Graphical abstract
The first step is Dataset preparation where sequences are extracted from the raw data. To make the data understandable for the network, the second step is sequence encoding where one-hot encoding is used. Further, as the third step CNN model training is carried out which is evaluated on the fourth step. At the last step, a webserver is generated for the researchers.
Keywords: Bioinformatics, 4mC modification, Computational Biology, Convolutional neural network, Deep learning
Abstract
DNA N4-methylcytosine (4mC) being a significant genetic modification holds a dominant role in controlling different biological functions, i.e., DNA replication, DNA repair, gene regulations and gene expression levels. The identification of 4mC sites is important to get insight information regarding different organics mechanisms. However, getting modification prediction from experimental methods is a challenging task due to high expenses and time-consuming techniques. Therefore, computational tools can be a great option for modification identification. Various computational tools are proposed in literature but their generalization and prediction performance require improvement. For this motive, we have proposed a neural network based tool named DCNN-4mC for identifying 4mC sites. The proposed model involves a set of neural network layers with a skip connection which allows to share the shallow features with dense layers. Skip connection have allowed to gather crucial information regarding 4mC sites. In literature, different models are employed on different species hence in many cases different datasets are available for a single species. In this research, we have combined all available datasets to create a single benchmark dataset for every species. To the best of our knowledge, no model in literature is employed on more than six different species. To ensure the generalizability of DCNN-4mC we have used 12 different species for performance evaluation. The DCNN-4mC tool has attained 2% to 14% higher accuracy than state-of-the-art tools on all available datasets of different species. Furthermore, independent test datasets are also engaged and DCNN-4mC have overall yielded high performance in them as well.
1. Introduction
Epigenetic modification is the heritable alteration that occurs in the gene expression keeping the original DNA sequence unchanged [1]. DNA methylation has been demonstrated in several studies to alter chromatin structure, DNA orientation, DNA integrity, and genetic code interactions [2], [3]. Furthermore, changes in DNA methylation pattern are considered to be a biological complication mechanism [4], leading to tumour formation [5] and other disorders [6].
N6-methyladenine (6 mA), 5-methylcytosine (5mC), and N4-methylcytosine (4mC) are all common forms of DNA methylation in genomics [7]. In both prokaryotes and eukaryotic genomes, these kinds of DNA methylation are predominantly found [8], [9]. 5mC is the most frequent DNA alteration in eukaryotes, and it is required for cell growth, transposon elimination, and gene imprinting [10], [11], [12]. Given their tiny size, 6 mA and 4mC can only be identified in eukaryotes using high sensitivity methods. 6 mA and 4mC are the most common in prokaryotes, and are primarily utilized to differentiate host DNA from foreign pathogenic DNA[13] and also 4mC regulates the replication process and fixes abnormalities in DNA replication [14]. Furthermore, as a segment of the restriction-modification system, 4mC inhibits restriction enzymes from damaging host DNA. 4mC is more prominent in mesophilic bacteria and is extremely hard to identify in eukaryotic genomes using conventional methods [15], [14].
Based on next-generation sequencing (NGS), bisulphite sequencing is a widely used method for detecting DNA methylation sites across the entire genome [16]. This experimental approach, however, is costly and prolonged method [17], and it can only detect 5mC [18]. Single-molecule real-time (SMRT) sequencing is a common method for detecting 4mC, 5mC and 6 mA sites from unknown DNA sequences [17]. However, the library preparation required in SMRT makes it a more expensive and time-consuming technique [19]. Furthermore, distinguishing 4mC from 5mC continues to be a significant problem for traditional experimental approaches. To overcome these issues, 4mC-Tet-assisted bisulfite-sequencing (4mC-TAB-seq), a 4mC-specific NGS-based technique for properly distinguishing 4mC from 5mC, has been suggested [19]. Another group recently used synthetic transcription activator-like effectors to differentiate between 4mC and 5mC sites [1]. Undoubtedly these experimental methods aid in the identification of 4mC sites, but they are too time-consuming and pricey techniques to be used for wide-range genome scanning. As a result, computational approaches for predicting DNA methylation sites are a valuable and compatible tool for high-throughput identification of DNA methylation sites, and they can tremendously aid experimental research.
Computational techniques, particularly machine-learning (ML) based approaches, have recently been successfully applied to a variety medical related issues [20], including 4mC site identification [21]. Chen et al. created first computational model iDNA4mC, for 4mC sites identification [21]. The iDNA4mC tool employs nucleotide chemical properties (NCP) and frequencies as features to create support vector machine (SVM) based prediction tool. In total six species which are Caenorhabditis elegans (C.elegans), Drosophila melanogaster (D.melanogaster), Arabidopsis thaliana (A.thaliana), Escherichia coli (E. coli), Geoalkalibacter subterraneus (G.subterraneus) and Geobacter pickeringii (G.pickeringii) were used to train and validate the iDNA4mC tool, and the results suggests that the tool is effective for differentiating 4mC sites from non-4mC sites. After iDNA4mC several other machine learning based tools were proposed like 4mCPred [22], 4mcPred-SVM [14] and 4mcPred-IFL [23] which improved the performance on same six species. Later a deep learning based approach, DeepTorrent was introduced which increased the performance and contributed more dataset for the similar species [24]. Some other deep learning based tools like 4mCCNN [25] were also proposed which provide improvement in performance for identification of 4mC site in these species. Further Rao et al. contributed an additional datatset for C.elegans, D.melanogaster and A.thaliana [26]. Subsequently Zeng et al. collected an additional dataset for C.elegans [27].
Recently an ensemble learning framework, 4mCpred-EL was proposed for Mus musculus [28]. Later a tool i4mC-ROSE was presented for 4mC identification in rosaceae genome [29]. The i4mC-ROSE was suggested for two species which are Fragaria vesca (F.vesca) and Rosa chinensis (R.chinensis). Another tool iDNA-MS was put forward for four species out of which one is F.vesca and other are Casuarina equisetifolia (C.equisetifolia), Saccharomyces cerevisiae (S.cerevisiae) and Tolypocladium sp. Sup5 [30]. Even though the aforementioned methods regularly perform efficiently, but they may lack generalizability, necessitating the creation of a new predictor for successful 4mC site detection with dependable transferability.
Machine learning based approaches have had a lot of success in predicting 4mC sites, and they have helped to speed up 4mC identification studies. The success of Machine learning based techniques (i.e., their predictive power) in differentiating 4mC sites from non-4mC sites, on the other hand, is highly dependent on the quality of features. Due to a paucity of research on 4mC, extracting useful characteristics with a significant discriminative capacity to forecast 4mC sites is difficult [1]. While on other side deep learning has emerged as a solution to such a problem with a capability of automatically learning deep features using several neural network layers [31]. Very few deep learning based techniques for 4mC sites identification have been proposed in literature while many hidden wonders of deep learning are still not explored for detecting 4mC sites. Further, the previously proposed deep learning based techniques still lack generalizability as none of the technique is proposed for more than six species.
In this work, we are proposing Densely Connected Neural Network Based N4-methylcytosine Site Prediction (DCNN-4mC), a general framework for twelve different species proposed in different studies. Further, in this study, we have combined all available datasets in literature and bring them under one umbrella, so that the research on computational models for 4mC can be carried out on common benchmark datasets, which will help in carrying out better comparative analysis. The proposed DCNN-4mC tool is a neural network based tool which employs multiple layers with a skip connection. The skip connection allows sharing the shallow features with the deeper layers, which results in great performance improvement. When compared to state-of-the-art techniques, extensive benchmarking studies on twelve distinct species indicate that DCNN-4mC obtains the greatest performance for 4mC site identification in all species. To facilitate the experts of the field, DCNN-4mC can be accessed freely at: http://nsclbio.jbnu.ac.kr/tools/DCNN-4mC/.
2. Materials and methods
2.1. Overall framework of DCNN-4mC
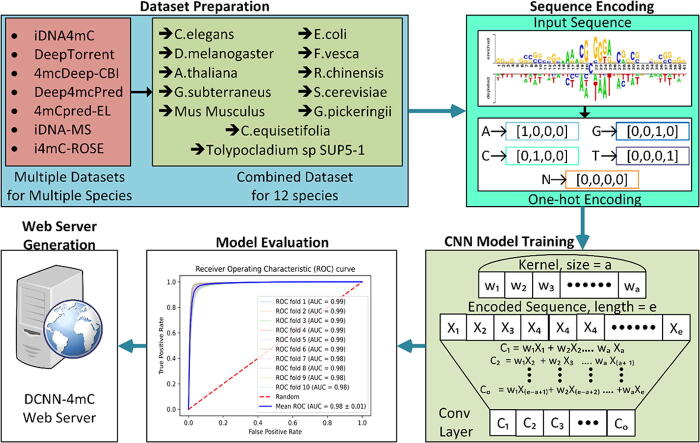
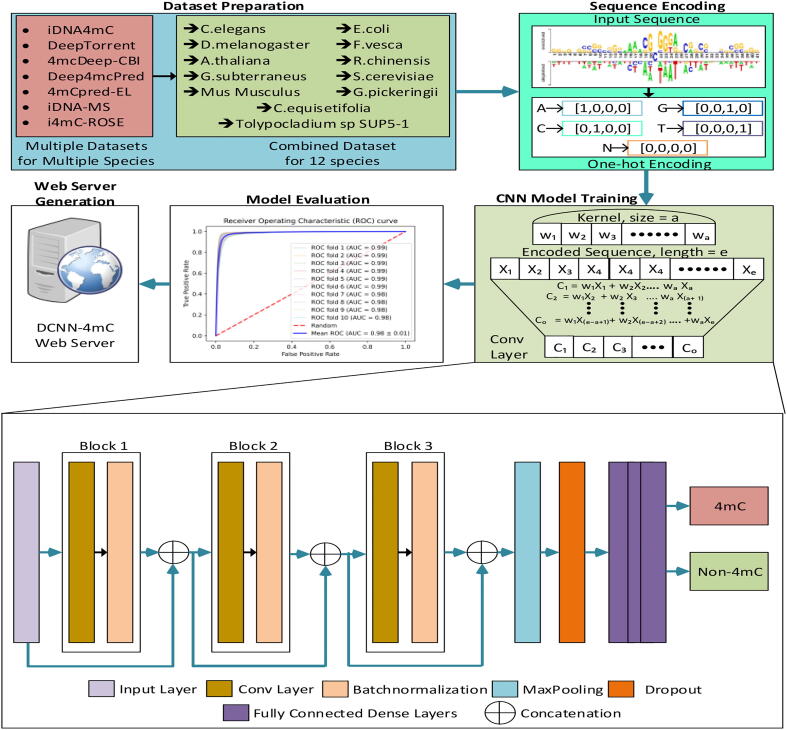
The overall framework of DCNN-4mC is depicted in Fig. 1. The development of the DCNN-4mC predictor consists of the following five major steps: Dataset Preparation; Sequence Encoding; CNN Model Training; Model Evaluation; (iv) WebServer Generation. In the first step, we collected all available datasets for different species from the literature after having an extensive literature review. Further, a single dataset for every species is prepared with the help of available datasets. At the second step, we carried out One-hot encoding for the input sequences. The third step involves the CNN model training from the encoded sequences. In the fourth step we evaluated the trained model using 10-fold cross-validation and by using an independent test dataset. The model evaluation is carried out based on the different figure of merits. The fifth step includes the construction of a webserver for the medical and bioinformatics experts.
Fig. 1.
The overall framework of DCNN-4mC. It entails the following steps: (i) Dataset Preparation; (ii) Sequence Encoding; (iii) CNN Model Training; (iv) Model Evaluation; (v) WebServer Generation.
2.2. Dataset preparation
In literature, two databases are used for constructing datasets for different species. These databases are MDR database [32] and MethSMRT database [10]. To the best of our knowledge, there are 12 different species for which the datasets are constructed in literature. Chen et al. constructed the dataset from MethSMRT for six species which are: C.elegans; D.melanogaster; A.thaliana; E. coli; G.subterraneus; G.pickeringii [21]. Liu et al. continued the work and collected more datasets for the aforementioned species from the MethSMRT database [24]. Rao et al. went on to further collect the dataset from MethSMRT for C.elegans, D.melanogaster and A.thaliana [27]. Zeng et al. further utilized the MethSMRT database to gather a dataset for C.elegans [27]. Hao et al. used SMRT and MDR databases to construct the dataset for four species which are: C.equisetifolia; S.cerevisiae; Tolypocladium sp. Sup5 [30]. In [28] authors collected a dataset for Mus musculus from the MethSMRT database. Further, Hasan et al. collected the dataset for F.vesca and R.chinensis from the MDR database [29].
All of the constructed datasets followed a similar procedure. The positive and negative sample sequences in the collection were all 41 bp long with cytosine (”C”) nucleotide at the centre. Positive samples that have been experimentally validated are confirmed using a relevant modification score (ModQV). In positive samples, the related cytosine is considered to be modified if the ModQV score >= 20. The CD-HIT software was used to remove the redundant sequences, which solves the bias problem in the curated sequences.
As for many species, there is more than one dataset, therefore, we combined them into a single dataset for every species, so that a single benchmark can be used by us and by future researchers. For all species, their training datasets are combined into one single benchmark dataset and the same is done with the testing dataset. As the origin of the datasets is the same which is either MethSMRT or MDR database, therefore redundant sequences are removed from benchmark training and testing datasets. It is also being ensured that there should be no similar sequence present in the training and testing dataset. Table 1 shows the statistical details regarding the dataset of every species.
Table 1.
Dataset Statistics.
| Species | Available Train Dataset | Train Dataset Size | Test Dataset Size | Updated TrainDataset | Updated Test Dataset | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Caenorhabditis elegans (C. elegans) | iDNA4mC (chen et al. [21]) | 4mC = 1554 | 4mC = 0 | 4mC = 7939 Non-4mC = 82033 | 4mC = 2352 Non-4mc = 2660 | |||||||||||||
| Non-4mC = 1554 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 55729 | 4mC = 2667 | ||||||||||||||||
| Non-4mC = 55729 | Non-4mC = 2667 | |||||||||||||||||
| Zeng et al. [27] | 4mC = 11173 | 4mC = 0 | ||||||||||||||||
| Non-4mC = 6635 | Non-4mC = 0 | |||||||||||||||||
| Rao et al. [26] | 4mC = 20000 | 4mC = 0 | ||||||||||||||||
| Non-4mC = 20000 | Non-4mC = 0 | |||||||||||||||||
| Drosophila melanogaster (D. melanogaster) | iDNA4mC (chen et al. [21]) | 4mC = 1769 | 4mC = 0 | 4mC = 72127 Non-4mC = 75460 | 4mC = 3332 Non-4mC = 3521 | |||||||||||||
| Non-4mC = 1769 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 53970 | 4mC = 3684 | ||||||||||||||||
| Non-4mC = 53970 | Non-4mC = 3684 | |||||||||||||||||
| Rao et al. [26] | 4mC = 20000 | 4mC = 0 | ||||||||||||||||
| Non-4mC = 20000 | Non-4mC = 0 | |||||||||||||||||
| Arabidopsis thaliana(A. thaliana) | iDNA4mC (chen et al. [21]) | 4mC = 1978 | 4mC = 0 | 4mC = 81143 Non-4mC = 85456 | 4mC = 10388 Non-4mC = 11172 | |||||||||||||
| Non-4mC = 1978 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 63720 | 4mC = 11 307 | ||||||||||||||||
| Non-4mC = 63720 | Non-4mC = 11 307 | |||||||||||||||||
| Rao et al. [26] | 4mC = 20000 | 4mC = 0 | ||||||||||||||||
| Non-4mC = 20000 | Non-4mC = 0 | |||||||||||||||||
| Escherichia coli (E. coli) | iDNA4mC (chen et al. [21]) | 4mC = 388 | 4mC = 0 | 4mC = 1959 Non-4mC = 2156 | 4mC = 126 Non-4mC = 126 | |||||||||||||
| Non-4mC = 388 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 1941 | 4mC = 126 | ||||||||||||||||
| Non-4mC = 1941 | Non-4mC = 126 | |||||||||||||||||
| Geoalkalibacter subterraneus (G. subterraneus) | iDNA4mC(chen et al. [21]) | 4mC = 905 | 4mC = 0 | 4mC = 10583 Non-4mC = 10780 | 4mC = 5263 Non-4mC = 5263 | |||||||||||||
| Non-4mC = 905 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 9934 | 4mC = 5263 | ||||||||||||||||
| Non-4mC = 9934 | Non-4mC = 5263 | |||||||||||||||||
| Geobacter pickeringii (G. pickeringii) | iDNA4mC (chen et al. [21]) | 4mC = 569 | 4mC = 0 | 4mC = 4703 Non-4mC = 4900 | 4mC = 1210 Non-4mC = 1210 | |||||||||||||
| Non-4mC = 569 | Non-4mC = 0 | |||||||||||||||||
| DeepTorrent (Liu et al. [24]) | 4mC = 4514 | 4mC = 1210 | ||||||||||||||||
| Non-4mC = 4514 | Non-4mC = 1210 | |||||||||||||||||
| Mus musculus | 4mCpred-EL [28] | 4mC = 800 | 4mC = 180 | 4mC = 800 Non-4mC = 800 | 4mC = 180 Non-4mC = 180 | |||||||||||||
| Non-4mC = 800 | Non-4mC = 180 | |||||||||||||||||
| Casuarina equisetifolia (C. equisetifolia) | iDNA-MS [30] | 4mC = 183 | 4mC = 183 | 4mC = 183 Non-4mC = 183 | 4mC = 183 Non-4mC = 183 | |||||||||||||
| Non-4mC = 183 | Non-4mC = 183 | |||||||||||||||||
| Saccharomyces cerevisiae (S. cerevisiae) | iDNA-MS [30] | 4mC = 990 | 4mC = 989 | 4mC = 990 Non-4mC = 990 | 4mC = 989 Non-4mC = 989 | |||||||||||||
| Non-4mC = 990 | Non-4mC = 989 | |||||||||||||||||
| Tolypocladium sp SUP5-1 (Tolypocladium) | iDNA-MS [30] | 4mC = 7664 | 4mC = 7663 | 4mC = 7664 Non-4mC = 7664 | 4mC = 7663 Non-4mC = 7663 | |||||||||||||
| Non-4mC = 7664 | Non-4mC = 7663 | |||||||||||||||||
| Fragaria vesca (F. vesca) | i4mC-ROSE [29] | 4mC = 4854 | 4mC = 1617 | 4mC = 12298 Non-4mC = 12152 | 4mC = 8819 Non-4mC = 9015 | |||||||||||||
| Non-4mC = 4854 | Non-4mC = 1617 | |||||||||||||||||
| iDNA-MS [30] | 4mC = 7899 | 4mC = 7898 | ||||||||||||||||
| Non-4mC = 7899 | Non-4mC = 7898 | |||||||||||||||||
| Rosa chinensis (R. chinensis) | i4mC-ROSE [29] | 4mC = 2337 | 4mC = 779 | 4mC = 2337 Non-4mC = 2337 | 4mC = 779 Non-4mC = 779 | |||||||||||||
| Non-4mC = 2337 | Non-4mC = 779 |
2.3. Sequence encoding
The input sequence to the proposed computational tool looks as follows,
where sequence ‘S’ is of length 41 and ’N’ represents the nucleotide and can be represented as, . The four nucleotides in a DNA sequence are adenine (A), cytosine (C), guanine (G) and thymine (T). For embedding these sequences to the neural network model. they first need to be represented as appropriate numerical data. As the neural networks extract the features from the numerical data only. For this reason, we have utilized a One-hot encoding scheme.
The one-hot encoding scheme is the simplest and efficient encoding algorithm used frequently in the field of bioinformatics [33], [34], [35], [36]. In this encoding scheme each nucleotide is mapped to integer values and further this integer value is assigned with a unique binary vector that includes all ‘0’ values apart from the index of the integer, which is kept as ‘1’. The one-hot encoding scheme is considered to be more expressive than the simple encoding scheme. The one-hot vector for four nucleotides present in a DNA sequence is represented as follows,
After one-hot encoding, the resultant matrix for a length ‘l’ input DNA sequence would be .
2.4. CNN model
The complete network architecture is illustrated at the bottom of Fig. 1. The network consists of single-dimensional (1-D) convolutional, max pooling, dropout and fully connected layers. After preprocessing the data is first passed through 1-D convolutional layers to extract robust and meaningful features for further processing. Each 1-D convolution layer is followed by a batch normalization (BN) layer and an activation layer unless specified explicitly. We use the rectified linear unit (ReLU) as an activation function throughout the network except for the last layer.
| (1) |
We further enhance the representational power of our network by incorporating skip connections. The skip connections follow the concept of identity mapping [37], which helps in the more efficient training of the network. In contrast to the original skip connections, instead of adding the input to the output of the convolutional layer, we concatenate both features and then pass them to the next convolutional layer for further processing. Concatenation operation is performed to combine the shallow features with the deeper features. As it allows the network to give importance to each feature map adaptively depending upon the input sequence, without distorting the extracted features of previous layers. Hyper-parameter tuning is done for the selection of finer parameters for the whole network. The hyper-parameters for tuning purposes are presented in Table 2. Whereas Table 3 shows the selected parameters for the CNN model. After performing three consecutive skip 1-D convolutions we perform max pooling operation followed by dropout layer to avoid overfitting and to increase the generalization of the network on unseen sequences. Finally, the features extracted from convolutional layers are flattened and passed on to the fully connected layers for the classification of the sequence into 4mC and Non-4mC. Sigmoid is used as an activation function for the output layer of the network.
| (2) |
The L2 regularization which is also known as ridge regression is used to prevent the network from over-fitting on training sequences. The loss function for L2 regularization is as follows,
| (3) |
where l is the true value and p is the predicted value. represents the loss of the model in which L2 regularization term () is added to prevent over-fitting. While is the regularization parameter which is tuned manually and must be greater than 0. Stochastic Gradient Descent (SGD) is used as an optimizer for training the network with the momentum of 0.8 and the initial learning is set to be 0.003. Loss function plays an important role in optimizing the neural network model. A single loss function sometimes is not capable enough to optimize the network at its best. Therefore we used a customized loss function for back-propagating errors and updating the network’s weights. The customized loss function is the sum of the Dice Loss Coefficient (DLC) and Weight Cross-Entropy (WCE). The formulation for these loss functions is as follows,
| (4) |
| (5) |
where Q is the total number of labels which in our case is 2 and is the label. The represents the Predicted class of the sequence, is the allotted weight and is the ground truth class of the pixel. The total loss function can be represented as,
| (6) |
Table 2.
Hyper-parameter tuning values.
| Parameters | Experiment Values |
|---|---|
| Number of Blocks/ Convolution Layers | [1,2,3,4,5] |
| Filters in convolution Layer | [8, 12, 16, 32, 64, 128] |
| Filter size | [1, 3, 5, 7, 11, 15] |
| MaxPooling Pool-size | [2, 4] |
| Dropout Ratio | [0.1, 0.2, 0.3, 0.4] |
Table 3.
Selected parameters for DCNN-4mC model.
| Parameters | Selected Values |
|---|---|
| Number of Filters (Block 1) | 64 |
| Filter Size (Block 1) | 11 |
| Number of Filters (Block 2) | 32 |
| Filter Size (Block 2) | 7 |
| Number of Filters (Block 3) | 32 |
| Filter Size (Block 3) | 5 |
| MaxPooling Pool-size | 4 |
| Dropout Ratio | 0.3 |
2.5. CNN model utilization for different datasets
The dataset of different species has different sizes. Therefore to go with 3 block architecture for all species generates the problem of over-fitting due to the limited dataset. The species with a good amount of data like C.elegans, D.melanogaster, A.thaliana, G.subterraneus and F.vesca uses all three blocks. While in the case of E.coli, G.pickeringii, Mus musculus, Tolypocladium and R.chinensis the block 1 is removed from the architecture due to the limited data and the encoded sequence is directly given to block2. The remaining architecture remains the same in this case. The dataset for C.equisetifolia is too small and for this purpose only block 3 is used in its architecture. The encoded sequence of C.equisetifolia species is directly given to block 3. This subtraction of blocks is performed due to the limitation of the dataset size used for training.
2.6. Figure of merits
We utilized four frequently used measures to assess the new method’s and existing techniques’ performance, including Sensitivity (also known as true positive rate), Specificity (also known as true negative rate), Accuracy (ACC), Precision (also known as positive predictive value), F1 score and Matthews correlation coefficient (MCC). Following are the mathematical expressions for these figure of merits,
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
where acronyms are,
TP: True Positive.
TN: True Negative.
FP: False Positive.
FN: False Negative.
Accuracy and MCC are two measures that assess the overall prediction performance of the prediction model. The ROC curve was also utilized to intuitively assess the overall performance of the model. The Area Under the ROC curve (AUC) is used to quantitatively validate the model’s overall prediction performance.
3. Results and discussion
In this part, we go through the DCNN-4mC tool performance evaluation results in depth. We ran performance assessment experiments on both the existing datasets and updated datasets in particular.
3.1. Performance comparison with the existing methods
To have the comparison with the existing models it is important to have similar datasets to get the quantitative results. For this purpose, we computed results on different existing datasets to have comparative analysis with the existing dataset-specific state-of-the-art techniques. Table 4 shows the performance comparison of DCNN-4mC on existing databases with state-of-the-art techniques of each database. All the results are computed using a 10-fold cross-validation process. For C.elegans results are computed on three different datasets, where the proposed model has achieved the highest performance concerning all metrics for all datasets. The results for species A.thaliana, D.melanogaster and F.vesca are calculated on two individual datasets for every specie while for the remaining species the results are evaluated on a single dataset for every species. The DCNN-4mC tool has outperformed in all datasets regardless of the species.
Table 4.
Performance Comparison of DCNN-4mC on existing databases with state-of-the-art techniques of each database. The bold values in the table shows high performance achieved for the particular database.
| Species | Dataset | Model | Sensitivity | Specificity | Accuracy | MCC | AUC |
|---|---|---|---|---|---|---|---|
| Caenorhabditis elegans (C. elegans) | Liu et al. [24] | DeepTorrent | 0.930 | 0.910 | 0.920 | 0.840 | 0.976 |
| DCNN-4mC | 0.971 | 0.968 | 0.969 | 0.938 | 0.992 | ||
| Zeng et al. [27] | 4mcDeep-CBI | 0.949 | 0.894 | 0.930 | 0.850 | 0.924 | |
| DCNN-4mC | 0.970 | 0.942 | 0.959 | 0.913 | 0.986 | ||
| Rao et al. [26] | Deep4mCPred | 0.915 | 0.872 | 0.893 | 0.787 | – | |
| DCNN-4mC | 0.955 | 0.951 | 0.953 | 0.906 | 0.982 | ||
| Drosophila melanogaster (D. melanogaster) | Liu et al. [24] | DeepTorrent | 0.939 | 0.899 | 0.919 | 0.838 | 0.971 |
| DCNN-4mC | 0.968 | 0.960 | 0.964 | 0.927 | 0.988 | ||
| Rao et al. [26] | Deep4mCPred | 0.876 | 0.866 | 0.871 | 0.742 | – | |
| DCNN-4mC | 0.952 | 0.939 | 0.945 | 0.890 | 0.977 | ||
| Arabidopsis thaliana (A. thaliana) | Liu et al. [24] | DeepTorrent | 0.879 | 0.844 | 0.862 | 0.723 | 0.929 |
| DCNN-4mC | 0.937 | 0.930 | 0.933 | 0.866 | 0.967 | ||
| Rao et al. [26] | Deep4mCPred | 0.860 | 0.829 | 0.844 | 0.689 | – | |
| DCNN-4mC | 0.934 | 0.928 | 0.931 | 0.863 | 0.967 | ||
| Escherichia coli (E. coli) | Liu et al. [24] | DeepTorrent | 0.937 | 0.878 | 0.908 | 0.816 | 0.967 |
| DCNN-4mC | 0.960 | 0.941 | 0.951 | 0.902 | 0.983 | ||
| Geoalkalibacter subterraneus (G. subterraneus) | Liu et al. [24] | DeepTorrent | 0.857 | 0.701 | 0.779 | 0.565 | 0.866 |
| DCNN-4mC | 0.920 | 0.917 | 0.919 | 0.837 | 0.967 | ||
| Geobacter pickeringii (G. pickeringii) | Liu et al. [24] | DeepTorrent | 0.895 | 0.788 | 0.842 | 0.687 | 0.923 |
| DCNN-4mC | 0.924 | 0.916 | 0.920 | 0.841 | 0.967 | ||
| Mus musculus | Manavalan et al. [28] | 4mCpred-EL | 0.804 | 0.787 | 0.795 | 0.591 | 0.874 |
| DCNN-4mC | 0.893 | 0.912 | 0.903 | 0.807 | 0.958 | ||
| Saccharomyces cerevisiae (S. cerevisiae) | Lv et al. [30] | iDNA-MS | 0.701 | 0.707 | 0.704 | 0.408 | 0.771 |
| DCNN-4mC | 0.877 | 0.896 | 0.886 | 0.774 | 0.947 | ||
| Casuarina equisetifolia (C. equisetifolia) | Lv et al. [30] | iDNA-MS | 0.717 | 0.705 | 0.711 | 0.422 | 0.780 |
| DCNN-4mC | 0.913 | 0.931 | 0.922 | 0.848 | 0.971 | ||
| Tolypocladium sp SUP5-1 (Tolypocladium) | Lv et al. [30] | iDNA-MS | 0.716 | 0.708 | 0.712 | 0.423 | 0.780 |
| DCNN-4mC | 0.850 | 0.858 | 0.854 | 0.708 | 0.915 | ||
| Fragaria vesca (F. vesca) | Lv et al. [30] | iDNA-MS | 0.830 | 0.818 | 0.824 | 0.648 | 0.900 |
| DCNN-4mC | 0.916 | 0.902 | 0.909 | 0.846 | 0.963 | ||
| Hasan et al. [29] | i4mC-ROSE | 0.635 | 0.899 | 0.767 | 0.545 | 0.883 | |
| DCNN-4mC | 0.951 | 0.939 | 0.945 | 0.860 | 0.978 | ||
| Rosa chinensis (R. chinensis) | Hasan et al. [29] | i4mC-ROSE | 0.668 | 0.900 | 0.784 | 0.563 | 0.889 |
| DCNN-4mC | 0.900 | 0.905 | 0.902 | 0.806 | 0.953 |
Liu et al. evaluated the DeepTorrent model on 6 different species whereas Lv et al. assess the iDNA-MS tool on 4 different species for 4mC identification. The proposed DCNN-4mC performed better than the DeepTorrent tool and iDNA-MS tool. To efficiently train higher-order feature representations, the DeepTorrent uses a multi-layer CNN model with an inception module coupled with bidirectional long short-term memory and four distinct feature encoding techniques to encode the sequence. The iDNA-MS tool uses multiple combinations of three encoding schemes to train a random forest classifier for the prediction. Another deep learning-based model Deep4mCPred uses multiple CNN layers to achieve high performing results on three species. While on other hand the proposed DCNN-4mC model uses a single and simple encoding scheme to train the densely connected neural network which uses skip connections to keep the track of the shallow features. An analysis from this comparison can be driven that the reason for the DCNN-4mC tool to perform higher than the other model is the skip connection. We have even tried to add a few processing units at the skip connections however that didn’t achieve better results. Therefore, this conceptualizes that the raw shallow information on the deeper layers of CNN plays an important role in the modification prediction.
3.2. Performance evaluation on updated datasets
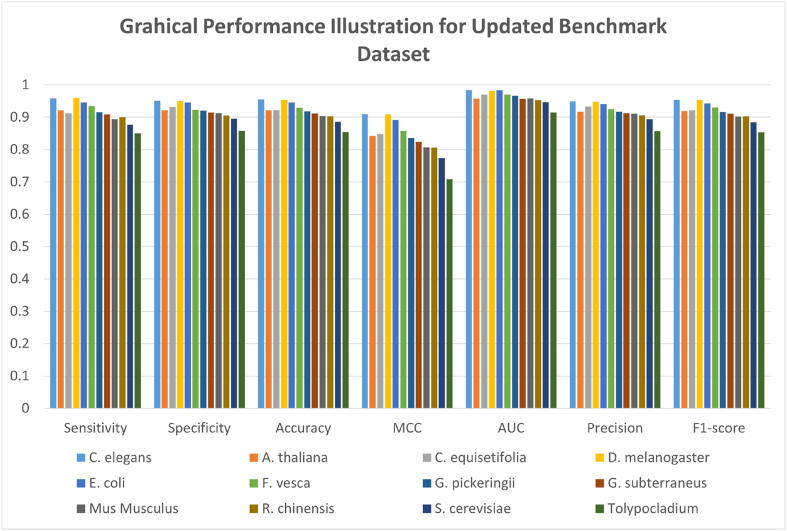
As this research presents the updated dataset for all the species taken into consideration, therefore, it is mandatory to evaluate the model for the updated training and independent dataset. This will help future researchers to use the updated benchmark dataset and have better comparative analysis with DCNN-4mC. Fig. 2 gives the graphical illustration of 10-fold cross-validation results achieved by the proposed architecture on 12 different species. Further Supplementary Table S1 shows the quantitative results in terms of sensitivity, specificity, ACC, MCC, AUC, precision and F1-score obtained by the proposed model. The results show that DCNN-4mC has attained good performance on the updated dataset for 10-fold cross-validation. The proposed tool attained accuracy of 0.954574, 0.921147, 0.922222, 0.954461, 0.945561, 0.928955, 0.917746, 0.911669, 0.903125, 0.902906, 0.886363 and 0.854032 for C.elegans, A.thaliana, C.equisetifolia, D.melanogaster, E.coli, F.vesca, G.pickeringii, G.subterraneus, Mus musculus, R.chinensis, S.cerevisiae and Tolypocladium, respectively. For all the species the obtained accuracy remained more than 85%. As suggested in literature the binary classification evaluation is better carried out by MCC rather than other [38], [39]. The MCC measurement suggests that the model is not biased towards one class. The high MCC values achieved by the proposed model suggests the high-quality prediction by it. Further ROC curve and AUC also represents the quality of the model. Therefore, Supplementary Figs. S1–S12 represents the ROC curves for all 10 folds for every species individually along with the computed AUC on every fold as well as the average. The AUC values achieved by the model are 0.984338, 0.957957, 0.970799, 0.981456, 0.983691, 0.970450, 0.966007, 0.956868, 0.958437, 0.953251, 0.946710 and 0.914519 for C.elegans, A.thaliana, C.equisetifolia, D.melanogaster, E.coli, F.vesca, G.pickeringii, G.subterraneus, Mus musculus, R.chinensis, S.cerevisiae and Tolypocladium, respectively.
Fig. 2.
The graphical illustration of 10-fold cross validation results on updated benchmark dataset of different species.
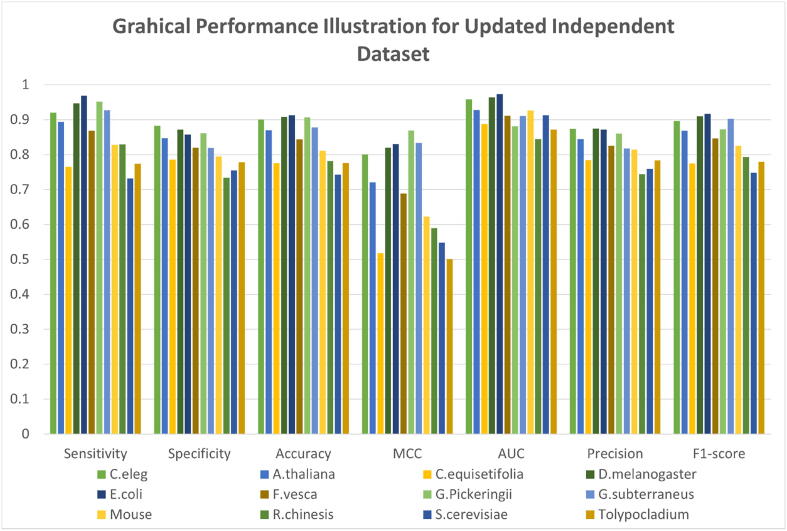
The proposed model is also assessed on the updated independent dataset. Fig. 3 shows the visual representation of the proposed model on an updated independent dataset while Supplementary Table S2 shows the numerical results of the same. The achieved F1-scores by the tool are 0.896252, 0.868546, 0.774721, 0.909667, 0.917293, 0.846171, 0.872471, 0.902786, 0.825847, 0.793190, 0.748015 and 0.779483 for C.elegans, A.thaliana, C.equisetifolia, D.melanogaster, E.coli, F.vesca, G.pickeringii, G.subterraneus, Mus musculus, R.chinensis, S.cerevisiae and Tolypocladium respectively. The DCNN-4mc tool exhibited good performance in this experiment as well.
Fig. 3.
The graphical illustration of results achieved on updated independent dataset of different species.
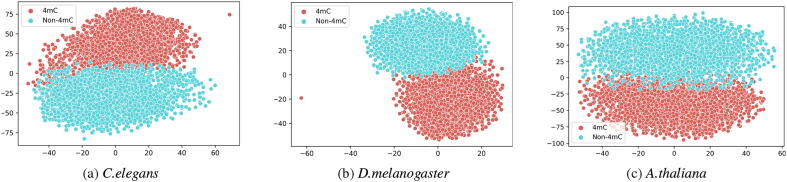
For the further experimental purpose we utilized t-SNE plots [40] to visualize the learned features by the proposed model. Fig. 4 represents the t-SNE plot for three different species which are C.elegans, D.melanogaster and A.thaliana. Each t-SNE plot illustrates the feature representation of 4mC and Non-4mC sites after the flattening layer. As showcased in the plots, the proposed framework is capable of learning distinct features which can efficiently discriminate 4mC sites from Non-4mC sites.
Fig. 4.
t-SNE plots of three different species, illustrating the feature representation after flattening layer.
3.3. Cross-species validation
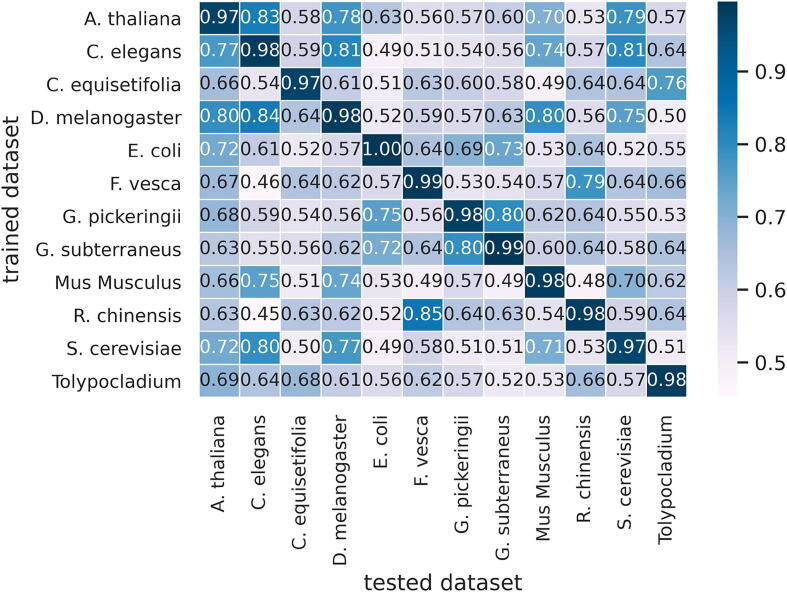
In bioinformatics, it is considered to be important that any artificial intelligence-based model should learn the genetic information rather than just learning the dataset. Therefore to evaluate the model in that perception, we have carried out cross-species validation. The computed cross-validation is compared with the phylogenetic tree which represents evolutionary relationships between numerous biological species. If any neural network based model learns the genetic information of the species so it would be an easy task for the network to perform the prediction of the closely related species.
In our case, we have some closely related biological species which can validate the model learning. Fig. 5 shows the cross-species validation heat map generated using the ACC values. The diagonal values of the heatmap show the result of species being trained and tested on the same dataset. While the neighbouring values represent the values of cross-species validation. The species A.thaliana, C.elegans, D.melanogaster, S.cerevisiae and Mus musculus are closely related species that belong to the same main branch of the phylogenetic tree. As can be seen in the heat map that the cross-species results are better in these species when compared to their results on other species. For instance, the model trained on A.thaliana gives good results when tested on C.elegans, D.melanogaster, Mus musculus and S.cerevisiae while the other species the model performance is not good. This shows that the proposed tool holds the capability to learn the insight genetic information of the biological species. Similarly, R.chinensis and F.vesca belong to the Rosace genome, which means they are highly related to each other. When the model is trained on F.vesca and tested on R.chinensis so the achieved accuracy is 0.79 and when the model is trained on R.chinensis and tested on F.vesca the achieved accuracy is 0.85. The model cross-species prediction results demonstrate that the proposed architecture is competent to be relied on.
Fig. 5.
Cross-species validation heat map.
4. Webserver
The proposed DCNN-4mC predictor has been implemented on PHP based user-friendly webserver, can be accessed freely at:http://nsclbio.jbnu.ac.kr/tools/DCNN-4mC/. The following is a set of instructions to use the webserver. Users can type FASTA format sequences into the text area or click the upload icon to upload a file containing FASTA format sequences. The sequences should be of length 41nt. Further in a single cycle maximum of 1000 sequences can be processed. By selecting the ‘Example’ button, an example of FASTA format sequences can be seen. Further, choosing the species must be specified during the process. The chosen species must be the same as that of the sequence belonging species, in order to achieve the expected prediction accuracy. Lastly, pressing the ’Submit sequences’ button will appear the anticipated outcomes.
5. Challenges and future work
The proposed tool has undoubtedly achieved good results on numerous biological species and holds the capability to be used by experts. But still, a gap of improvement in 4mC sites classification is there. Here we have discussed some of the challenges that can be addressed in future work. Dataset is considered to be the backbone of any artificial intelligence model. The same is the case with this research problem. Some of the species have a very limited amount of datasets that restricts the artificial intelligence experts to propose an effective model. The same case happened in this research, due to the limited amount of datasets, we reduced the number of blocks for few species as discussed in the methodology section. The increase in dataset size will allow the researchers to have complex computational models which can give good classification performance. In this research, we have tried to cover all available species datasets. Still, the authors hold an opinion that the dataset for new species needs to be explored. This will allow the tools to learn distinct insight information from different species. Moreover, the techniques of neural networks need to be explored which are not yet used for the purpose of DNA modification identification. One such effort is made in this research where the role of skip connection in the neural networks is explored for the said research problem.
6. Conclusion
In this research, a neural network based tool known as DCNN-4mC is proposed for 4mC site prediction. This tool is a CNN-based framework with skip connections which uses a one-hot encoding scheme to encode the raw DNA sequence. The DCNN-4mC tool has contributed towards addressing the issue of generalizability that lacks in the previously proposed frameworks. In this study, we collected all the available datasets of different species under a single umbrella. Where different datasets for similar species are efficiently combined into a single dataset so that future researchers can have a single benchmark dataset. So far, in bio-informatics dataset for 12 different species are explored for 4mC site classification. The proposed model has exhibited state-of-the-art results and has outperformed all existing architectures. The skip connection in the proposed tool helped to learn the insight genomics features of different species and the results of cross-species validation prove that. The proposed approach not only achieved high results on existing databases but also performed well on the updated dataset. For the ease of the research community, we have made a freely accessible webserver of this powerful tool for high-throughput 4mC site classification from DNA sequences.
CRediT authorship contribution statement
Mobeen Ur Rehman: Conceptualization, Methodology, Software, Writing-original-draft, Writing-review-editing. Hilal Tayara: Conceptualization, Software, Validation, Supervision, Writing-review-editing. Kil To Chong: Conceptualization, Validation, Supervision, Writing-review-editing, Funding-acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by the National Research Foundation of Korea(NRF) grant funded by the Korea government(MSIT) (No. 2020R1A2C2005612) and in part by the Brain Research Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No. NRF-2017M3C7A1044816).
Footnotes
Supplementary data associated with this article can be found, in the online version, athttps://doi.org/10.1016/j.csbj.2021.10.034.
Contributor Information
Mobeen Ur Rehman, Email: cmobeenrahman@jbnu.ac.kr.
Hilal Tayara, Email: hilaltayara@jbnu.ac.kr.
Kil To Chong, Email: kitchong@jbnu.ac.kr.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rathi P., Maurer S., Summerer D. Selective recognition of n 4-methylcytosine in dna by engineered transcription-activator-like effectors. Philos Trans R Soc B: Biol Sci. 2018;373(1748):20170078. doi: 10.1098/rstb.2017.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., Cai J., Lu H., Mao S., Dai S., Hu J., Wang L., Hua X., Xu H., Tian B., et al. N4-cytosine dna methylation is involved in the maintenance of genomic stability in deinococcus radiodurans. Front Microbiol. 2019;10:1905. doi: 10.3389/fmicb.2019.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen-wen W., Li-hua Q. Current review on dna methylation in ovarian cancer. J Int Reprod Health/Family Plann. 2012;31(4):312. [Google Scholar]
- 4.Santos K., Mazzola T., Carvalho H. The prima donna of epigenetics: the regulation of gene expression by dna methylation. Braz J Med Biol Res. 2005;38(10):1531–1541. doi: 10.1590/s0100-879x2005001000010. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich M. Dna methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 6.Robertson K.D. Dna methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 7.Cheng X. Dna modification by methyltransferases. Curr Opin Struct Biol. 1995;5(1):4–10. doi: 10.1016/0959-440x(95)80003-j. [DOI] [PubMed] [Google Scholar]
- 8.Liang Z., Shen L., Cui X., Bao S., Geng Y., Yu G., Liang F., Xie S., Lu T., Gu X., et al. Dna n6-adenine methylation in arabidopsis thaliana. Develop Cell. 2018;45(3):406–416. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Ratel D., Ravanat J.-L., Berger F., Wion D. N6-methyladenine: the other methylated base of dna. Bioessays. 2006;28(3):309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye P., Luan Y., Chen K., Liu Y., Xiao C., Xie Z. Methsmrt: an integrative database for dna n6-methyladenine and n4-methylcytosine generated by single-molecular real-time sequencing. Nucl Acids Res. 2016:gkw950. doi: 10.1093/nar/gkw950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyko F. The dna methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki M.M., Bird A. Dna methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 13.Heyn H., Esteller M. An adenine code for dna: a second life for n6-methyladenine. Cell. 2015;161(4):710–713. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Wei L., Luan S., Nagai L.A.E., Su R., Zou Q. Exploring sequence-based features for the improved prediction of dna n4-methylcytosine sites in multiple species. Bioinformatics. 2019;35(8):1326–1333. doi: 10.1093/bioinformatics/bty824. [DOI] [PubMed] [Google Scholar]
- 15.Schweizer H.P. Bacterial genetics: past achievements, present state of the field, and future challenges. Biotechniques. 2008;44(5):633–641. doi: 10.2144/000112807. [DOI] [PubMed] [Google Scholar]
- 16.Lister R., Ecker J.R. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19(6):959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W. Direct detection of dna methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7(6):461. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Z., Li J., Zhang J.-R., Zhang X. qdnamod: a statistical model-based tool to reveal intercellular heterogeneity of dna modification from smrt sequencing data. Nucl Acids Res. 2014;42(22):13488–13499. doi: 10.1093/nar/gku1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M., Ji L., Neumann D.A., Chung D.-H., Groom J., Westpheling J., He C., Schmitz R.J. Base-resolution detection of n 4-methylcytosine in genomic dna using 4mc-tet-assisted-bisulfite-sequencing. Nucl Acids Res. 2015;43(21):e148. doi: 10.1093/nar/gkv738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehman M.U., Akhtar S., Zakwan M., Mahmood M.H. Novel architecture with selected feature vector for effective classification of mitotic and non-mitotic cells in breast cancer histology images. Biomed Signal Process Control. 2022;71 [Google Scholar]
- 21.Chen W., Yang H., Feng P., Ding H., Lin H. idna4mc: identifying dna n4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics. 2017;33(22):3518–3523. doi: 10.1093/bioinformatics/btx479. [DOI] [PubMed] [Google Scholar]
- 22.He W., Jia C., Zou Q. 4mcpred: machine learning methods for dna n4-methylcytosine sites prediction. Bioinformatics. 2019;35(4):593–601. doi: 10.1093/bioinformatics/bty668. [DOI] [PubMed] [Google Scholar]
- 23.Wei L., Su R., Luan S., Liao Z., Manavalan B., Zou Q., Shi X. Iterative feature representations improve n4-methylcytosine site prediction. Bioinformatics. 2019;35(23):4930–4937. doi: 10.1093/bioinformatics/btz408. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q., Chen J., Wang Y., Li S., Jia C., Song J., Li F. Deeptorrent: a deep learning-based approach for predicting dna n4-methylcytosine sites. Briefings Bioinform. 2021;22(3):bbaa124. doi: 10.1093/bib/bbaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khanal J., Nazari I., Tayara H., Chong K.T. 4mccnn: Identification of n4-methylcytosine sites in prokaryotes using convolutional neural network. IEEE Access. 2019;7:145455–145461. [Google Scholar]
- 26.Zeng R., Liao M. Developing a multi-layer deep learning based predictive model to identify dna n4-methylcytosine modifications. Front Bioeng Biotechnol. 2020;8:274. doi: 10.3389/fbioe.2020.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng F., Fang G., Yao L. A deep neural network for identifying dna n4-methylcytosine sites. Front Genet. 2020;11:209. doi: 10.3389/fgene.2020.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manavalan B., Basith S., Shin T.H., Lee D.Y., Wei L., Lee G., et al. 4mcpred-el: an ensemble learning framework for identification of dna n4-methylcytosine sites in the mouse genome. Cells. 2019;8(11):1332. doi: 10.3390/cells8111332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasan M.M., Manavalan B., Khatun M.S., Kurata H. i4mc-rose, a bioinformatics tool for the identification of dna n4-methylcytosine sites in the rosaceae genome. Int J Biolog Macromolecules. 2020;157:752–758. doi: 10.1016/j.ijbiomac.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Lv H., Dao F.-Y., Zhang D., Guan Z.-X., Yang H., Su W., Liu M.-L., Ding H., Chen W., Lin H. idna-ms: an integrated computational tool for detecting dna modification sites in multiple genomes. Iscience. 2020;23(4) doi: 10.1016/j.isci.2020.100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z.-Y., Xing J.-F., Chen W., Luan M.-W., Xie R., Huang J., Xie S.-Q., Xiao C.-L. Mdr: an integrative dna n6-methyladenine and n4-methylcytosine modification database for rosaceae. Horticulture Res. 2019;6(1):1–7. doi: 10.1038/s41438-019-0160-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehman M.U., Chong K.T. Dna6ma-mint: Dna-6ma modification identification neural tool. Genes. 2020;11(8):898. doi: 10.3390/genes11080898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbas Z., Tayara H., Chong K. Spinenet-6ma: a novel deep learning tool for predicting dna n6-methyladenine sites in genomes. IEEE Access. 2020;8:201450–201457. [Google Scholar]
- 35.Alam W., Ali S.D., Tayara H., Chong K. A cnn-based rna n6-methyladenosine site predictor for multiple species using heterogeneous features representation. IEEE Access. 2020;8:138203–138209. [Google Scholar]
- 36.Shujaat M., Lee S.B., Tayara H., Chong K.T. Cr-prom: A convolutional neural network-based model for the prediction of rice promoters. IEEE Access. 2021 [Google Scholar]
- 37.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition, in; pp. 770–778. [Google Scholar]
- 38.Chicco D., Tötsch N., Jurman G. The matthews correlation coefficient (mcc) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Mining. 2021;14(1):1–22. doi: 10.1186/s13040-021-00244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chicco D., Jurman G. The advantages of the matthews correlation coefficient (mcc) over f1 score and accuracy in binary classification evaluation. BMC Genom. 2020;21(1):1–13. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Maaten L., Hinton G. Visualizing data using t-sne. J Mach Learn Res. 2008;9(11) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.