Abstract
Background
Autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is a cerebrovascular disease closely related to the NOTCH3 gene. More than 200 mutations in this gene have been reported to be associated with this disease.
Methods
The NOTCH3 gene from CADASIL patient was screened for mutations by whole‐exome sequencing (WES). PCR amplification and direct Sanger sequencing were used to verify the suspicious gene mutation sites detected by WES.
Results
We performed second‐generation sequencing on a sample of the patient's genome and found a heterozygous deletion‐insertion mutation c.512_605delinsA in exon 4 of NOTCH3, which resulted in amino acid changes p.G171_A202delinsE. This variation was confirmed by the direct Sanger sequencing. It may be rated as a CADASIL clinical variation.
Conclusion
Discovery of this mutation site provides an important theoretical basis for specific gene‐based diagnosis and treatment of CADASIL.
Keywords: CADASIL, gene mutation, leukoencephalopathy, microbleeds, NOTCH3
1. INTRODUCTION
Autosomal‐dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is a monogenic inherited cerebrovascular disease. The disease is caused by the mutations in NOTCH3 on chromosome 19. 1 , 2 CADASIL usually starts in the middle age, which is different from the traditional cerebrovascular disease, and it usually has no clear high‐risk factors for cerebrovascular disease, with recurrent stroke as the main manifestation, which can be accompanied by cognitive impairment, dementia, mental and emotional disorders, and migraine with aura. 3 The presence of granular osmiophilic material (GOM) in close proximity to smooth muscle cells, pericytes, and endothelial cells is critical for the diagnosis of CADASIL. 4 , 5
Analysis of all the exons of the NOTCH3 gene is crucial to determine the mutation that causes the pathology. 6 , 7 The NOTCH3 gene, located on chromosome 19p13, encodes a single‐pass transmembrane receptor, which is composed of a large extracellular domain (ECD) with 34 tandem epidermal growth factor‐like (EGF) repeats encoded by exons 2–24, where NOTCH3 mutations are commonly found, a transmembrane domain, and an intracellular domain (ICD). 8 , 9 , 10 In all, 33 exons and 2321 amino acids constitute NOTCH3. 11 CADASIL is caused by the presence of only one mutation in one of the two alleles of NOTCH3 because pathogenic mutations are dominant in this disease. 12
Here, we report a case of sporadic cerebral small vessel disease in CADASIL. Gene detection revealed a heterozygous mutation in exon 4 of NOTCH3, c.512_605delinsA, and no pathogenic report was found for the mutation in the Human Gene Mutation Database (HGMD). Hence, the case has been reported here.
2. MATERIALS AND METHODS
2.1. Patients and families
We present a patient with CADASIL who was admitted to the Department of Neurology, First Affiliated Hospital of Dalian Medical University, China. Peripheral blood samples were collected for investigation. The study was approved by the Ethics Committee of the First Affiliated Hospital of Dalian Medical University and was performed in accordance with the recommendations of the Declaration of Helsinki. Written informed consent was obtained from the patient. No family history of CADASIL was provided.
Venipuncture was used to obtain the whole blood in tubes containing EDTA. Routine techniques were used to harvest genomic DNA from the peripheral blood. Spectrophotometric analysis was used to measure the concentration and purity of DNA samples. 13 , 14
2.2. Mutation analysis
Whole‐exome sequencing (WES) was performed on the DNA extracted from the peripheral blood samples. PCR amplification and direct Sanger sequencing were used to verify the suspicious gene mutation sites detected by WES. The amplified PCR products of the NOTCH3 gene were visualized using a 2% agarose gel. To discover harmful mutations, BLAST (https://blast.ncbi.nlm.nih.gov/) was used to align the sequence data with the NOTCH3 reference DNA sequence. 15 , 16
3. CASE DESCRIPTION
3.1. Disease history
A 58‐year‐old woman complained of weakness in the right lower limb for 4 days. Four days prior, the patient developed right lower limb weakness with dizziness and persistent symptoms, but no headache, nausea, vomiting, unclear speech, blurred vision, dysdipsia and dysphagia, limb twitch, dysuria, or unconsciousness.
3.2. Past history
The patient had a history of cerebral infarction for 9 years, no history of hypertension, diabetes, coronary heart disease, or smoking.
3.3. Physical examination of the nervous system
The patient was conscious, fluent in the language, and had normal intelligence. Left central facial paralysis was detected. The muscle strength of the right lower limb was scored as 4, muscle tension of the right limb was found to be increased, and the right Babinski sign was positive. No abnormalities were found in the other nervous systems.
3.4. Additional examinations
Head MRI/DWI/MRA/SWI: (1) Acute lacunar infarction of the left thalamus. (2) Multiple old lacunar cerebral infarctions. (3) Demyelination of the white matter (Figure 1A). (4) Multiple microbleeding foci were found in the bilateral semioval center, paraventricular horn, right basal ganglia, bilateral thalamus, and brainstem (Figure 1B). (5) The M1 segment of the right middle cerebral artery narrowed slightly (Figure 1C).
FIGURE 1.
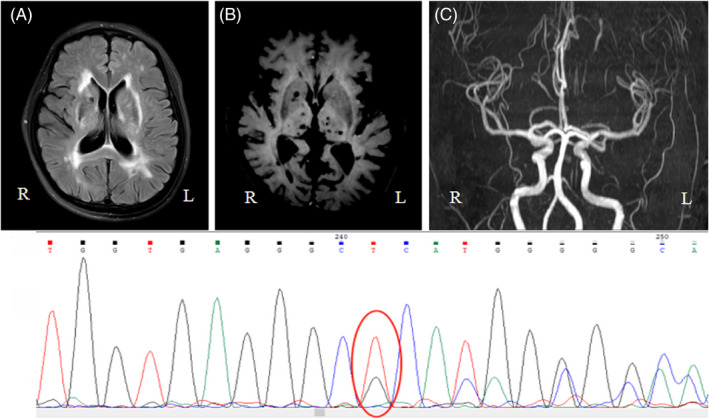
Brain MR exam and genotype result of the proband. (A) Severe diffuse leukoencephalopathy in deep white matter. (B) Microbleeds on MRI‐SWI. (C) MRA. (D) Sequence of the heterozygous deletion‐insertion mutation c.512_605delinsA in exon 4 of NOTCH3
3.5. Genetic analysis
Genomic DNA was extracted from the peripheral blood samples of the patient. One deletion insertion heterozygous variation, c.512_605delinsA, was identified in exon 4 of the NOTCH3 gene by WES. The quality control data for WES are shown in Table 1. In order to further verify the gene test results, PCR amplification and direct Sanger sequencing were used to detect the variant in the sequence exons 2–8, 11, 12, 18, and 19 (both coding sections and intron/exon borders), which are mutational hotspots in the NOTCH3 gene. The sequences of primers used are as follows: F‐5′‐TGGCGACCTCACTTACGACT‐3′ and R‐5′‐CACTGGCAGTTATAGGTGTTGAC‐3′. It was verified that there was a definite gene mutation of c.512_605delinsA in exon 4 of NOTCH3 (Figure 1D). No pathogenic gene mutations were found in other hotspots in this gene.
TABLE 1.
Quality control data of WES
| Total | |
|---|---|
| Raw_data (Mb) | 2026.02 |
| Clean_data (Mb) | 1971.61 |
| Aligned (%) | 99.97 |
| Initial bases on target | 1,929,086 |
| Base covered on target | 1,924,904 |
| Coverage of target region | 99.80% |
| Total effective yield (Mb) | 1435.79 |
| Effective sequence on target (Mb) | 797.63 |
| Fraction of effective bases on target | 55.60% |
| Average sequencing depth on target | 413.48 |
| Fraction of target covered with at least 4X | 99.40% |
| Fraction of target covered with at least 10X | 98.60% |
| Fraction of target covered with at least 20X | 97.00% |
| Duplication rate (%) | 26.81 |
4. DISCUSSION
CADASIL, a hereditary small artery disease, is the most common hereditary vascular disease in adults. Typical clinical features include migraine, subcortical ischemic events, cognitive impairment, and mental symptoms. The disease aggravates progressively, eventually leading to severe disability and dementia. 17 The head MRI findings of CADASIL are correlated with age and disease stage. It showed symmetrical and extensive hyperintensity on T2‐weighted images and T2 flair images. Hyperintensity lesions with limited diffusion appeared on DWI images, indicating acute or subacute cerebral infarction. SWI revealed microbleeds, white matter lesions are often involved in the anterior temporal lobe, external capsule, superior frontal gyrus, and temporal pole. These special parts of the lesions have high sensitivity and specificity for diagnosis.
The pathophysiological mechanism of CADASIL is mainly due to the deposition of mutated Notch3 protein aggregates on the surface of smooth muscle cells of the medium‐sized artery wall, resulting in vascular stenosis and occlusion, which then lead to a cerebral ischemic attack. The deposition is usually systemic and systemic, but deposition on the wall of cerebral vessels is more significant. Granular osmiophilic substances can also be found in areas outside the cerebral vessels, such as the skin and muscle vessels.
Mutation in the NOTCH3 gene is the main genetic mechanism of CADASIL. To date, the hot spots of NOTCH3 mutations in Chinese mainland CADASIL families have been mainly located in exons 3, 4, 11, 12, 13, and 14. For example, Fan Weiming et al described a 100 pare base fragment deletion (ENST 00000263388, c.512‐611del) with CADASIL. 18 Moreover, the Notch3 gene mainly encodes a highly conserved transmembrane receptor with 2321 amino acids, which is specifically expressed in vascular smooth muscle cells and plays an important role in the stability of vascular smooth muscle cells. 19 , 20 From a microscopic point of view, the core pathogenesis of CADASIL is that NOTCH3 gene mutations interfere with the expression of receptors, affect the production of vascular smooth muscle cells, and cause the degeneration of vascular smooth muscle cells. At present, most researchers believe that in CADASIL families, NOTCH3 mutations lead to the duplication or loss of cysteine residues in the repeat region of the epidermal growth factor; thus, the transmembrane protein encodes changes in structure and function. Macroscopically, the pathogenesis of this disease is caused by the deposition of mutated NOTCH3 protein aggregates (granular osmiophilic substances), resulting in vascular stenosis and occlusion, which then leads to cerebral ischemic attack. The deposition is usually systemic and systemic, but deposition on the wall of cerebral vessels is more significant. Whether from the macro or micro point of view, the etiology and pathogenesis of CADASIL are still unclear and require further study. 21 , 22 In terms of treatment, no drugs have been proven to be beneficial for the treatment and prevention of CADASIL, including antiplatelet drugs, lipid‐lowering drugs, homocysteine‐lowering drugs, and antihypertensive drugs. 23 Therefore, only symptomatic treatment can be offered. Identifying the potential risk factors leading to the mutation of the CADASIL gene may more accurately guide the treatment and prevention, and help to identify new drug targets in the future.
Gene examination is the gold standard for the diagnosis of this disease. 6 , 24 In this report, we detected a heterozygous deletion‐insertion mutation, c.512_605delinsA, in exon 4 of NOTCH3, resulting in amino acid changes in P. G171_A202delinsE. Although there was no association between this heterozygous mutation and CADASIL in HGMD, according to ACMG guidelines, 25 the variation can be rated as a pathogenic variation. 20 , 26 If the variation is pathogenic, it may theoretically be pathogenic. If this is further confirmed in family samples, the clinical diagnosis and treatment rate of CADASIL may be further improved.
5. CONCLUSION
In conclusion, we found a new heterozygous deletion‐insertion mutation c.512_605delinsA in exon 4 of NOTCH3 that may be associated with CADASIL. This new mutation expands the genetic spectrum of NOTCH3‐related diseases, which will contribute to further study of this disease in the future.
CONFLICT OF INTERESTS
The present study does not have any conflict of interest.
ACKNOWLEDGMENTS
The authors thank the patient for the participation.
Liu J, Zhang Q, Wang Q, et al. A case of CADASIL caused by NOTCH3 c.512_605delinsA heterozygous mutation. J Clin Lab Anal. 2021;35:e24027. 10.1002/jcla.24027
Jiahui Liu and Qiaoyu Zhang contributed equally to this work and should be considered as equal first coauthors.
Contributor Information
Huijie Dong, Email: Donghuijie1978@163.com.
Xiaofei Ji, Email: jixiaofei1979@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Joutel A, Bousser M‐G, Biousse V, et al. A gene for familial hemiplegic migraine maps to chromosome 19. Nat Genet. 1993;5(1):40‐45. [DOI] [PubMed] [Google Scholar]
- 2. Tournier‐Lasserve E, Joutel A, Melki J, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy maps to chromosome 19q12. Nat Genet. 1993;3(3):256‐259. [DOI] [PubMed] [Google Scholar]
- 3. Chen S, Ni W, Yin X‐Z, et al. Clinical features and mutation spectrum in Chinese patients with CADASIL: a multicenter retrospective study. CNS Neurosci Ther. 2017;23(9):707‐716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muiño E, Gallego‐Fabrega C, Cullell N, et al. Systematic review of cysteine‐sparing NOTCH3 missense mutations in patients with clinical suspicion of CADASIL. Int J Mol Sci. 2017;18(9):1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishiko A, Shimizu A, Nagata E, et al. Notch3 ectodomain is a major component of granular osmiophilic material (GOM) in CADASIL. Acta Neuropathol. 2006;112(3):333‐339. [DOI] [PubMed] [Google Scholar]
- 6. Joutel A, Corpechot C, Ducros A, et al. Notch3 mutations in CADASIL, a hereditary adult‐onset condition causing stroke and dementia. Nature. 1996;383(6602):707‐710. [DOI] [PubMed] [Google Scholar]
- 7. Dieu JH, Veyckemans F. Perioperative management of a CADASIL type arteriopathy patient. Br J Anaesth. 2003;91(3):442‐444. [DOI] [PubMed] [Google Scholar]
- 8. Dziewulska D, Lewandowska E. Pericytes as a new target for pathological processes in CADASIL. Neuropathology. 2012;32(5):515‐521. [DOI] [PubMed] [Google Scholar]
- 9. Meng HE, Zhang X, Yu G, et al. Biochemical characterization and cellular effects of CADASIL mutants of NOTCH3. PLoS One. 2012;7(9):e44964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duering M, Karpinska A, Rosner S, et al. Co‐aggregate formation of CADASIL‐mutant NOTCH3: a single‐particle analysis. Hum Mol Genet. 2011;20(16):3256‐3265. [DOI] [PubMed] [Google Scholar]
- 11. Wesołowski W, Dziewulska D, Koziarska M, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL)—literature review apropos an autopsy case. Pol J Pathol. 2015;66(3):323‐329. [DOI] [PubMed] [Google Scholar]
- 12. Opherk C, Duering M, Peters N, et al. CADASIL mutations enhance spontaneous multimerization of NOTCH3. Hum Mol Genet. 2009;18(15):2761‐2767. [DOI] [PubMed] [Google Scholar]
- 13. Han P, Wei G, Cai K, et al. Identification and functional characterization of mutations in LPL gene causing severe hypertriglyceridaemia and acute pancreatitis. J Cell Mol Med. 2020;24(2):1286‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang R, Chen S, Han P, et al. Whole exome sequencing identified a homozygous novel variant in CEP290 gene causes Meckel syndrome. J Cell Mol Med. 2020;24(2):1906‐1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai Y, Liang S, Dong X, et al. Whole exome sequencing identified a novel DAG1 mutation in a patient with rare, mild and late age of onset muscular dystrophy‐dystroglycanopathy. J Cell Mol Med. 2019;23(2):811‐818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng Y, Xu J, Liang S, et al. Whole exome sequencing identified a novel heterozygous mutation in HMBS gene in a Chinese patient with acute intermittent porphyria with rare type of mild anemia. Front Genet. 2018;9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Machowska‐ Sempruch K, Bajer‐ Czajkowska A, Makarewicz K, et al. A novel NOTCH3 gene mutation in a polish CADASIL family. J Stroke Cerebrovasc Dis. 2019;28(3):574‐576. [DOI] [PubMed] [Google Scholar]
- 18. Weiming F, Yuliang W, Youjie LI, et al. A novel Notch3 deletion mutation in a Chinese patient with cerebral autosomal dominant arteriopathy with subcortical infarcts and leucoencephalopathy (CADASIL). J Clin Neurosci. 2013;20(2):322‐323. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z, Yuan Y, Zhang W, et al. NOTCH3 mutations and clinical features in 33 mainland Chinese families with CADASIL. J Neurol Neurosurg Psychiatry. 2011;82(5):534‐539. [DOI] [PubMed] [Google Scholar]
- 20. Rutten JW, Haan J, Terwindt GM, et al. Interpretation of NOTCH3 mutations in the diagnosis of CADASIL. Expert Rev Mol Diagn. 2014;14(5):593‐603. [DOI] [PubMed] [Google Scholar]
- 21. Muller K, Courtois G, Ursini MV, Schwaninger M. New insight into the pathogenesis of cerebral small‐vessel diseases. Stroke. 2017;48(2):520‐527. [DOI] [PubMed] [Google Scholar]
- 22. Hedera P. NOTCHing toward better understanding of CADASIL. Neurology. 2020;95(13):565‐566. [DOI] [PubMed] [Google Scholar]
- 23. Bersano A, Bedini G, Oskam J, et al. CADASIL: treatment and management options. Curr Treat Options Neurol. 2017;19(9):31. [DOI] [PubMed] [Google Scholar]
- 24. Hung LY, Ling TK, Lau NKC, et al. Genetic diagnosis of CADASIL in three Hong Kong Chinese patients: a novel mutation within the intracellular domain of NOTCH3. J Clin Neurosci. 2018;56:95‐100. [DOI] [PubMed] [Google Scholar]
- 25. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferrante EA, Cudrici CD, Boehm M. CADASIL: new advances in basic science and clinical perspectives. Curr Opin Hematol. 2019;26(3):193‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


