Abstract
Background
Circulating long non‐coding RNAs (lncRNAs) are emerging as promising biomarkers for non‐small cell lung cancer (NSCLC). This study aimed to detect serum exosomal lncRNA SNHG15 expression in NSCLC and evaluate its potential clinical value.
Methods
A total of 238 serum samples were collected from 118 patients with NSCLC, 40 patients with benign pulmonary lesions and 80 healthy volunteers. The expression levels of serum exosomal lncRNA SNHG15 were measured by quantitative real‐time polymerase chain reaction (qRT‐PCR). Then, the relationship between serum exosomal lncRNA SNHG15 expression and clinical parameters was analyzed.
Results
The serum exosomal lncRNA SNHG15 expression was markedly higher in NSCLC patients compared to patients with benign pulmonary lesions and normal controls. As expected, serum exosomal lncRNA SNHG15 was greatly decreased after surgery. High serum exosomal lncRNA SNHG15 expression was closely associated with poor differentiation (p=0.035), positive lymph node metastasis (p=0.009) and advanced TNM stage (p<0.001). Receiver operating characteristic (ROC) curve analysis demonstrated that serum exosomal lncRNA SNHG15 well differentiated all stage NSCLC, stage I/II NSCLC patients or stage III/IV NSCLC patients from controls, and the combination of serum exosomal lncRNA SNHG15 and CEA showed an elevated AUC for distinguishing NSCLC from healthy individuals. In univariate and multivariate analyses, serum exosomal lncRNA SNHG15 was confirmed as an independent prognostic predictor for overall survival.
Conclusion
In conclusion, our findings suggest that serum exosomal lncRNA SNHG15 might be a potential biomarker for early diagnosis and prognosis prediction of NSCLC.
Keywords: diagnosis, lncRNA, non‐small cell lung cancer, prognosis, SNHG15
Kaplan‐Meier survival curves demonstrated that NSCLC patients with higher serum exosomal lncRNA SNHG15 expression had significant shorter overall survival than those with lower serum exosomal lncRNA SNHG15 expression.

1. INTRODUCTION
Lung cancer is the leading cause of cancer‐associated deaths around the world and is one of the most global health problems. 1 , 2 Non‐small cell lung cancer (NSCLC) accounts for about 85% of lung cancer cases. 3 Despite great effort has been made in the treatment of NSCLC over the past decades, the clinical outcome of NSCLC patients remains quite unfavorable. 4 Since NSCLC shows unobvious incipient symptom, most of patients are at advanced stages or suffer metastasis at initial diagnosis. 5 Therefore, identification of novel biomarkers for early diagnosis and prognosis prediction are urgently required to improve the survival rate of NSCLC patients.
Exosomes are 30–150 nm sized membranous vesicles and contain different molecules, such as proteins, lipids and nucleic acids. 6 Exosomes can be extracted from multiple body fluids such as blood, saliva and urine in a highly stable form. 7 , 8 Long non‐coding RNAs (lncRNAs) are non‐coding RNAs with more than 200 nucleotides in length. 9 Growing evidence has demonstrated that lncRNAs are abnormally expressed in various cancer types and involve in cancer progression. 10 Exosomal lncRNAs could be used as diagnostic and prognostic indicators for NSCLC. For instance, Zhang et al found that exosomal lncRNA DLX6‐AS1 was highly expressed in NSCLC tissues and associated with poor clinical variables. 11 Lv et al showed that lncRNA LINC00662 was significantly higher in NSCLC patients, and its overexpression greatly enhanced NSCLC cell proliferation, migration and invasion. 12
To the best of our knowledge, the diagnostic power of serum exosomal lncRNA SNHG15 and its associations with clinical parameters of NSCLC have not yet been explored. Thus, the aim of this study was to measure the expression levels of serum exosomal lncRNA SNHG15 in NSCLC patients and assess its potential clinical value for the early detection and prognosis prediction of NSCLC.
2. MATERIALS AND METHODS
2.1. Blood samples
Blood samples from patients diagnosed with NSCLC (n=118), patients with benign pulmonary lesions (n=40) and healthy volunteers (n=80) were collected in EDTA‐tubes. Patients were excluded if they had received any chemotherapy or radiotherapy before blood collection. Patients were staged by the classification of the 7th lung cancer TNM classification. A total of 52 stage I/II NSCLC patients and 7 stage III NSCLC patients underwent curative surgery. The clinical characteristics of all NSCLC patients included gender, age, histology, smoking status, differentiation, lymph node metastasis and TNM stage, which were summarized in Table 1 . This study was approved by the Ethics Committee of the First People's Hospital of Yulin City. All participants provided signed written informed consents before the study. Clinical follow‐up was available for all NSCLC patients. Overall survival (OS) was defined as the time from the date of diagnosis to the date of death or the last follow‐up. Peripheral blood samples were centrifuged at 3,000 × g for 15 min at 4℃ and then stored at –80℃ until further use.
TABLE 1.
Demographic characteristics of 118 patients with NSCLC
| Variables | All patients | Serum exosomal lncRNA SNHG15 | P‐value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | ||||
| Women | 47 | 22 | 25 | 0.572 |
| Men | 71 | 37 | 34 | |
| Age | ||||
| <60 | 67 | 31 | 36 | 0.353 |
| ≥60 | 51 | 28 | 23 | |
| Histology | ||||
| Adenocarcinoma | 53 | 25 | 28 | 0.579 |
| Squamous cell carcinoma | 65 | 34 | 31 | |
| Smoking status | ||||
| No | 42 | 19 | 23 | 0.441 |
| Yes | 76 | 40 | 36 | |
| Differentiation | ||||
| Well/moderate | 75 | 32 | 43 | 0.035 |
| Poor | 43 | 27 | 16 | |
| Lymph node metastasis | ||||
| No | 50 | 18 | 32 | 0.009 |
| Yes | 68 | 41 | 27 | |
| TNM stage | ||||
| I/II | 52 | 16 | 36 | <0.001 |
| III/IV | 66 | 43 | 23 | |
2.2. Isolation of exosomes
The exosomes were extracted from serum samples using the ExoQuick kit (SBI, Mountain View, CA, USA) according to the manufacturer's instructions. Briefly, the serum sample was thawed on ice and centrifuged at 3000 g for 15 min at 4℃ to remove cells and cell debris. Then, one‐fourth volume of ExoQuick solution was added to the supernatants. The mixture was incubated at 4℃ for 30 min and centrifuged at 1500 g for 30 min. The final exosome‐containing pellets were collected for characterization and RNA isolation.
2.3. Western blotting
The proteins were size‐fractionated by electrophoresis on 4–20% SDS‐PAGE gels. Proteins were transferred to a polyvinylidene fluoride membrane. The membrane was blocked with nonfat dry milk for 30 min at room temperature and probed with a primary antibody (anti‐CD63, CD81 and TSG101) at 4℃ overnight. Following incubating secondary antibodies, bands were visualized using an Amersham ECL Western Blotting Detection Kit (GE Healthcare, Chicago, IL, USA).
2.4. RNA extraction and quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA was isolated from the exosomes using MirVana™ miRNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). 5 μL of synthetic Caenorhabditis elegans miR‐39 (cel‐miR‐39) was spiked into each sample. cDNA was synthesized from total RNAs using a TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). The qRT‐PCR was conducted on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Each experiment was repeated in triplicate. The relative expression levels of serum exosomal lncRNA SNHG15 were normalized against cel‐miR‐39 using the comparative 2–ΔΔCt method. The primer sequences of SNHG15 and cel‐miR‐39 are as follows: SNHG15 forward: 5′‐GCTGAGGTGACGGTCTCAAA‐3′, SNHG15 reverse: 5′‐GCCTCCCAGTTTCATG GACA‐3′; cel‐miR‐39 forward: 5′‐UCACCGGGUGUAAAU CAGCUUG‐3′, cel‐miR‐39 reverse: 5′‐TCACCGGGTGTAAAT CAGCTTG‐3′.
2.5. Statistical analysis
Statistical differences of serum exosomal lncRNA SNHG15 expression among groups were calculated using Kruskal‐Wallis test. The relationship between serum exosomal lncRNA SNHG15 expression and clinicopathological factors was analyzed by the chi‐square test. Receiver operating characteristic (ROC) curve was carried out to analyze the diagnostic accuracy of serum exosomal lncRNA SNHG15 and CEA. Kaplan‐Meier method was used to calculate cumulative survival rates, and the log‐rank test was used to assess the differences between the subgroups. Univariate and multivariate analyses were performed to evaluate the independent factors for OS. GraphPad Prism 6.01 (GraphPad Software, Inc., San Diego, CA, USA) was used for data analyses. Statistically significant level was defined as p< 0.05.
3. RESULTS
3.1. Serum exosomal lncRNA SNHG15 was significantly upregulated in NSCLC
Our results showed that the exosomes isolated from the serum samples were positive for CD63, CD81 and TSG101 (Figure 1A). However, no or weak signal was detected in the supernatant samples, indicating that the extracellular vesicles we isolated were exosomes. QRT‐PCR was performed to investigate the expression levels of serum exosomal lncRNA SNHG15 in NSCLC patients, patients with benign pulmonary lesions and healthy controls. Our PCR results demonstrated that the expression level of serum exosomal lncRNA SNHG15 was significantly higher in NSCLC patients than in patients with benign pulmonary lesions and healthy controls. However, no significant difference was found for the serum exosomal lncRNA SNHG15 level between patients with benign pulmonary lesions and healthy controls. In addition, serum exosomal lncRNA SNHG15 expression was not significantly different between stage III/IV patients and stage I/II patients. (Figure 1B).
FIGURE 1.
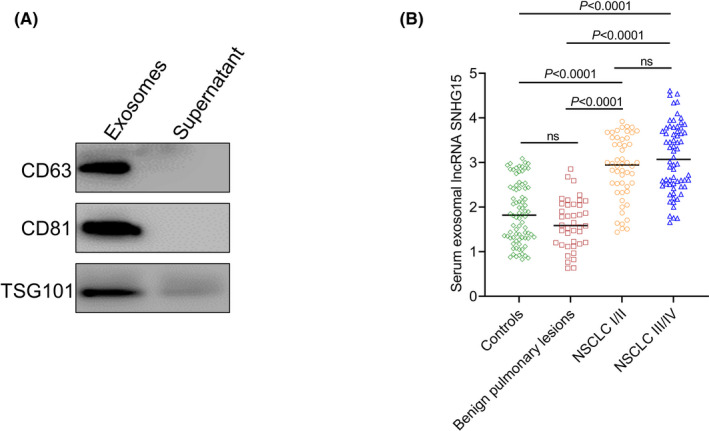
Serum exosomal lncRNA SNHG15 was significantly higher in NSCLC. (A) The exosomes we isolated from serum samples were positive for CD63, CD81 and TSG101. (B) The level of serum exosomal lncRNA SNHG15 was significantly higher in NSCLC patients than in patients with benign pulmonary lesions and healthy controls
3.2. Serum exosomal lncRNA SNHG15 was a potential non‐invasive marker for NSCLC
Next, ROC curves were generated to assess the potential diagnostic accuracy of serum exosomal lncRNA SNHG15 for NSCLC. The results revealed that serum exosomal lncRNA SNHG15 was robust in differentiating NSCLC patients, early‐stage (I/II) NSCLC and advanced stage (III/IV) NSCLC from control subjects, with an AUC value of 0.856 (Figure 2A), 0.838 (Figure 2B) and 0.870 (Figure 2C), respectively. Carcinoembryonic antigen (CEA) is a clinical biomarker that is currently used for NSCLC detection. In this study, CEA differentiated all NSCLC patients, early‐stage NSCLC and advanced stage NSCLC from normal controls with an AUC of 0.812 (Figure 2D), 0.792 (Figure 2E) and 0.828 (Figure 2F), respectively. When serum exosomal lncRNA SNHG15 was combined with CEA, ROC analysis revealed an increased AUC value of 0.915, with 92.3% sensitivity and 76.2% specificity for the detection of NSCLC (Figure 2G). The AUC was 0.869, with 75.0% sensitivity and 90.0% specificity for the detection of early‐stage NSCLC (Figure 2H). Moreover, the AUC was 0.900, with 77.3% sensitivity and 87.5% specificity for the detection of advanced stage NSCLC (Figure 2I).
FIGURE 2.
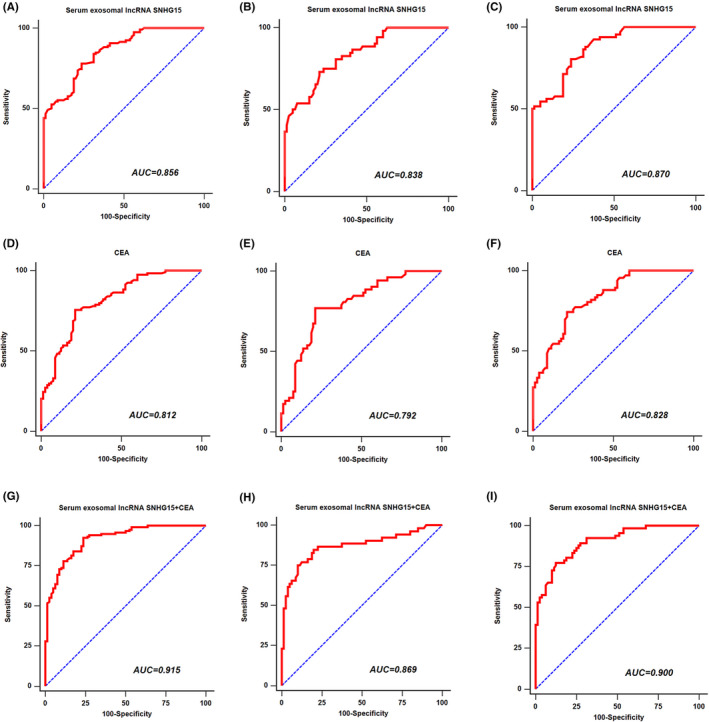
The diagnostic value of serum exosomal lncRNA SNHG15 for HNSCC. (A) ROC curve of serum exosomal lncRNA SNHG15 for all stage NSCLC patients. (B) ROC curve of serum exosomal lncRNA SNHG15 for early‐stage NSCLC patients. (C) ROC curve of serum exosomal lncRNA SNHG15 for advanced stage NSCLC patients. (D) ROC curve of CEA for all stage NSCLC patients. (E) ROC curve of CEA for early‐stage NSCLC patients. (F) ROC curve of CEA for advanced stage NSCLC patients. (G) ROC curve of the combination of two markers for all stage NSCLC patients. (H) ROC curve of two markers for early‐stage NSCLC patients. (I) ROC curve of two markers for advanced stage NSCLC patients
3.3. Relationship between serum exosomal lncRNA SNHG15 level and clinical features
As shown in Table 1, all 118 patients with NSCLC were classified into the two subgroups according to serum exosomal lncRNA SNHG15 expression: The high serum exosomal lncRNA SNHG15 group (n=59) and the low serum exosomal lncRNA SNHG15 group (n=59). The correlations between clinical features and serum exosomal lncRNA SNHG15 levels were analyzed. The results demonstrated that serum exosomal lncRNA SNHG15 expression was significantly higher in NSCLC patients with poor differentiation (p=0.035), or with positive lymph node metastasis (p=0.009) or at the advanced TNM stage (p<0.001). However, serum exosomal lncRNA SNHG15 levels were not correlated with gender, age, histology and smoking status (all p>0.05).
Thereafter, serum samples were collected from 59 NSCLC patients 3 months after their surgical resection. Compared to the paired pre‐operative serum samples, it was interesting to note that serum exosomal lncRNA SNHG15 levels were significantly decreased in post‐operative serum samples (p<0.0001, Figure 3).
FIGURE 3.
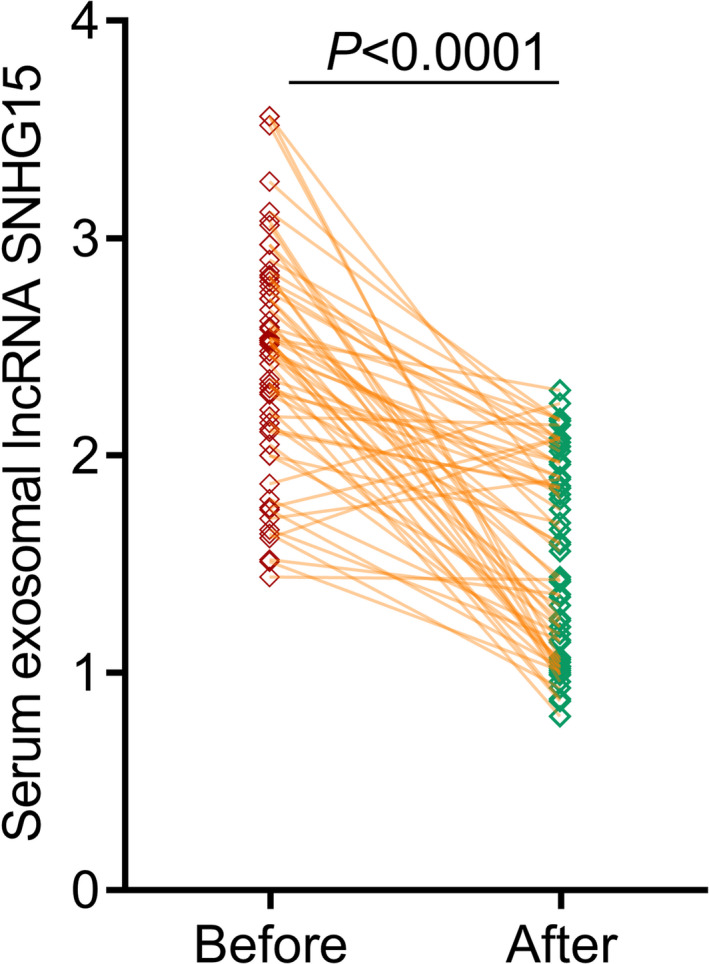
Compared to pre‐operative blood samples, serum exosomal lncRNA SNHG15 levels were significantly decreased
3.4. Survival analyses
Kaplan‐Meier survival analyses were performed to analyze the prognosis of NSCLC patients according to serum exosomal lncRNA SNHG15 expression, differentiation, lymph node metastasis and TNM stage. As anticipated, NSCLC patients with higher serum exosomal lncRNA SNHG15 expression, with poor differentiation, with positive lymph node metastasis or with advanced TNM stage had shorter OS (p=0.0153, Figure 4A; p=0.0006, Figure 4B; p=0.0164, Figure 4C; p<0.0001, Figure 4D, respectively). In addition, univariate analysis indicated that serum exosomal lncRNA SNHG15 (HR=4.13, 95% CI: 1.94–7.58, p=0.013), differentiation (HR=2.36, 95% CI: 1.17–4.69, p=0.041), lymph node metastasis (HR=3.18, 95% CI: 1.44–5.83, p=0.027) and TNM stage (HR=4.75, 95% CI: 2.32–8.46, p=0.003) were significantly correlated with OS. Moreover, multivariate analysis demonstrated that serum exosomal lncRNA SNHG15 (HR=3.71, 95% CI: 1.67–6.94, p=0.020), lymph node metastasis (HR=2.52, 95% CI: 1.25–4.84, p=0.036) and TNM stage (HR=4.34, 95% CI: 2.16–7.73, p=0.009) were independent indicators for OS (Table 2).
FIGURE 4.
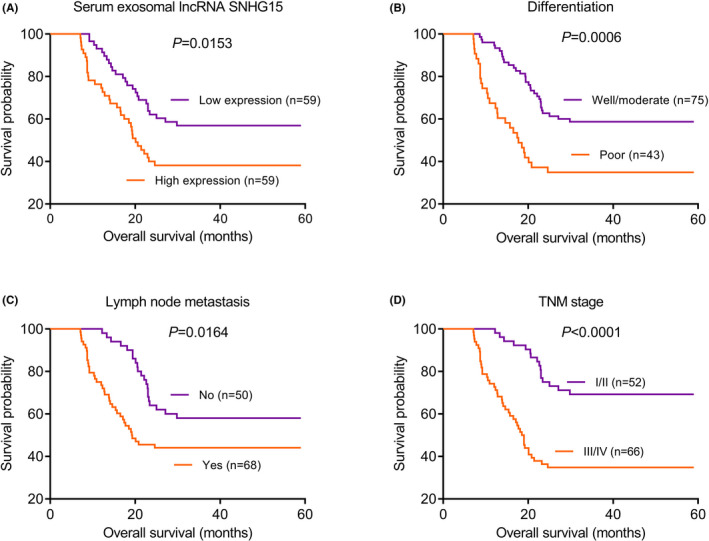
Kaplan‐Meier survival curves of NSCLC patients. (A) Patients with high serum exosomal lncRNA SNHG15 expression had shorter OS. (B) Patients with poor differentiation had shorter OS. (C) Patients with lymph node metastasis had shorter OS. (D) Patients with advanced TNM stage had shorter OS
TABLE 2.
Univariate and multivariate Cox regression analyses of overall survival
| Variables | Overall Survival | ||
|---|---|---|---|
| Hazard Ratio | 95% CI | P‐value | |
| Univariate analysis | |||
| Serum exosomal lncRNA SNHG15 | 4.13 | 1.94–7.58 | 0.013 |
| Differentiation | 2.26 | 1.17–4.69 | 0.041 |
| Lymph node metastasis | 3.18 | 1.44–5.83 | 0.027 |
| TNM stage | 4.75 | 2.32–8.46 | 0.003 |
| Multivariate analysis | |||
| Serum exosomal lncRNA SNHG15 | 3.71 | 1.67–6.94 | 0.020 |
| Differentiation | 1.54 | 0.86–3.89 | 0.056 |
| Lymph node metastasis | 2.52 | 1.25–4.84 | 0.036 |
| TNM stage | 4.34 | 2.16–7.73 | 0.009 |
4. DISCUSSION
The NSCLC is the main cause of cancer‐related deaths around the world. The high mortality rate of the malignancy can be prevented if NSCLC patients could be diagnosed at early stage. Recent studies have demonstrated that SNHG15 expression levels were significantly upregulated in cancerous tissues and cell lines. In vitro analysis showed that SNHG15 knockdown markedly suppressed cancer cell proliferation, invasion, metastasis and induced cell apoptosis. The xenograft model indicated that downregulation of SNHG15 dramatically inhibited NSCLC cell growth by targeting miR‐486 and miR‐211‐3p. NSCLC patients with high SNHG15 expression was positively associated with poor prognosis. 13 , 14 , 15 The results were in line with our findings. In this study, elevated serum exosomal lncRNA SNHG15 levels were observed in patients with NSCLC. ROC analysis demonstrated that serum exosomal lncRNA SNHG15 could well differentiate all stage NSCLC, stage I/II patients or stage III/IV patients from normal controls. The combination of serum exosomal lncRNA SNHG15 and CEA exhibited higher accuracy for the early diagnosis of NSCLC. In addition, high serum exosomal lncRNA SNHG15 levels were positively associated with differentiation, lymph node metastasis and TNM stage. Expression levels of serum exosomal lncRNA SNHG15 in the pre‐operative serum markedly declined following surgical removal of the tumors. Moreover, NSCLC patients with higher serum exosomal lncRNA SNHG15 expression levels showed a poorer survival rate compared with patients with lower lncRNA SNHG15 expression levels. Furthermore, serum exosomal lncRNA SNHG15 maintained the significance as an independent prognostic indicator for OS. Collectively, this study suggested that serum exosomal lncRNA SNHG15 might be a promising diagnostic and prognostic maker in NSCLC.
Recently, the oncogenic role of SNHG15 was also reported in various cancer types. For instance, SNHG15 overexpression was observed in both breast cancer (BC) tissues and cells and predicted unfavorable prognosis. Decreased SNHG15 expression significantly restrained the proliferation, migration and promoted cisplatin sensitivity of BC cells as well as remarkably suppressed the tumor growth in vivo. 16 , 17 Compared to adjacent normal tissues, SNHG15 expression levels were overexpressed in hepatocellular carcinoma (HCC). SNHG15 downregulation greatly attenuated the tumorigenesis. High SNHG15 expression was closely correlated with aggressive clinical parameters and poor survival of HCC. 18 , 19 In addition, upregulation of SNHG15 significantly promoted osteosarcoma cell proliferation, invasion, migration and autophagy by silencing miR‐141 expression, while SNHG15 downregulation showed the opposite effects. 20 Colorectal cancer (CRC) patients with increased SNHG15 expression had shorter survival. SNHG15 overexpression not only significantly stimulated CRC cell proliferation and inhibited cell apoptosis in vitro, but also promoted tumorigenicity in vivo. 21 Moreover, Ma and colleagues revealed that SNHG15 expression levels were markedly elevated in tissues of pancreatic cancer (PC). SNHG15 upregulation was significantly associated with worse clinical variables of PC patients. In vitro and in vivo evidence showed that decreased SNHG15 expression markedly suppressed oncogenic activities via interacting with P15 and KLF2. 22
In summary, serum exosomal lncRNA SNHG15 showed good performance in discriminating NSCLC cases from normal controls, and its level was significantly reduced following surgical treatment. In addition, serum exosomal lncRNA SNHG15 might be used to predict the prognosis of NSCLC. Further validation is required to analyze the exact efficiency of serum exosomal lncRNA SNHG15 as the tumor marker.
CONFLICTS OF INTEREST
We declared no competing interests exist.
AUTHOR CONTRIBUTIONS
Pengfei Han, Jia Zhao, and Lun Gao designed the study, conducted the experiments, analyzed the data, and wrote the study. All authors have prepared, edited, reviewed, and approved the study.
ETHICAL APPROVAL
This study was approved by the Ethics Committee of the First People's Hospital of Yulin City. All participants provided signed written informed consents before the study (approval no. 20200215X).
Han P, Zhao J, Gao L. Increased serum exosomal long non‐coding RNA SNHG15 expression predicts poor prognosis in non‐small cell lung cancer. J Clin Lab Anal. 2021;35:e23979. 10.1002/jcla.23979
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non‐small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor‐microenvironment in lung cancer‐metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558‐566. [DOI] [PubMed] [Google Scholar]
- 5. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679‐695. [DOI] [PubMed] [Google Scholar]
- 6. Bang C, Thum T. Exosomes: new players in cell‐cell communication. Int J Biochem Cell Biol. 2012;44(11):2060‐2064. [DOI] [PubMed] [Google Scholar]
- 7. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255‐289. [DOI] [PubMed] [Google Scholar]
- 8. Nicholas J. A new diagnostic tool with the potential to predict tumor metastasis. J Natl Cancer Inst. 2013;105(6):371‐372. [DOI] [PubMed] [Google Scholar]
- 9. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kondo Y, Shinjo K, Katsushima K. Long non‐coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927‐1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non‐coding RNA DLX6‐AS1 as a potential diagnostic biomarker for non‐small cell lung cancer. Oncol Lett. 2019;18(5):5197‐5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lv X, Lian Y, Liu Z, Xiao J, Zhang D, Yin X. Exosomal long non‐coding RNA LINC00662 promotes non‐small cell lung cancer progression by miR‐320d/E2F1 axis. Aging (Albany NY). 2021;13(4):6010‐6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma XR, Xu YL, Qian J, Wang Y. Long non‐coding RNA SNHG15 accelerates the progression of non‐small cell lung cancer by absorbing miR‐211‐3p. Eur Rev Med Pharmacol Sci. 2019;23(4):1536‐1544. [DOI] [PubMed] [Google Scholar]
- 14. Dong YZ, Meng XM, Li GS. Long non‐coding RNA SNHG15 indicates poor prognosis of non‐small cell lung cancer and promotes cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2018;22(9):2671‐2679. [DOI] [PubMed] [Google Scholar]
- 15. Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non‐coding RNA SNHG15 promotes CDK14 expression via miR‐486 to accelerate non‐small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233(9):7164‐7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu LB, Jiang ZJ, Jiang XL, Wang S. Up‐regulation of SNHG15 facilitates cell proliferation, migration, invasion and suppresses cell apoptosis in breast cancer by regulating miR‐411‐5p/VASP axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1899‐1912. [DOI] [PubMed] [Google Scholar]
- 17. Mi H, Wang X, Wang F, et al. SNHG15 contributes to cisplatin resistance in breast cancer through sponging miR‐381. Onco Targets Ther. 2020;13:657‐666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang JH, Wei HW, Yang HG. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20(9):1720‐1724. [PubMed] [Google Scholar]
- 19. Dai W, Dai JL, Tang MH, Ye MS, Fang S. lncRNA‐SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR‐490‐3p/ histone deacetylase 2 axis. World J Gastroenterol. 2019;25(38):5789‐5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu K, Hou Y, Liu Y, Zheng J. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR‐141. J Biomed Sci. 2017;24(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M, Bian Z, Jin G, et al. LncRNA‐SNHG15 enhances cell proliferation in colorectal cancer by inhibiting miR‐338‐3p. Cancer Med. 2019;8(5):2404‐2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma Z, Huang H, Wang J, et al. Long non‐coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2‐mediated H3K27me3. Oncotarget. 2017;8(48):84153‐84167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.


