Abstract
Objective
This investigation devoted to lncRNA FGF14 antisense RNA 2 (FGF14‐AS2) in prostate carcinoma progression.
Methods
The levels of lncRNA FGF14‐AS2, miR‐96‐5p, and Adherens junction‐associated protein‐1 (AJAP1) in prostate carcinoma were tested by Western blot and qRT‐PCR. How these two genes interacted was confirmed by RNA immunoprecipitation and dualluciferase gene methods. The effect of FGF14‐AS2/miR‐96‐5p/AJAP1 axis in prostate carcinoma progression was determined by MTT, Transwell, and nude mice tumor model.
Results
FGF14‐AS2 was a downregulated lncRNA in prostate carcinoma tissue and cells. FGF14‐AS2 could restrain miR‐96‐5p expression while miR‐96‐5p hampered AJAP1. FGF14‐AS2 could effectively decrease the biological behaviors of prostate carcinoma cells, while knock‐down of FGF14‐AS2 triggered opposite results. Moreover, miR‐96‐5p mimic presented a cancer promoter role in prostate carcinoma cells. AJAP1 expression level could affect levels of proteins related to epithelial‐mesenchymal transition. In vivo experiment suggested that overexpressing FGF14‐AS2 could reverse the promotion of silenced AJAP1 on prostate carcinoma cell metastasis, thus to inhibit tumor growth.
Conclusion
lncRNA FGF14‐AS2 was a downregulated lncRNA in prostate carcinoma and influenced cell proliferation and metastasis. The influence relied on modulating miR‐96‐5p and its target gene AJAP1.
Keywords: AJAP1, FGF14‐AS2, miR‐96‐5p, progression, prostate carcinoma
For in vitro cellular functional assay, FGF14‐AS2 expression was decreased in prostate carcinoma cells and the sponging effect of this gene on miR‐96‐5p was attenuated. As a result, miR‐96‐5p bound with its target gene AJAP1, resulting in the degradation of AJAP1. In this way, the proliferation, migration, invasion, and epithelial‐mesenchymal transition of prostate carcinoma cells were regulated.For in vivo assay, a xenograft transplantation nude mouse tumor model was constructed, suggesting that overexpressing FGF14‐AS2 reversed the promoting effect of silencing AJAP1 on the tumor growth of prostate carcinoma.
![]()
1. INTRODUCTION
Prostate carcinoma is the second largest factor of death relevant to cancers. 1 In China, bad diet habits like excessive fat consumption and cutting fiber intake gradually increase prostate carcinoma patients. The morbidity of prostate carcinoma ranks 7th, and the mortality ranks 10th in the malignant tumors in China. 1 Early prostate carcinoma is often ignored because of the lack of early evident symptoms, leading to a decreased cure rate. In addition, the molecular mechanism of prostate carcinoma metastasis remains unclear, and relevant molecular markers that effectively predict prostate carcinoma progression are lacking. Hence, it is urgent to identify a novel biomarker and make a corresponding therapeutic strategy.
Long non‐coding RNAs (lncRNAs) participate in many biological processes, including tumor progression. 2 , 3 So far, many lncRNAs are recognized as biomarkers of prostate carcinoma, like prostate cancer antigen 3 (PCA3). 3 , 4 LncRNA maternally expressed gene 3 (MEG3) inhibits prostate carcinoma progression via regulating miR‐9‐5p/Quaking‐5 (QKI‐5) axis. 5 Li et al. 6 elaborated that prostate cancer‐associated lncRNA on chromosome 7 (lncRNA PCAL7) aggravates prostate cancer. Li et al. 7 also mined some lncRNAs which might be relevant to autophagy. Differential expression analysis here showed downregulated lncRNA FGF14‐AS2 level in prostate carcinoma. Yang et al. 8 illuminated that FGF14‐AS2 functions as an inhibitor gene in breast cancer. Nonetheless, the effect and mechanism of FGF14‐AS2 in prostate carcinoma remain to be further explored.
Abundant evidence illustrated that various miRNAs participate in tumor initiation, development, and metastasis of prostate carcinoma. 9 For example, Mazzu et al. 10 demonstrated that silence of miR‐193b may release prostate cancer subtype 1 (PCS1) gene inhibition to promote prostate carcinoma progression. Chen et al. 11 discovered that miR‐9‐5p hastens epithelial‐mesenchymal transition (EMT) and prostate carcinoma cell growth. Here, we discovered that FGF14‐AS2 could competitively bind miR‐96‐5p. However, studies about the mechanism of FGF14‐AS2 in human prostate carcinoma remain uncertain; therefore, we are interested in investigating the effect of FGF14‐AS2 in prostate carcinoma progression.
Here, we researched the interplay between FGF14‐AS2, miR‐96‐5p, and Adherens junctions‐associated protein‐1 (AJAP1) based on the downregulation of FGF14‐AS2 in prostate carcinoma and probed their effect in cancer progression. FGF14‐AS2 overexpression could sponge miR‐96‐5p and modulate AJAP1 levels in prostate carcinoma to obstruct cancer progression. Hence, it was concluded that lncRNA FGF14‐AS2 modulated prostate carcinoma cell progression via miR‐96‐5p/AJAP1 axis. We offer a reference for finding a biomarker of prostate carcinoma treatment.
2. MATERIALS AND METHODS
2.1. Bioinformatics analysis
Firstly, expression data of lncRNA and mRNA (normal: 52, tumor: 499) and mature miRNA (normal: 52, tumor: 499) were downloaded from TCGA database (https://portal.gdc.cancer.gov/), along with corresponding clinical data. Differential expression analysis (|logFC|>1.5, padj<0.05) was performed on lncRNAs in the normal group and tumor group by using “edgeR” package to obtain differential lncRNAs. The researched lncRNA of the study was determined by combining references. Differential analysis (|logFC|>1.5, padj<0.05) was also undertaken on miRNA and mRNA expression data in the normal group and tumor group by using R package “edgeR.” Then, miRNAs that interact with FGF14‐AS2 were predicted by lncBase (http://carolina.imis.athena‐innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex). Downstream regulatory target genes of target miRNA were predicted by miRWalk (http://mirwalk.umm.uni‐heidelberg.de), mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp), and TargetScan (http://www.targetscan.org/vert_72/). The predicted mRNAs were intersected with differentially downregulated mRNAs to obtain potential target mRNA.
2.2. Cell culture and treatment
Normal human prostate epithelial cell line RWPE‐1 (BNCC100292) and human prostate carcinoma cell lines DU145 (BNCC338240), PC‐3 (BNCC100267), PC‐3 M (BNCC340074), and LNCaP (BNCC342627) were purchased from BeNa Culture Collection (Beijing, China). RWPE‐1 cell line was cultured in keratinocyte serum‐free medium (K‐SFM) (Invitrogen, GIBCO). DU145 cell line was cultured in Dulbecco's modified Eagle medium (DMEM) (BNCC351841, BNCC, China) containing 10% fetal bovine serum (FBS). PC‐3 M cell line was cultured in F‐12K medium (BNCC341829, BNCC, China) containing 10% FBS. PC‐3 and LNCaP cell lines were cultured in roswell park memorial institute (RPMI)‐1640 (BNCC341471, BNCC, China) containing 10% FBS. All cell lines were cultured with 5% CO2 at 37℃.
Cell transfection was conducted with LipofectamineTM 3000 (Invitrogen). Vectors used in the experiments were pcDNA3.1 (Thermo Fisher Scientific, Waltham, MA, USA). Cells were usually allocated into different groups as follows: (1) NC group and pcDNA3.1‐FGF14‐AS2 group (FGF14‐AS2 group); (2) sh‐NC group and sh‐FGF14‐AS2 group; (3) pcDNA group, FGF14‐AS2+mimic NC group, FGF14‐AS2+miR‐96‐5p mimic group, FGF14‐AS2+si‐NC group, and FGF14‐AS2+si‐AJAP1 group; (4) NC+si‐NC group, NC+si‐AJAP1 group, and FGF14‐AS2+si‐AJAP1 group.
2.3. RNA isolation and qRT‐PCR
Total RNA was isolated from cancer cells by TaKaRa MiniBEST universal RNA extraction kit (Ambion lnc., Austin, TX, USA). Quantitative reverse transcription kit (QIAGEN, FSQ‐101, Japan) was used to synthesize cDNA. Real‐time quantitative polymerase chain reaction kit (qRT‐PCR) was provided by Kapa Biosystems Company (Boston, USA). Relative gene expression level was calculated by 2−ΔΔCt method, and data were analyzed by StepOne software. GAPDH and U6 were taken as the internal reference. See Table 1 for primer sequences.
TABLE 1.
Primer sequences
| Target gene | Primer (5′–3′) |
|---|---|
| miR−96‐5p | F: TCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCAAAAA |
| R: ACACTCCAGCTGGGTTTGGCACTAGCACATT | |
| U6 | F: CTCGCTTCGGCAGCACATA |
| R: AACGATTCACGAATTTGCGT | |
| FGF14‐AS2 | F: AGTTCCAGTTACCATCTTCA |
| R: AGGTTCATAGTTGCCAGAC | |
| AJAP1 | F: TCTGAGGCCCCGCTCCCCGAAACGTGA |
| R: GGCGTCTGCCCTGCCCCCAGGAGGTAAA | |
| GAPDH | F: AAATGGTGAAGGTCGGTGTGAAC |
| R: CAACAATCTCCACTTTGCCACTG |
2.4. MTT assay
LNCaP and DU145 cell suspension (5 × 104 cells/well) were cultured in 96‐well plates. Each well was added with 10 μl MTT reagent at specific times for cell incubation at 37 ℃ for 4 h. Afterward, spectrophotometry of each sample was determined at 490 mm. All experiments were repeated 3 times with 3 parallel wells to calculate their average value.
2.5. Transwell assay
In vitro invasive and migratory abilities of prostate carcinoma cells were evaluated by Transwell (Costar, NY, USA). After being treated under assorted culture conditions, 1 × 105 cells in serum‐free medium were inoculated in the upper chamber coated (invasion assay) or uncoated (migration assay) with growth factor reduced (GFR) basement membrane matrix®. The lower chamber was added with medium containing 10% FBS as chemical attractant. Q‐tip was applied to remove cells on the upper surface of the membrane in due time. Migrating and invading cells were fixed with formaldehyde and stained with 0.5% crystal violet (Sigma). Lastly, invading and migrating cells were counted in 5 random fields under a microscope.
2.6. Fluorescence in situ hybridization (FISH) assay
FGF14‐AS2 expression in prostate carcinoma cells was detected by lncRNA FISH probe and FISH kit (RIBO Bio, China). Cells were washed with phosphate buffer saline (PBS) and fixed with 4% formaldehyde at room temperature for 10 min. Then, cells were permeated with 0.5% Triton X‐100 at 4℃ and hybridized with target probes according to the manufacturer's instruction. FGF14‐AS2 expression in prostate carcinoma cells was detected by a confocal microscope. Cell nucleus was stained by DAPI (Thermo Fisher Scientific).
2.7. Nucleus and cytoplasm separation assay
PARIS kit (AM1921, Life Technologies, Carlsbad, CA, USA) was employed to separate nucleus and cytoplasm of prostate carcinoma cells.
2.8. Dual‐luciferase reporter gene assay
FGF14‐AS2 fragments containing predicted binding sites of miR‐96‐5p were firstly PCR amplified. Subsequently, the amplified FGF14‐AS2 fragments were cloned to dual‐luciferase miRNA target expression vector (Promega, Madison, WI, USA) to construct wild‐type FGF14‐AS2 (FGF14‐AS2‐WT). Mutant FGF14‐AS2 (FGF14‐AS2‐MUT) was constructed by the same method. Similarly, AJAP1‐WT and AJAP1‐MUT were established. Next, luciferase activity of cells in each group was detected by dual‐luciferase reporter gene detection system (Promega, Madison, WI, USA).
2.9. RNA immunoprecipitation (RIP) assay
RIP was detected with Imprint RIP kit (Sigma‐Aldrich, St. Louis, MO, USA). The transfected cells were collected. Then, cells were suspended with RIP lysis buffer (Solarbio) and centrifuged under 12,000 g condition for 5 min. Later, cell lysis products were incubated with anti‐argonaute2 (anti‐Ago2) or anti‐IgG (negative control) at 4 ℃ overnight. Next, protein A magnetic beads were added to obtain immunoprecipitate. Total RNA in cells was extracted with GenEluteTM Total RNA Purification Kit (Sigma‐Aldrich). Relative enrichment of FGF14‐AS2 and miR‐96‐5p was analyzed by qRT‐PCR.
2.9.1. Western blot assay
Cancer cells were lysed in radio immunoprecipitation assay (RIPA) buffer. Proteins were separated in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and electro‐transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with primary antibodies at 4 ℃ overnight and then incubated with horseradish peroxidase (HRP)‐labeled secondary antibody IgG (Abcam, UK) at room temperature for 3 h. Immunolabeling was visualized by enhanced chemiluminescence (ECL) system (Amersham, Bucks, UK). Primary antibodies were rabbit polyclonal antibodies: anti‐AJAP1 antibody (Abcam, UK), anti‐E‐cadherin (Abcam, UK), anti‐N‐cadherin (Abcam, UK), anti‐Vimentin (Abcam, UK), and anti‐GAPDH (Abcam, UK).
2.10. Nude mice tumor assay
2 × 107 DU145 cells were subcutaneously injected into 18 male BALB/c nude mice (5 weeks). Before transfection, DU145 cells were transfected with NC+si‐NC, NC+si‐AJAP1, and FGF14‐AS2+si‐AJAP1, respectively, and divided into 3 groups with each group 6 randomly allocated nude mice. Tumor volume and weight were measured at specific time point. Mice were killed 4 weeks later. Tumors were resected and measured for immunohistochemical assay and qRT‐PCR assay.
2.11. Immunohistochemical (IHC) assay
Paraffin‐embedded xenograft tumor tissue was fixed, dewaxed, and washed with PBS. Samples were detected with AJAP1 (Abcam, UK), N‐cadherin (Abcam, UK), E‐cadherin (Abcam, UK), GAPDH (Abcam, UK), and Vimentin (Abcam, UK) antibodies at 4 ℃ overnight. Afterward, samples were incubated with secondary antibody goat anti‐rabbit IgG (Abcam, UK) at 37 ℃ for 60 min. Then, 0.5 mg/mL DAPI was applied to incubate cells at 37 ℃. All cover glasses were detected under a microscope (Nikon C1‐Si, Mississauga, Canada). EZ‐C1 3.20 FreeViewer software was utilized for image analysis.
2.12. Data analysis
GraphPad Prism 6.0 (GraphPad Prism 6.0, San Diego, CA, USA) was employed for data analysis. Each experiment was repeated at least 3 times. Results were presented as mean ±standard deviation (SD). Two‐tailed Student's t test or one‐way analysis of variance was applied to determine difference between groups. p < 0.05 was significantly different.
3. RESULTS
3.1. FGF14‐AS2 is lowly expressed in prostate carcinoma tissue
Differentially expressed lncRNAs were screened through edgeR differential analysis, and the screened 786 differential lncRNAs were shown in volcano plot (Figure 1A). Among them, FGF14‐AS2 was stably expressed in normal samples and tumor samples, and it was highly significantly lowly expressed in tumor samples (Figure 1B). Meanwhile, FGF14‐AS2 expression level was lower in 4 prostate carcinoma cell lines in comparison with normal cells (Figure 1C). Additionally, FGF14‐AS2 level was the highest in DU145 cells while the lowest in LNCaP cells. Thus, these two cell lines were used in the experiments.
FIGURE 1.
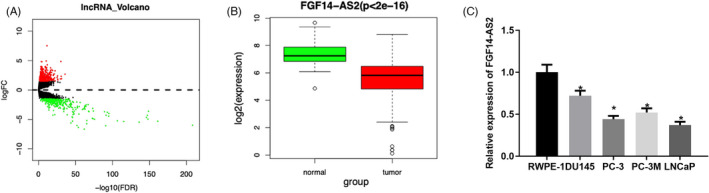
FGF14‐AS2 is lowly expressed in prostate carcinoma. (A) Volcano plot showed differential lncRNAs in prostate carcinoma tissue and adjacent tissue (red: upregulated expression; green: downregulated expression). (B) Boxplot of FGF14‐AS2 expression in normal group and tumor group. (C) qRT‐PCR exhibited that FGF14‐AS2 was downregulated in human prostate carcinoma cell lines. *Denotes p < 0.05
3.2. FGF14‐AS2 expression level affects biological functions of prostate carcinoma cells
We overexpressed FGF14‐AS2 in LNCaP cells and silenced FGF14‐AS2 in DU145 cells, respectively. FGF14‐AS2 expression was elevated after LNCaP cells were transfected with FGF14‐AS2 vector while declined after DU145 cells were transfected with sh‐FGF14‐AS2 (Figure 2A). MTT assay illuminated that FGF14‐AS2 suppressed cell proliferation while sh‐FGF14‐AS2 facilitated cell proliferation (Figure 2B). Transwell assay also bared that FGF14‐AS2 overexpression repressed cancer cell migration and invasion. However, silenced FGF14‐AS2 expression accelerated cancer cell migration and invasion (Figure 2C). Therefore, FGF14‐AS2 may work as a tumor‐inhibiting lncRNA in prostate carcinoma progression.
FIGURE 2.
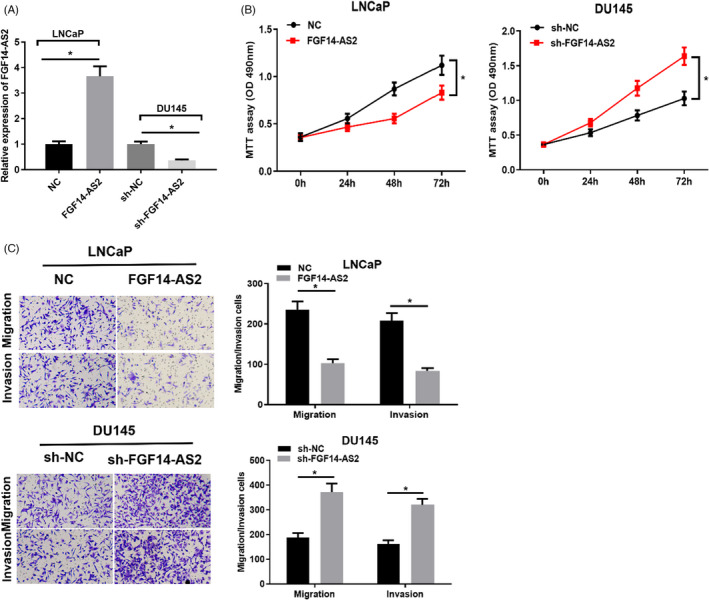
FGF14‐AS2 inhibits functions of prostate carcinoma cells. (A) FGF14‐AS2 level in NC, FGF14‐AS2, sh‐NC, and sh‐FGF14‐AS2 groups. (B) MTT assay detected LNCaP and DU145 cell proliferation. (C) Cell migratory and invasive abilities in each group (100×). *Denotes p < 0.05
3.3. FGF14‐AS2 directly represses miR‐96‐5p expression
It was reported that lncRNA plays a role in tumor progression by adsorbing miRNA. 12 The location of FGF14‐AS2 in subcellular fractions was determined by FISH and qRT‐PCR assays. The two assays both suggested that FGF14‐AS2 was mostly localized in the cytoplasm in prostate carcinoma cells (Figure 3A, 3B). Afterward, 51 differentially expressed miRNAs were obtained from TCGA (Figure 3C). Based on the negative regulation of lncRNA‐miRNA in ceRNA, miRNAs that interact with FGF14‐AS2 were predicted on lncBase, and the predicted results were intersected with the 36 upregulated miRNAs obtained from differential analysis. Then, 3 differential miRNAs with binding sites with FGF14‐AS2 were obtained (Figure 3D). MiR‐96‐5p was significantly negatively correlated with FGF14‐AS2 as presented by Pearson correlation analysis (Figure 3E) and remarkably highly expressed in prostate carcinoma tissue (Figure 3F). Putative binding sites between FGF14‐AS2 and miR‐96‐5p were shown in Figure 3G. Dual‐luciferase reporter gene assay exhibited that miR‐96‐5p mimic could markedly decline the luciferase activity of cells with FGF14‐AS2‐WT (Figure 3H ). RIP analysis unmasked that FGF14‐AS2 and miR‐96‐5p both mainly enriched in Ago2 immunoprecipitate (Figure 3I). qRT‐PCR bared that FGF14‐AS2 overexpression remarkably declined miR‐96‐5p expression in prostate carcinoma cells, while inhibiting FGF14‐AS2 expression conspicuously upregulated miR‐96‐5p expression (Figure 3J). Besides, miR‐96‐5p expression in prostate carcinoma cells was detected. miR‐96‐5p expression was remarkably upregulated in prostate carcinoma cells (Figure 3K). Given the above, FGF14‐AS2 works as a ceRNA for miR‐96‐5p in prostate carcinoma progression.
FIGURE 3.
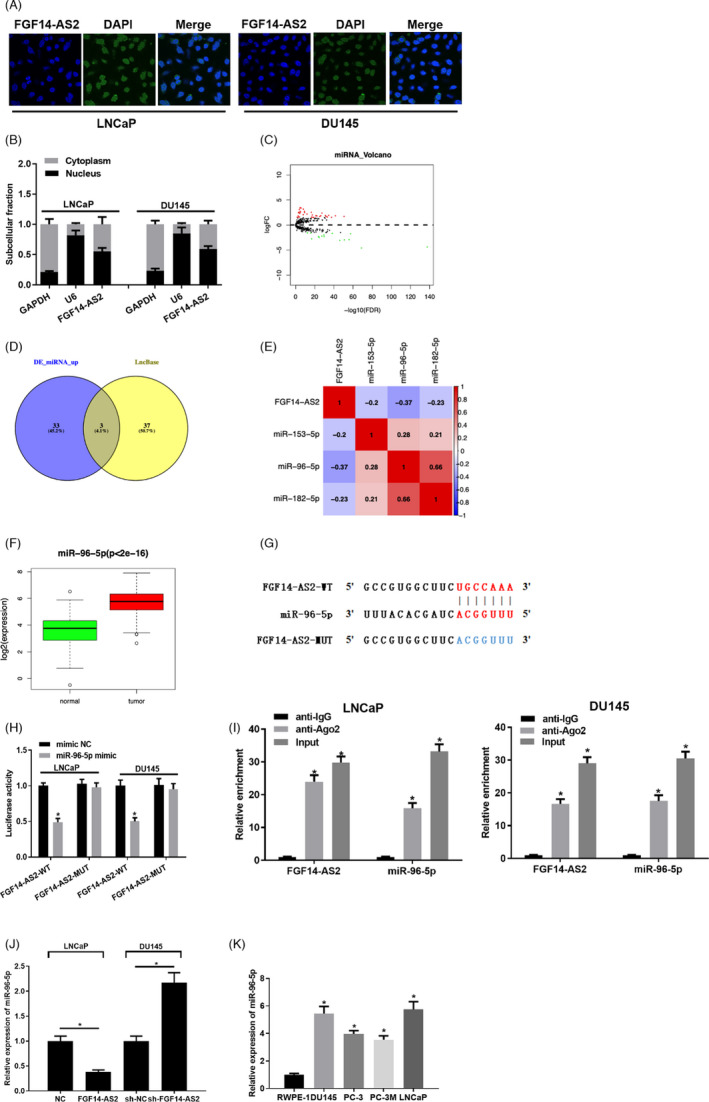
FGF14‐AS2 directly inhibits miR‐96‐5p expression. (A) Results of FISH assay of LNCaP and DU145 cells. (B) qRT‐PCR detected FGF14‐AS2, U6, and GAPDH expression level in cell nucleus and cytoplasm in LNCaP and DU145 cells after separation of nucleus and cytoplasm. (C) 51 differentially expressed miRNAs obtained from TCGA database (red: upregulated expression; green: downregulated expression). (D) Venn diagram of differentially expressed miRNAs and predicted miRNAs. (E) Association between miRNA candidates and FGF14‐AS2. (F) Boxplot of miR‐96‐5p levels. (G) Binding sites between FGF14‐AS2 and miR‐96‐5p. (H‐I) The binding relationship between miR‐96‐5p and FGF14‐AS2. (J) qRT‐PCR detected the effect of FGF14‐AS2 expression level on miR‐96‐5p expression in NC, FGF14‐AS2, sh‐NC, and sh‐FGF14‐AS2 groups. (K) Level of miR‐96‐5p. *Denotes p < 0.05
3.4. MiR‐96‐5p binds AJAP1 and modulates EMT pathway
mRNA expression data (normal: 52, tumor: 499) of the cancer were downloaded from TCGA database. Altogether, 1412 differentially expressed mRNAs were obtained by differential analysis using edgeR (Figure 4A). Next, potential target genes of miR‐96‐5p were predicted by databases. The predicted genes were intersected with differentially downregulated mRNAs, and 6 potential downstream target genes of miR‐96‐5p were obtained (Figure 4B). Pearson correlation analysis was performed on these 6 mRNAs and FGF14‐AS2. It was found that AJAP1 had the most significant positive correlation with FGF14‐AS2 (Figure 4C). TCGA data showed markedly low level of AJAP1 in prostate carcinoma tissue (Figure 4D). Similar results were examined by qRT‐PCR in cancer cells (Figure 4E). Hence, AJAP1 was taken as the downstream gene to research in this study.
FIGURE 4.

MiR‐96‐5p directly affects AJAP1 and regulates EMT signaling pathway. (A) Volcano plot of differential mRNAs in prostate carcinoma data (red: upregulated expression; green: downregulated expression). (B) Venn diagram of differentially downregulated mRNAs and predicted target mRNAs. (C) Heat map of Pearson correlation analysis between FGF14‐AS2 and mRNA candidates. (D) Boxplot of AJAP1 level (green: normal; red: tumor) in TCGA. (E) qRT‐PCR detected AJAP1 mRNA expression level in cell lines. (F) Binding region of miR‐96‐5p on AJAP1 mRNA 3’UTR. (G) The binding relationship between miR‐96‐5p and AJAP1. (H) The impacts of miR‐96‐5p overexpression on E‐cadherin, AJAP1, Vimentin, and N‐cadherin protein expression level. *Denotes p < 0.05
TargetScan predicted that miR‐96‐5p had binding relationship with AJAP1 (Figure 4F), and the interplay was verified by dual‐luciferase reporter gene assay. It was discovered that miR‐96‐5p mimic declined the luciferase activity of LNCaP and DU145 cells with AJAP1‐WT (Figure 4G). Moreover, a study manifested that AJAP1 can affect EMT and metastasis of hepatocellular carcinoma (HCC). 13 Hence, the expression level of EMT markers in prostate carcinoma cell lines LNCaP and DU145 was also detected. It was discovered that miR‐96‐5p overexpression evidently declined AJAP1 protein expression and E‐cadherin expression while upregulated N‐cadherin and Vimentin expression (Figure 4H). Given the above results, it was concluded that miR‐96‐5p could bind AJAP1 and thus regulate EMT pathway.
3.5. FGF14‐AS2 inhibits malignant behaviors of prostate carcinoma cells through modulating miR‐96‐5p/AJAP1
Later, 5 groups of the LNCaP cell line were settled to research whether FGF14‐AS2 modulates AJAP1 through miR‐96‐5p, thereby mediating EMT pathway activation. In Figure 5A, we detected FGF14‐AS2, miR‐96‐5p, and AJAP1 mRNA expression in LNCaP cells in 5 groups, respectively, and it was presented that FGF14‐AS2 overexpression could downregulate miR‐96‐5p level while upregulate AJAP1 expression. Nevertheless, AJAP1 expression was decreased after simultaneously overexpressing FGF14‐AS2 and miR‐96‐5p, and silencing AJAP1 upon FGF14‐AS2 overexpression could also reverse the promotion of FGF14‐AS2 on AJAP1 expression. Western blot exhibited the same results. Besides, E‐cadherin protein expression trend was basically consistent with AJAP1, while Vimentin and N‐cadherin protein expressions were increased opposite to AJAP1 expression (Figure 5B). Moreover, cell functional experiments were also conducted for testification. It was displayed that the proliferative, migratory, and invasive abilities were weakened in high AJAP1 expression group. It was presented that over‐expression of miR‐96‐5p or silencing AJAP1 could reverse the tumor‐inhibitory effects of FGF14‐AS2 in the prostate carcinoma cells (Figure 5C–5E).
FIGURE 5.
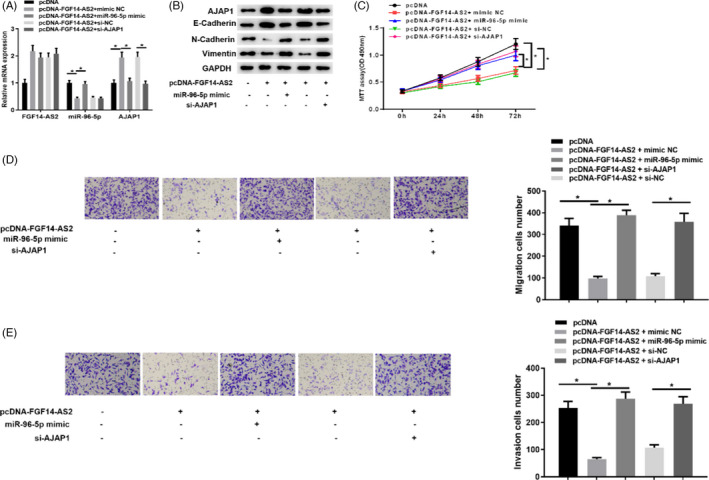
FGF14‐AS2 represses prostate carcinoma cell functions through FGF14‐AS2/miR‐96‐5p/AJAP1 axis. (A) qRT‐PCR detected FGF14‐AS2, miR‐96‐5p, and AJAP1 mRNA expression level in pcDNA, pcDNA‐FGF14‐AS2+mimic NC, pcDNA‐FGF14‐AS2+miR‐96‐5p mimic, pcDNA‐FGF14‐AS2+si‐NC, and pcDNA‐FGF14‐AS2+si‐AJAP1 groups. (B) Western blot detected AJAP1 and key proteins of EMT markers in each group. (C) MTT assay detected cell viability in 5 transfection groups. (D‐E) Cell migration and invasion in each group (100×). * Denotes p < 0.05
3.6. FGF14‐AS2 overexpression declines tumorigenesis of prostate carcinoma cells in vivo
To verify the carcinogenesis of FGF14‐AS2 in prostate carcinoma progression, we constructed a xenograft transplantation nude mice tumor model. After 4 weeks of subcutaneous injection, tumor formation in NC+si‐AJAP1 group was significantly quicker than NC+si‐NC group. Besides, tumor volume was evidently decreased in FGF14‐AS2+si‐AJAP1 group than that in NC+si‐AJAP1 group (Figure 6A–6C). Next, qRT‐PCR displayed that FGF14‐AS2 overexpression remarkably downregulated miR‐96‐5p expression while upregulated AJAP1 expression (Figure 6D). Immunohistochemical assay also indicated that AJAP1 expression was related to EMT. Knock‐down of AJAP1 could upregulate Vimentin N‐and cadherin expression while downregulate E‐cadherin expression. After overexpressing FGF14‐AS2 which led to promoted AJAP1 expression, E‐cadherin expression was strengthened while Vimentin and N‐cadherin expression were weakened (Figure 6E). The above results confirmed that AJAP1 could suppress EMT, and FGF14‐AS2 overexpression could inhibit the malignant progression and EMT signaling pathway of prostate carcinoma cells.
FIGURE 6.
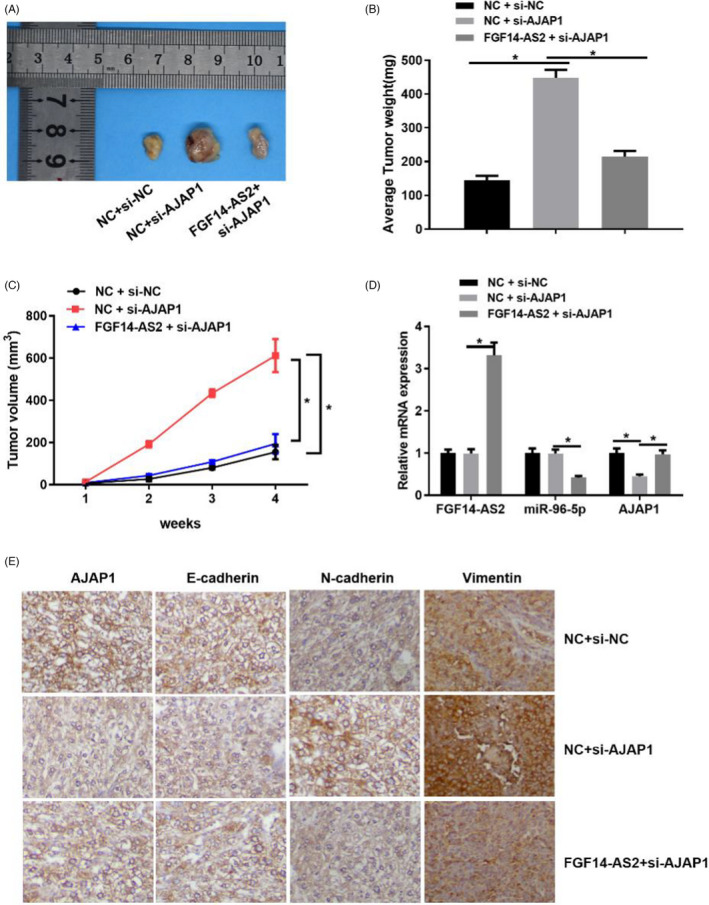
FGF14‐AS2 overexpression declines tumorigenesis of prostate carcinoma cells in vivo (A) Images of tumors in mice. (B)–(C) Average tumor weight and volume. (D) qRT‐PCR detected FGF14‐AS2, miR‐96‐5p, and AJAP1 mRNA expression in tissue. (E) Immunohistochemical staining showed Vimentin, E‐cadherin, AJAP1, N‐and cadherin expression in tissue (400×). *Denotes p < 0.05
4. DISCUSSION
The morbidity and mortality of prostate carcinoma are soaring worldwide. 14 , 15 Therefore, comprehensive understanding of the pathogenesis of prostate carcinoma is an important basis for developing new treatment strategies. Recently, abundant newly found lncRNAs were proved to be vital in human diseases, especially cancers(16). For instance, Wan et al. 16 researched that ADAM Metallopeptidase with Thrombospondin Type 1 Motif 9 Antisense RNA 1 (ADAMTS9‐AS1) is a molecular sponge of miR‐96 as a ceRNA and modulates PRDM16 expression, providing evidence for the treatment strategy of prostate carcinoma. You et al. 17 found that lncRNA colon cancer‐associated transcript 1 (CCAT1) facilitates prostate carcinoma cell proliferation through miR‐28‐5p/DEAD‐Box Helicase 5 (DDX5) interaction. Han et al. 18 found that downregulated lncRNA small nucleolar RNA host gene 7 (SNHG7) represses prostate carcinoma cell EMT through miR‐324‐3p/Wnt family member 2B (WNT2B) axis. This investigation focused on FGF14‐AS2/miR‐96‐5p/AJAP1 axis in prostate carcinoma proliferation and metastasis. It was believed that FGF14‐AS2 represses prostate carcinoma progression through modulating miR‐96‐5p/AJAP1 axis.
FGF14‐AS2 level was found to be low in prostate carcinoma. A study revealed that FGF14‐AS2 is relevant to cancer progression and prognosis. 8 Yang et al. 19 demonstrated that FGF14‐AS2 represses breast cancer proliferation, migration, and invasion and induces apoptosis through sponging miR‐205‐5p. Besides, FGF14‐AS2 serves as a regulator of colorectal cancer by downregulating miR‐1288‐3p. 20 The effect of FGF14‐AS2 in our study was similar to that on other tumors. According to in vitro experiments, FGF14‐AS2 overexpression remarkably decreased biological functions of prostate carcinoma cells, while inhibiting FGF14‐AS2 expression markedly strengthened cell proliferation, migration, and invasion in prostate carcinoma. Altogether, FGF14‐AS2 may be a cancer inhibitor in prostate carcinoma.
To determine the molecular mechanism of FGF14‐AS2 as ceRNA in prostate carcinoma cells, we intersected potential interacted genes of FGF14‐AS2 predicted on lncBase and differentially upregulated miRNAs in TCGA. It was discovered that miR‐96‐5p may be a potential regulatory target of FGF14‐AS2. Yu et al. 21 manifested that miR‐96 facilitates prostate carcinoma cell proliferation and clonogenicity through targeting FOXO1. Hong et al. 22 found that miR‐96 accelerates breast cancer cell proliferation, migration, and invasion via binding tyrosine‐protein phosphatase non‐receptor type 9 (PTPN9). Liu et al. 23 elaborated that miR‐96 aggravates ovarian cancer via binding Caveolae1 and may be an effective target for treating ovarian cancer. In our study, FGF14‐AS2 may directly impact miR‐96‐5p in prostate carcinoma.
This investigation discovered that AJAP1 was a target of miR‐96‐5p. AJAP1 is a posited tumor suppressor gene located in chromosome 1 region 1p36 which is frequently deleted in human cancers. 24 AJAP1 expression is downregulated in various malignant tumors. For instance, Yang et al. 25 discovered that AJAP1 is lowly expressed in glioblastoma and positively correlated with patient's poor survival. AJAP1 can inhibit the adhesive and migratory ability of oligodendroglioma cells. 26 Ezaka et al. 27 elaborated that the lack of AJAP1 in HCC cell lines and tissue strengthens its inhibition on HCC and its intermediation in EMT. Low levels of AJAP1 in hepatocellular cancer are relevant to tumor metastasis. 28 Here, it was found that AJAP1 expression evidently declined in prostate carcinoma cell lines. We confirmed that AJAP1 was a potential target of miR‐96‐5p in prostate carcinoma progression. Besides, functional assay and in vivo experiment indicated that FGF14‐AS2 could regulate AJAP1 expression, to hinder prostate carcinoma progression and modulate EMT pathway. FGF14‐AS2/miR‐96‐5p/AJAP1 axis is essential for prostate carcinoma progression.
Viewed in toto, it was illustrated that FGF14‐AS2 upregulation adjusted AJAP1 and EMT pathway via sponging miR‐96‐5p, thereby mediating cell behaviors in prostate carcinoma. It was also proved that FGF14‐AS2 had a value as a potential target for treating prostate carcinoma.
CONFLICT OF INTEREST
All authors declare no conflicts of interest in this work.
AUTHORS CONTRIBUTIONS
RBL designed the study. YCC performed the experiment. JWW acquired and analyzed the data. XBC drafted the first version of the article. SNZ and HQY contributed significantly on software and prepared manuscript. YMW and FW revised the article. FW decided the final version of the manuscript for submission.
Li R, Chen Y, Wu J, et al. LncRNA FGF14‐AS2 represses growth of prostate carcinoma cells via modulating miR‐96‐5p/AJAP1 axis. J Clin Lab Anal. 2021;35:e24012. 10.1002/jcla.24012
Funding information
None.
DATA AVAILABILITY STATEMENT
The data and materials in the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5‐29. [DOI] [PubMed] [Google Scholar]
- 2. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629‐641. [DOI] [PubMed] [Google Scholar]
- 3. de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62(9):2695‐2698. [PubMed] [Google Scholar]
- 4. Sartori DA, Chan DW. Biomarkers in prostate cancer: what's new? Curr Opin Oncol. 2014;26(3):259‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu M, Huang Y, Chen T, et al. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR‐9‐5p/QKI‐5 axis. J Cell Mol Med. 2019;23(1):29‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z, Teng J, Jia Z, Zhang G, Ai X. The long non‐coding RNA PCAL7 promotes prostate cancer by strengthening androgen receptor signaling. J Clin Lab Anal. 2021;35(2):e23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J, Du H, Chen W, Qiu M, He P, Ma Z. Identification of potential autophagy‐associated lncRNA in prostate cancer. Aging. 2021;13(9):13153‐13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang F, Liu YH, Dong SY, et al. A novel long non‐coding RNA FGF14‐AS2 is correlated with progression and prognosis in breast cancer. Biochem Biophys Res Commun. 2016;470(3):479‐483. [DOI] [PubMed] [Google Scholar]
- 9. Wu X, Xiao Y, Zhou Y, Zhou Z, Yan W. LncRNA FOXP4‐AS1 is activated by PAX5 and promotes the growth of prostate cancer by sequestering miR‐3184‐5p to upregulate FOXP4. Cell Death Dis. 2019;10(7):472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mazzu YZ, Yoshikawa Y, Nandakumar S, et al. Methylation‐associated miR‐193b silencing activates master drivers of aggressive prostate cancer. Mol Oncol. 2019;13(9):1944‐1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L, Hu W, Li G, Guo Y, Wan Z, Yu J. Inhibition of miR‐9‐5p suppresses prostate cancer progress by targeting StarD13. Cell Mol Biol Lett. 2019;24:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710‐718. [DOI] [PubMed] [Google Scholar]
- 13. Qu W, Wen X, Su K, Gou W. MiR‐552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression. J Cell Mol Med. 2019;23(2):1541‐1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133‐1145. [DOI] [PubMed] [Google Scholar]
- 15. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan J, Jiang S, Jiang Y, et al. Data mining and expression analysis of differential lncRNA ADAMTS9‐AS1 in prostate cancer. Front Genet. 2019;10:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. You Z, Liu C, Wang C, et al. LncRNA CCAT1 promotes prostate cancer cell proliferation by interacting with DDX5 and MIR‐28‐5P. Mol Cancer Ther. 2019;18(12):2469‐2479. [DOI] [PubMed] [Google Scholar]
- 18. Han Y, Hu H, Zhou J. Knockdown of LncRNA SNHG7 inhibited epithelial‐mesenchymal transition in prostate cancer though miR‐324‐3p/WNT2B axis in vitro. Pathol Res Pract. 2019;215(10):152537. [DOI] [PubMed] [Google Scholar]
- 19. Yang Y, Xun N, Wu JG. Long non‐coding RNA FGF14‐AS2 represses proliferation, migration, invasion, and induces apoptosis in breast cancer by sponging miR‐205‐5p. Eur Rev Med Pharmacol Sci. 2019;23(16):6971‐6982. [DOI] [PubMed] [Google Scholar]
- 20. Hou R, Liu Y, Su Y, Shu Z. Overexpression of long non‐coding RNA FGF14‐AS2 inhibits colorectal cancer proliferation via the RERG/Ras/ERK signaling by sponging microRNA‐1288‐3p. Pathol Oncol Res. 2020;26(4):2659‐2667. [DOI] [PubMed] [Google Scholar]
- 21. Yu JJ, Wu YX, Zhao FJ, Xia SJ. miR‐96 promotes cell proliferation and clonogenicity by down‐regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31(4):910. [DOI] [PubMed] [Google Scholar]
- 22. Hong Y, Liang H, Uzair Ur R, et al. miR‐96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu B, Zhang J, Yang D. miR‐96‐5p promotes the proliferation and migration of ovarian cancer cells by suppressing Caveolae1. J Ovarian Res. 2019;12(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68(8):2551‐2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang C, Li YS, Wang QX, et al. EGFR/EGFRvIII remodels the cytoskeleton via epigenetic silencing of AJAP1 in glioma cells. Cancer Lett. 2017;403:119‐127. [DOI] [PubMed] [Google Scholar]
- 26. Zeng L, Kang C, Di C, et al. The adherens junction‐associated protein 1 is a negative transcriptional regulator of MAGEA2, which potentiates temozolomide‐induced apoptosis in GBM. Int J Oncol. 2014;44(4):1243‐1251. [DOI] [PubMed] [Google Scholar]
- 27. Ezaka K, Kanda M, Sugimoto H, et al. Reduced expression of adherens junctions associated protein 1 predicts recurrence of hepatocellular carcinoma after curative hepatectomy. Ann Surg Oncol. 2015;22(Suppl 3):S1499‐S1507. [DOI] [PubMed] [Google Scholar]
- 28. Han J, Xie C, Pei T, et al. Deregulated AJAP1/beta‐catenin/ZEB1 signaling promotes hepatocellular carcinoma carcinogenesis and metastasis. Cell Death Dis. 2017;8(4):e2736. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials in the current study are available from the corresponding author on reasonable request.


