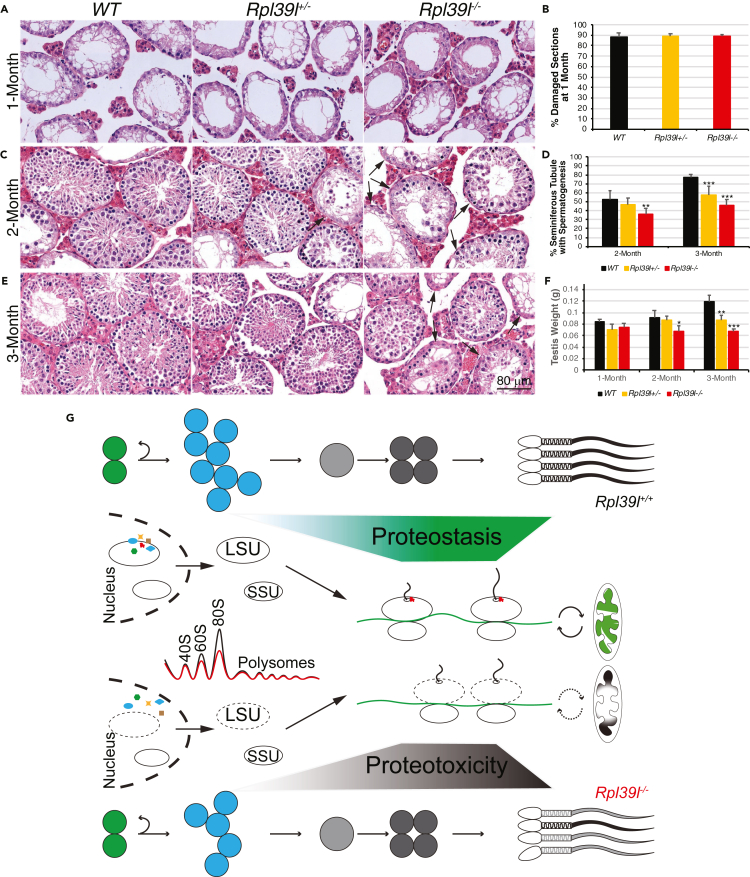
Reduced regenerative spermatogenesis in Rpl39l null testes
Loss of proteostasis causes cell degeneration and aging. The global proteotoxic stress caused by Rpl39l deletion would reduce functionality of SSCs and differentiating spermatogenic cells. To test this, we applied chemotoxic insult on testes using busulfan. Busulfan treatment in testes causes damage to most mitotic cells and degeneration of spermatogenesis. However, surviving SSCs could replenish testes with newly established stem cell population and regenerate spermatogenesis in time (Bucci and Meistrich, 1987; Kitadate et al., 2019). Following busulfan injection, progress of testis regeneration was compared in mice with different genotypes. As shown in Figures 7A and 7B, most of the spermatogenic cells in testes were eliminated similarly after 1 month following a single dose of busulfan injection (20 mg/kg). However, both wild type and Rpl39l+/– mice regained spermatogenesis within 2 months, whereas spermatogenesis in Rpl39l–/– mice only partially recovered (Figures 7C and 7D). The deleterious effect of Rpl39l deletion on regenerative spermatogenesis was more evident at 3 months following tissue damage when wild type testes completely recovered, testes from Rpl39l–/– mice still contained substantial fractions of damaged tubules (Figures 7D–7F). Furthermore, testes from 8-month-old Rpl39l–/– mice contained more degenerative seminiferous tubules, compared with their wild type and Rpl39l+/– counterparts (Figure S7). These data suggest that spermatogenic cells are less effective in regenerating spermatogenesis and more prone to degenerate with age in the absence of RPL39L.
Figure 7.
Reduced regenerative spermatogenesis in Rpl39l null testes
(A) Hematoxylin/Eosin (H/E) staining of testis sections at 1-month post-busulfan injection.
(B) Percent of cross-sections containing mainly empty lumens at 1-month post-busulfan injection. N = 6 testes.
(C) H/E staining of testis sections at 2 months after busulfan injection. Arrows indicate damaged seminiferous tubules.
(D) Percent of seminiferous tubules containing developing spermatogenic cells after 2 and 3 months after busulfan injection. N = 6 testes.
(E) H/E staining of testis sections at 3 months after busulfan injection. Arrows indicate damaged seminiferous tubules.
(F) Testis weight measurements at different times after busulfan injection. N = 3 testes. One-way ANOVA Tukey’s test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
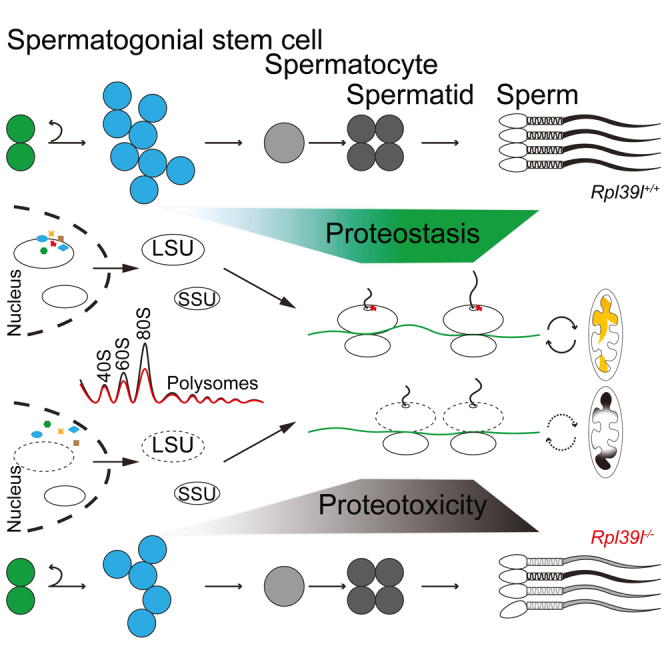
(G) Proposed model of proteostasis regulated by RPL39L maintains normal spermatogenesis, in which Rpl39l deletion may interrupt biogenesis of ribosome subunits, leading to reduced protein synthesis and aberrant protein quality control. The poor quality of proteins synthesized may introduce proteotoxicity and compromise normal spermatogenesis, including decreased SSC proliferation and abnormal spermiogenesis, thus producing sperm with poor qualities of morphology and motility.
Discussion
The present study shows that ribosomal protein RPL39L is required for mouse spermatogenesis. Deletion of Rpl39l caused subfertility in male mice, likely a cumulative effect of reduced proliferation of differentiating SSCs, compromised mitochondrial functions and morphogenesis of sperm. On the molecular level, lack of RPL39L affected biogenesis of ribosomal large subunits, causing reduced protein synthesis and aberrant PQC, during which protein ubiquitination plays an important role. Consequently, the global proteotoxicity occurring in spermatogenic cells is deleterious to spermatogenesis (Figure 7G).
Several lines of evidence indicate the ablation of RPL39L in Rpl39l–/– mice: 1) RT-PCR showed that only mutant Rpl39l mRNAs containing a frame-shift mutation were expressed in Rpl39l–/– mice (Figure S1E); 2) immunostaining using a pan-RPL39/RPL39L antibody showed elimination of protein signals in elongating spermatids and residue bodies of Rpl39l–/– testis, where the expression of Rpl39 is presumably turned off by MSCI (Figure S1G); and 3) deletion of Rpl39l caused reduction of proliferating SSCs by nearly 50% comparing to wild type mice, consistent with its expression in SSCs (Figure 2). Although it is not clear what regulates the expression of paralogous RP genes, the ubiquitous expression of Rpl36al (Uechi et al., 2002) and the expression of Rpl39l in embryonic stem cells and cancerous cells (Wong et al., 2014) suggest broader regulatory mechanisms than MSCI, such as the compensatory roles proposed for Rpl10l (Jiang et al., 2017; Wang, 2004). It will be of interest to find what regulates the expression of Rpl39 and Rpl39l, including putative transcription factors and their differential 5′-cis elements.
How ribosome homeostasis and protein synthesis are differentially influenced by individual RPs is not fully understood. Cells maintain balanced ribosome biogenesis and protein synthesis in order to meet demands of cellular growth (Emmott et al., 2019). To support this, GST pull-down assays revealed several ribosome AFs, including SSU processome factors IMP3 (U3 small nucleolar ribonucleoprotein) and EMG1 (N1-specific pseudouridine methyltransferase), suggesting that assembly of LSU integrates with SSU formation. In addition, transcriptional and epigenetic regulators of ribosomal genes were found to change in Rpl39l–/– mice, including elevated ENY2 and ATX7L3B, known to facilitate nuclear acetylation and de-ubiquitination of histones H2A/B during ER stress responses (Li et al., 2016; Schram et al., 2013). Whether changes of ribosome biogenesis regulate transcription of RP genes via feedback signals warrants further investigation.
How protein synthesis is coordinated with cellular metabolism has attracted attention in recent years (Rangaraju et al., 2019; Ruan et al., 2020). Several signaling pathways are known to modify cellular growth according to environmental cues, including mTORC, AMPK, and PKA pathways (Gonzalez et al., 2020; Herzig and Shaw, 2018; Lempiainen and Shore, 2009; Meng et al., 2018). mTORC2 responds to cellular energetic states and regulate the expression of ribosomal genes (Morita et al., 2017; Zinzalla et al., 2011). Intriguingly, translation of 5′-TOP mRNAs regulated by CNBP is also the target of mTORC signaling (Calcaterra et al., 2010). Whether RPL39L preferentially regulates the translation of mitochondrial proteins is not clear (Segev and Gerst, 2018), however, a distinct group of mitochondrial proteins were reduced in Rpl39l–/– mice, including mitochondrial RPs and metabolic enzymes, e.g., CISD3, of which mRNA contains putative 5′-TOP sequences. Understanding whether ribosome homeostasis signals cellular growth through mitochondrial translation machinery and activities will be critical to fully appreciate the functional roles of proteostasis during animal development.
Crystallographic structures of eukaryotic ribosomes indicate that RPL39 is located in the ribosomal LSU near the exit tunnel of nascent polypeptide chain (Ben-Shem et al., 2011; Deuerling et al., 2019). Presumably, RPL39L occupies the same location when assembled into ribosomal LSU, providing a way of co-translational PQC. Decreased PQC that fails to sense and clear aberrant proteins often leads to formation of misfolded protein aggregates, hallmarks of degenerative diseases and aging (Balch et al., 2008; Lopez-Otin et al., 2013). This is in line with the increased protein aggregation and decreased molecular chaperones in Rpl39l–/– mice, including DNAJB6/8 and mitochondria-specific DNAJC30 (Gillis et al., 2013; Tebbenkamp et al., 2018). It was recently shown that the DNAJ family of HSP40 co-chaperones recognize substrates in cell-type specific manner (Piette et al., 2021; Thiruvalluvan et al., 2020), reminiscent of our findings that DNAJB6 presents in pre-meiotic cells and DNAJB8 is mainly expressed in post-meiotic cells. The decreased protein ubiquitination further supports the notion of aberrant PQC in the absence of RPL39L (An and Harper, 2020; Inada, 2017; Sung et al., 2016; Takehara et al., 2021). Although largely unexplored, molecular chaperones and ubiquitin ligases have been implicated in facilitating normal spermatogenesis (Grad et al., 2010; Tokuhiro et al., 2012). Collectively, the presented study suggests that maintaining protein homeostasis underlies potential pathways to regulate spermatogenesis and improve reproductive health for humans.
Limitations of the study
Although the presented data indicate that the ribosomal large subunit protein RPL39L participates in the regulation of proteostasis that is important for the progression and maintenance of mouse spermatogenesis, little is known about how spermatogenic cells regulate the expression of paralogous RPL39 and RPL39L. It appears that the expression of Rpl39 may not compensate for the loss of Rpl39l in spermatogenic cells, a phenomenon that usually would occur for some other RP paralogs. In addition, whether RPL39 and RPL39L could functionally complement each other in spermatogenic cells if their expressions are accommodated should also be investigated in the future. In this regard, generating RPL39L specific antibodies and a germline specific Rpl39/Rpl39l duel deletion mouse model should be helpful to further elucidate the roles of these ribosomal proteins during mouse spermatogenesis. Furthermore, due to the limited numbers of endogenous SSCs that could be obtained, stage-specific roles of RPL39L were difficult to dissect. An in vitro culture system of SSCs may be applied to solve this issue.
Lack of RPL39L disrupted 60S ribosomal large subunit assembly, suggesting that ribosome homeostasis is an intrinsic part of gene expression network that regulates spermatogenesis. However, we did not apply in vitro biochemical systems to directly test how RPL39L participates in the regulation of fidelity and efficiency of protein synthesis and whether RPL39L-less ribosomes differentially synthesize proteins. The ubiquitination proteomes during ribosome biogenesis and nascent protein synthesis should be identified so that their roles during spermatogenesis could be further studied. Using quantitative mass spectrometry, numerous proteins that are affected by or interact with RPL39L were identified. Whether they elicit similar regulatory roles during spermatogenesis await to be established. Phenotypes of mitochondrial abnormality in sperm provide interesting links between cytoplasmic translation and mitochondrial functions, a phenomenon that should be further investigated. Answers to these questions should provide us more comprehensive understanding of the relationships between ribosomal control of proteostasis and spermatogenesis. In addition, it will be of great importance to establish whether RPL39/RPL39L and their associated proteins that participate in ribosome biogenesis and protein synthesis are correlated with human infertility and reproductive health.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| eFluor 780-conjugated anti-cKIT | eBioscience | Cat#47117180 |
| PE-conjugated anti-PLZF | eBioscience | Cat#12932080 |
| Rabbit anti-KI67 | Abcam | Cat#ab15580; RRID: AB_443209 |
| Goat anti-GFRα1 | R&D systems | Cat#AF560 |
| Rabbit anti-PLZF | Santa Cruz | Cat#sc-22839; RRID: AB_2304760 |
| Rabbit anti-UCHL1 | Abcam | Cat#ab108986 |
| Rabbit anti-SYCP3 | Abcam | Cat#ab15093 |
| Mouse anti-γH2AX | Millipore | Cat#05-636 |
| Rabbit anti-SOX9 | Millipore | Cat#AB5535 |
| Alexa Fluor 488-conjugated Lectin PNA | Molecular Probes | Cat#L-21409 |
| Rabbit anti-RPL39 | Proteintech | Cat#14990-1-AP; RRID: AB_2238670 |
| Mouse anti-Puromycin | Millipore | Cat#MABE343; RRID: AB_2566826 |
| Rabbit anti-CNBP | Proteintech | Cat#14717-1-AP; RRID: AB_2081548 |
| Rabbit anti-RPL28 | Proteintech | Cat#16649-1-AP; RRID: AB_2254021 |
| Rabbit anti-RPS19 | Bethyl | Cat#A304-002A; RRID: AB_2620351 |
| Rabbit anti-CREM | Santa Cruz | Cat#sc-440; RRID: AB_673599 |
| Rat anti-FUBI | R&D systems | Cat#MAB9036 |
| Rabbit anti-DNAJB6 | Proteintech | Cat#11707-1-AP; RRID: AB_2230757 |
| Rabbit anti-DNAJB8 | Proteintech | Cat#17071-1-AP; RRID: AB_2094743 |
| Mouse anti-DNAJC30 | Abnova | Cat#H00084277-B01P |
| Rabbit anti-COX4 | Proteintech | Cat#11242-1-AP |
| Rabbit anti-Ubiquitin | Abcam | Cat#ab7780; RRID: AB_306069 |
| Rabbit anti-MRPL2 | Proteintech | Cat#16492-1-AP; RRID: AB_2145746 |
| Rabbit anti-MRPL17 | Bethyl | Cat#A305-670A-T; RRID: AB_2891579 |
| Rabbit anti-SPATA22 | Proteintech | Cat#16989-1-AP; RRID: AB_2286640 |
| Mouse anti-Lamin B1 | Proteintech | Cat#66095-1-lg; RRID: AB_2721256 |
| Mouse anti-GST | Abmart | Cat#M20007; RRID: AB_2864360 |
| Rabbit anti-NOP10 | Abcam | Cat#ab134902; RRID: AB_10674253 |
| Mouse anti-αTubulin | Proteintech | Cat#66031-1-lg |
| Alexa Fluor 488-conjugated goat anti-rabbit IgG | Invitrogen | Cat#A-11034 |
| Alexa Fluor 568-conjugated goat anti-rabbit IgG | Invitrogen | Cat#A-11036 |
| Alexa Fluor 488-conjugated goat anti-mouse IgG | Invitrogen | Cat#A-11001 |
| Alexa Fluor 568-conjugated goat anti-mouse IgG | Invitrogen | Cat#A-11004 |
| Alexa Fluor 488-conjugated donkey anti-goat IgG | Invitrogen | Cat#A-11055 |
| Alexa Fluor 488-conjugated goat anti-rat IgG | Abcam | Cat#Ab150165 |
| HRP-conjugated goat anti-mouse IgG | Multi Sciences | Cat#GAM007 |
| HRP-conjugated goat anti-rabbit IgG | Multi Sciences | Cat#GAR007 |
| HRP-conjugated goat anti-rat IgG | Multi Sciences | Cat#GRT007 |
| Rabbit IgG | GeneTex | Cat#GTX35035 |
| Bacterial and virus strains | ||
| E.coli BL-21 | Tiangen | Cat#CB105 |
| Chemicals, peptides, and recombinant proteins | ||
| PMSG | San Sheng Biotech. | N/A |
| HCG | Merck | Cat#230734 |
| Taq DNA polymerase | Tiangen | Cat#ET101 |
| Reverse Transcriptase XL, AMV | Tanaka | Cat#2620A |
| TB Green Premix ExTaq | Takara | Cat#PR420A |
| HTF | Millipore | Cat#MR-070-D |
| Paraformaldehyde | Sigma-Aldrich | Cat#P6148 |
| Glutaraldehyde | Sigma-Aldrich | Cat#G5882 |
| DMEM basic media | Gibco | Cat#C11995500CP |
| Collagenase IV | Worthington | Cat#LS004188 |
| DNase I | Worthington | Cat#LS002139 |
| Trypsin | Worthington | Cat#LS003703 |
| HBSS | Gibco | Cat#14025090 |
| Fetal Bovine Serum | Gibco | Cat#16140071 |
| Hoechst 33342 | Sigma-Aldrich | Cat#14533 |
| DAPI (4’, 6-diamidino-2-phenylindole dihydrochloride) | Sigma-Aldrich | Cat#D8419-1MG |
| Propidium Iodide | Sigma-Aldrich | Cat#P4170 |
| Giemsa | Sigma-Aldrich | Cat#GS500 |
| Photo-Flo 200 solution | Kodak Professional | Cat#1464510 |
| MitoTrcker Red CMXRos | Invitrogen | Cat#M7512 |
| Protease Inhibitor cocktail | Roche | Cat#4693132001 |
| Puromycin | Gibco | Cat#A1113803 |
| RRI | Takara | Cat#2313B |
| Iodoacetamide (IAA) | Sigma-Aldrich | Cat#I1149 |
| Ammonium bicarbonate (ABC) | Sigma-Aldrich | Cat#09830 |
| Trypsin (for protein mass spectrometry samples) | Promega | Cat#V5280 |
| Trifluoroacetic Acid (TFA) | Sigma-Aldrich | Cat#1002641000 |
| Glutathione agarose beads | GenScript | Cat#L00206 |
| Protein G agarose beads | GenScript | Cat#L00209 |
| RIPA buffer | Beyotime | Cat#P0013E |
| 5X loading buffer | Cwbio | Cat#CW0027 |
| Busulfan | Supelco | Cat#B2635 |
| Critical commercial assays | ||
| RNAsimple ToTal RNA Kit | Tiangen | Cat#DP419 |
| TIANgel Midi Purification Kit | Tiangen | Cat#DP209 |
| MAXIscript T7 Transcription Kit | Invitrogen | Cat#AM1312 |
| mirVana miRNA Isolation Kit | Invitrogen | Cat#AM1561 |
| mMESSAGE mMACHINE SP6 Transcription Kit | Invitrogen | Cat#AM1340 |
| RNeasy Micro Kit | Qiagen | Cat#74004 |
| PrimeScript RT Reagent Kit | Takara | Cat#RR047A |
| Eponate 12 Embedding Kit | Ted Pella | Cat#18012 |
| OxiSelect in vitro ROS/RNS Assay Kit | Cell Biolabs | Cat#STA-347 |
| Mitochondrial Membrane Potential Indicator JC-1 Kit | Beyotime | Cat#C2006 |
| Enhanced ATP Assay Kit | Beyotime | Cat#S0027 |
| Enhanced BCA Protein Assay Kit | Beyotime | Cat#P0010 |
| Proteostat Protein Aggregation Assay Kit | Enzo Lifescience | Cat#ENZ-51023-KP050 |
| One Step TUNEL Apoptosis Assay Kit | Beyotime | Cat#C1088 |
| Tissue Mitochondria Isolation Kit | Beyotime | Cat#C3606 |
| ECL Detection Kit | Tanon | Cat#180-501 |
| Deposited data | ||
| Raw protein mass spectrometry data | This paper | ProteomeXchange Consortium: PXD029274 |
| Original uncropped western blots | This paper | Mendeley Data: https://doi.org/10.17632/53sdh5s4bz.1 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse: B6;CBA-Tg(Pou5f1-EGFP)2Mnn/J (OG2) | The Jackson Laboratory | JAX: 004654 |
| Oligonucleotides | ||
| 5’-CACCGCAGGAATCGCTTGATCCTGA-3’ | This study | Rpl39l-1-S |
| 5’-AAACTCAGGATCAAGCGATTCCTGC-3’ | This study | Rpl39l-1-AS |
| 5’-CACCGACATTGGAGACGAACCAAAT-3’ | This study | Rpl39l-2-S |
| 5’-AAACATTTGGTTCGTCTCCAATGTC-3’ | This study | Rpl39l-2-AS |
| 5’-CAGGAATCGCTTGATCCTGA-3’ | This study | sgRNA-1 |
| 5’-ACATTGGAGACGAACCAAAT-3’ | This study | sgRNA-2 |
| Recombinant DNA | ||
| pGEX-4T-2 vector | Life Sciences | Cat#27458101 |
| pGST-Rpl39 | This study | N/A |
| pGST-Rpl39l | This study | N/A |
| pT7-gRNA vector | Addgene | Cat#46759 |
| pCS2-3XFLAG-NLS-SpCas9-NLS | Addgene | Cat#51307 |
| Software and algorithms | ||
| CRISPR Finder | WTSI Genome Editing (WGE) | http://www.sanger.ac.uk |
| MaxQuant (Version 1.6.17.0) | MaxQuant | https://www.maxquant.org |
| DEP Package | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DEP.html |
| DAVID Bioinformatics Resources 6.8 | LHRI | https://david.ncifcrf.gov |
| ImageJ | NIH | https://imagej.nih.gov/ij/download.html |
Resource availability
Lead contact
Further information and requests for reagents may be directed to, and will be fulfilled by, the lead contact, Huayu Qi (qi_huayu@gibh.ac.cn).
Materials availability
This study did not generate new unique reagents. Rpl39l gene null mouse line will be available upon request.
Experimental model and subject details
Animals
Gene targeting was done using C57BL/6J mouse strain, CD1 female mice were used as foster mice following superovulation and mated with ligated males. All experiments utilizing animals are proved by IACUC of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, permit number: 2020240. Mice were housed in an environment controlled for light (12 hours on/off) at ambient temperature (21-23°C) with ad libitum access to water and food (normal mouse chow). Mouse strains used are listed in key resources table.
Collection and preparation of tissues
No human samples were used in this study.
Method details
Animal handling
C57BL/6J mice at various ages were used for dissection of testes, epididymides, isolation of spermatogenic cells, and preparation of tissue lysates. Before dissection, mice were sacrificed by anesthetization with CO2 followed by cervical dislocation. Testes and epididymides were then extracted, weighed, and photographed or used for tissue sectioning and cell purification. To obtain 2-cell embryos for microinjection, 4- to 6-week-old female mice were super-ovulated by subcutaneous injection of 100 μl PMSG (Sansheng Biotech) (5 IU in 0.9% NaCl for each mouse), followed by injection of 100 μl HCG (Merck) (5 IU in 0.9% NaCl for each mouse) after 46–48 hrs. Female mice were mated right after HCG injection with 2-month-old male mice and two-cell embryos were isolated from oviduct after 40–42 hrs. All animal housing and handling were carried out according to IACUC guidelines and that of Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences (Permit No. 2020240).
RT-PCR
To examine the mRNA expression of wild type and mutant Rpl39l, total RNAs were extracted from the testes of wild type, Rpl39l+/– and Rpl39l–/– mice using RNAsimple ToTal RNA Kit (Tiangen). 2 μg of total RNA was reverse transcribed using Reverse Transcriptase XL (AMV, Tanaka) and oligo-dT, followed by PCR reactions using gene specific primers with Taq DNA polymerase (Tiangen). PCR reactions were run at 94°C for 2 min (minutes) first, followed by 30 cycles of 94°C, 30 sec (seconds), 60°C, 30 sec and 72°C, 30 sec. Amplified cDNAs were examined using agarose gel electrophoresis and imaged with a gel imaging system (Tanon, 1600). Gapdh was used as a control. All primers used are listed in Table S1.
Real-time quantitative RT-PCR
To examine the expression levels of Rpl39 and Rpl39l in various mouse tissues and the relative expressions of various genes in mouse testes, quantitative RT-PCR was applied. Following extraction of total RNAs from mouse tissues and testes at various ages using RNAsimple ToTal RNA Kit, first strand cDNAs were reverse transcribed with oligo-dT primer. Real-time quantitative PCR was then performed using TB Green Premix ExTaq (Takara) and gene-specific primers. PCR reactions were run on a CFX96 Real-Time system (Bio-Rad, 1855195) with 40 cycles of 95°C, 5 sec, 60°C, 30 sec following a denaturation step of 95°C, 30 sec. Gapdh was used as an internal control. Relative levels of mRNA expression were calculated using 2–ΔCT (ΔCT = CTTarget gene – CTInternal gene) for different tissues and 2–ΔΔCT (ΔΔCT = ΔCTTreated sample – ΔCTControl sample) for testes at various ages using testes at 3-dpp as the control. For quantitative RT-PCR of various genes, testes from wild type and Rpl39l–/– adult mice were used for total RNA extraction and first strand cDNA amplification. Differential expression was compared using 2–ΔΔCT.
To compare the relative expression levels of Rpl39 and Rpl39l in mouse spermatogonial stem cells (SSCs), SSCs were first sorted from 10-day-old mice by FACS using GFP and cKIT expression (see section of Fluorescence activated cell sorting). Total RNAs from sorted SSCs were extracted and reverse transcribed to cDNAs using method described above. cDNA fragments of Rpl39 and Rpl39l were then amplified using gene specific primers, resolved by agarose gel electrophoresis and purified from agarose gels using TIANgel Midi Purification Kit (Tiangen). Concentrations of cDNAs were measured by spectrometry (OD260) and copy numbers of Rpl39 and Rpl39l cDNAs were calculated according to their molecular weights in order to construct standard curves of expression levels. A series dilution of cDNAs (2 × 10(7, 6, 5, 4, 3, 2, 1, 0) copies/μl) were made and used for PCR. First strand cDNAs of Rpl39 and Rpl39l from SSCs were amplified using PCR with TB Green Premix Ex Taq on the CFX96 Real-Time system using 40 cycles of 95°C for 5 sec and 60°C for 30 sec after pre-denaturation at 95°C for 30 sec. Relative copy numbers of Rpl39 and Rpl39l mRNAs were calculated using the respective equations of standard curves (copy number of Rpl39 = 2 × 10(38.03–CT)/3.692; copy number of Rpl39l = 2 × 10(37.46–CT)/3.595).
To examine the changes of mRNA distributions in different fractions collected from sucrose gradient sedimentation of testis lysates, RNAs from fractions representing RNPs, monosomes and polysomes were extracted using RNAsimple ToTal RNA kit (Tiagen). Concentrations of extracted RNAs were measured by spectrophotometry. RNA solutions at the same volume were used to reverse transcribe cDNAs using PrimeScript RT Reagent kit (Takara), followed by quantitative PCR using TB Green Premix ExTaq (Takara) and gene specific primers. PCR reactions were run using 40 cycles of 95°C, 5 sec, 60°C, 30 sec after a pre-denaturation step at 95°C for 30 sec. Relative mRNA contents of target genes in each fraction were normalized to those of Gapdh (2–ΔCT = CT(Gene)–CT(Gapdh)) and changes of mRNA contents in corresponding fractions of Rpl39l–/– and wild type mice were calculated as the ratio of relative mRNA contents (R = 2–ΔCT–Rpl39l–/–/2–ΔCT–Wild type). Changes of mRNAs in each fraction caused by Rpl39l deletion (see below) were designated as R < 1: decreased, R = 1: no change and R > 1: increased. All primers used are listed in Table S1.
Generation of Rpl39l gene null mice
Rpl39l gene null mice were generated using CRISPR/Cas9 method. Two sgRNAs targeting Rpl39l genomic regions near the start codon in Exon3 was selected using the online CRISPR Finder from WTSI Genome Editing (WGE) (www.sanger.ac.uk). Oligonucleotides encoding sgRNAs were synthesized and sub-cloned into pT7-gRNA vector (Addgene). sgRNAs were in vitro transcribed using MAXIscript T7 Transcription Kit (Invitrogen) and purified with mirVana miRNA Isolation Kit (Invitrogen). The mRNA of spCAS9 endonuclease was in vitro transcribed from pCS2-3XFLAG-NLS-SpCas9-NLS plasmid (Addgene) using mMESSAGE mMACHINE SP6 Transcription Kit (Invitrogen) and purified with RNeasy Micro Kit (Qiagen). Concentrations of RNAs were determined by spectrometry and RNAs were stored at −80°C. To generate gene mutation in mice, RNA mixture (SpCas9:sgRNA = 1:3 μg:μg, with sgRNAs mixed at 1:1 ratio) were microinjected into the cytoplasm of 2-cell embryos isolated from superovulated mice. Injected 2-cell embryos were then transplanted into oviduct of pseudo-pregnant foster CD-1 female mice. Genomic DNAs of tail clips from live pups were genotyped using PCR with gene specific primers flanking the sgRNA target sites. Sequences of PCR products were determined by direct DNA sequencing. Various mutations were found at the Rpl39l loci in 8 pups obtained. One female mouse carrying a 19-bp nucleotide insertion near the start codon was chosen to breed further in order to establish the Rpl39l mutant mouse line. Homozygous mutant mice obtained were examined for putative off-target mutations predicted by the CRISPR Finder for the two Rpl39l sgRNAs using PCR and DNA sequencing. No off-targeting mutations were found (Table S2). For subsequent experiments, Rpl39l mutant mice were genotyped using one pair of primers flanking the mutation site. Oligonucleotides encoding sgRNAs and primers used for genotyping are listed in Table S1.
Mouse fertility test
To examine the fertility of Rpl39l mutant male mice, each male mouse of different genotypes (5 wild type, 5 Rpl39l+/– and 10 Rpl39l–/– mice) was mated with two wild-type female mice; the litter size and number of pups in each litter were recorded for a period of three months. Results were calculated and compared using Excel.
Computer assisted sperm analysis (CASA)
The number and motility of sperm were analyzed using sperm extracted from cauda epididymides of 2-3-month-old male mice. Cauda epididymides isolated from mice were briefly washed in PBS and then placed in pre-warmed HTF medium (Millipore) (1 ml/per mouse). Sperm were squeezed out with a fine needle and collected into 15-ml Corning tubes and centrifuged for 10 min at 1,000 rpm, RT (room temperature) using a table-top centrifuge (Eppendorf, 5427R). After removing supernatant, sperm were re-suspended in same volumes of pre-warmed HTF medium. Numbers of sperm were determined using a hemacytometer (Hausser Scientific, 1492). Sperm motility was analyzed using a computer-assisted sperm analysis system (Microptic, Sperm Class Analyzer). Briefly, 10 μl of sperm suspension was loaded into the test chamber, observed using 10X phase contrast objective and recorded via a CCD camera attached to the microscope. Total sperm motility, progressive motility, hyperactivity and various parameters, including VCL, VSL, VAP, ALH, LIN, WOB and BCF were analyzed using SCA (Sperm Class Analyzer) software. Data for sperm from wild type, Rpl39l+/– and Rpl39l–/– mice were calculated and compared using Excel.
Histology of mouse testis
Isolated testes and cauda epididymides were fixed in 4% paraformaldehyde (PFA) (Sigma-Aldrich) for overnight at 4°C, then embedded in paraffin after dehydration through a series of increasing concentrations of ethanol (30%, 50%, 70%, 80%, 90%, 95%, and 100%), 5 min each, and cut into 5-μm thin sections with a microtome (Leica, RM2255). Tissue sections were stained with Hematoxylin and Eosin and imaged using a digital slide scanning system (Motic, BA600Mot).
Morphology of sperm
To examine the morphology of mature sperm, sperm isolated from cauda epididymides of wild type, Rpl39l+/– and Rpl39l–/– mice were spread on glass slides and air-dried. They were then mounted in 50% glycerol/PBS and photographed using an inverted light microscope (Olympus, IX71) equipped with a CCD camera. Total sperm and sperm with bend at the neck region were counted from images taken. Percentages of abnormal sperm with bend head were calculated and compared using excel. About 1,000 sperm were analyzed for each genotype using 4–5 mice in three independent experiments.
Transmission electron microscopy (TEM)
The ultra-structure of sperm was examined using transmission electron microscopy (TEM) as described previously (Xu et al., 2020). Briefly, sperm isolated from cauda epididymides were washed in 1X PBS, pH7.4, pelleted by centrifugation at 1,000 rpm for 10 min and fixed in 2.5% glutaraldehyde (Sigma-Aldrich) overnight at 4°C. They were then washed in 1X PBS and pelleted and fixed again in 1% OsO4 for 2 hrs at 4°C. Fixed sperm were dehydrated through a series of increasing concentrations of ethanol (30%, 50%, 70%, 80%, 90%, 95% and 100%), 5 min each, infiltrated and embedded with Eponate 12 using Eponate 12 Embedding Kit (Ted Pella). Embedded sperm pellets were then cut into 100-nm ultrathin sections with an ultramicrotome (Leica, EM UC7) and mounted onto copper grids. The grids were double-stained with 10% uranyl acetate/methanol for 20 min and then in lead citrate for 15 min. Stained sections were then examined and photographed with a Tecnai G2 Spirit transmission electron microscope (FEI).
Gravity sedimentation of spermatogenic cells
To isolate spermatocytes, round and elongating spermatids, testicular cells were first dispersed into single cells using a two-step enzyme digestive method (Bellvé, 1993). Testes extracted from adult mice were first removed of tunica albuginea and seminiferous tubules were washed and cut into small pieces in DMEM basic media (Gibco) containing 2 mg/ml Collagenase IV (Worthington) and 5 μg/ml DNase I (Worthington). Minced seminiferous tubules were incubated in a humidified incubator with 5% CO2 at 37°C for 10 min with occasional pipetting. Trypsin (Worthington) was then added to a final concentration of 0.5 mg/ml and incubation continued for another 15 min. Cells were completely dissociated with gentle pipetting every 5 min. The resulting single cell suspension was filtered through a 70-μm cell strainer (Falcon, 352350) and collected by centrifugation for 10 min at 1,000 rpm, 4°C, using a table-top centrifuge (Eppendorf, 5427R). Cell pellets were washed twice and re-suspended in 10 ml 1X PBS. They were then laid on top of a prepared 2–4% BSA gradient and sedimented for 2.5 hrs. 5-ml fractions were collected manually into 15-ml Corning tubes. Spermatocytes, round and elongating spermatids were examined using EVOSfl microscope (Advanced Microscopy Group, AMF5000) according to their cellular morphologies. Cells were also stained with Hoechst 33342 and cell types verified by their nuclear morphology using the EVOSfl microscope. Fractions containing the same cell types were pooled together and used for further experiments.
Flow cytometry
Populations of spermatogenic cells at various developmental stages were analyzed using flow cytometry of testicular cells dispersed from testes of mice at different ages (Bastos et al., 2005). Briefly, testes were dissected and dispersed using the two-step enzyme digestive method as described above. After washing in 1X PBS briefly, spermatogenic cells were resuspended in HBSS (Gibco) containing 5% fetal bovine serum (Gibco) at a concentration of 1 X 106 cells/ml. Cells were then stained with 10 μg/ml Hoechst 33342 (Sigma-Aldrich) for 20 min at 32°C. Before flow cytometry analysis, 2 μg/ml propidium iodide (PI) (Sigma-Aldrich) was added and used for discriminating live and dead cells. Cells were run on a flow cytometer (BD Biosciences, LSR Fortessa) and gated by Hoechst red and Hoechst blue parameters. Moreover, forward scattering (FSC) was also applied to separate round and elongating spermatids.
To analyze spermatogonial stem cells, dispersed testicular cells were fixed in 4% PFA (2 X 106 cells/ml) on ice for 30 min and permeabilized by 0.2% Triton X-100 for 15 min. After washing with 1X PBS containing 2% FBS, fixed and permeabilized cells were stained with PE-conjugated anti-PLZF (1:200) and rabbit anti-Ki67 (1:100) or eFluor 780-conjugated anti-cKIT (1:500) antibodies in 1X PBST (1X PBS containing 0.01% Triton X-100) on ice for 60 min. Stained cells were washed for 3 times, 10 min each, and then incubated with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:500) in 1X PBST. After washing again three times in 1X PBST, 10 min each, stained cells were analyzed using LSR Fortessa. Sources of all antibodies are listed in the key resources table.
Fluorescence activated cell sorting (FACS)
For fluorescence activated cell sorting of spermatogonial stem cells, testes from 10-day-old OG2 mice were digested enzymatically as described above and dispersed cells were resuspended in HBSS containing 5% FBS to a concentration of 2 X 106 cells/ml. The cell suspension was then incubated with eFluor 780-conjugated anti-cKIT antibody (1:500) for 45 min on ice. Stained cells were washed and run on an accelerated cell sorting system (Beckman Coulter, MoFlo Astrios) in order to sort out GFPHcKIT–, GFPHcKIT+, and GFPLcKIT+ cells. Sources of antibodies are listed in the key resources table.
Analysis of meiotic cell cycle progression
Meiosis progression was examined using air-drying method as described before (Evans et al., 1964). Briefly, 6-cm long dissociated seminiferous tubules from the testes of 3-month-old mice were transferred into 1.5 ml isotonic sodium citrate solution (2.2%) and spermatogenic cells were squeezed out with forceps. Dispersed cells were pelleted at 750 rpm for 5 min using a table-top centrifuge. Cell pellets were resuspended in 1.5 ml hypotonic sodium citrate solution (1%) and incubated for 12 min at RT. Cells were then span down and pellets resuspended in 0.5 ml fixative containing 74% ethanol, 25% acetic acid and 1% chloroform and incubated for 5 min at RT. Cells were washed twice by resuspension and pelleting with the fixative before resuspended in 200 μl fixative. Cells were then dropped onto glass slides, air-dried and stained with Giemsa (Sigma-Aldrich) (1:20) for 30 min at RT. The slides were examined under a light microscope and the number of nuclei from cells at first and second meiotic metaphase were counted in defined areas that contain 1,000 mid-pachytene nuclei.
The progression of first meiotic prophase was further detected using a drying-down method for spermatocyte spreading combined with immunofluorescent staining as described previously (Peters et al., 1997). Briefly, dismantled seminiferous tubules from testes of 3-month-old mice were incubated in 1 ml hypotonic solution containing 30 mM Tris (pH 8.2), 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 1 mM DTT, 1mM PMSF and 1Xprotease inhibitor cocktail for 60 min at RT. 40 μl cell suspension was made in 100 mM sucrose solution (pH8.2) and spread onto glass cover slides that were wetted by a fixative containing 1% PFA, 0.15% Triton X-100 and 10 mM sodium borate (pH9.2) in order to expose cell nuclei. The slides were air-dried for 2 hrs and then washed twice with 0.4% Photo-Flo 200 solution (Kodak Professional), 3 min each, followed by immunofluorescent staining with antibodies against SYCP3 (1:400) and γH2AX (1:500). The number of nuclei at leptotene, zygotene, pachytene, and diplotene stages defined by distinct staining patterns of SCYP3 and γH2AX were counted from images taken using laser confocal scanning microscopy (LCSM) (Zeiss, LSM800). Results were analyzed using Excel. Sources of antibodies used are listed in the key resources table.
Immunocytochemistry
Testis sections were permeabilized using 0.3% Triton X-100 in PBS for 15 min and then blocked in 2% BSA in PBST (PBS containing 0.01% Triton X-100) for 2 hrs at RT. Blocked sections were incubated with primary antibodies diluted in 1% BSA/PBST for overnight at 4°C and then washed three times in PBST, 10 min each. They were then incubated with appropriate secondary antibodies diluted in 1% BSA/PBST containing 1 μg/ml DAPI for 1 hr at RT. After washing three times in PBST, 10 min each, sections were mounted in 50% glycerol/PBS and examined using LCSM. Antibodies used as: goat polyclonal anti-GFRα1 (1:50); rabbit polyclonal antibodies: anti-PLZF (1:100), anti-UCHL1 (1:200), anti-SOX9 (1:500), anti-RPL39 (1:100), anti-CNBP (1:100), anti-RPL28 (1:100), anti-FUBI (1:50), anti-DNAJB6 (1:100), anti-DNAJB8 (1:100), anti-COXIV (1:200); mouse monoclonal antibodies: anti-Puromycin (1:1,000), anti-DNAJC30 (1:50); Alexa Fluor 488-conjugated Lectin PNA (1:1,000); secondary antibody conjugates: Alexa Fluor 488-conjugated goat-anti-rabbit IgG (1:500), Alexa Fluor 568-conjugated goat-anti-rabbit IgG (1:500), Alexa Fluor 488-conjugated goat-anti-mouse IgG (1:500), Alexa Fluor 568-conjugated goat-anti-mouse IgG (1:500), Alexa Fluor 488-conjugated donkey-anti-goat IgG (1:500) and Alexa Fluor 488-conjugated goat-anti-rat IgG (1:500). Sources of all antibodies are listed in the key resources table.
Measurements of mitochondrial functions
Sperm mitochondria were measured using MitoTracker staining of mature sperm isolated from cauda epididymides. About 1 X 106 sperm were stained with 100 nM MitoTracker Red CMXRos (Invitrogen) in 0.5 ml HTF media for 30 min in a humidified incubator with 5% CO2 at 37°C. The sperm were then harvested and resuspended in 0.5 ml HBSS and MitoTracker Red fluorescent signals were analyzed using Fortessa.
Mitochondria membrane potential in spermatogenic cells and mature sperm were measured using mitochondrial membrane potential indicator JC-1 Kit (Beyotime). Briefly, 1 X 106 spermatogenic cells separated from BSA gradient or mature sperm were collected, washed, and resuspended in 0.5 ml DMEM media (DMEM basic media supplemented with 10% FBS, 1X NEAA, 1X Penicillin/Streptomycin) and HTF media, respectively. 0.5 ml 1X JC-I staining solution (200X JC-1 stock solution diluted with 1X washing buffer provided in the kit) was then used to incubate cells for 20 min in a 37°C incubator, with 5% CO2. Stained sperm and spermatogenic cells were washed twice with 1 ml 1X washing buffer, 5 min each time, resuspended in 0.5 ml 1X washing buffer and analyzed using Fortessa. Fluorescent signals of JC-1 red aggregates were used as the indicator for mitochondrial membrane potential. Measured data were then calculated and compared using Excel.
The level of ROS (reactive oxygen species) in testicular cells was measured using OxiSelect in vitro ROS/RNS Assay Kit (Cell Biolabs). Testes of adult mice were dissected and weighed. They were then homogenized in 1X PBS at 50 mg/ml on ice. Insoluble aggregates were removed by centrifugation for 10 min at 10,000 rpm, 4°C, using a table-top centrifuge. Supernatants were then transferred into fresh Eppendorf tubes. 50 μl supernatant was mixed with 50 μl catalyst provided in the kit, added into each well of a 96-well plate and incubated for 5 min. 100 μl DCFH solution (provided in the kit) was added into each well and incubation continued for 30 min. Fluorescent signals were measured using a multimode microplate reader (Berthold, LB943) with excitation/emission wavelengths at 480/530 nm, respectively.
ATP contents in spermatogenic cells and mature sperm were measured using an Enhanced ATP Assay Kit (Beyotime). About 2 X 106 collected cells were first lysed with the 200 μl lysis buffer provided in the kit on ice for 5 min. Cell lysates were centrifuged for 5 min at 12,000 g, 4°C, and supernatants were transferred to fresh Eppendorf tubes. Protein concentrations were determined using an Enhanced BCA Protein Assay Kit (Beyotime). 20 μl cell lysates were used for ATP measurements on a microplate reader using chemiluminescence method. The intracellular ATP level was calculated as nmol ATP per mg proteins.
Measurement of protein aggregation
Protein aggregation in testes was determined using the Proteostat Protein Aggregation Assay Kit (Enzo Lifescience). Extracted testes of adult mice were weighed and homogenized using glass homogenizer in lysis buffer containing 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.01% Triton X-100, 5% glycerol and 1X Protease Inhibitor cocktail (Roche) (pH 7.4) at 100 mg/ml on ice for 5 min. Testis lysates were centrifuged for 5 min at 6000 g, 4°C. Supernatants were transferred into fresh Eppendorf tubes. 98 μl of the homogenates was mixed with 2 μl 1X Detection Reagent (diluted from 20X Detection Reagent with 1X assay buffer provided in the kit) and added into each well of a 96-well plate and incubated for 15 min in dark at RT. Fluorescent signals were read using the multimode microplate reader with excitation/emission wavelengths at 550/600 nm, respectively.
TUNEL assay
Apoptotic testicular cells were detected in frozen testis sections using One-step TUNEL Apoptosis Assay Kit (Beyotime) according to the manufacturer’s instruction. Briefly, frozen sections fixed in ice-cold acetone were washed twice with 1X PBS, then permeabilized with 0.5% Triton X-100 in PBS for 5 min at RT. After washing with PBS twice, 5 min each time, each sample was incubated with 50 μl TUNEL detecting reagent for 60 min at 37°C in the dark, followed by DAPI staining (2 μg/ml) for 30 min. Testis sections were then washed three times with 1X PBS, 10 min each and mounted in 50% glycerol/PBS. Immunofluorescent signals were examined and images taken using LCSM. Apoptotic rates were calculated by counting TUNEL positive cross-sections and TUNEL positive cells.
Nascent protein labeling
Nascent protein synthesis in testicular cells was detected using puromycin labeling method. For in vivo labeling, 3-4-month-old male mice were intraperitoneally injected with puromycin (Gibco) (65 mg/kg body weight). After 1.5 hrs, mice were sacrificed and testes were dissected out. For each mouse, one testis was used for tissue lysate preparation and the other one was fixed in 4% PFA and used for paraffin section preparation. For in vitro labeling, 2 X 106 dispersed testicular cells were incubated with 5 μg/ml puromycin in 1 ml DMEM complete media for 1 hr in a humidified 37°C incubator, 5% CO2. Cells were then collected and washed twice in 1X PBS. A portion of cells was then fixed in 4% PFA, whereas the remaining cells were used for cell lysate preparation. Fixed paraffin testis sections and testicular cells were immunostained with anti-puromycin antibody and Alexa Fluor 568-goat-anti-mouse secondary antibody conjugates, using protocols for immunocytochemistry as described before. Samples were then examined and fluorescent images taken using LCSM. Antibodies used are listed in the key resources table.
Sucrose gradient sedimentation
Sucrose gradient sedimentation was done using a 10–50% sucrose gradient. Extracted testes from adult mice were homogenized in cell lysis buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 5 mM MgCl2, 1% NP40, 1 mM DTT, 1X protease inhibitor cocktail, 0.2 U/μl recombinant RNase inhibitor (RRI) (Takara), 1 ml per testis, with a glass homogenizer. Testes were briefly grinded and incubated on ice for 30 min. Crude lysates were centrifuged at 12,000 g for 10 min at 4°C. 0.5 ml supernatant was layered on top of a 10–50% sucrose gradient prepared with Gradient Master 108 (BioComp Instrument Inc.) in a 12.5 ml ultracentrifuge tube (Beckman Coulter, 344059). Samples were centrifuged at 210,000 g for 2 hrs at 4°C using an Optima L-80 XP Ultracentrifuge (Beckman Coulter). 24 0.5-ml fractions were collected using BioComp Fractionator (BioComp Instrument Inc.) and absorbance at 260 nm was monitored with UV detector.
Protein tandem mass spectrometry
For protein mass spectrometry, testes extracted from adult mice with different genotypes were lysed in 8 M urea containing 0.1 M Tris-HCl (pH8.5) and 10 mM DTT. Protein concentrations of testis lysates were measured using the BCA method. Whole proteome peptides were prepared using the Filter Aided Sample Preparation (FASP) protocol as described before (Wisniewski et al., 2009). Briefly, 50 μg soluble lysates were added onto a filter with 30-KDa cut-off and centrifuged at 11,000 rpm for 15 min at 20°C. 50 mM iodoacetamide (IAA, Sigma-Aldrich) in 8 M urea was used to alkylate proteins at 20°C for 15 min. After a few washes with 8 M urea and 50 mM ammonium bicarbonate (ABC, Sigma-Aldrich), 100 ng Trypsin (Promega) in 50 mM ABC was used to digest proteins in a wet chamber overnight at 37°C. Tryptic peptides were extracted by 50 mM ABC and acidified by 10% Trifluoroacetic acid (TFA, Sigma-Aldrich). Stage tipping was performed as described previously (Zhang et al., 2018). Tryptic peptides were then separated using an Easy-nLC 1200, connected online to a Fusion Lumos mass spectrometer equipped with FAMIS Pro (Thermo Fisher Scientific), with 240 min total data collection time. Scans were collected in data-dependent top-speed mode with dynamic exclusion at 90 sec. Raw data were analyzed using MaxQuant (Version 1.6.17.0) searching against mouse FastA database, with label-free quantification (LFQ) and match between runs functions enabled. The output protein list was analyzed and visualized using DEP package as described before (Zhang et al., 2018).
To compare mitochondrial proteomes, mitochondria were first purified from testes using Tissue Mitochondria Isolation Kit (Beyotime). Briefly, testes extracted from mice with different genotypes were weighed and homogenized in mitochondria isolation reagent provided in the kit at 100 mg/ml on ice. Homogenates were then centrifuged at 1000 g, for 5 min at 4°C. Supernatants were transferred into fresh Eppendorf tubes and centrifuged again at 3500 g, for 10 min at 4°C. After discard supernatants, pellets containing mitochondria were washed twice with mitochondria isolation reagent by centrifugation under the same condition. Pellets of mitochondria were lysed with 0.1 M Tris buffer containing 8 M urea and 10 mM DTT (pH 8.5). Protein concentrations of dissolved mitochondria were determined by BCA method and subjected to LC-MS/MS using method as described above.
GST pull-down assay
GST pull-down assays were done using bacterially expressed GST-RPL39 and GST-RPL39L fusion proteins with testis lysates. cDNAs encoding RPL39 and RPL39L were synthesized and sub-cloned into pGEX-4T-2 vector (Life Sciences) at BamHI and NotI sites. Plasmids were then transformed into BL21 E. coli (Tiangen). Transformed bacteria were inoculated into 40 ml LB broth and grown at 37°C to log phase (OD600 = 0.5). 0.1 ml 200 mM IPTG was then added to induce expression of fusion proteins and bacteria were continued to grow at 18°C for overnight. Overnight cultures were centrifuged for 5 min at 6000 g, 4°C and bacteria pellets were resuspended in 5 ml lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP40, 5% glycerol and 1X protease inhibitor cocktail), homogenized briefly in a glass homogenizer and incubated for 30 min on ice. After centrifugation at 12,000 g for 15 min at 4°C, supernatants were transferred into fresh tubes, mixed with 0.25 ml glutathione agarose beads (GenScript) and rotated for 4 hrs at 4°C. Protein-bound agarose beads were then pelleted and washed twice with lysis buffer and once with 1X PBS, 15 min each. They were then mixed with testis lysates prepared using lysis buffer (50 mM Tris, pH8.0, 150 mM NaCl, 0.5% NP40, 10% glyceral and 1X protease inhibitor cocktail) and incubated overnight at 4°C. Glutathione agarose beads were then washed three times with testis lysis buffer and 1 time with PBS, 15 min each. After complete removal of PBS, proteins pulled down were digested using on-bead digestion method as described before (Hubner and Mann, 2011). Briefly, washed beads were incubated with 100 μl elution buffer (2 M urea, 10 mM DTT and 100 mM Tris, pH 8.5) for 20 min. Then IAA was added to a final concentration of 50 mM and incubated for 10 min in dark. Beads were then partially digested with 250 ng trypsin (Promega) for 2 hrs and supernatants collected into fresh tubes. Beads were then incubated with 100 μl elution buffer for another 5 min and supernatants pooled with previous ones. All procedures were conducted at RT on a thermoshaker at 1500 rpm. Combined eluates were further digested with 100 ng trypsin overnight at RT. Tryptic peptides were then acidified to pH < 2 by adding 10 ml 10% TFA and desalted using homemade C18 StageTips prior to being analyzed by LC-MS/MS as described above with data collection of 140 min.
Co-immunoprecipitation
Co-immunoprecipitation of testis lysates was done according to the procedure described previously (Xu et al., 2014). Briefly, testis lysates were prepared by homogenizing the tissue in ice-cold lysis buffer (50 mM Tris, pH8.0, 150 mM NaCl, 0.5% NP40, 10% glycerol and 1X protease inhibitor cocktail). Homogenates were incubated for 30 min at 4°C with rotating. After centrifugation at 12,000 g for 10 min at 4°C, the supernatant was first precleared with Protein G agarose beads (GenScript) and then incubated with anti-RPL39 (1:100) or anti-NOP10 (1:100) by rotating at 4°C for overnight. Isotype rabbit IgG (GeneTex) was used as the control. Immunocomplexes were then incubated with Protein G agarose beads at 4°C for 2 hrs. Immunoprecipitates were collected by brief centrifugation, washed three times with ice-cold lysis buffer, then dissolved in denaturing sample buffer for Western blotting.
Western blotting
Western blotting was used to determine the expression of various proteins. Total proteins of dispersed testicular cells or testes were extracted using ice-cold RIPA buffer (Beyotime), supplemented with 1X protease inhibitor cocktail. Protein concentrations were determined using BCA method using known quantity of BSA as standard. Protein samples were mixed with 5X loading buffer (Cwbio), boiled for 5 min, and separated by SDS-PAGE. After transfer onto nitrocellulose membrane (Millipore, HATF00010), the membrane was blocked with 5% BSA in TBST (Tris-buffered saline containing 0.1% Tween20) for 2 hrs at RT, then incubated with primary antibodies diluted in 1% BSA/TBST overnight at 4°C. After washing three times in PBST, 10 min each, membranes were further incubated with appropriate secondary antibodies diluted in 2% BSA/PBST for 1 hr at RT. Protein signals were then developed using the ECL Detection Kit (Tanon). Relative quantitation of protein band intensities was done using ImageJ. Antibodies were used as: rabbit polyclonal antibodies: anti-RPL39 (1:1,000), anti-CNBP (1:1,000), anti-RPL28 (1:1,000), anti-CREM (1:1,000), anti-DNAJB6 (1:2,000), anti-DNAJB8 (1:2,000), anti-COXIV (1:5,000), anti-UBIQUITIN (1:1,000), anti-MRPL2 (1:1,000), anti-MRPL17 (1:1,000), anti-SPATA22 (1:1,000), anti-NOP10 (1:2,000), anti-RPS19 (1:1,000); rat monoclonal anti-FUBI (1:1,000); mouse monoclonal antibodies: anti-Puromycin (1:10,000), anti-DNAJC30 (1:1,000), anti-LaminB1 (1:3,000), anti-GST (1:5,000), anti-α-TUBULIN (1:5,000); secondary antibody conjugates: HRP-conjugated goat-anti-rabbit IgG (1:5,000), HRP-conjugated goat-anti-mouse IgG (1:5,000) and HRP-conjugated goat-anti-rat IgG (1:5,000). Sources of antibodies are listed in the key resources table.
Busulfan treatment of mouse testis
To evaluate the recovery of spermatogenesis after germ cell depletion caused by busulfan administration, 3-month-old male mice were intraperitoneally injected once with 20 mg/kg busulfan (Supelco) dissolved in DMSO, and deionized water at 1:1 volume ratio. Testes were then dissected out from mice at different times after busulfan injection (1 month, 2 months, and 3 months). Extracted testes were fixed in 4% PFA and paraffin sections prepared. Testis sections were then stained with Hematoxylin and Eosin, spermatogenic cells and contents of testis were examined using images taken from Motic digital slide scanning system.
Quantification and statistical analysis
For quantitative protein mass spectrometry, the differentially expressed proteins were selected using Log2FC > 1, p < 0.05. Heatmaps and Volcano plots of differentially enriched proteins identified were generated using DEP package based on LFQ intensity as described previously (Zhang et al., 2018). DEP groups of Heatmaps were clustered by k-means clustering. Putative hits of GST pull-down assays were selected according to the number of identified unique peptides (GST-tag alone < 1, GST-RPL39 and GST-RPL39L > 3), protein sequence coverage (> 20%) and iBAQ intensity (>1,00,000). GO analyses were done using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov). Intensities of protein bands from Western blotting were measured using ImageJ. All statistical analyses were performed using two-tailed unequal Student’s t-test for paired samples and One-way ANOVA Tukey’s multiple comparison test for N ≥ 3 samples; significances were set as ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001. Significances shown in the graph are those of mutants versus wild type. Data are presented as mean ± S.D. and plotted using Excel software.
Acknowledgments
We would like to thank our lab members for their discussion and criticism during the course of this study, the Common Facility Center at Guangzhou Institutes of Biomedicine and Health for the help with flow cytometry and imaging, Liman Guo from Metabolomics and Preoteomics Center, Bioland Laboratory, for the quantitative proteomics. We also thank Dr. Wenliang Zhou for sharing his CASA apparatus. This work is supported by the National Key Research and Development Program of China, Ministry of Science and Technology of the People’s Republic of China, 2018YFC1004700 (to H.Q.), the Frontier Research Program of Guangzhou Regenerative Medicine and Health, Bioland Laboratory, 2018GZR110105021 (to H.Q.), Basic Research grant, Guangdong Department of Science and Technology, 2020A1515010882 (to H.Q.), and is partly supported by the Guangdong Provincial Key Laboratory of Stem Cell and Regenerative Medicine grant 2020B1212060052 and the Chinese Academy of Sciences.
Author contributions
Conceptualization and design: Q.Z., H.Q.; Investigation: Q.Z., L.Y.; Data analysis: Q.Z., L.Y., H.Q.; MS and data analysis: Y.Q., R.S., X.Z., Q.Z.; Writing: Q.Z., H.Q.; Supervision: H.Q.; Funding acquisition: H.Q. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103396.
Supplemental information
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (www.iprox.org) (Ma et al., 2019) with the dataset identifier PXD029274. The processed LC-MS/MS data are included in Table S3. Proteins identified from GST pulled-down assays, related to Figure 4, Table S4. Differentially expressed proteins between Rpl39l null and wild type testes identified by LC-MS/MS, related to Figure 5, Table S5. Differentially expressed proteins in enriched mitochondria of Rpl39l null and wild type testes (FC > 2, p < 0.05), related to Figure 6. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This study does not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Albert B., Kos-Braun I.C., Henras A.K., Dez C., Rueda M.P., Zhang X., Gadal O., Kos M., Shore D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife. 2019;8:e45002. doi: 10.7554/eLife.45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H., Harper J.W. Ribosome abundance control via the ubiquitin-proteasome system and autophagy. J. Mol. Biol. 2020;432:170–184. doi: 10.1016/j.jmb.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badertscher L., Wild T., Montellese C., Alexander L.T., Bammert L., Sarazova M., Stebler M., Csucs G., Mayer T.U., Zamboni N., et al. Genome-wide RNAi screening identifies protein modules required for 40s subunit synthesis in human cells. Cell. Rep. 2015;13:2879–2891. doi: 10.1016/j.celrep.2015.11.061. [DOI] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Bastos H., Lassalle B., Chicheportiche A., Riou L., Testart J., Allemand I., Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- Bellvé A.R. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bezzi G., Piga E.J., Binolfi A., Armas P. CNBP binds and unfolds in vitro G-quadruplexes formed in the sars-cov-2 positive and negative genome strands. Int. J. Mol. Sci. 2021;22:2614–2635. doi: 10.3390/ijms22052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar R.B., Makley L.N., Tsonis P.A. The other lives of ribosomal proteins. Hum. Genomics. 2010;4:327–344. doi: 10.1186/1479-7364-4-5-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci L.R., Meistrich M.L. Effects of busulfan on murine spermatogenesis: cytotoxicity, sterility, sperm abnormalities, and dominant lethal mutations. Mutat. Res. 1987;176:259–268. doi: 10.1016/0027-5107(87)90057-1. [DOI] [PubMed] [Google Scholar]
- Calcaterra N.B., Armas P., Weiner A.M., Borgognone M. CNBP: a multifunctional nucleic acid chaperone involved in cell death and proliferation control. IUBMB Life. 2010;62:707–714. doi: 10.1002/iub.379. [DOI] [PubMed] [Google Scholar]
- Casteels D., Poirier C., Guenet J.L., Merregaert J. The mouse Fau gene: genomic structure, chromosomal localization, and characterization of two retropseudogenes. Genomics. 1995;25:291–294. doi: 10.1016/0888-7543(95)80140-h. [DOI] [PubMed] [Google Scholar]
- D'Amours O., Calvo E., Bourassa S., Vincent P., Blondin P., Sullivan R. Proteomic markers of low and high fertility bovine spermatozoa separated by Percoll gradient. Mol. Reprod. Dev. 2019;86:999–1012. doi: 10.1002/mrd.23174. [DOI] [PubMed] [Google Scholar]
- Dave B., Granados-Principal S., Zhu R., Benz S., Rabizadeh S., Soon-Shiong P., Yu K.D., Shao Z., Li X., Gilcrease M., et al. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc. Natl. Acad. Sci. U S A. 2014;111:8838–8843. doi: 10.1073/pnas.1320769111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Gomez-Herreros F., Rodriguez-Galan O., Begley V., de la Cruz Munoz-Centeno M., Chavez S. Feedback regulation of ribosome assembly. Curr. Genet. 2018;64:393–404. doi: 10.1007/s00294-017-0764-x. [DOI] [PubMed] [Google Scholar]
- Deuerling E., Gamerdinger M., Kreft S.G. Chaperone interactions at the ribosome. Cold Spring Harb. Perspect. Biol. 2019;11:a033977. doi: 10.1101/cshperspect.a033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N., Coux O., Rodriguez M.S., Barrio R., Core Group Members of PROTEOSTASIS PROTEOSTASIS: a european network to break barriers and integrate science on protein homeostasis. Trends Biochem. Sci. 2019;44:383–387. doi: 10.1016/j.tibs.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Dresios J., Derkatch I.L., Liebman S.W., Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- Egan E.D., Collins K. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol.Cell. Biol. 2012;32:2428–2439. doi: 10.1128/MCB.00286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E., Jovanovic M., Slavov N. Ribosome stoichiometry: from form to function. Trends Biochem. Sci. 2019;44:95–109. doi: 10.1016/j.tibs.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E.P., Breckon G., Ford C.E. An air-drying method for meiotic preparations from mammalian testes. Cytogenetics. 1964;3:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
- Fang X., Lee C.S. Proteome characterization of mouse brain mitochondria using electrospray ionization tandem mass spectrometry. Methods Enzymol. 2009;457:49–62. doi: 10.1016/S0076-6879(09)05003-4. [DOI] [PubMed] [Google Scholar]
- Farley-Barnes K.I., McCann K.L., Ogawa L.M., Merkel J., Surovtseva Y.V., Baserga S.J. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell. Rep. 2018;22:1923–1934. doi: 10.1016/j.celrep.2018.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Yamazaki T., Kroemer G. Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol.Cell Biol. 2018;19:731–745. doi: 10.1038/s41580-018-0068-0. [DOI] [PubMed] [Google Scholar]
- Genuth N.R., Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol.Cell. 2018;71:364–374. doi: 10.1016/j.molcel.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth N.R., Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 2018;19:431–452. doi: 10.1038/s41576-018-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J., Schipper-Krom S., Juenemann K., Gruber A., Coolen S., van den Nieuwendijk R., van Veen H., Overkleeft H., Goedhart J., Kampinga H.H., et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J. Biol. Chem. 2013;288:17225–17237. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Hall M.N., Lin S.C., Hardie D.G. AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell. Metab. 2020;31:472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- Grad I., Cederroth C.R., Walicki J., Grey C., Barluenga S., Winssinger N., De Massy B., Nef S., Picard D. The molecular chaperone Hsp90alpha is required for meiotic progression of spermatocytes beyond pachytene in the mouse. PLoS One. 2010;5:e15770. doi: 10.1371/journal.pone.0015770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.D., Ma Q., Manske G.L., Shami A.N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S.J., Moore B.B., et al. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq. Dev.Cell. 2018;46:651–667.e610. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernstroer B., et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell. Rep. 2018;25:1650–1667.e1658. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermo L., Pelletier R.M., Cyr D.G., Smith C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 2010;73:241–278. doi: 10.1002/jemt.20783. [DOI] [PubMed] [Google Scholar]
- Herzig S., Shaw R.J. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol.Cell. Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R.M., Seandel M., Falciatori I., Rafii S., Pandolfi P.P. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N.C., Mann M. Extracting gene function from protein-protein interactions using quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Hwang J., Qi L. Quality control in the endoplasmic reticulum: crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018;43:593–605. doi: 10.1016/j.tibs.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T. The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem. Sci. 2017;42:5–15. doi: 10.1016/j.tibs.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Jayaraj G.G., Hipp M.S., Hartl F.U. Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol. 2020;12:a033951. doi: 10.1101/cshperspect.a033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Li T., Zhang X., Zhang B., Yu C., Li Y., Fan S., Jiang X., Khan T., Hao Q., et al. RPL10L is required for male meiotic division by compensating for rpl10 during meiotic sex chromosome inactivation in mice. Curr. Biol. 2017;27:1498–1505 e1496. doi: 10.1016/j.cub.2017.04.017. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol.Cell Biol. 2019;20:368–383. doi: 10.1038/s41580-019-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas K., Michiels L., Merregaert J. Genomic structure and expression of the human fau gene: encoding the ribosomal protein S30 fused to a ubiquitin-like protein. Biochem. Biophys. Res. Commun. 1992;187:927–933. doi: 10.1016/0006-291x(92)91286-y. [DOI] [PubMed] [Google Scholar]
- Kitadate Y., Jorg D.J., Tokue M., Maruyama A., Ichikawa R., Tsuchiya S., Segi-Nishida E., Nakagawa T., Uchida A., Kimura-Yoshida C., et al. Competition for mitogens regulates spermatogenic stem cell homeostasis in an open niche. Cell Stem Cell. 2019;24:79–92 e76. doi: 10.1016/j.stem.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S., Woolford J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol.Cell Biol. 2019;20:116–131. doi: 10.1038/s41580-018-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C.R., Shimizu K., Hsieh A.C., Xue S., Ishijima J., Shiroishi T., Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C., Riera-Escamilla A. Genetics of male infertility. Nat. Rev. Urol. 2018;15:369–384. doi: 10.1038/s41585-018-0003-3. [DOI] [PubMed] [Google Scholar]
- Lempiainen H., Shore D. Growth control and ribosome biogenesis. Curr. Opin.Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li W., Atanassov B.S., Lan X., Mohan R.D., Swanson S.K., Farria A.T., Florens L., Washburn M.P., Workman J.L., Dent S.Y.R. Cytoplasmic ATXN7L3B interferes with nuclear functions of the SAGA deubiquitinase module. Mol.Cell. Biol. 2016;36:2855–2866. doi: 10.1128/MCB.00193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton J.M., Ellis S.R. Diamond Blackfan anemia 2008-2009: broadening the scope of ribosome biogenesis disorders. Curr. Opin. Pediatr. 2010;22:12–19. doi: 10.1097/MOP.0b013e328334573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes A.M., Miguel R.N., Sargent C.A., Ellis P.J., Amorim A., Affara N.A. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol. Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Guo S. Mechanisms linking mitochondrial dysfunction and proteostasis failure. Trends Cell. Biol. 2020;30:317–328. doi: 10.1016/j.tcb.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., et al. iProX: an integrated proteome resource. Nucl. Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V.P., Edelman G.M. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U S A. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D., Frank A.R., Jewell J.L. mTOR signaling in stem and progenitor cells. Development. 2018;145:dev152595. doi: 10.1242/dev.152595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.W., Green R. Ribosomopathies: there's strength in numbers. Science. 2017;358:6363. doi: 10.1126/science.aan2755. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Prudent J., Basu K., Goyon V., Katsumura S., Hulea L., Pearl D., Siddiqui N., Strack S., McGuirk S., et al. mTOR controls mitochondrial dynamics and cell survival via MTFP1. Mol.Cell. 2017;67:922–935 e925. doi: 10.1016/j.molcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Nadano D., Notsu T., Matsuda T., Sato T. A human gene encoding a protein homologous to ribosomal protein L39 is normally expressed in the testis and derepressed in multiple cancer cells. Biochim. Biophys. Acta. 2002;1577:430–436. doi: 10.1016/s0167-4781(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Netherton J.K., Hetherington L., Ogle R.A., Velkov T., Baker M.A. Proteomic analysis of good- and poor-quality human sperm demonstrates that several proteins are routinely aberrantly regulated. Biol. Reprod. 2018;99:395–408. doi: 10.1093/biolre/iox166. [DOI] [PubMed] [Google Scholar]
- Oakberg E.F. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- Pena C., Hurt E., Panse V.G. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 2017;24:689–699. doi: 10.1038/nsmb.3454. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Plug A.W., van Vugt M.J., de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Petersen C., Aumuller G., Bahrami M., Hoyer-Fender S. Molecular cloning of Odf3 encoding a novel coiled-coil protein of sperm tail outer dense fibers. Mol. Reprod. Dev. 2002;61:102–112. doi: 10.1002/mrd.1136. [DOI] [PubMed] [Google Scholar]
- Piette B.L., Alerasool N., Lin Z.Y., Lacoste J., Lam M.H.Y., Qian W.W., Tran S., Larsen B., Campos E., Peng J., et al. Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol.Cell. 2021;81:2549. doi: 10.1016/j.molcel.2021.04.012. [DOI] [PubMed] [Google Scholar]
- Rangaraju V., Lauterbach M., Schuman E.M. Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell. 2019;176:73–84 e15. doi: 10.1016/j.cell.2018.12.013. [DOI] [PubMed] [Google Scholar]
- Rashid R., Liang B., Baker D.L., Youssef O.A., He Y., Phipps K., Terns R.M., Terns M.P., Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol.Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Ravanelli S., den Brave F., Hoppe T. Mitochondrial quality control governed by ubiquitin. Front.Cell. Dev. Biol. 2020;8:270. doi: 10.3389/fcell.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A., Bertolotti A. An evolutionarily conserved pathway controls proteasome homeostasis. Nature. 2016;536:184–189. doi: 10.1038/nature18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L., Wang Y., Zhang X., Tomaszewski A., McNamara J.T., Li R. Mitochondria-associated proteostasis. Annu. Rev. Biophys. 2020;49:41–67. doi: 10.1146/annurev-biophys-121219-081604. [DOI] [PubMed] [Google Scholar]
- Schafer M., Nayernia K., Engel W., Schafer U. Translational control in spermatogenesis. Dev. Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Schram A.W., Baas R., Jansen P.W., Riss A., Tora L., Vermeulen M., Timmers H.T. A dual role for SAGA-associated factor 29 (SGF29) in ER stress survival by coordination of both histone H3 acetylation and histone H3 lysine-4 trimethylation. PLoS One. 2013;8:e70035. doi: 10.1371/journal.pone.0070035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Gerst J.E. Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J.Cell. Biol. 2018;217:117–126. doi: 10.1083/jcb.201706059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.Y., Qian S.B. Less is more: improving proteostasis by translation slow down. Trends Biochem. Sci. 2013;38:585–591. doi: 10.1016/j.tibs.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Sitron C.S., Brandman O. Detection and degradation of stalled nascent chains via ribosome-associated quality control. Annu. Rev. Biochem. 2020;89:417–442. doi: 10.1146/annurev-biochem-013118-110729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N., Semrau S., Airoldi E., Budnik B., van Oudenaarden A. Differential stoichiometry among core ribosomal proteins. Cell. Rep. 2015;13:865–873. doi: 10.1016/j.celrep.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara Y., Honda H., Iida T., Morinaga T., Hino S., Okajima T., Matsuda T., Nadano D. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- Sugihara Y., Sadohara E., Yonezawa K., Kugo M., Oshima K., Matsuda T., Nadano D. Identification and expression of an autosomal paralogue of ribosomal protein S4, X-linked, in mice: potential involvement of testis-specific ribosomal proteins in translation and spermatogenesis. Gene. 2013;521:91–99. doi: 10.1016/j.gene.2013.02.040. [DOI] [PubMed] [Google Scholar]
- Sung M.K., Reitsma J.M., Sweredoski M.J., Hess S., Deshaies R.J. Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol. Biol.Cell. 2016;27:2642–2652. doi: 10.1091/mbc.E16-05-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi S., Sonenberg N., Hershey J.W.B., Mathews M.B. Protein synthesis and translational control: a historical perspective. Cold Spring Harb. Perspect. Biol. 2019;11:a035584. doi: 10.1101/cshperspect.a035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara Y., Yashiroda H., Matsuo Y., Zhao X., Kamigaki A., Matsuzaki T., Kosako H., Inada T., Murata S. The ubiquitination-deubiquitination cycle on the ribosomal protein eS7A is crucial for efficient translation. iScience. 2021;24:102145. doi: 10.1016/j.isci.2021.102145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Song H.W., Wilkinson M.F. Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development. 2020;147:dev183251. doi: 10.1242/dev.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp A.T.N., Varela L., Choi J., Paredes M.I., Giani A.M., Song J.E., Sestan-Pesa M., Franjic D., Sousa A.M.M., Liu Z.W., et al. The 7q11.23 protein DNAJC30 interacts with ATP synthase and links mitochondria to brain development. Cell. 2018;175:1088–1104.e1023. doi: 10.1016/j.cell.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiruvalluvan A., de Mattos E.P., Brunsting J.F., Bakels R., Serlidaki D., Barazzuol L., Conforti P., Fatima A., Koyuncu S., Cattaneo E., et al. DNAJB6, a key factor in neuronal sensitivity to amyloidogenesis. Mol.Cell. 2020;78:346–358.e349. doi: 10.1016/j.molcel.2020.02.022. [DOI] [PubMed] [Google Scholar]
- Tokuhiro K., Ikawa M., Benham A.M., Okabe M. Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility [corrected] Proc. Natl. Acad. Sci. U S A. 2012;109:3850–3855. doi: 10.1073/pnas.1117963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi T., Maeda N., Tanaka T., Kenmochi N. Functional second genes generated by retrotransposition of the X-linked ribosomal protein genes. Nucl. Acids Res. 2002;30:5369–5375. doi: 10.1093/nar/gkf696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Waterbeemd M., Fort K.L., Boll D., Reinhardt-Szyba M., Routh A., Makarov A., Heck A.J. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat. Methods. 2017;14:283–286. doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- Visser L., Repping S. Unravelling the genetics of spermatogenic failure. Reproduction. 2010;139:303–307. doi: 10.1530/REP-09-0229. [DOI] [PubMed] [Google Scholar]
- Wang P.J. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Wolff S., Weissman J.S., Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Wong Q.W., Li J., Ng S.R., Lim S.G., Yang H., Vardy L.A. RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol. 2014;11:33–41. doi: 10.4161/rna.27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Yang L., Zhang L., Qi H. Lack of AKAP3 disrupts integrity of the subcellular structure and proteome of mouse sperm and causes male sterility. Development. 2020;147:dev181057. doi: 10.1242/dev.181057. [DOI] [PubMed] [Google Scholar]
- Xu K., Yang L., Zhao D., Wu Y., Qi H. AKAP3 synthesis is mediated by RNA binding proteins and PKA signaling during mouse spermiogenesis. Biol. Reprod. 2014;90:1–14. doi: 10.1095/biolreprod.113.116111. [DOI] [PubMed] [Google Scholar]
- Yang L., Wu W., Qi H. Gene expression profiling revealed specific spermatogonial stem cell genes in mouse. Genesis. 2013;51:83–96. doi: 10.1002/dvg.22358. [DOI] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Scholer H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K., Nishikawa S., Ogawa M., Hayashi S., Kunisada T., Fujimoto T., Nishikawa S. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–699. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- Zhang X., Smits A.H., van Tilburg G.B., Ovaa H., Huber W., Vermeulen M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018;13:530–550. doi: 10.1038/nprot.2017.147. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wolfle T., Rospert S. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J. Biol. Chem. 2013;288:33697–33707. doi: 10.1074/jbc.M113.508283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W., Hall M.N. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (www.iprox.org) (Ma et al., 2019) with the dataset identifier PXD029274. The processed LC-MS/MS data are included in Table S3. Proteins identified from GST pulled-down assays, related to Figure 4, Table S4. Differentially expressed proteins between Rpl39l null and wild type testes identified by LC-MS/MS, related to Figure 5, Table S5. Differentially expressed proteins in enriched mitochondria of Rpl39l null and wild type testes (FC > 2, p < 0.05), related to Figure 6. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This study does not generate original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.