Highlights
-
•
Extracellular Vesicles/EVs can be tested in small clinically relevant serum samples.
-
•
Antibody-coated immunobeads is a simple, fast, user-friendly method for this.
-
•
EVs may help diagnose breast cancer patients versus those with a benign mass.
-
•
Gremlin-1 cargo of EVs may be a diagnostic biomarker for breast cancer.
Keywords: Extracellular vesicles, breast cancer, Clinical utility, standard-of-care, Gremlin-1
Abbreviations: EVs, Extracellular vesicles; DIFF UC, Differential ultracentrifugation; PEG, PEG precipitation; SEC, Size-exclusion chromatography; NBI, Nickel-based isolation; BCA, Bicinchoninic acid assay; NTA, Nanoparticle tracking analysis; TEM, Transmission electron microscopy
Abstract
Extracellular vesicles (EVs) have potential as minimally invasive biomarkers. However, the methods most commonly used for EV retrieval rely on ultracentrifugation, are time-consuming, and unrealistic to translate to standard-of-care. We sought a method suitable for EV separation from blood that could be used in patient care. Sera from breast cancer patients and age-matched controls (n = 27 patients; n = 36 controls) were analysed to compare 6 proposed EV separation methods. The EVs were then characterised on 8 parameters. The selected method was subsequently applied to independent cohorts of sera (n = 20 patients; n = 20 controls), as proof-of-principle, investigating EVs’ gremlin-1 cargo. Three independent runs with each method were very reproducible, within each given method. All isolates contained EVs, although they varied in quantity and purity. Methods that require ultracentrifugation were not superior for low volumes of sera typically available in routine standard-of-care. A CD63/CD81/CD9-coated immunobead-based method was most suitable based on EV markers' detection and minimal albumin and lipoprotein contamination. Applying this method to independent sera cohorts, EVs and their gremlin-1 cargo were at significantly higher amounts for breast cancer patients compared to controls. In conclusion, CD63/CD81/CD9-coated immunobeads may enable clinical utility of blood-based EVs as biomarkers.
Introduction
Extracellular vesicles (EVs) are nanosized lipid bilayer-surrounded particles released by cells into their surrounding biofluid [1]. Blood-based EVs have potential as minimally invasive diagnostic, prognostic and/or predictive tools. Proteins used for breast cancer diagnosis include uPA and PAI-1, but these require invasive biopsies [2]. Serum biomarkers CA15–3, carcinoembryonic antigen, and tissue polypeptide antigen are not sensitive enough to detect early-stage breast cancer [3]. Jesneck et al. [4] investigated if a panel of 98 serum freely-circulating biomarkers could decipher breast cancer from healthy controls, but found that they could not.
As we recently reviewed, a number of studies have investigated the potential of blood based EVs as biomarkers for breast cancer [5]. Typically, those studies selected one EV separation method, applied it, and sometimes did no EV characterisation; often not adhering to MISEV2018 guidelines [6]. Our group found that miR-134 was at significantly lower levels in EVs from serum of breast cancer patients compared to heathy controls, directly reflecting comparative tissue-based miR-134 [7]. A limitation of our study was that it employed the popular differential ultracentrifugation method of Thery et al. [8,9]. Although appropriate in a research setting, that method is not translatable to standard-of-care clinical settings.
Unrelated to cancer, EV separation techniques have been compared using conditioned media or blood. Andreu et al. [10] compared EV enrichment methods using four sera samples, but with the specific focus of analysing EV-encapsulated miRNAs. EVs' structure was not analysed. Also focusing on profiling miRNAs, Buschmann et al. [11] compared 5 methods on serum from septic shock patients and controls. Karimi et al. [12] compared 3 methods of EV separation from serum/plasma of healthy subjects. A similar study of 4 EV separation methods focused on healthy donor sera only [13]. A 3-step protocol to isolate EVs from blood of healthy humans was developed. However, as this requires PEG-precipitation, iohexol gradient ultracentrifugation, and size-exclusion chromatography it, too, will never be translated to routine clinical utility [14]. Furthermore, many such studies use volumes of serum (e.g. 1 mL) that are too large to routinely take from cancer patients. When we consider the patients’ perspective, 1 mL of serum is the yield from approximately 3 mL of blood. From cancer patients – already ill and under-going multiple longitudinal investigations- the equivalent of 3 mL of blood is a quite a large volume to use for any one test type, as multiple other routine tests are typically needed to be performed.
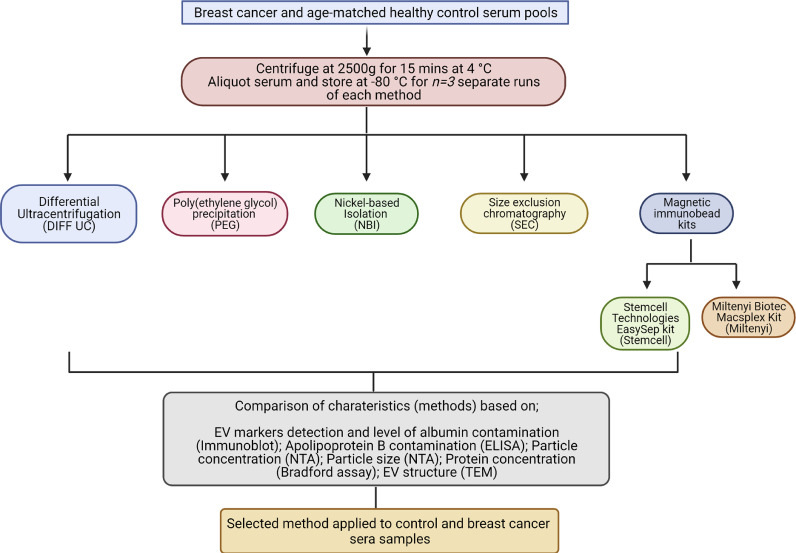
Thus, with a specific focus on clinical ulility rather than fundamental research, we first aimed to compare the separation of EVs from sera of breast cancer patients’ and aged-matched controls, using 6 different methods. Specifically, we wished to identify a method that is effective at separating the purest EVs possible, while also being a quick, cost-effective technique that realistically could be translated to a hospital setting. Although we do not claim -or, indeed, believe- that isolates obtained following all methods are free of co-isolated “contaminants” from within the preparations, we use the term EVs here as there is no suitable term yet selected to cover EVs with or without possible co-isolated non-EV material (initially we used the term EVs/isolates here, but it is considered too cumbersome and confusing). Our second aim was to apply the selected method to the separation of EVs from independent cohorts of breast cancer patients’ and controls’ sera and then compare the EVs quantities and gremlin-1 cargo. Gremlin-1 was selected as its cellular expression is emerging as an important molecule in breast cancer development and metastasis, but its blood-/EV-based presence and relevance are unknown [15], [16], [17], [18]. Our study design is summarised in Fig. 1.
Fig. 1.
Study design overview
BC patients’ and healthy controls’ sera pools were prepared to establish if an optimal method for clinical utility could be identified, when comparing six proposed EV separation methods. Following extensive characterisation of EVs, the selected method was applied to an independent cohort of sera samples (n = 20 from patients; n = 20 from controls). Estimates of EVs amounts, and a cargo protein of interest, were evaluated.
Materials and methods
Serum collection
The work described has been performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). Ethics approval was obtained at St. Vincent's University Hospital and Trinity College Dublin. Blood samples (9 mL) were donated by individuals (n = 63) referred to Breast Care Clinic at SVUH due to a breast mass/abnormal growth that was relatively small (≤ 1.5 cm). Blood was procured immediately upon arrival prior to examination and intervention. This was because any intervention could stimulate release of cellular material, including EVs, into the blood. A useful diagnostic would be present in blood without any intervention. After discarding the first 5 mL, blood was collected in non-heparinised tubes, clotted for 30–60mins., and centrifuged at 1000 g for 10mins. Serum was collected, leaving ∼100μL to avoid non-sera components. It was later on that day established which individuals had breast cancer (“BC patients”) and which did not i.e. breast mass was benign (“controls”).
For the first aim, to use exactly the same sera for comparison of 6 EV separation methods, 14.5 mL volumes of sera from patients and controls, respectively, were pooled. These were spun at 2500 g for 15mins. at 4 °C, aliquoted, and stored at −80 °C. Each EV separation used 500µL aliquots of pooled cancer and control sera, in n = 3 separate experiments. All steps were performed at room temperature, unless indicated. For the second aim, individual aliquots of sera from patients (n = 20) and controls (n = 20), that were not used in the pools, were analysed.
Differential ultracentrifugation (DIFF-UC)
Based on the method reported by Thery et al. [10], sera were ultracentrifuged at 100,000 g for 60mins. in polyallomer tubes (Beckman Coulter;Cat.#:326823), SW32Ti swinging rotor, at 4 °C. Supernatant was removed, pellets washed with PBS, and spun at 100,000 g for 60mins. Final EVs were resuspended in 125µL PBS.
PEG precipitation (PEG)
Based on the previously described methods [14,19], sera were mixed 1:1 with PEG buffer, to a final 10% w/v of PEG. PEG buffer contains: 20% w/v PEG6000 (Sigma;Cat.#:81260) in 200 mM NaCl (Sigma;Cat.#:376), 10 mM EDTA (Invitrogen;Cat.#:15,575–038), 200 mM Tris-HCL (Sigma,Cat.#:T3253), pH7.0. Samples were rotated for 1h. at 4 °C and centrifuged at 4000 g, 15mins., at 4 °C. Supernatant was removed; pellets washed with PBS; and centrifuged at 120,000 g, 2 h., at 4 °C. Final EVs were resuspended in 125uL PBS.
Nickel-based isolation (NBI)
NBI was performed as previously reported [20]. Final EVs were brought to 125μL with PBS.
Size-exclusion chromatography (SEC)
SEC was performed as the manufacturer of Izon qEV original 70 nm (Izon;Cat.#:SP1) columns recommended. Pooled fractions 7–9 were concentrated using 10 kDa Pierce protein concentrator, PES10MWCO (Sigma;Cat.#:88513) to 125µL.
Stemcell Technologies EasySep Pan extracellular vesicles kit
The principle is that EVs (Stemcell Technologies/Stemcell;Cat.#:17,891) are pulled out by anti-CD9/CD81/CD63 antibodies-coated spheres/immunobeads. Following the manufacturers' instructions, final EVs were resuspended in 125µL PBS.
Miltenyi Biotec Macsplex Exosome Isolation kit
Principle, as for Stemcell, this (Miltenyi Biotec;Cat.#:130–110–912) was performed following the manufacturers' instructions. The EVs were adjusted to 125µL with PBS.
Protein quantification
Bradford assay using the bio-rad protein assay dye reagent (Bio-Rad;Cat.#:500–0006).
Nanoparticle tracking analysis (NTA)
EVs samples were automatically injected into a NS500 (NanoSight, UK) under constant flow conditions (flow rate=50). Videos were analysed using NTA3.1.54 software.
Transmission electron microscopy (TEM)
EVs (10µL) were placed on formvar carbon-coated nickel grids (Ted Pella Inc;Cat.#:01813-F) and settled for 10mins. A droplet of paraformaldehyde (2%) was placed on parafilm, the grid placed on top for 10mins., contrasted in 2% uranyl acetate (BDH;Cat.#:230550), and imaged by JEOL TEM-2100.
Immunoblotting
Performed as we previously described [21], using 10 µg EVs or cell lysates. Primary antibodies: anti-CD63 (1:500;Abcam;Cat.#:ab68418), anti-CD9 (1:1000;Abcam;Cat.#:ab92726), anti-CD81 (1:200;Santa Cruz;Cat.#:sc-23,962), anti-albumin (1:1000;Abcam;Cat.#:ab190806). HRP-linked secondary antibodies: anti-mouse (1/1000 in 5% BSA/PBS-T;Cell Signalling;Cat.#:7076) or anti-rabbit (1/1000 in 5% BSA/PBS-T;Cell Signalling;Cat.#:7074).
Apolipoprotein B ELISA
Apo B, indicative of “contamination” with lipoproteins, was investigated using 500 ng of EVs or Hs578Ts(i)8 lysate as control, according to (Abcam;Cat.#:ab190806) manufacturers’ instructions.
Gremlin-1 ELISA
Assay Genie (Cat.#:HUFI01783) was performed according to manufacturers’ instructions using 5 µg lysed EVs.
Statistical analysis
Using GraphPad Prism9, paired t-test compared two groups and one-way ANOVA comparing more than two groups.
Results
Immunoblotting for EV markers and contaminating albumin
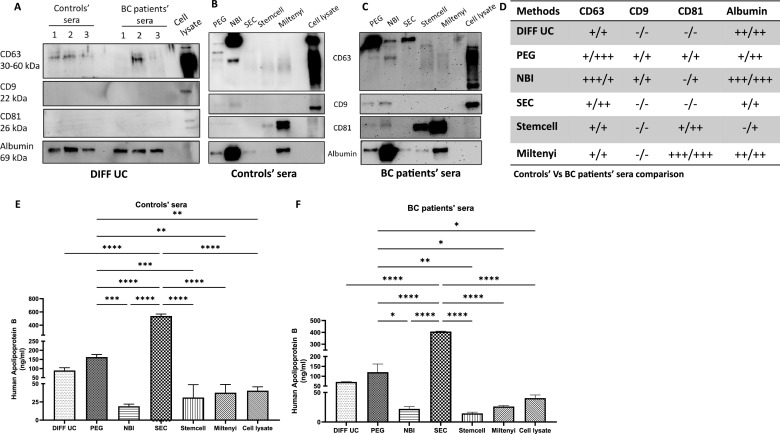
Initially considering the commonly used DIFF-UC approach, the presence of established EV markers i.e. tetraspanins CD63, CD9 and CD81 were investigated. With both control and patients’ EVs, CD63 was occasionally detected, but CD9 and CD81 were not (Fig. 2A,D). With the other 5 methods, CD63 was detected in both control and patients’ sera EVs (Fig. 2B–D). CD9 was detected at low levels in PEG and NBI EVs only. CD81 was detected in both control and patients’ EVs following Stemcell and Miltenyi methods, and only in patients’ EVs with NBI. Overall, with the exception of DIFF-UC and SEC, 2 of 3 EV markers analysed were detected.
Fig. 2.
Immunoblotting for EV biomarkers and potential albumin contamination and ELISAs for potential ApoB contamination
EV biomarkers and albumin contamination in EVs from the traditional DIFF-UC method, n = 3 separations (A). Representative immunoblots following the other 5 methods i.e. controls (B) and BC (C), summarised in (D). ELISA of ApoB contamination of controls (E) and BC (F) EVs. Hs578Ts(i)8 cell lysate was the control. Graphs are mean of n = 3±SEM experiments. One-way ANOVA; *P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Albumin (Fig. 2A–D) and ApoB (Fig. 2E,F), representing key serum components that should not be in pure EV preparations, were also investigated. Stemcell and NBI EVs had very low levels of ApoB contamination. However, NBI EVs contained a substantial amount of albumin. This was not so for Stemcell which, based on albumin and ApoB, produced the cleanest EVs.
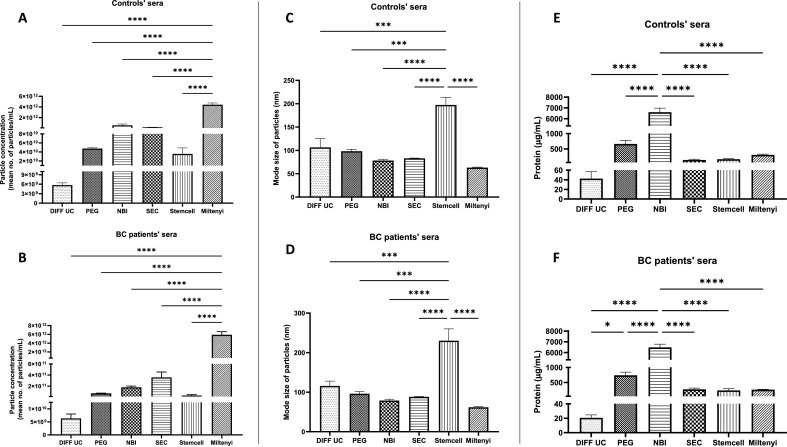
Particle concentration, size and protein content
NTA estimates particles (EVs) quantities/concentration and sizes. With both control and cancer sera, DIFF-UC gave the lowest yields (Fig. 3A,B). Miltenyi apparently produced significantly higher yields when compared to other methods (Fig. 3A,B). However, this may be due to remaining immunobeads that have not captured EVs but are also detected as particles by NTA.
Fig. 3.
Particle concentration, size and protein concentration of EVs
Concentration (particles/mL) (A, B), size (C, D), and protein quantification (E, F) of EVs of controls and patients, respectively. Graphs represent mean±SEM of n = 3 independent experiments. One-way ANOVA was used as statistical test. *P < 0.05, ***P < 0.001, ****P < 0.0001.
All methods resulted in particles of 60–142 nm in size, except Stemcell method where the particles were 197–230 nm (Fig. 3C,D). As EVs remain attached to Stemcell immunobeads, this difference is likely due to the EV-bead complexes being sized as single particles. Overall, despite the limitations of NTA in deciphering EVs from other free- or bound- particles, it is noteworthy that all 3 runs of each NTA experiment were highly reproducible.
Total protein quantification (e.g. by Bradford as used here, or BCA assay) is sometimes used as a surrogate for EV quantities. Its disadvantage is that it quantifies all proteins present and is not specific to those that are integral components of EVs i.e. other proteins precipitated/spun-down with EVs will also be quantified. However, unlike NTA, the Bradford assay only quantifies biological material and -importantly here- is a simple, routine technique. Like NTA, all 3 runs of each Bradford assay experiment had a high level of reproducibility. Furthermore, as with NTA, DIFF-UC isolates from both control and cancer sera had lowest protein concentration. This was only significant when compared to NBI for control and cancer sera (Fig. 3E,F) and when compared to PEG for cancer sera (Fig. 3F). NBI resulted in significantly higher amounts of protein compared to all other methods, with both control and cancer sera (Fig. 3E,F). This was unsurprising given that NBI EVs had substantially the most albumin contamination (Fig. 2B–D). Overall, NBI (and, to a lesser extent, PEG) seemed to carry over most non-EV proteins.
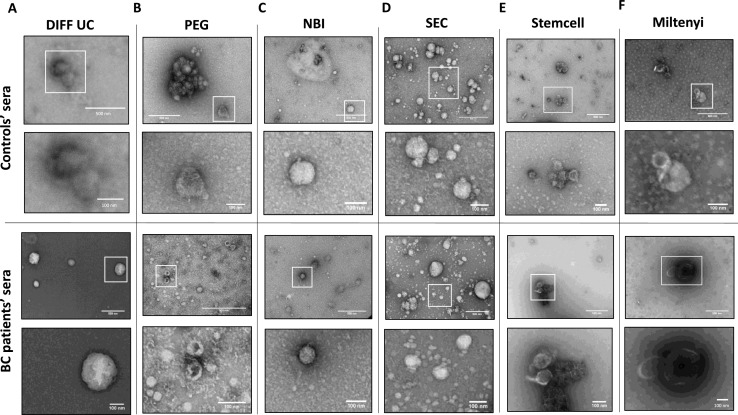
TEM analysis of separated EVs
TEM showed heterogeneous populations of EVs, with characteristic EV shapes and sizes, resulting from each method of separation and both control and patients’ sera. Although TEM is qualitative, it was apparent that DIFF-UC EVs were fewest in numbers, in keeping with the NTA data (Fig. 4A). PEG EVs had aggregates present (Fig. 4B). NBI EVs were rounded, vesicular structures (Fig. 4C). SEC products had structures of typical EV sizes, but also numerous small vesicular structures (Fig. 4D), aligned with our observation of significantly more ApoB/lipoprotein contamination. Stemcell and Miltenyi EVs had typical EVs structures. Doublets/clumps present were likely due to free- or EV-attached immunobeads (Fig. 4E,F). Overall, all isolates contained some structures indicative of EVs, no substantial differences were noted between controls and cancer EVs, but SEC produced a predominance of particularly small structures. Thus, with the exception of SEC, TEM would not contribute to any method being either favoured or ruled out.
Fig. 4.
TEM analysis of separated EVs
TEM analysis of EVs from control (top panel) and patient (bottom panel) sera using DIFF-UC (A), PEG (B), NBI (C), SEC (D), Stemcell (E), and Miltenyi (F) methods. Scale bars are 500 nm (top images of each panel) and 100 nm (bottom images of each panel).
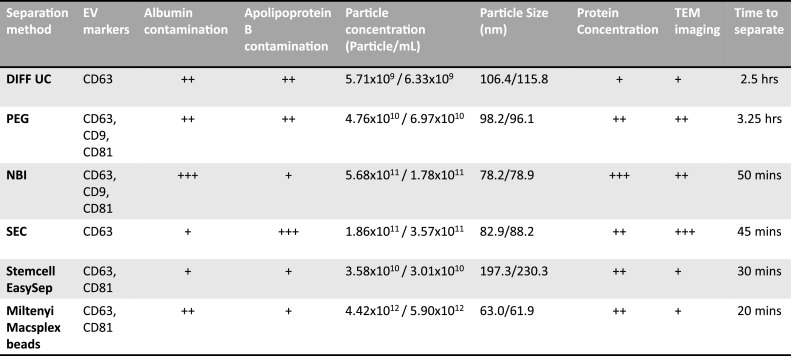
Overview of characteristics of the methods and resulting EVs
Fig. 5 summarises the results from the 8 comparisons made from the 6 methods. Clearly no method -suitable for working with small volumes of sera that would be available on a routine basis from cancer patients- is perfect; but also, no method was particularly worse for EVs from cancer patients compared to controls. Stemcell method's EVs had the least contaminating albumin and ApoB, while also having comparable amounts of particles (although still attached to immunobeads, so apparently bigger) and protein when compared to most other methods. Although, CD9 was undetected on the Stemcell EVs, 2 established EV markers, CD63 and CD81, were confirmed. The short time (∼30 mins.) needed, ease of use, relatively low levels of contamination, suggest that of the methods tested, this may be most suitable for routine use in a clinical setting.
Fig. 5.
Characteristics of the methods and resulting EVs
A summary of the results of each of the 8 characteristics used to compare the 6 separation methods. The results are qualitatively graded by low (+), moderate (++), high (+++). Particle concentration and size are divided into two columns, representing control and patients separately.
Gremlin-1 in EVs
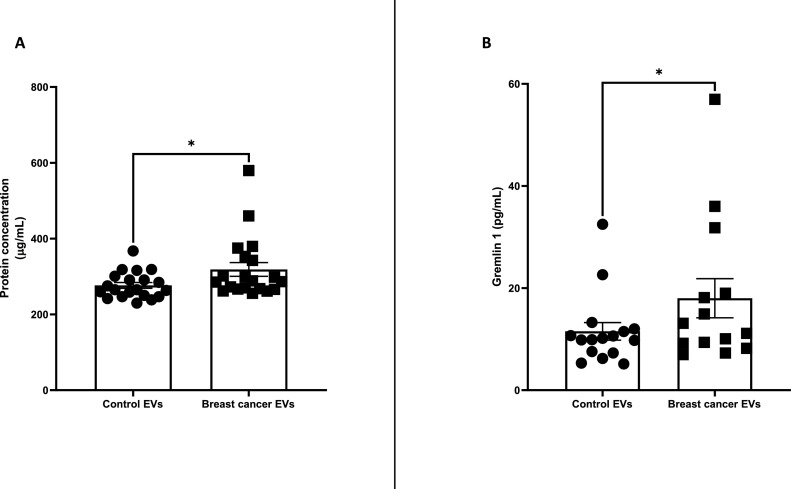
Following selection of Stemcell immunobeads as possibly the best of the methods for progressing to clinical samples and applying it to an independent cohort of control (n = 20) and patients’ (n = 20) sera samples, the protein quantities of the EVs -as surrogate for EVs- were significantly higher with breast cancer compared to controls (Fig. 6A). Gremlin-1 cargo of the lysed EVs was also significantly higher in breast cancer EVs compared to control EVs (Fig. 6B).
Fig. 6.
Gremlin-1 in EVs separated using Stemcell method
EVs from sera of BC patients (n = 20) and age-matched controls (n = 20) were separated using the Stemcell kit. Protein concentration of lysed EVs (A). Gremlin-1 content of lysed EVs (B). Graphs represent mean±SEM of n = 3 independent experiments. µg/mL reports the amount of EV protein per millilitre (mL) of lysed EV suspension. Paired t-test was used as statistical test. *P < 0.05.
Discussion
After extensive investigations of EVs produced by 6 different EV separation methods, from cancer and controls sera, and characterised on 8 parameters, evidently no method is perfect at producing pure EVs from small volumes of sera. Granted, achieving pure EVs is not necessarily a priority for clinical utility as biomarkers. Additional to the fact that DIFF-UC is unsuitable for routine clinical utility based on the need for ultracentrifugation, it did not produce adequate EVs from clinically relevant sera volumes to detect any EV markers investigated. In agreement, when DIFF-UC was used on larger serum volumes (1 mL), no EV markers were detected [11]. Arguments could be made in favour of other methods tested but as we aimed to select and apply a method that could realistically be translated to a routine care, the strongest case could be made for Stemcell CD63/CD81/CD9 immunobeads.
Stemcell method-separated EVs met most of the MISEV2018 criteria needed for an acceptable EV isolation. Furthermore, aside from the fact that it can be performed in ∼30 min; requires no high-end expensive equipment; could routinely be performed by trained laboratory personnel; we found that the Stemcell EVs contained 2 of 3 EV markers investigated and were most pure of both contaminating albumin and ApoB. The particle numbers and associated protein quantities were not significantly different to those obtained by DIFF-UC or, indeed, most of the other methods tested. Stemcell EVs were apparently larger than those obtained with all other methods, likely due to the fact that the immunobeads remain attached to the EVs. So, as evidenced by the actual NTA particle concentration results and also their reproducibility, the presence of the EV-attached immunobeads did not significantly affect EV counts. It did affect their apparent size, as both the EV and attached immunobead would have been sized together. Thus, we suggest that NTA is not a very suitable method for analysis of EVs generated from patient or controls sera by Stemcell. However, as NTA would not be typically available in a hospital laboratory to use in decisions on patient care, this is a moot point.
As proof-of-principle this method was applied to independent cohorts of sera samples (n = 40), as could be done in a hospital laboratory. Using protein quantification as surrogate for EV quantities (a simple method that would be available in a hospital laboratory), it was observed that significantly more EVs are present in sera of cancer patients compared to age-matched controls who have a similarly sized, benign breast mass. By ELISA (also a routine assay), this was also found to be so for gremlin-1. Gremlin-1 has been shown to be overexpressed in human cancers. In breast cancer, gremlin-1 has been shown to promote the metastasis to the lungs [18]. Specifically, gremlin-1 was present in EVs from both patients’ and controls’ sera, but at significantly higher levels for patients. Although there is data to suggest that gremlin-1 is mechanistically involved in cancer development and metastasis, this is the first study to investigate it as a blood-based biomarker and even more specifically associated with EVs.
In conclusion, a few methods evaluated may have relevance in separating EVs from sera of cancer patients and individuals who have a benign breast growth/mass. However, due to their complexity, specialised equipment needed, and length of time to perform, some of those could never be advanced to clinical utility. Considering all 6 methods and 8 characteristics evaluated, when aiming to select one method, the Stemcell CD63/CD81/CD9 immunobeads seems the most optimal. Arguably with any EV separation technique there is a compromise between separating pure EVs and having a method that is clinically relevant. However, this does not detract from EVs’ potential clinical relevance. Although independent validation studies are needed, our application of this immunobead approach followed by ELISA for gremlin-1 suggests that EVs carrying gremlin-1 may have relevance as a breast cancer biomarker. Maybe even more relevant, we believe we have identified a method that has potential for advancing other blood-based biomarkers studies.
CRediT authorship contribution statement
Niamh McNamee: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Roisin Daly: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. John Crown: Investigation, Resources, Writing – review & editing. Lorraine O'Driscoll: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
CRediT authorship contribution statement
Niamh McNamee: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Róisín Daly: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. John Crown: Investigation, Resources, Writing – review & editing. Lorraine O'Driscoll: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was supported by an Irish Research Council Advanced Laureate Award to LOD [IRCLA/2019/49] and a philanthropic donation by Carrick Therapeutics [TCDLOD001]. Thanks also to Clinical Cancer Research Trust (Ireland) for their support of this research. The funding sources had no such involvement in any part of the design, execution, analysis of the study or in the drafting of this manuscript.
References
- 1.O'Driscoll L. Expanding on exosomes and ectosomes in cancer. N. Engl. J. Med. 2015;372:2359–2362. doi: 10.1056/NEJMcibr1503100. [DOI] [PubMed] [Google Scholar]
- 2.Duffy M.J., McGowan P.M., Harbeck N., Thomssen C., Schmitt M. uPA and PAI-1 as biomarkers in breast cancer: validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res. 2014;16:428. doi: 10.1186/s13058-014-0428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3.Duffy M.J. Serum tumor markers in breast cancer: are they of clinical value? Clin. Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 4.Jesneck J.L., Mukherjee S., Yurkovetsky Z., Clyde M., Marks J.R., Lokshin A.E., et al. Do serum biomarkers really measure breast cancer? BMC Cancer. 2009;9:164. doi: 10.1186/1471-2407-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly R., O’Driscoll L. Extracellular vesicles in blood: are they viable as diagnostic and predictive tools in breast cancer? Drug Discov. Today. 2021;26:778–785. doi: 10.1016/j.drudis.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien K., Lowry M.C., Corcoran C., Martinez V.G., Daly M., Rani S., et al. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget. 2015;6:32774–32789. doi: 10.18632/oncotarget.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner C., Di Vizio D., Sahoo S., Théry C., Witwer K.W., Wauben M., et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J. Extracell. Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreu Z., Rivas E., Sanguino-Pascual A., Lamana A., Marazuela M., González-Alvaro I., et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles. 2016;5:31655. doi: 10.3402/jev.v5.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buschmann D., Kirchner B., Hermann S., Märte M., Wurmser C., Brandes F., et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimi N., Cvjetkovic A., Jang S.C., Crescitelli R., Hosseinpour Feizi M.A., Nieuwland R., et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cellular and molecular life sciences. CMLS. 2018;75:2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Romero N., Madurga R., Rackov G., Palacín-Aliana I., Núñez-Torres R., Asensi-Puig A., et al. Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation. J. Transl. Med. 2019;17:75. doi: 10.1186/s12967-019-1825-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Borg E.G.F., Liaci A.M., Vos H.R., Stoorvogel W. A novel three step protocol to isolate extracellular vesicles from plasma or cell culture medium with both high yield and purity. J. Extracell. Vesicles. 2020;9 doi: 10.1080/20013078.2020.1791450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neckmann U., Wolowczyk C., Hall M., Almaas E., Ren J., Zhao S., et al. GREM1 is associated with metastasis and predicts poor prognosis in ER-negative breast cancer patients. Cell Commun. Signal. CCS. 2019;17:140. doi: 10.1186/s12964-019-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S.A., Sung N.J., Choi B.J., Kim W., Kim S.H., Surh Y.J. Gremlin-1 augments the oestrogen-related receptor α signalling through EGFR activation: implications for the progression of breast cancer. Br. J. Cancer. 2020;123:988–999. doi: 10.1038/s41416-020-0945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim N.H., Sung N.J., Youn H.S., Park S.A. Gremlin-1 activates Akt/STAT3 signaling, which increases the glycolysis rate in breast cancer cells. Biochem. Biophys. Res. Commun. 2020;533:1378–1384. doi: 10.1016/j.bbrc.2020.10.025. [DOI] [PubMed] [Google Scholar]
- 18.Sung N.J., Kim N.H., Surh Y.J., Park S.A. Gremlin-1 promotes metastasis of breast cancer cells by activating STAT3-MMP13 signaling pathway. Int. J. Mol. Sci. 2020;21:9227. doi: 10.3390/ijms21239227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig A.K., De Miroschedji K., Doeppner T.R., Börger V., Ruesing J., Rebmann V., et al. Precipitation with polyethylene glycol followed by washing and pelleting by ultracentrifugation enriches extracellular vesicles from tissue culture supernatants in small and large scales. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1528109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notarangelo M., Ferrara D., Potrich C., Lunelli L., Vanzetti L., Provenzani A., et al. Rapid nickel-based isolation of extracellular vesicles from different biological fluids. Bio Protoc. 2020;10:e3512. doi: 10.21769/BioProtoc.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien K., Rani S., Corcoran C., Wallace R., Hughes L., Friel A.M., et al. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur. J. Cancer. 2013;49:1845–1859. doi: 10.1016/j.ejca.2013.01.017. [DOI] [PubMed] [Google Scholar]