Abstract
The J domain of simian virus 40 (SV40) large T antigen is required for efficient DNA replication and transformation. Despite previous reports demonstrating the promiscuity of J domains in heterologous systems, results presented here show the requirement for specific J-domain sequences in SV40 large-T-antigen-mediated activities. In particular, chimeric-T-antigen constructs in which the SV40 T-antigen J domain was replaced with that from the yeast Ydj1p or Escherichia coli DnaJ proteins failed to replicate in BSC40 cells and did not transform REF52 cells. However, T antigen containing the JC virus J domain was functional in these assays, although it was less efficient than the wild type. The inability of some large-T-antigen chimeras to promote DNA replication and elicit cellular transformation was not due to a failure to interact with hsc70, since a nonfunctional chimera, containing the DnaJ J domain, bound hsc70. However, this nonfunctional chimeric T antigen was reduced in its ability to stimulate hsc70 ATPase activity and unable to liberate E2F from p130, indicating that transcriptional activation of factors required for cell growth and DNA replication may be compromised. Our data suggest that the T-antigen J domain harbors species-specific elements required for viral activities in vivo.
The large and small tumor antigens of simian virus 40 (SV40) are multifunctional regulatory proteins that play vital roles at many steps in the virus life cycle. In particular, the large tumor antigen (T antigen) is necessary for diverse functions, including viral DNA replication and transcriptional regulation; T antigen is also necessary and often sufficient for tumorigenesis. The ability of T antigen to elicit neoplastic transformation depends upon its capacity to alter a number of multiprotein complexes, including those containing members of the retinoblastoma (Rb) tumor suppressor family (p107, p130, and pRb) and those containing p53 (12, 13, 14, 24, 26; reviewed in reference 5).
Essential T-antigen functions have been shown to depend completely or in part on activities mapping within the first 80 amino acids of T antigen (29). This amino-terminal segment of T antigen is homologous to and functions both in vivo and in vitro as a J domain (6, 21, 36). J domain-containing proteins constitute a family of molecular chaperones known as DnaJ homologues or hsp40s (40-kDa heat shock proteins). Hsp40s associate with a partner chaperone, known as a DnaK homologue or hsc70 (70-kDa heat shock cognate proteins), and stimulate their ATPase activity, consequently modulating the ability of the hsc70 to bind and release polypeptides or protein complexes. As a result, hsc70-hsp40 chaperone pairs may remodel the conformations and regulate the activities of cellular protein machines (reviewed in reference 19). Because T antigen also acts on several protein complexes (see above), we proposed that the chaperone activity of T antigen is required to modulate the activities of these multiprotein assemblies (5).
Experiments demonstrating that the amino terminus of T antigen is a functional J domain also indicated that the SV40 T-antigen J domain is functionally promiscuous. First, the J domain from SV40 T antigen substitutes functionally for the J domain of the Escherichia coli DnaJ protein, as assessed by the ability of the chimeric protein to support both phage lambda plaque formation and bacterial growth at a restrictive temperature (21). Second, the J domains from two human hsp40s could replace the J domain in T antigen and support SV40 replication (6). Third, a 136-amino-acid N-terminal fragment of T antigen that includes the J domain stimulates the ATPase activities of both mammalian and yeast hsc70s (36). Fourth, the T-antigen J domain, when inserted into the J domain of the cytoplasmic yeast protein, Ydj1p, rescues a temperature-sensitive growth defect when expressed in yeast containing mutations in YDJ1 (S. W. Fewell and J. L. Brodsky, unpublished data). In accordance with these data, promiscuous hsc70-hsp40 interactions have been observed both in vivo and in vitro in a number of experimental systems (22, 25, 32, 33). In contrast, a significant body of data indicate that only specific interactions between an hsc70 and a unique hsp40 can promote chaperone-dependent functions both in vitro and in vivo (for examples, see references 2, 4, 10, 11, 27, 28, and 41). The determinants of this specificity remain unknown and, because of the results presented above, it is unclear if T antigens similarly possess such determinants.
To address this question, we substituted the J domain of SV40 T antigen with the yeast Ydj1p and E. coli DnaJ J domains and examined viral growth and infection, DNA replication, and the ability of the hybrid T antigens to transform mammalian cells. In addition, because the J domain from T antigens in the JC virus and SV40 are highly homologous (31), the T-antigen J domain from JC virus was used to replace the J domain of the SV40 T antigen. Although the J domain from JC virus complemented each activity to some extent, we found that the bacterial and yeast J domains could not substitute for the T-antigen J domain. In accordance with these results, only the SV40 T antigen chimera containing the JCV J domain was able to dissolve an Rb family-EF2 transcription factor complex. However, both the SV40 T antigen and a defective T antigen containing the DnaJ J domain bound hsc70, but only wild-type SV40 T antigen stimulated proficiently the ATPase activity of hsc70. These data suggest that the T-antigen J domain harbors amino acid sequence elements required for it to engineer virus-specific activities in vivo and that hsc70 binding is not sufficient for T-antigen J domain activities.
MATERIALS AND METHODS
Chimera construction.
The starting point for these constructions was pSV-B3 (30), a plasmid which contains the SV40 genome ligated into pBR322 at the BamHI sites. A new plasmid (pSV40*) was made by replacing the HindIII restriction site 7 bp upstream of the T-antigen translation start site with a BglII site. This was accomplished by two-step PCR with two flanking primers: SV-JD Taq (CTA AAG CAT TCG AAG CAG TAG CAA TC) and SV-Ori (CAT TCT CCG CCC CAT GGC TGA C) and an internal primer SV-JD Bgl II (GGC TTT TGC AA AGA TCT TGC AAA GAT GGA TAA AG) (1). The resulting fragment was cut with BglI and BstXI and ligated into the corresponding sites of pSV-B3. No differences in viral growth were observed between pSV40* and wild-type SV40 (J. D. Tremblay, J. A. Lewis, and J. M. Pipas, unpublished observations). The J domain for each chimera was PCR amplified and cloned into the BglII and ApaI sites in the polylinker of pSKB, a Bluescript (Stratagene)-derived vector in which the PstI site was replaced with a BglII site. The first 210 nucleotides (nt) of yeast YDJ1 were amplified from plasmid pET9d.Ydj1p (8) using YDJ-Bgl II (GCG AGA TCT TGC AAA GAT GGT TAA AGA AAC TAA) and YDJ-Apa I (GCC GGG CCC AAA TTG GTC ATA TAT ATC TCT C). The first 225 nt of the JC virus T-antigen gene (16) were amplified using primers JCV-Bgl II (GCG AGA TCT TGC AAA GAT GGA CAA AGT GCT GAA TAG) and JCV-Apa I (TCC GGG CCC AAA ATC AGG CTG ATG AGC). The first 210 nt of E. coli DNA J (21) were amplified using DNAJ-Bgl II (GCG AGA TCT TGC AAA GAT GGC TAA GCA AGA TTA TTA) and DNAJ-Apa I (TCC GGG CCC ATA CTG ATC GTA TGC CGC). DNA from SV40 T antigen was PCR amplified to introduce an NaeI site at the J-domain border using primers SV-JD Nae I (CAA CCT GAC TTT GCC GGC TTC TGG GAT G) and a ClaI site using SV-JD Taq (see above) and then cloned into pSKB at the NaeI and ClaI sites to create pSKB Nae.
To complete the construction of the chimeras (SV-JCV, SV-YDJ, and SV-DNAJ), each J-domain plasmid was cut with BglII and NlaIV, the SV40 fragment in pSKB Nae was removed with BstXI and NaeI, and the two fragments were ligated into pSV40* that was digested with BglII and BstXI. NlaIV and NaeI are both blunt-end cutters whose cleaved sites when ligated in these clones maintain the open reading frame but result in the loss of both restriction sites. All constructs were verified by nucleotide sequence analysis. For plaque and replication assays, the viral genomes were excised from pBR322 using BamHI and recircularized with T4 DNA ligase as described elsewhere (30).
The intron in pSV40* and pSV-DNAJ was removed by inverse PCR using oligonucleotides SWF5 (ATC TCG AGC TCA GTT GCA TCC CAG) and SWF6 (ATC TCG AGA TTC CAA CCT ATG GAA CTG) and the Expand Long Template PCR System (Roche Molecular Biochemicals). Amplified DNAs were digested with XhoI and religated to generate pSV40*-Xho and pSV-DNAJ-Xho. Baculovirus expression plasmids were constructed by subcloning BglII-BamHI fragments from pSV40*-Xho and pSV-DNAJ-Xho into the BamHI site in pFastBac1 (Gibco-BRL). Proteins were expressed in and purified from Sf9 insect cells as described previously (7).
Plaque assays.
Freshly confluent 6-cm plates of BSC40 cells were transfected using 0.5 mg of DEAE-dextran (Sigma) per ml in minimum essential medium (MEM; Gibco) with 15 ng of DNA in a total volume of 200 μl. This mixture was carefully added to the center of each plate after removal of the old medium. After 15 min, plates were overlaid with 4 ml of a mixture containing one part melted 1.8% Bacto-Agar (Difco) cooled to 45°C and one part 2× modified Eagle medium without phenol red (Gibco) but supplemented with 10% fetal bovine serum (FBS; HyClone) at 37°C. An additional 3 ml of this mixture was added every 3 days (35). On the sixth day, Neutral Red (Sigma) was added at a final concentration of 0.05 mg/ml to the mixture in order to visualize plaques. Plaques were scored by number, size, and time of appearance.
In vivo replication assay.
Freshly confluent 75-cm2 flasks of BSC40 cells were transfected with 45 ng of DNA containing or lacking 45 ng of dl1007 DNA (control dishes) using DEAE-dextran in a total volume of 800 μl. After 15 min, dishes were fed with 10 ml of MEM–2% FBS and incubated at 37°C for 2 to 3 days. Cells were trypsinized from the plates, and DNA was extracted with the Qiagen Plasmid Mini Kit. Extracted DNA was linearized with BclI and quantified. The DNA was also treated with DpnI to digest methylated input template DNA, while leaving newly replicated DNA intact (30). DNA extracts were then separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with a 32P-labeled probe synthesized using random hexamer primers with SV40 genomic DNA (30) as a template.
Transformation assay.
Transformation activity was measured using a dense focus formation assay in REF52 cells (36). Subconfluent plates of REF52 cells were transfected with 2 μg of DNA using Lipofectamine as described by the manufacturer (Gibco). After 24 h, plates were split 1:3 and fed twice weekly with MEM–10% FBS. After 4 to 6 weeks, plates were stained with crystal violet (Sigma), and foci were counted.
Expression of T-antigen hybrids.
In order to verify expression of each chimera, cells were transfected as described above but with the addition of 0.2 μg of pRSV.neo (36) per dish. After being split 1:3, plates were either used in the transformation assay as described above or selected for drug resistance using 300 μg of G418 (BioWhittaker) per ml. Drug-resistant colonies were picked and expanded into lines. Lysates were prepared by resuspending cell pellets in 300 μl of lysis buffer (50 mM Tris-HCl [pH 8]–5 mM EDTA–150 mM NaCl–0.5% Nonidet P-40 supplemented with the following protease inhibitors to the respective final concentrations: leupeptin, 1 μg/ml; pepstatin, 0.7 μg/ml; E64, 1 μg/ml; phenylmethylsulfonyl fluoride, 50 ng/ml; aprotinin, 1 μg/ml; trypsin inhibitor, 10 μg/ml; and tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK], 10 μg/ml) with rocking for 30 min at 4°C. Clarified extracts were prepared by centrifuging the extract at 13,000 rpm in a microcentrifuge for 15 min at 4°C. Equal amounts of total protein (determined by measuring the absorbance of a diluted aliquot of the sample at 280 nm in a spectrophotometer) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (Schleicher & Schuell). PAb901, an antibody specific to the C terminus of large T antigen (kindly provided by Judith Tevethia, Pennsylvania State University Medical School, Hershey, Pa.), was used to detect each of the T-antigen chimeras, and anti-mouse actin antibody (Boehringer Mannheim) was used to detect actin levels as a loading control. Both antibodies were decorated using horseradish peroxidase-conjugated sheep anti-mouse antiserum (Amersham) and the SuperSignal West Chemiluminescence system from Pierce.
Gel shift assay.
Sf9 insect cells and their handling have been described previously (7). Baculoviruses expressing human E2F4 and a truncated version of human p130(Δ2–371) were kindly provided by Peter Whyte (Institute for Molecular Biology and Biotechnology, McMaster University, Hamilton, Ontario, Canada). Baculoviruses expressing human DP1 and E2F1 were generously provided by Helen Piwnica Worms (Washington University Medical School, St. Louis, Mo.).
Lysates enriched for E2F4-Dp1 or p130-E2F4-Dp1 were generated either by double infecting Sf9 cells with multiple viruses encoding E2F4 and DP1 or triple infecting them with multiple viruses encoding E2F4, DP1, and p130. Cells were incubated at 27°C for 43 h. Parental and transfected REF52 cells or insect cells were harvested 10-fold in buffer B (50 mM HEPES, pH 7.9; 400 mM KCl; 0.5 μM EDTA; 10% glycerol; 0.1% Nonidet P-40), 1 μg of pepstatin per ml, and the recommended dilution of Complete EDTA-free Protease Inhibitor Cocktail as recommended by the supplier (Boehringer Mannheim). Cells were lysed for 25 min on ice and centrifuged at 16,000 × g in a microcentrifuge for 30 min at 4°C. The pellets were discarded, and protein in the supernatant was quantified with the Bio-Rad protein assay reagent using bovine serum albumin as a standard.
For the gel shift reaction, 10 μg of REF52 or insect cell lysate was incubated with 1 ng of DNA probe containing an E2F binding site (5′-ATTTAAGTTTCGCGCCCTTTCTCAA-3′) end labeled with 32P (U.S. Biochemical T4 protocol). Reactions (20 μl, total volume) were incubated for 10 min on ice and then for 20 min at room temperature in 20 mM HEPES (pH 7.4), 50 mM KCl, 8.5% glycerol, 1 mM EDTA, and 1 mM MgCl2. Reactions were loaded onto a 0.25 × TBE–4.5% acrylamide gel and run at 200 V for 3 h at 4°C. Where indicated, competitor DNA (5′-ATTTAAGTTTCGATCCCTTTCTCAA-3′) was also included at a 500-fold molar excess (underlined nucleotides denote mutated sequence). The following antibodies were added to identify components of the shifted DNA binding complexes: anti-p130-C20, p107-SD9, E2F1-C20, E2F2-C20, E2F3-C18, E2F4-C20, E2F5-C20, and pRb IF8 (Santa Cruz Biotechnologies); anti-pRb AB2 (Calbiochem); and anti-pRb 14001A (Pharmingen).
Hsc70 binding and ATPase assays.
REF52 lysate (prepared as described above) was incubated with anti-T-antigen antibody (PAb901) for 2 h on ice. The complexes were captured via a 20-s spin in a microcentrifuge at 16,000 × g and washed three times with 1 ml of buffer I (20 mM HEPES, pH 7.8; 40 mM KCl; 6 mM MgCl2; 0.1% Nonidet P-40). The pellets were resuspended in 100 μl of buffer I, in the presence of an ATP regeneration system (50 μM GDP mannose, 40 μM creatine phosphate, 0.2 mg of creatine phosphokinase per ml), and 1 μg of purified bovine hsc70 (StressGen) for 30 min at room temperature. The complexes were recaptured via a 20-s spin in a microcentrifuge at 16,000 × g and washed two times with 1 ml of phosphate-buffered saline (Gibco) and two times with 1 ml of buffer I. The final pellets were resuspended in 2× SDS-PAGE sample buffer containing dithiothreitol and β-mercaptoethanol and resolved by electrophoresis on an 8% polyacrylamide gel. Proteins were transferred to an Immobilon polyvinylidene difluoride membrane (Millipore) and immunoblots were performed using the PAb901 or anti-hsc70 antibodies (StressGen) as described above.
Ssa1p-ATP complexes were formed essentially as described previously for Ssc1p (12a). Ssa1p (25 μg) was mixed with 100 μCi of [α-32P]ATP (NEN) and 25 μM ATP in complex buffer (25 mM HEPES-KOH, pH 7.5; 100 mM KCl; 11 mM magnesium acetate) and incubated for 30 min on ice. Complexes were purified from free ATP on a NICK spin column (Amersham/Pharmacia Biotech) preequilibrated with complex buffer. Fifteen fractions were collected (two drops per fraction) and monitored with a Geiger counter. Peak complex fractions were pooled, adjusted to 10% glycerol, and frozen in liquid nitrogen. Single turnover assays were performed at 30°C by mixing 25 μl of thawed complex and 25 μl of complex buffer with or without 1.6 μg of the indicated T antigen. At specified time points, a 6-μl aliquot of the reaction was removed and added to 2 μl of stop solution (36 mM ATP; 2 M LiCl; 4 M formic acid) on ice. Triplicate 2-μl aliquots of this mixture were spotted onto a thin-layer chromatography plate and developed as described previously (36).
RESULTS
Yeast and E. coli J-domain chimeric SV40 viruses fail to produce plaques.
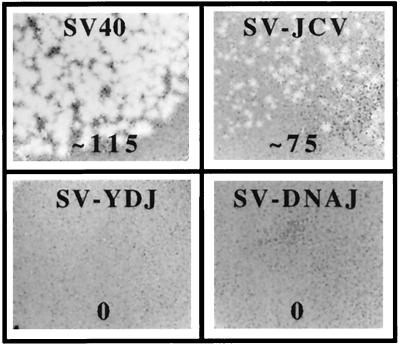
To test the ability of various J domains to functionally substitute for the SV40 T-antigen J domain in vivo, chimeric SV40 DNAs containing the J domains of JC virus T antigen, Saccharomyces cerevisiae Ydj1p, or E. coli DnaJ were constructed (Fig. 1). We first examined whether these recombinant viruses formed plaques after being transfected into monkey BSC40 cells. The wild-type SV40 control produced visible plaques after 7 days, as expected (Fig. 2). The JC virus construct (SV-JCV) produced plaques 2 to 3 days later and with a much smaller size (Fig. 2). In contrast, the cells transfected with the yeast (SV-YDJ)- and E. coli (SV-DNAJ)-derived constructs failed to produce plaques, even after 21 days of incubation. To establish that the inability of these constructs to produce plaques arose specifically from a defective T antigen, each construct was cotransfected with dl1007, an SV40 mutant defective for viral coat protein production (23). We found that dl1007 complemented both defective hybrid T antigens (Tremblay et al., unpublished). Thus, the failure of SV-YDJ and SV-DNAJ to produce plaques was due to inactive chimeric T antigens.
FIG. 1.
(A) Sequence alignment of the relevant J domains. The aligned sequences of SV40 large T-antigen (residues 1 to 80), JCV large T-antigen (residues 1 to 75), Ydj1p (residues 1 to 70), and DnaJ (residues 1 to 70) J domains are shown (5, 6, 36). Helices in the E. coli DnaJ J domain are underlined. (B) Construction of the chimeric T-antigen J-domain proteins. The J-domain sequences of the JCV T antigen, Ydj1p, and DnaJ used to replace the J domain of SV40 large T antigen are depicted as shaded boxes. The p53 and Rb family binding site (LXCXE) is also shown.
FIG. 2.
The Ydj1p and DnaJ J domains cannot functionally replace the J domain of SV40 large T antigen. The ability of wild-type SV40 and three SV40 mutants containing chimeric T-antigen constructs to form infectious virions was assessed by a plaque assay: SV-JCV, the JCV J domain in T antigen; SV-YDJ, the Ydj1p J domain in T antigen; and SV-DNAJ, the DnaJ J domain in T antigen. The number of plaques per nanogram of input DNA is shown below each dish. Wild-type SV40 plaques were detected 7 days after infection, while plaques on the SV-JCV plate appeared 9 to 10 days after infection and were noticeably smaller than those on the wild-type SV40 dish. Plaques were absent on plates infected with the SV-YDJ or SV-DNAJ chimeric viruses. A portion of each plaque assay dish is shown.
SV40 T-antigen chimeras containing yeast or E. coli J domains do not support viral DNA replication.
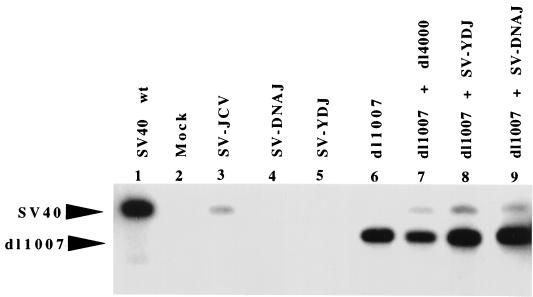
Because the J domain has been shown to be required for efficient viral DNA replication (6, 29), the ability of each construct to support in vivo replication was examined by extracting DNA from BSC40 cells that had been transfected with each of the chimeric virus constructs. Southern blot analysis of DpnI-treated DNA extracts showed that SV-YDJ and SV-DNAJ failed to exhibit replication activity in vivo. In contrast, the SV-JCV hybrid replicated DNA but was only ∼10% as proficient as wild-type SV40 (Fig. 3), a finding consistent with the compromised ability of this chimera to form plaques (Fig. 2). When wild-type large T antigen (dl1007) was transfected with the defective chimeras in trans, replication of the chimeric DNA was recovered (Fig. 3), providing evidence that the defect is not related to the condition of the input DNA and is specific to the J domain in the T-antigen constructs.
FIG. 3.
DNA replication in defective SV40 containing the Ydj1p and DnaJ J domains. DNA was extracted from BSC40 cells infected with wild-type, control, or chimeric SV40 viral DNA (lanes 1 to 5) and assayed by Southern blot analysis using a SV40 probe. In addition, wild-type T-antigen DNA (dl1007) was transfected alone (lane 6) or in combination with DNA encoding SV40 coat proteins (dl4000; lane 7) or the Ydj1p (lane 8) or DnaJ (lane 9) chimeric SV40 genomes and analyzed similarly. The positions of replicated SV40 and dl1007 DNA were indicated.
SV40 T-antigen chimeras containing yeast or E. coli J domains are transformation defective.
The transforming ability of each chimeric virus was measured by its ability to form dense foci on a monolayer of REF52 cells. Subconfluent plates of cells were transfected with a vector containing the complete SV40 genome encoding either the SV40, JC virus, Ydj1p, or DnaJ J domains. After transfection, plates were split and maintained for 4 to 6 weeks before being stained with crystal violet. Foci were scored as darkly stained piles of cells emerging from the monolayer. As shown in Fig. 4A and Table 1, constructs expressing the yeast and E. coli J-domain chimeric T antigens failed to produce foci, whereas the JC virus J-domain construct was able to produce foci, but at a somewhat reduced efficiency.
FIG. 4.
(A) The Ydj1p and DnaJ chimeric T antigens fail to transform REF52 cells. Transformation of REF52 cells was measured by a dense focus assay. Cells were fixed and stained 4 weeks after transfection with SV40 large T antigen, SV-JCV chimeric T antigen, SV-DNAJ T antigen, SV-YDJ T antigen, or vector DNA. Photographs of a representative portion of tissue culture dishes are shown. (B) Steady-state levels of chimeric T antigens are similar to that of wild-type T antigen. REF52 cells were cotransfected with pRSV. Neo and SV40, SV-JCV, SV-YDJ, or SV-DNAJ T-antigen DNAs. G418-resistant colonies were expanded into clonal lines, and the steady-state levels of each chimeric large T antigen in cell extracts were measured by immunoblot analysis using PAb901 (see Materials and Methods). The level of actin was also determined as a loading control.
TABLE 1.
Characteristics of SV40 J-domain chimerasa
| DNA source | No. of focib | Replication (% wild type) | No. of plaques/ng | Disruption of p130-E2F |
|---|---|---|---|---|
| Mock | 0, 0, 0 | 0 | 0 | No |
| SV40 | 49, 45, 42 | 100 | 115 | Yes |
| SV-JCV | 23, 20, 25 | 9.6 | 75 | Yes |
| SV-YDJ | 0, 0, 0 | 0 | 0 | No |
| SV-DNAJ | 0, 0, 0 | 0 | 0 | No |
A summary of wild-type and chimeric T-antigen activities in plaque (Fig. 2) and of DNA replication (Fig. 3), transformation (Fig. 4), and p130-E2F complex disruption (Fig. 6) assays, performed as described in Materials and Methods, is presented.
The number of foci from three independent experiments is shown.
To verify that the chimeric T antigens were expressed efficiently in REF52 cells, replicate plates were transfected as described in the Materials and Methods but with the addition of a pRSV.neo plasmid. After being split, cells were tested either for focus formation or selected for drug resistance using G418. Extracts from drug-resistant colonies were prepared and then screened by immunoblot analysis using an anti-T-antigen-specific antibody. As presented in Fig. 4B, the level of each chimeric T antigen was reduced compared to the wild-type level. However, the level of SV-JCV (which was transformation and replication competent) was identical to the levels of the defective hybrids. Therefore, the inability of SV-YDJ and SV-DNAJ and compromised ability of SV-JCV to form foci was not solely due to lower steady-state levels of the protein.
SV40 T-antigen chimeras containing yeast or E. coli J domains are defective for abolishing a p130-E2F DNA binding complex.
One function of the T-antigen J domain is to act in cis with the Rb binding motif to disable the growth inhibitory functions of the Rb family of proteins (18, 36, 37, 37a; 42). When bound by the Rb family of proteins, such as pRb, p107, or p130, E2Fs are unable to transactivate genes that induce DNA synthesis and cellular division; T antigen disrupts this complex, thereby activating DNA synthesis and cell division (12, 14, 40). Thus, we wished to determine if the chimeric T antigens were capable of disrupting Rb-E2F family complexes.
Lysates of stable REF52 cell lines expressing SV40 large T antigen were incubated in a gel shift reaction using an oligonucleotide containing an E2F consensus-binding site. To demonstrate the relative migration of “free” E2F versus Rb family-E2F complexes, a gel shift of lysate from insect cells expressing only E2F4-DP1 or p130-E2F4-DP1 was performed (Fig. 5A, lanes 7 and 8). When incubated in the gel shift reaction, 2-day-postconfluent parental REF52 cell lysate yielded bands that migrated as free E2F as well as Rb family-E2F complexes (Fig. 5A, lane 1). As expected, the Rb family-E2F complexes were absent from lysates of 2-day-postconfluent wild-type T-antigen REF52 cell lines but free E2F-DNA binding complexes were present (Fig. 5A, lane 2). The E2F-DNA binding complexes were specific since coincubation of an excess of a 500-fold molar ratio of specific but not mutated nonradioactive oligonucleotide abolished DNA binding (Fig. 6A, lanes 3 to 6).
FIG. 5.
Analysis of Rb family-E2F complexes in T-antigen-transfected REF52 cells. Lysates from parental cells (P) and cells transfected with SV40 T antigen (T) were subjected to gel shift analysis using radiolabeled nucleotide probe containing a consensus E2F binding site. (A) A 500-fold molar excess of specific (lanes 3 and 4) or mutated (lanes 5 and 6) competitor DNA was included in the gel shift reactions. Arrows indicate the migration of E2F4, and p130-E2F4-Dp1 complexes from doubly E2F4-Dp1 baculovirus-infected insect cells (lane 7) and triply p130-E2F4-Dp1 infected cells (lane 8). (B) Parental or T-antigen-transfected REF52 cell lysates were incubated alone (lanes 1 and 2) or in the presence of antibody against p107 (lanes 3 and 4), p130 (lanes 5 and 6), or pRb (lanes 7 and 8). Lane 8 appears overexposed as a result of incubation with multiple anti-pRb antibodies. Multiple antibodies are required to effect efficient binding to pRb (data not shown). (C) Parental or T-antigen-transfected REF52 cell lysates were incubated alone (lanes 1 and 2) or in the presence of antibodies against E2F1 (lanes 3 and 4), E2F2 (lanes 5 and 6), E2F3 (lanes 7 and 8), E2F4 (lanes 9 and 10), or E2F5 (lanes 11 and 12).
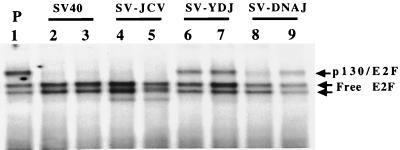
FIG. 6.
The Ydj1p and DnaJ chimeric T antigens cannot fully abolish a p130-E2F DNA binding complex. Lysates from parental REF52 cells (P; lane 1) and two clones from REF52 cells transfected with SV40 large T antigen (SV40, lanes 2 and 3), SV-JCV (lanes 4 and 5), SV-YDJ (lanes 6 and 7), and SV-DNAJ (lanes 8 and 9) were subjected to gel shift analysis using an E2F consensus site-containing probe.
To identify the Rb family member(s) present, antibodies either to p107, p130, or pRb were included in the gel shift reaction. Anti-p130 antibody shifted a majority of the parental REF52 complex that migrates at the Rb family-E2F position (Fig. 5B, lane 5). Incubation with anti-p107 or anti-pRb had no effect on the E2F-DNA complexes (Fig. 5B, lanes 3 and 4 and lanes 7 and 8). We conclude that the Rb family complex present in postconfluent parental REF52 cells but absent in postconfluent T-antigen-expressing cells includes p130-E2F.
Similar supershift experiments were performed with antibodies against the various E2F family members. Anti-E2F4 antibody supershifted a majority of the free E2F complexes present in the T-antigen-expressing cells (Fig. 5C, lane 10). In contrast, antibodies directed against E2Fs 1 to 3 and E2F5 had little effect in this assay. Thus, the free E2F induced by T antigen and the E2F bound to p130 is primarily E2F4.
Finally, we tested whether the chimeric T antigens disrupted p130-E2F4 DNA binding complexes. Two cell lines derived from independent clones were grown to 2 days postconfluency and then lysed. E2F gel shift reactions conducted with these lysates demonstrated that the SV-JCV chimera completely abolished the p130-E2F4 DNA binding complex (Fig. 6, lanes 4 and 5), whereas the SV-YDJ and SV-DNAJ chimeras did not (Fig. 6, lanes 6 to 9). Therefore, only SV-JCV, like wild-type T antigen disrupts a p130-E2F-DNA binding complex in postconfluent cells.
Wild-type and chimeric T antigens bind mammalian hsc70, but only wild-type T antigen significantly enhances hsc70 ATPase activity.
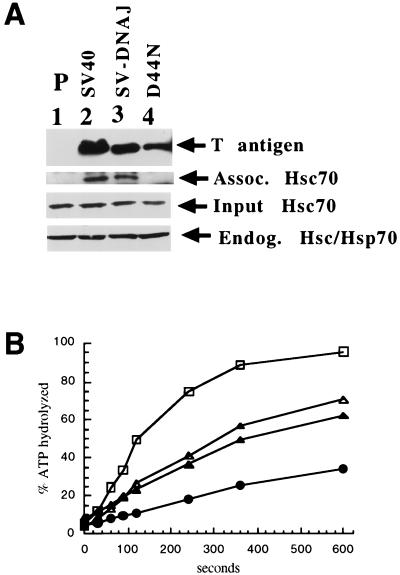
The J domain of SV40 T antigen has been shown to bind hsc70, and mutations in T antigen that disrupt this association abrogate SV40 DNA replication (6). Thus, it was possible that the SV-YDJ and SV-DNAJ chimeras were simply unable to complex with hsc70. To test this hypothesis, we developed an in vitro assay to measure hsc70 binding to T antigen prepared from transformed cell lysates. As a positive control, we prepared lysate from cells transformed with wild-type SV40 T antigen and, as a negative control, parental (untransformed) cell lysate was used. In addition, because it had been shown that mutations in the conserved HPD sequence in the J domain of T antigen abolished T-antigen function and hsc70 binding (reference 6; see also the introduction), lysates from cells transformed with SV40 T antigen containing the D44N mutation (5110, [29]) were prepared. Finally, to examine whether a defective chimera bound hsc70, we made lysate from the SV-DNAJ-transformed cells. These lysates were incubated with purified bovine brain hsc70. T antigen was then immunoprecipitated using a C-terminal-specific antibody, and the pelleted complexes were examined via immunoblot analysis. Approximately equal levels of all T-antigen constructs were immunoprecipitated (Fig. 7A, lanes 2 to 4). Control reactions demonstrated that equal amounts of endogenous and exogenous hsc70 were also present in each assay. As expected, wild-type T antigen, but not the T-antigen J-domain point mutant (D44N), bound hsc70 (Fig. 7A, lanes 2 versus lane 4); lysate from parental cells also failed to display hsc70 in the immunoprecipitate. However, T antigen from cells transformed with the SV-DNAJ T antigen were hsc70-binding proficient (lane 3).
FIG. 7.
T-antigen binding to hsc70 and stimulation of hsc70 ATPase activity. (A) Lysates containing various T antigens were incubated with hsc70 and immunoprecipitated for T antigen. T antigen and hsc70 associated with the pellets from the parental (lane 1), wild-type (lane 2), SV-DNAJ hybrid (lane 3), and the SV40 J-domain mutant D44N (5110, lane 4) cell lysates are shown. Input and endogenous hsc70 levels were also measured. (B) ATPase assays were performed as described in Materials and Methods. Stimulation of Ssa1p ATPase activity by wild-type T antigen (□), SV-DnaJ (▵), D44N (▴), and Ssa1p alone (●) are shown.
The result presented above indicating that a nonfunctional T-antigen hybrid (SV-DNAJ) bound to hsc70 was surprising, given that other T antigens containing a mutated J domain fail to interact with hsc70 (D44N, Fig. 7A) (6). However, it is possible that the interaction between the cochaperones may be unproductive, i.e., the J domain of SV-DNAJ may be unable to enhance the ATPase activity of hsc70, an activity that is required to promote viral replication and cellular transformation (36). To test this hypothesis, we purified wild-type T antigen, the D44N T antigen mutant, and SV-DNAJ from baculovirus-infected cells as previously described (7). To discount the endogenous ATPase activity of T antigen, we preformed ATP-hsc70 complexes (see Materials and Methods) and then added T antigen. Such single-turnover ATPase assays measure the ability of a J-domain-containing protein to stimulate specifically the hydrolysis of hsc70-bound ATP (12a). When we performed this assay multiple times, we observed that wild-type T antigen stimulated endogenous hsc70 ATP hydrolysis by 4.6- to 5.5-fold, while D44N and SV-DNAJ stimulated hydrolysis by only 2.3- and 2.2- to 2.9-fold, respectively (Fig. 7B displays a representative experiment). As a control, we found that purified T antigen completely lacking the J domain (12b) stimulated hsc70 ATP turnover by 0.4-fold (data not shown). These results indicate that the defective T-antigen J-domain mutant and SV-DNAJ hybrid proteins are compromised for their ability to interact productively with hsc70.
DISCUSSION
Our results show that the S. cerevisiae Ydj1p and E. coli DnaJ J domains fail to substitute functionally for the SV40 T-antigen J domain for one or more activities necessary for a complete virus life cycle (Fig. 2), including the ability to support viral DNA replication (Fig. 3). In addition the J domains from Ydj1p and DnaJ cannot substitute functionally for the SV40 T-antigen J domain in activities required for dense foci formation in REF52 cells (Fig. 4). In contrast, the JC virus J domain can replace the SV40 T antigen J domain for all of the activities necessary for a complete SV40 life cycle in cell culture, for viral DNA replication in an established in vivo system, and for the ability to transform REF52 cells in a dense focus assay. In each of these assays, however, the JC virus J domain is less efficient than the wild-type SV40 T-antigen J domain. This result is not unexpected, since the decreased efficiency of JC virus compared with SV40 has been seen in the results from other SV40-JC virus chimeras studied (9). Nevertheless, these results provide further evidence that not all J domains are interchangeable. Most striking, reciprocal domain-swapping experiments yielded opposing results: the yeast Ydj1p protein containing the T-antigen J domain (Fewell and Brodsky, unpublished) and the bacterial DnaJ protein harboring the T-antigen J domain (21) are active in yeast and bacteria, respectively, but the DnaJ and Ydj1p J domains fail to function in the context of SV40.
One role of the J domain is to interact with a cognate hsc70 molecular chaperone. The J domain forms a four-bundle α-helix, in which helices I and IV occupy the base of the structure and helices II and III form a finger-like projection, with the conserved amino acid sequence HPD located at the tip of the projection. Nuclear magnetic resonance studies have indicated that hsc70 interacts primarily with amino acid residues in helix II and the HPD sequence (17). Consistent with these data, mutations in the HPD motif in bacterial (39) and yeast J domains (15, 38) are known to abolish the activities of DnaJ homologue-hsc70 complexes. Because the T-antigen J domain can substitute functionally for the Ydj1p (Fewell and Brodsky, unpublished) and DnaJ (21) J domains, we suggest that the T-antigen J domain is proficient for binding to the bacterial and yeast hsc70 partner proteins. Thus, one explanation for the inability of the DnaJ and Ydj1p J domains to replace the T-antigen J domain is that mammalian hsc70 cannot bind to the other J domains. We found, however, that the bacterium-derived chimera could bind to hsc70 (Fig. 7). Because SV-DNAJ, which is defective for viral DNA replication, focus formation, and disrupting p130-E2F complexes, associates with hsc70 as well as wild-type T antigen, the simplest conclusion from these data is that hsc70 association alone is not sufficient to support J-domain function in DNA replication and cellular transformation. In accordance with this hypothesis, we found that both the D44N and SV-DnaJ proteins displayed a reduced ability to enhance hsc70 ATPase activity.
Although the ability of D44N and SV-DNAJ to enhance the ATPase activity of hsc70 was compromised with respect to wild-type T antigen, we note that the level of stimulation (∼2- to 3-fold) was higher than that obtained when a T-antigen construct lacking the J domain was incubated with hsc70 (0.4-fold). Thus, these defective proteins can interact productively to some extent with hsc70. For D44N, we suggest that the productive interaction is transient, since stable complexes with hsc70 were not observed (Fig. 7A). In contrast, SV-DNAJ formed a stable complex with hsc70 but could not fully stimulate the ATPase activity of hsc70.
E2F dissociated from Rb family members (pRb, p107, or p130) induces transactivation of the genes required for DNA replication and cellular division, whereas binding of E2F by Rb family members inhibits the transactivation activity of E2F (40). The J domain of T antigen is required in cis with the Rb binding motif to induce cellular division (36), and is also required to free p130 from E2F4 and to decrease the half life of p130 (37a, 42). Here we show that a p130-E2F DNA binding complex is present in postconfluent REF52 cells and that the action of T antigen or the SV-JCV chimera disrupts this complex, thereby liberating E2F4 (Fig. 5). In contrast, neither SV-YDJ nor SV-DNAJ disrupts the p130-E2F DNA binding complex. This result provides a mechanism to explain the inability of the T-antigen chimeras that contain other J domains to induce focus formation and to successfully complete viral DNA replication.
In conclusion, our work suggests that there is a specificity element within some J domains that is not present in yeast or E. coli J domains but that is required for viral DNA replication and transformation. This element is not restricted to viral J domains, since two mammalian J domains are functional in the SV40 T antigen (6). Furthermore, this specificity element is not simply an inability of a defective J domain to bind to hsc70. One possibility is that there is another T-antigen function in amino acids 1 to 75 other than the J-domain chaperone activity. For example, the T-antigen J domain may contain binding sites for cellular targets other than hsc70. In support of this idea, it has been shown that the binding site for the Tst-1/Oct-6/SCIP transcription factor maps to the T-antigen J domain (34). Alternatively, it is possible that although the T-antigen chimeras containing nonfunctional J domains can bind to hsc70, they may fail to regulate its activity, as suggested by the experiments presented in Fig. 7B. It is also possible that the association between a cochaperone or modulator of chaperone action, such as BAG-1 (3, 20), and the T-antigen J domain in the chimera is absent. In these last scenarios, the net effect is that T antigen fails to induce hsc70 to act on some cellular protein, such as the transcriptional repressor p130; thus, the cells would not be transformed. Future experiments will aim to identify the specificity element(s) in the T-antigen J domain and decipher its role in inducing DNA replication and transformation.
ACKNOWLEDGMENTS
This work was supported by grant CA40586 to J.M.P. from the National Institutes of Health and by a grant from the American Cancer Society (RPG-99-267-01-MBC) to J.L.B. S.W.F. was supported by a National Research Service Award from the National Institutes of Health.
We thank Paul Cantalupo for the expert technical assistance and protein purification and Thomas Harper for help with the figures.
REFERENCES
- 1.Barik S. Site-directed mutagenesis by double polymerase chain reaction: megaprimer method. Methods Mol Biol. 1993;15:277–286. doi: 10.1385/0-89603-244-2:277. [DOI] [PubMed] [Google Scholar]
- 2.Becker J, Walter W, Yan W, Craig E A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bimston, Song J, Winchester D, Takayama S, Reed J C, Morimoto R I. BAG-1, a negative regulator of hsc70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky J L, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky J L, Pipas J M. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J Virol. 1998;72:5329–5334. doi: 10.1128/jvi.72.7.5329-5334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 7.Cantalupo P, Sáenz-Robles M T, Pipas J M. Expression of SV40 large T antigen in Baculovirus systems and purification by immunoaffinity chromatography. Methods Enzymol. 1999;306:297–307. doi: 10.1016/s0076-6879(99)06019-x. [DOI] [PubMed] [Google Scholar]
- 8.Caplan A J, Tsai J, Casey P J, Douglas M G. Farnesylation of Ydj1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 9.Chuke W F, Walker D L, Peitzman L B, Frisque R J. Construction and characterization of hybrid polyomavirus genomes. J Virol. 1986;60:960–971. doi: 10.1128/jvi.60.3.960-971.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyr D M. Cooperation of the molecular chaperone Ydj1p with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- 11.Cyr D M, Douglas M G. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- 12.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 12a.Davis J E, Voisine C, Craig E. Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase- and peptide-binding domains. Proc Natl Acad Sci USA. 1999;16:9269–9276. doi: 10.1073/pnas.96.16.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b.Dornreiter I, Hoss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson N, Buchkovich K, Whyte P, Harlow E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 14.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein p120. Cell. 1989;2:405–415. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 15.Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene M K, Maskos K, Landry S J. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large T antigen: requirement of both the pRB-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 20.Höhfeld J. Regulation of the heat shock cognate hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol Chem. 1998;379:269–274. [PubMed] [Google Scholar]
- 21.Kelley W L, Georgopoulos C. The T/t common exon of SV40, JCV, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King C, Eisenberg E, Greene L. Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J Biol Chem. 1995;270:22535–22540. doi: 10.1074/jbc.270.38.22535. [DOI] [PubMed] [Google Scholar]
- 23.Lai C-J, Nathans D. Deletion mutants of simian virus 40 generated by enzymatic excision of DNA segments from the viral genome. J Mol Biol. 1974;89:179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- 24.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 25.Levy E J, Bukau B, Chirico W J. Conserved ATPase and luciferase refolding activities between bacteria and yeast hsp70 chaperones and modulators. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- 26.Linzer D I, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 27.Lu Z, Cyr D M. Protein folding activity of hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 28.McClellan A J, Endres J, Vogel J P, Palazzi D, Rose M D, Brodsky J L. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peden K W C, Pipas J M. Simian virus 40 mutants with amino acid substitutions near the amino-terminus of large T antigen. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 30.Peden K W C, Pipas J M, Pearson-White S, Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980;209:1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- 31.Pipas J M. Common and unique features of T antigens encoded by the polyoma group. J Virol. 1992;66:979–985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlenstedt G, Harris S, Risse B, Lill R, Silver P A. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher R J, Hansen W J, Freeman B C, Alnemri E, Litwack G, Toft D O. Cooperative action of hsp70, hsp90 and DnaJ proteins in protein renaturation. Biochemistry. 1996;35:14889–14898. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]
- 34.Sock E, Enderich J, Wegner M. The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol Cell Biol. 1999;19:2455–2464. doi: 10.1128/mcb.19.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spence S L, Pipas J M. SV40 large T antigen functions at two distinct steps in virion assembly. Virology. 1994;204:200–209. doi: 10.1006/viro.1994.1524. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of SV40 large and small T antigens function as a J-domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 38.Tsai J, Douglas M G. A conserved HPD sequence of the J domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 39.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for λ replication. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 40.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Yan W, Craig E A. The glycine-phenylalanine-rich region determines the specificity of the yeast hsp40, Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]