Abstract
Background
Brain‐derived neurotrophic factor (BDNF) is considered to be one of the best candidate genes for depression. However, whether sertraline treatment affects the methylation level of this gene remains unknown.
Methods
Fifty‐three patients with depression and 51 healthy controls were included in the study. The methylation level of BDNF exon I was determined in blood samples from these subjects. The Hamilton Depression Scale was used to evaluate the depression status of patients. Single nucleotide polymorphism detection was used for genotyping, and a receiver operating characteristic (ROC) curve was used to evaluate the predictive value of the methylation level of this locus in patients with depression.
Results
There was a significant difference in the methylation level of BDNF exon I between the control and depression groups. No effect of sertraline monotherapy on BDNF methylation was found in subjects with depression. Moreover, no interaction was found between BDNF genotype and the per cent methylation of BDNF exon I. However, methylation at this site was positively correlated with diurnal variation and retardation scores. Blood homocysteine concentrations were significantly reduced by sertraline treatment. No influence of genotype on serum BDNF concentration was found in subjects with depression. The ROC curve showed that methylation of BDNF exon I may be used to distinguish patients from healthy people, to a certain extent.
Conclusion
Methylation of BDNF exon I may be used as a biomarker of depression and may be a therapeutic target for previously untreated depression.
Keywords: 5‐hydroxytryptamine, BDNF, depression, gene methylation, sertraline
The Brain‐derived neurotrophic factor (BDNF) exon I methylation level in Depression patients was significantly lower than that in the healthy controls,and Receiver operation characteristics (ROC) curve showed that the BDNF promoter region I methylation could be used for distinguishing patients from healthy people to a certain extent.

1. INTRODUCTION
Depression is one of the 10 most common diseases worldwide, and it is estimated that it will become the second most common disease by 2030. 1 , 2 Depressive patients are mainly characterized by depression, a decline in interest and fatigue, accompanied by varying degrees of impaired social function. 1 , 2 Thus far, no specific genetic, biological, biochemical or neuropathological changes have been identified as objective biological indicators of depression. 3 At present, treatment of depression is mainly based on the empirical use of antidepressants to control the main symptoms. However, there is insufficient evidence to prove that different antidepressants have significant differences in efficacy. In addition, depression, like many diseases, often has obvious individual treatment differences due to heterogeneity in age, depression type and genetic background. 4 Exploring the aetiological mechanism and specific disease indicators of depression to further optimize diagnosis and identify new targets for antidepressants is very important for the future diagnosis and treatment of depression.
Early genetic studies found a significant relationship between genetic factors and the pathogenesis of depression. 5 Previous studies have shown that there may be a large number of depression susceptibility loci in humans. 6 In recent years, with the development of molecular genetic techniques, these susceptibility genes have been found to be involved in a variety of cellular metabolic pathways and neurotransmitter transmission pathways, and their corresponding variations may be involved in the occurrence and development of disease by causing abnormal levels of neurotransmitters (5‐hydroxytryptamine, norepinephrine, monoamine oxidase inhibitors) in the brain and even brain pathology and structure in patients with depression. 7 , 8 Brain‐derived neurotrophic factor (BDNF), a member of the neurotrophic factor family, is considered to be one of the best candidate genes for depression. 9 It can stimulate and promote the growth and differentiation of nerve cells, maintain their survival and normal function and prevent neuronal death. 10
DNA methylation is a common mechanism of epigenetic regulation and has been found to be involved in the pathophysiological mechanism of depression. DNA methylation refers to the selective addition of a methyl group (‐CH3) to the cytosine nucleotide of CpG sites in DNA to form 5´‐CH3 cytosine. 11 , 12 The methylation status of functional genes can lead to the silencing of gene expression and can be inherited. 12 The effects of diet, genetic variation, social and psychological stimulation and environmental chemicals can lead to changes in the level of gene methylation, which can lead to a variety of mental illnesses, including depression. 13 , 14 , 15 The methylation level of some BDNF gene regions has been found to be associated with certain phenotypes of depression. 8 , 9 , 16
A single nucleotide polymorphism (SNP) mainly refers to a single nucleotide variation at the genome level, due to the insertion, deletion or substitution of a base. 17 SNPs may lead to disorders through changes in protein function and thus may provide some reference for genetic disease mechanisms. 17 Based on this, some scholars believe that BDNF gene polymorphisms may be closely associated with depression. 18 For example, Lee et al. found that the Val/Met and Met/Met genotypes of the Val66Met polymorphism are associated with 1.67 times and 2.58 times the risk of depression, respectively, compared with the Val/Val genotype. 18 In addition, Pei et al. 19 found that Met allele carriers have a higher risk of senile depression. These studies confirmed that BDNF polymorphisms affect BDNF protein expression and that the Met allele is a risk factor for depression. However, Duncan et al. 20 found that the Val/Val genotype is associated with a higher risk of depression. Moreover, some studies have shown that the Val66Met polymorphism of the BDNF gene has no obvious relationship with depression. 21 , 22 The inconsistencies of the above results indicate that gene polymorphisms may also be affected by other genetic or environmental factors. Epigenetic regulation of the BDNF gene is thought to be affected by environmental factors. Methylation is one of the most common epigenetic regulatory mechanisms of the BDNF gene.
The mechanism involved in the response of BDNF to different antidepressants is unknown. More in‐depth exploration of the relationship between antidepressants and BDNF will help us to explore the aetiological mechanism of depression and develop improved treatments. Sertraline is a 5‐hydroxytryptamine (5‐HT) reuptake inhibitor commonly used as an antidepressant in the clinic. In this study, sertraline‐treated patients with depression were selected as the case group, and age‐ and gender‐matched healthy adults were selected as the control group. Genotypes at the rs7103411 locus of BDNF were assessed. Sequencing was used to compare the methylation level of exon I of BDNF between the case and control groups and explore the correlations between the methylation level and sertraline use, genotype and clinical symptoms of depression to identify epigenetic markers that may facilitate diagnosis and antidepressant treatment guidance.
2. MATERIALS AND METHODS
2.1. Subjects
Fifty‐three patients with depression (27 males and 26 females), including outpatients and inpatients, were recruited from December 2015 to July 2019 from Ningbo Kangning Hospital. Healthy subjects (25 males and 26 females) with no family history of mental illness and no major physical disease who were willing to participate in this study were recruited from among healthy volunteers at Ningbo Kangning Hospital from April 2016 to July 2019. The patients with depression ranged in age from 18 to 68 years, with an average age of 47.41 years. Patients were included if they (i) were first‐episode, drug‐free depressive patients who voluntarily gave informed consent to participate in this study; (ii) were diagnosed with depressive disorder according to the diagnostic criteria of the International Classification of Diseases‐10; (iii) had a Hamilton Depression Scale (HAMD) score greater than or equal to 18; (iv) were independently diagnosed with depression by two senior professional psychiatrists; (v) had no history of chronic disease or severe physical disease; (vi) had no history of electroconvulsive therapy; (vii) had no history of psychoactive substance abuse; and (viii) were able to complete an assessment using the relevant scale. On the day before commencing sertraline treatment, whole blood samples were collected. A second blood sample was collected after 4 weeks of sertraline monotherapy. The blood homocysteine (Hcy) of subjects was measured by Chemistry Analyzer (AU2700, Olympus) at the Clinical Chemistry Laboratory of Ningbo Kangning Hospital. All study procedures were performed in accordance with the Declaration of Helsinki and were approved by the Medical Ethics Committee of Ningbo Kangning Hospital.
2.2. Collection of blood samples and determination of serum BDNF concentrations
After fasting, a 5 ml venous peripheral blood sample was collected from each patient with depression and centrifuged at 1500× g for 12 min. The supernatant was transferred to a new centrifuge tube, while avoiding the transfer of any cellular components. The serum samples and blood cellular components were stored at −80°C. Serum BDNF concentrations were determined by enzyme‐linked immunosorbent assay according to the manufacturer's instructions (Abcam). BDNF concentrations were measured before and after sertraline treatment.
2.3. Detection of methylation in exon I of the BDNF gene
The BDNF gene sequence was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). Genomic DNA was extracted from blood samples using a TIANamp Genomic DNA Kit (DP304) according to the manufacturer's instructions (Tiangen). The primers were designed using PyroMark Assay Design software (Qiagen). PCR was performed in a total volume of 10 μl, containing 1 μl of bisulphite‐converted DNA, 3 μl of double‐distilled (dd) H2O, 5 μl of enzyme and 0.5 μl of each primer. The thermal cycling conditions were as follows: 95°C for 10 min, followed by 45 cycles of 95°C for 20 s, 45°C for 20 s, and 72°C for 30 s, followed by one cycle of 42°C for 5 min (Eppendorf). Quantitative methylation‐specific PCR (qMSP) was performed in a total volume of 10 μl containing 5 μl of 2× SYBR Green I, 3.5 μl of ddH2O, 1 μl of DNA template and 0.25 μl of each primer.
2.4. BDNF genotyping at rs7103411
The literature was searched for relevant SNPs, and the polymorphic site, rs7103411, was selected from the dbSNP database of the National Center for Biotechnology Information. A MassARRAY system (Agena Bioscience) was used for SNP identification in this study. This technology uses matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry. Mass spectrometry can be used to identify SNPs of interest, and the method is flexible, with an accuracy greater than 99.7%. Moreover, this method does not require fluorescent labelling, which further reduces the cost. The primer sequences used for the identification of the BDNF polymorphism (rs7103411) were as follows: forward primer, 5′‐GTAGTTTTCGTAGGATGAGGAAGC‐3′; reverse primer, 5′‐AATATAAATTAACAACCCCGATACG‐3′. The amplification products were sequenced for SNP detection using the Sanger method.
2.5. Depression scale assessment
The HAMD scale is a classic rating scale for clinical depression. It is often used in parallel with other new scales to test their validity. It has good consistency, reflects the severity of clinical symptoms and has a moderate length and a clear operational evaluation standard. The most widely used version of this scale consists of 24 items that are scored using a 5‐point system with grades of 0–4. The total score reflects the severity of depression, and the evolution of the disease can be evaluated by monitoring changes in the total score. Factor analysis reflects the psychopathological characteristics of patients with depression and the effect of treatment on symptoms of depression. In this study, 53 depressive patients were selected for HAMD score evaluation, and their peripheral blood samples were taken for BDNF methylation detection. The patients were assessed once before sertraline treatment and once after sertraline treatment, corresponding to the times of blood sample collection. The assessments were administered by senior psychiatrists. The correlations between the scores for each factor and methylation status were analysed to explore the correlation between depressive symptoms and BDNF methylation before sertraline treatment.
2.6. Statistical analysis
Statistical Program for Social Sciences 17.0 software (IBM) was used for statistical analysis. After logarithmic transformation, the measurement data met the normality and homogeneity of variance requirements. Student's t test was used for comparisons between the two groups, and a one‐way analysis of variance was used for comparisons between multiple groups. A chi‐square test was used to compare categorical variables between groups. A receiver operating characteristic (ROC) curve was used to evaluate the specificity and effectiveness of methylation sites for diagnosing depression. A p value <0.05 was considered statistically significant.
3. RESULTS
Fifty‐three cases and 51 normal controls were included in this study. The demographic characteristics of the participants are presented in Table 1. There were no significant differences in age, gender, educational level or frequency of smoking between the case and control groups. The HAMD score was significantly higher in the case group than the healthy control group before sertraline treatment. After sertraline treatment, the scores for both male and female patients significantly improved (Table 2). The sequence information of CpG sites in BDNF exon I and rs7103411 location was shown in Figure 1. There was a significant difference in the methylation level of BDNF exon I between the case and control groups before sertraline treatment (Figures 1 and 2). However, no significant difference was found in the methylation level of BDNF exon I before and after sertraline monotherapy in subjects with depression (Figure 2). We also explored the association between the per cent of BDNF exon I methylation and BDNF genotype in subjects with depression but found no interaction between these factors (Table 3). Further analysis of the relationship between the 24 symptom clusters of the HAMD scale and methylation showed that that methylation at this site was not significantly correlated with probing mood, feelings of guilt, suicide ideation or insomnia, but was positively correlated with diurnal variation and retardation scores (Figure 3).
TABLE 1.
Clinical characteristics of subjects with depression and healthy controls
| Characteristics | HC | Depression | p value |
|---|---|---|---|
| Age (mean ± SD years) | 46.32 ± 5.36 | 47.41 ± 6.31 | ns |
| Gender (M/F) | |||
| Male | 25 | 27 | ns |
| Female | 26 | 26 | |
| Education level | |||
| Primary school | 11 | 10 | ns |
| Middle school | 30 | 29 | |
| Graduate/postgraduate | 10 | 14 | |
| BMI (kg/m2) | 26.60 ± 3.26 | 25.16 ± 3.19 | ns |
| Smoker/Nonsmoker | 19/32 | 20/33 | ns |
| HAMD−24 (±SD) | 7.94 ± 1.82 | 23.65 ± 3.30 | *** |
| Marital status | |||
| Single | 8 | 9 | ns |
| Married | 35 | 38 | |
| Divorced/widowed | 8 | 6 | |
| Occupation | |||
| Employee | 41 | 41 | ns |
| Housewife/Unemployed | 10 | 12 | |
| Location | |||
| Urban | 35 | 37 | ns |
| Rural | 16 | 16 | |
Abbreviation: ns, not significant.
*:p < 0.05; **p < 0.01;*** p < 0.001.
TABLE 2.
HAMD scores of patients with depression before and after sertraline treatment
| Item | Pre‐treatment | Post‐treatment | p value |
|---|---|---|---|
| Total | 23.65 ± 3.30 | 5.92 ± 2.72 | *** |
| Male | 23.57 ± 2.48 | 6.36 ± 1.53 | *** |
| Female | 23.74 ± 3.35 | 5.46 ± 2.67 | *** |
Abbreviation: ns, not significant.
p < 0.001.
FIGURE 1.
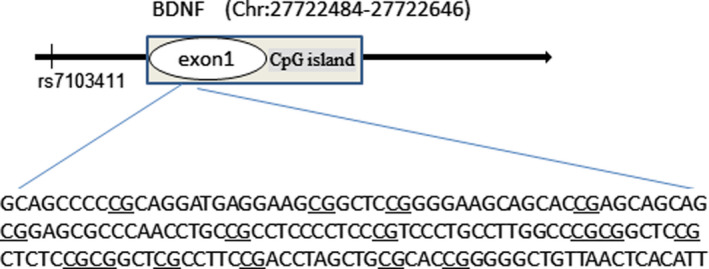
The sequence of CpG sites in BDNF exon I from the UCSC genome browser and rs7103411 location information (Chr11: 27678578)
FIGURE 2.
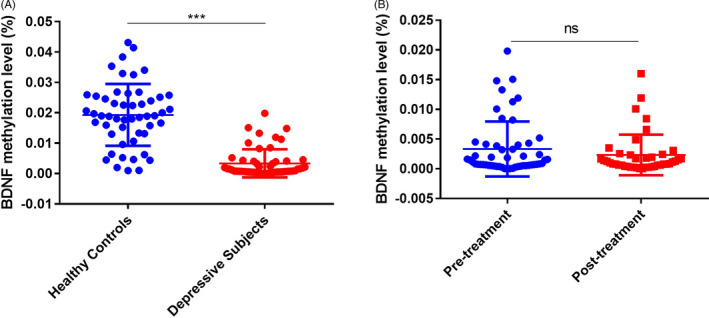
Methylation levels of BDNF exon I in healthy controls and subjects with depression. (A) The methylation level was significantly lower in patients with depression than in healthy controls. (B) No significant difference in the methylation level was found in subjects with depression before and after sertraline monotherapy. ns, not significant;*p < 0.05; **p < 0.01;*** p < 0.001
TABLE 3.
Methylation level of BDNF according to genotype
| Group | rs7103411 | p value | ||
|---|---|---|---|---|
| CC | CT | TT | ||
| Total (%) | 0.29 ± 0.12 | 0.39 ± 0.07 | 0.31 ± 0.15 | ns |
| Male (%) | 0.31 ± 0.15 | 0.36 ± 0.16 | 0.29 ± 0.10 | ns |
| Female (%) | 0.27 ± 0.16 | 0.40 ± 0.17 | 0.30 ± 0.09 | ns |
Abbreviation: ns, not significant.
FIGURE 3.
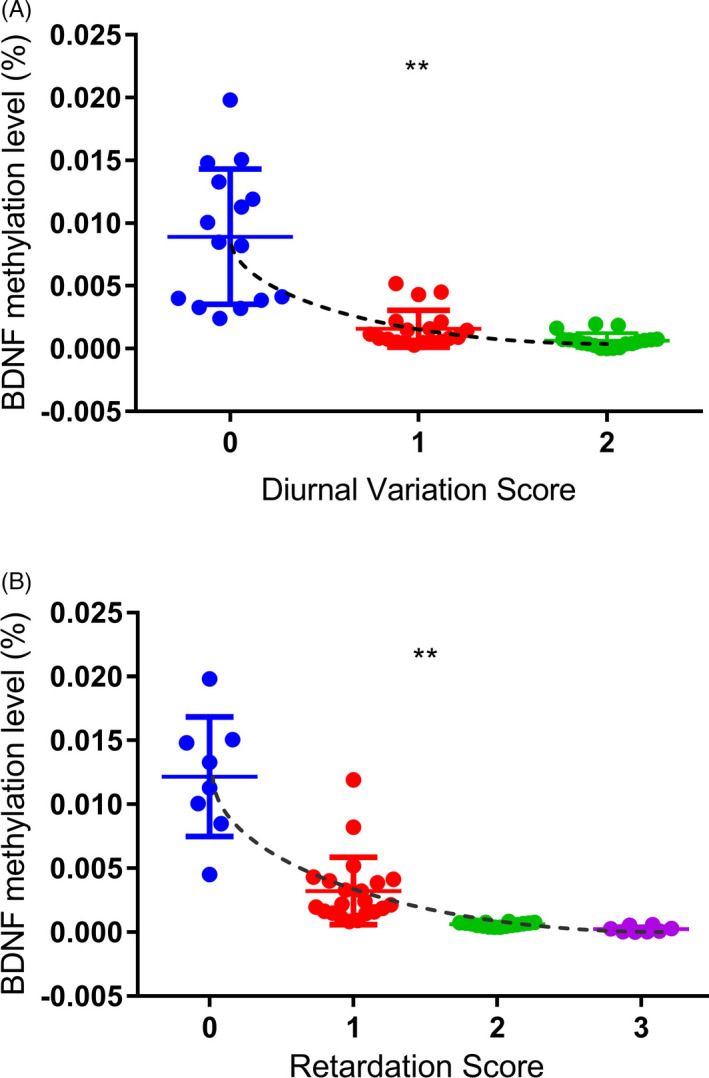
Association between the methylation level of BDNF exon I and HAMD factor scores in subjects with depression before sertraline treatment. (A) Correlation between BDNF exon I methylation and diurnal variation score in subjects with depression. (B) Correlation between BDNF exon I methylation and the retardation score in subjects with depression. ns, not significant; *p < 0.05; **p < 0.01; *** p < 0.001
Previous studies have shown that there is a correlation between homocysteine (Hcy) metabolism and methylation. In this study, blood Hcy concentrations were measured before and after sertraline treatment. The blood Hcy concentration was found to be significantly reduced by sertraline treatment (Figure 4). However, sertraline treatment had no significant effect on the serum BDNF concentration in subjects with depression (Table 4). There was also no significant difference in the serum BDNF concentration according to gender (Table 4). Subjects with depression were divided into three groups according to BDNF genotype (CC, CT and TT) to investigate the influence of genotype on the serum BDNF concentration. The results showed that there were no significant differences in the BDNF concentration between the three genotypes before or after sertraline treatment (Table 5). ROC curve analysis showed that the level of BDNF exon I methylation before sertraline treatment had good specificity and accuracy for distinguishing between patients and healthy individuals (Figure 5).
FIGURE 4.
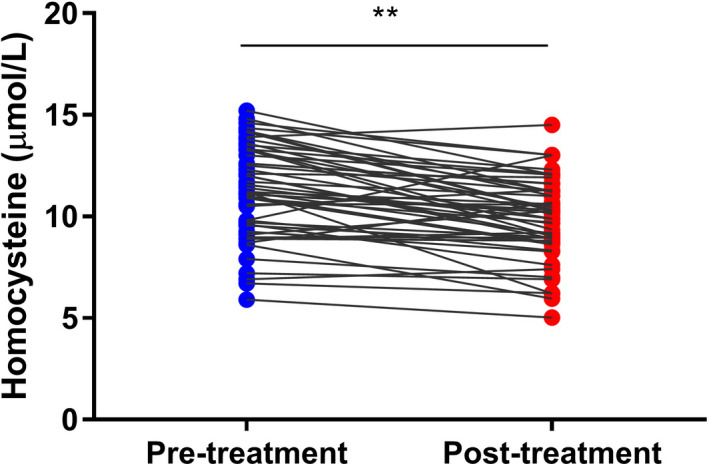
Homocysteine concentrations in the blood of patients with depression before and after sertraline monotherapy (n = 53, **p < 0.01)
TABLE 4.
Serum BDNF concentrations in patients with depression before and after sertraline treatment
| BDNF level (ng/ml) | Mean value | Male (n = 27) | Female (n = 26) |
|---|---|---|---|
| Pre‐treatment | 305.20 ± 95.86 | 281.55 ± 37.38 | 329.76 ± 48.38 |
| Post‐treatment | 296.94 ± 83.11 | 263.11 ± 26.04 | 332.07 ± 42.00 |
| p value | ns | ns | ns |
Abbreviation: ns, not significant.
TABLE 5.
Serum BDNF concentration according to BDNF genotype
| BDNF level (ng/ml) | rs7103411 | ||
|---|---|---|---|
| CC (n = 18) | CT (n = 19) | TT (n = 16) | |
| Pre‐treatment | 309.61 ± 48.46 | 305.96 ± 49.97 | 299.34 ± 18.25 |
| Post‐treatment | 313.30 ± 31.85 | 296.24 ± 43.65 | 279.37 ± 31.15 |
| p value | ns | ns | ns |
Abbreviation: ns, not significant.
FIGURE 5.
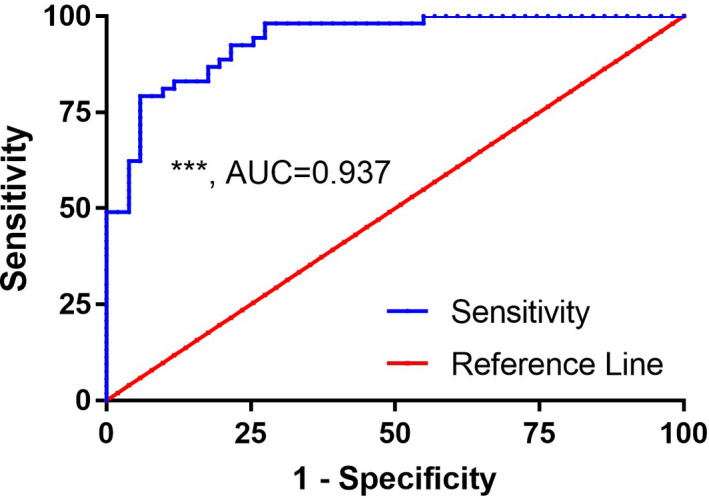
Receiver operating characteristic (ROC) curve of BDNF exon I methylation in subjects with depression. BDNF exon I methylation was able to predict depression to a certain degree
4. DISCUSSION
Depression is a common chronic mental disorder that has high recurrence and disability rates. However, the exact pathologic gene and mechanism associated with depression have not been identified. However, there are many relatively specific changes observed in patients with depression, especially in neurotransmitter levels. These changes are an important part of the disease mechanism. There is evidence that an early change in the plasma BDNF concentration, together with early disease progression, may be a predictive peripheral marker of treatment efficacy in patients with depression. 23 Further, studies have found that the peripheral blood BDNF concentration is decreased in patients with depression but increases after antidepressant treatment. 24 , 25 In this study, no significant differences in the serum BDNF concentration were found after sertraline treatment in subjects with depression.
SNPs in the BDNF gene are a common component of the genetic mechanism of depression. In China, rs6265 is the most frequently studied BDNF polymorphism. However, the results of studies investigating the aetiological mechanism of BDNF polymorphisms in depression have not been consistent. First, rs6265 and rs7103411 polymorphisms in the BDNF gene are not associated with depression in the Chinese Han population, 26 and this has been confirmed in other studies. 27 , 28 However, studies from other countries have found that the rs6265 polymorphism of the BDNF gene is associated with depression. Hwang et al. studied this polymorphism in patients with senile depression and concluded that the polymorphism may be involved in the occurrence of depression. 14 In addition, Strauss et al. 29 found that childhood‐onset depression is significantly associated with the T allele of rs7103411. To determine whether the T allele of rs7103411 is involved in modifying BDNF gene methylation in patients with depression in the Ningbo area, we selected rs7103411 as the polymorphic site for this study. The results indicated no significant difference in the methylation of BDNF among the CC, CT and TT genotypes in depressive patients. These results suggest that this BDNF gene polymorphism may not be a factor influencing BDNF gene methylation in the Han population. We note that rs7103411 is located outside the CpG island region, which may be one of the reasons for this result. In addition, studies have confirmed that the methylation level of BDNF changes after 8 weeks of antidepressant treatment. 30 , 31 , 32 In this study, BDNF methylation was assessed after 4 weeks of sertraline treatment. The potential association between methylation status and gene polymorphisms may need a longer period of antidepressant treatment to manifest, because gene polymorphisms are not the main factor affecting short‐term antidepressant efficacy.
Compared with the effect of polymorphisms on gene stability, DNA methylation is more flexible in terms of regulating gene expression. Antidepressants may increase BDNF promoter methylation and perform divergent actions, compared with mood stabilizers. 31 , 33 Animal experiments have shown that the dynamic expression of BDNF is related to the methylation of CpG island dinucleotides in the mouse BDNF gene. 34 A number of studies have shown that the concentration of BDNF in the peripheral blood is reduced in patients with depression, and animal experiments have shown that methylation of CpG in exon IV is significantly lower in the hippocampus, where BDNF is expressed at high levels, than in the cerebral cortex and striatum, which have low levels of BDNF expression. 26 , 34 Previous studies have found that there is a significant internal correlation between the methylation levels of exon I of the BDNF gene in the brain, peripheral blood and muscle tissue. 27 The BDNF gene and 5‐HT neurons can interact with each other, regulate each other and maintain a certain dynamic balance. Therefore, we hypothesize that methylation of the exon I region of BDNF may be a diagnostic biomarker for depression and that sertraline is involved in the regulation of depression by affecting BDNF gene methylation. The results showed that there was a significant difference in the methylation level of BDNF exon I between the control and depression groups before treatment. This result is consistent with the results of a study of 20 depressive patients by Fuchikami et al. 28 We further compared the methylation levels before and after sertraline treatment in the depression group and found no significant difference. This result indicated that modification of BDNF methylation may not be the mechanism responsible for the antidepressant effect of sertraline and that baseline methylation levels cannot be used as a predictor of drug treatment efficacy during follow‐up. However, some reports have suggested that the methylation of BDNF exon I may be a biomarker of the response to electroconvulsive therapy. 29 We analysed the correlation between the level of BDNF gene methylation and HAMD factor scores in the depression group before sertraline treatment. The results showed that diurnal variation and retardation scores were positively correlated with the methylation status of BDNF exon I. The ROC mode indicated that methylation at this site had a potential diagnostic value in the differential diagnosis of depression.
In this study, sertraline was selected as the treatment for depression, but it did not yield the expected results. This may be because depression is a polygenic disease, and interactions between many genes may weaken the correlation between BDNF methylation and clinical drug efficacy. In addition, qMSP was selected as the sequencing method due to its advantages of convenience and cost‐effectiveness. However, the accuracy of this method is relatively low compared with bisulphite sequencing PCR and high‐throughput sequencing technology. Hcy participates in energy metabolism and is indispensable to the process of methylation. Studies have shown that Hcy can affect the overall and promoter‐region methylation of genes involved in susceptibility to depression, thus participating in the regulation of depression. 35 Serum Hcy concentrations are higher in individuals with depression than in healthy subjects. 36 In this study, we found that the serum Hcy concentrations in patients with depression decreased significantly after sertraline treatment. However, we found no correlation between the Hcy concentration and BDNF methylation in further analysis. These results suggest that Hcy may not participate in the pathophysiological mechanism of depression through methylation of the exon I region of BDNF.
5. CONCLUSIONS
In conclusion, this study found that the methylation level of exon I of the BDNF gene may be used as a marker for the diagnosis of depression and that methylation at this site is correlated with HAMD factor scores in subjects with depression. Sertraline treatment significantly reduced the Hcy concentration in patients with depression, but did not affect the methylation status of the exon I region of BDNF. This methylation site is a potential therapeutic target for previously untreated depression.
CONFLICT OF INTERESTS
The authors declare that there are no conflicts of interest.
Xing Y, Sun T, Li G, Xu G, Cheng J, Gao S. The role of BDNF exon I region methylation in the treatment of depression with sertraline and its clinical diagnostic value. J Clin Lab Anal. 2021;35:e23993. 10.1002/jcla.23993
Yuhua Xing and Ting Sun are co‐first authors of this paper.
Funding information
This work was supported by the Zhejiang Provincial Medical Technology Project (No. 2015KYB351 and 2019KY628), the Natural Science Foundation of Ningbo (No. 2013A610249 and 2015A610196), the Life and Health Technology Innovation Team Fund of Ningbo (No. 2015C110026) and the Natural Science Foundation of Zhejiang Province (LY21H090003)
Contributor Information
Jia Cheng, Email: drchengjia@163.com.
Shugui Gao, Email: gaoshugui@sina.com.
DATA AVAILABILITY STATEMENT
The data sets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Malhi GS, Mann JJ. Depression. Lancet. 2018;392(10161):2299‐2312. [DOI] [PubMed] [Google Scholar]
- 2. Tian H, Li G, Xu G, et al. Inflammatory cytokines derived from peripheral blood contribute to the modified electroconvulsive therapy‐induced cognitive deficits in major depressive disorder. Eur Arch Psychiatry Clin Neurosci. 2021;271(3):475‐485. [DOI] [PubMed] [Google Scholar]
- 3. Riquin E, Lamas C, Nicolas I, et al. A key for perinatal depression early diagnosis: the body dissatisfaction. J Affect Disord. 2019;245:340‐347. [DOI] [PubMed] [Google Scholar]
- 4. Penner‐Goeke S, Binder EB. Epigenetics and depression. Dialogues Clin Neurosci. 2019;21(4):397‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mullins N, Lewis CM. Genetics of depression: progress at last. Curr Psychiatry Rep. 2017;19(8):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glanville KP, Coleman JRI, Howard DM, et al. Multiple measures of depression to enhance validity of major depressive disorder in the UK Biobank. BJPsych Open. 2021;7(2):e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buch AM, Liston C. Dissecting diagnostic heterogeneity in depression by integrating neuroimaging and genetics. Neuropsychopharmacology. 2021;46(1):156‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi S, Han KM, Won E, Yoon BJ, Lee MS, Ham BJ. Association of brain‐derived neurotrophic factor DNA methylation and reduced white matter integrity in the anterior corona radiata in major depression. J Affect Disord. 2015;172:74‐80. [DOI] [PubMed] [Google Scholar]
- 9. Januar V, Ancelin ML, Ritchie K, Saffery R, Ryan J. BDNF promoter methylation and genetic variation in late‐life depression. Transl Psychiat. 2015;5:e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu X, Ji H, Liu G, et al. A significant association between BDNF promoter methylation and the risk of drug addiction. Gene. 2016;584(1):54‐59. [DOI] [PubMed] [Google Scholar]
- 11. Cheng J, Wang Y, Zhou K, et al. Male‐specific association between dopamine receptor D4 gene methylation and schizophrenia. PLoS One. 2014;9(2):e89128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao S, Cheng J, Li G, et al. Catechol‐O‐methyltransferase gene promoter methylation as a peripheral biomarker in male schizophrenia. Eur Psychiatry. 2017;44:39‐46. [DOI] [PubMed] [Google Scholar]
- 13. Zhou J, Li M, Wang X, et al. Drug response‐related DNA methylation changes in Schizophrenia, bipolar disorder, and major depressive disorder. Front Neurosci. 2021;15:674273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Starnawska A, Demontis D. Role of DNA methylation in mediating genetic risk of psychiatric disorders. Front Psychiatry. 2021;12:596821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA‐methylation changes associated with major psychosis. Am J Hum Genet. 2008;82(3):696‐711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kang HJ, Kim JM, Bae KY, et al. Longitudinal associations between BDNF promoter methylation and late‐life depression. Neurobiol Aging. 2015;36(4):1764. [DOI] [PubMed] [Google Scholar]
- 17. Gao S, Hu Z, Cheng J, et al. Impact of catechol‐o‐methyltransferase polymorphisms on risperidone treatment for schizophrenia and its potential clinical significance. Clin Biochem. 2012;45(10–11):787‐792. [DOI] [PubMed] [Google Scholar]
- 18. Hwang JP, Tsai SJ, Hong CJ, Yang CH, Lirng JF, Yang YM. The Val66Met polymorphism of the brain‐derived neurotrophic‐factor gene is associated with geriatric depression. Neurobiol Aging. 2006;27(12):1834‐1837. [DOI] [PubMed] [Google Scholar]
- 19. Pei Y, Smith AK, Wang Y, et al. The brain‐derived neurotrophic‐factor (BDNF) val66met polymorphism is associated with geriatric depression: a meta‐analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(5):560‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duncan LE, Hutchison KE, Carey G, Craighead WE. Variation in brain‐derived neurotrophic factor (BDNF) gene is associated with symptoms of depression. J Affect Disord. 2009;115(1–2):215‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X, Xu Y, Jiang S, Cui D, Qian Y, Jiang K. Family‐based association study between brain‐derived neurotrophic factor gene and major depressive disorder of Chinese descent. Psychiatry Res. 2009;169(2):169‐172. [DOI] [PubMed] [Google Scholar]
- 22. Schumacher J, Jamra RA, Becker T, et al. Evidence for a relationship between genetic variants at the brain‐derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58(4):307‐314. [DOI] [PubMed] [Google Scholar]
- 23. Dreimuller N, Schlicht KF, Wagner S, et al. Early reactions of brain‐derived neurotrophic factor in plasma (pBDNF) and outcome to acute antidepressant treatment in patients with Major Depression. Neuropharmacology. 2012;62(1):264‐269. [DOI] [PubMed] [Google Scholar]
- 24. Karege F, Bondolfi G, Gervasoni N, Schwald M, Aubry JM, Bertschy G. Low brain‐derived neurotrophic factor (BDNF) levels in serum of depressed patients probably results from lowered platelet BDNF release unrelated to platelet reactivity. Biol Psychiatry. 2005;57(9):1068‐1072. [DOI] [PubMed] [Google Scholar]
- 25. Gervasoni N, Aubry JM, Bondolfi G, et al. Partial normalization of serum brain‐derived neurotrophic factor in remitted patients after a major depressive episode. Neuropsychobiology. 2005;51(4):234‐238. [DOI] [PubMed] [Google Scholar]
- 26. Lee HY, Kim YK. Plasma brain‐derived neurotrophic factor as a peripheral marker for the action mechanism of antidepressants. Neuropsychobiology. 2008;57(4):194‐199. [DOI] [PubMed] [Google Scholar]
- 27. Stenz L, Zewdie S, Laforge‐Escarra T, et al. BDNF promoter I methylation correlates between post‐mortem human peripheral and brain tissues. Neurosci Res. 2015;91:1‐7. [DOI] [PubMed] [Google Scholar]
- 28. Fuchikami M, Morinobu S, Segawa M, et al. DNA methylation profiles of the brain‐derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS One. 2011;6(8):e23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kleimann A, Kotsiari A, Sperling W, et al. BDNF serum levels and promoter methylation of BDNF exon I, IV and VI in depressed patients receiving electroconvulsive therapy. J Neural Transm. 2015;122(6):925‐928. [DOI] [PubMed] [Google Scholar]
- 30. Lisoway AJ, Zai CC, Tiwari AK, Kennedy JL. DNA methylation and clinical response to antidepressant medication in major depressive disorder: A review and recommendations. Neurosci Lett. 2018;669:14‐23. [DOI] [PubMed] [Google Scholar]
- 31. Webb LM, Phillips KE, Ho MC, Veldic M, Blacker CJ. The Relationship between DNA Methylation and Antidepressant Medications. A systematic review. Int J Mol Sci. 2020;21(3):826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang P, Zhang C, Lv Q, et al. Association of DNA methylation in BDNF with escitalopram treatment response in depressed Chinese Han patients. Eur J Clin Pharmacol. 2018;74(8):1011‐1020. [DOI] [PubMed] [Google Scholar]
- 33. Carlberg L, Scheibelreiter J, Hassler MR, et al. Brain‐derived neurotrophic factor (BDNF)‐epigenetic regulation in unipolar and bipolar affective disorder. J Affect Disord. 2014;168:399‐406. [DOI] [PubMed] [Google Scholar]
- 34. Dennis KE, Levitt P. Regional expression of brain derived neurotrophic factor (BDNF) is correlated with dynamic patterns of promoter methylation in the developing mouse forebrain. Brain Res Mol Brain Res. 2005;140(1–2):1‐9. [DOI] [PubMed] [Google Scholar]
- 35. Wan L, Li Y, Zhang Z, Sun Z, He Y, Li R. Methylenetetrahydrofolate reductase and psychiatric diseases. Transl Psychiatry. 2018;8(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moradi F, Lotfi K, Armin M, Clark CCT, Askari G, Rouhani MH. The association between serum homocysteine and depression: A systematic review and meta‐analysis of observational studies. Eur J Clin Invest. 2021;51(5):e13486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analysed during the current study are available from the corresponding author on reasonable request.


