Highlights
-
•
Tregs lead immune-evasive TME of early lung cancer of never smoker.
-
•
Depletion of γδT and NK cells and infiltration of B cells begins in early TME.
-
•
Early lung cancer cells were characterized by dysregulated surfactant pathway.
-
•
CAFs show enrichment of gene sets that inhibit vascular formation.
In tumor tissue, tip-like endothelial cells begin to be replaced with immature ones.
Keywords: Never smoker, Early lung cancer, Single-cell RNA sequencing, Tumor microenvironment
Abbreviations: BC, B cell; CA, cancer cell; CAF, cancer-associated fibroblast; DC, dendritic cell; DEG, differentially expressed gene; EC, endothelial cell; EP, epithelial cell; FB, fibroblast; GEM, gel bead in emulsion; GGN, Ground-glass nodule; HRCT, high-resolution computed tomography; IHC, immunohistochemical; MA, mast cell; MALT, mucosa-associated lymphoid tissue; MDSC, myeloid-derived suppressor cell; MY, myeloid cell; NSCLC, non-small cell lung cancer; NK, natural killer; NL, normal lung tissue; TAM, tumor-associated macrophage; TC, T cell; TIM, tissue-infiltrating macrophage; Treg, regulatory T cell; Tu, tumor
Abstract
With the increasing interest in health screening with chest CT Ground-glass nodule (GGN) has become one of the common lung lesions encountered in daily medical practice. Because lung adenocarcinoma in the form of GGN is an ideal model for studying early lung carcinogenesis, 11 GGN and normal lung specimens from 6 never smoker patients were studied by single-cell RNA sequencing. Lung cancer cells showed enrichment of gene sets related to small vesicle processing and surfactant homeostasis compared to non-malignant lung epithelial cells, suggesting the dysregulation of surfactant pathway may be involved in early lung carcinogenesis. Along with cancer-associated fibroblasts showing enrichment of gene sets involved in negative regulation of protein kinase activity and negative regulation of endothelial cell proliferation, tumor microenvironment (TME) was dominated by infiltration of TNFRSF4+/TNFRSF18+/CTLA4+ regulatory T cells (Treg) and depletion of CD8+ cytotoxic T cells (TC) and γδTC. Majority of mucosa-associated lymphoid tissue B cells (BCs) and follicular BCs were detected within tumor tissue, which was associated with CXCL13 overexpressed in intratumoral Tregs and CD4+ memory TCs. Coordination of components of the TME towards immune evasion is governed by Tregs from the onset of lung cancer, requiring unremitting efforts to target and overcome them. This provision of information on changes in cancer cell-specific biomarkers and TME using early lung cancer from never smokers will provide new insight into early lung carcinogenesis and useful targets for treatment.
Introduction
Lung cancer is the leading cause of cancer-related death, and the majority of lung cancers are directly or indirectly related to smoking [1,2]. In recent years, lung cancer in never smokers has ranked among the top 10 causes of cancer deaths in both men and women and significantly differs in pathogenesis and clinical aspects from lung cancer in smokers [2,3]. Adenocarcinoma is the most dominant histology in lung cancer in never smokers, and early lung adenocarcinoma in never smokers often presents in the form of ground-glass nodules (GGNs) on chest CT scans.
GGN is defined as pulmonary parenchymal blurred opacity seen on chest high-resolution computed tomography (HRCT) that does not obscure the underlying bronchi and pulmonary vascular structures [4,5]. Increasing interest in health screening has resulted in the widespread application of low-dose chest CT scans; thus, the detection rate of GGN is increasing, as is the diagnosis rate of early lung cancer. As GGNs are radiologically defined lesions, they encompass benign lesions, including lesions that result from inflammation and parenchymal hemorrhage. However, persistent GGN, which remains unchanged for more than 3 months, indicates potential malignancy and pathologically encompasses a spectrum of lesions ranging from atypical adenomatous hyperplasia to invasive lung adenocarcinoma. As early lung adenocarcinomas present in the form of GGN, it is a good model investigating early lung carcinogenesis [6].
Single-cell transcriptomic analysis is a powerful tool, enabling assessment of the heterogeneity of the tumor microenvironment (TME) and progression of lung cancer even in early lesions such as GGNs. To study early lung carcinogenesis and changes in the TME, early lung adenocarcinomas in the form of GGNs were explored by single-cell transcriptomic analysis and compared with corresponding adjacent normal-appearing nonmalignant lung tissues (referred to as normal lung tissues hereafter). When lung cancer cells were projected onto the trajectories of normal lung epithelial cell clusters, they were observed around the trajectories of differentiated type I or type II alveolar cells. Compared with normal lung tissue, the differentially expressed gene (DEG) set of lung cancer cells showed enrichment of gene sets related to vacuole/vesicle processing and surfactant homeostasis. Even in this early-stage lung cancer microenvironment, we observed depletion of CD8+ T cells (TCs), γδ TCs, and NK cells whereas enrichment of regulatory T cells (Tregs), and B cell (BC) subclusters comparing to the normal lung tissue. The emergence of cancer associated fibroblast (CAF) and immature endothelial cell (EC) populations with decrease in tip–like EC population facilitate the formation of TME with immune evasion properties.
Materials and methods
Details on the materials and methods are provided in an online data supplement entitled ‘Detailed experimental methods’
Study cases and ethical approval
Samples were obtained from patients who visited affiliated hospitals of Yonsei University from 2019 to 2020 for treatment for persistent pure or part-solid GGN. The detailed inclusion criteria were as follows: (1) part-solid or pure GGN on the chest CT lung window setting, (2) no evidence of metastasis on additional staging tests, PET or brain MRI, (3) no prior history of cancer, (4) never smoker according to the CDC's National Health Interview Survey Glossary, and (5) patients who consented to provide residual tumor and adjacent normal lung tissue samples. Patients who received adjuvant chemotherapy or neoadjuvant chemotherapy were excluded (Table 1). This study was approved by the IRB of our institution (IRB #3-2017-5509).
Table 1.
Characteristics of the study cases.
| Case_01 | Case_02 | Case_03 | Case_04 | Case_05 | Case_06* | |
|---|---|---|---|---|---|---|
| Age | 60 | 76 | 71 | 73 | 68 | 67 |
| Gender | F | F | F | M | F | F |
| Smoking | Never | Never | Never | †Never | Never | Never |
| Stage (AJCC 8th Ed) | pT2aN0M0 | pT1aN0M0 | pT1bN0 | pTmiNx | pT1bN0 | pT1aN0M0 |
| Total size of tumor (cm) | 4.0 × 2.5 × 2.9 | 2.0 × 1.7 × 0.9 | 1.9 × 1.2 × 1.2 | 1.8 × 1.2 × 1.0 | 2.3 × 1.8 × 1.5 | 1.4 × 1.0 × 0.7 |
| Diagnosis | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma |
| Histologic component | Acinar 85%, lepidic 10%, micropapillary 5% | Lepidic 60%, acinar 40% | Lepidic 70%, acinar 30% | Lepidic 90%, acinar 10% | Lepidic 80%, acinar 20% | Lepidic 70%, acinar 30% |
| ⁎⁎ EGFR mutation | L858R | L858R | L858R | Wild type | Wild type | E19del |
Localization with lipiodol was performed prior to the lung resection.
PANAMutyper™ R EGFR mutation detection kit (real time PCR).
Had smoked temporarily in his early 20 s.
Sample preparation, gel bead in emulsion (GEM) and library construction, and sequencing
Paired samples of tumor and adjacent normal lung tissue were obtained from 5 patients, and only lung tumor tissue was obtained from 1 patient, resulting in a total of 11 samples for the study. The specimens obtained in the operating room were transferred to the pathology laboratory and examined by frozen sectioning to confirm lung cancer. Fresh tumor tissue and normal lung tissue more than 2 cm from the tumor were cut into 0.5 × 0.5 × 0.5 cm3-sized cubes and processed by a pathologist (YJ Cha). The specimens were placed in a MACS Tissue Storage Solution at 4 °C, transferred macrogen Korea® ( Seoul, Republic of Korea, https://www.macrogen.com/en/company/summary.php), and then processed according to companies standardized protocol based on the single cell expression protocol of 10X GENOMICS®. Briefly, the specimens were processed into a single-cell suspension as described elsewhere using a gentleMACS Octo Dissociator with Heater and a Multi Tissue Dissociation Kit 1 (Miltenyi Biotec) [7]. GEMs and the library were created by barcoding up to 10,000 cells from each sample using the Chromium Single Cell 3′ GEM, Library & Gel Bead Kit v3 Protocol. After capturing polyadenylated mRNA using a poly (dT) primer, barcoded full-length cDNA was generated, and paired-end sequencing was performed with the Illumina sequencing system. Using the Cellranger mkfastq, count, and aggr modules, raw base call (BCL) files were demultiplexed into a FASTQ file, and sorting, filtering, barcode counting, and UMI counting were performed.
Analysis tools
The data obtained in the laboratory were analyzed using the Seurat R package version 3.2.2. The barcodes of the cells with perturbation of chromosomal gene expression were secured through InferCNV package version 1.6.0. Briefly, (i) the cells obtained from tumor and adjacent normal lung tissue in a patient were independently clustered. (ii) We extracted epithelial cell barcodes obtained from tumor tissues, (iii) designated the cell clusters obtained from normal lung tissues and the nonepithelial cell clusters obtained from tumor tissues as the reference, (iv) applied a nonsupervised clustering method in the InferCNV package, and (v) designated cells with clear chromosomal gene expression perturbation as lung cancer cells. Details on the annotation of individual clusters are described in the ‘Annotation of cluster and subclusters’ part of DETAILED EXPERIMENTAL METHODS. Selected gene sets were classified and analyzed according to functional gene ontology (GO) biological process gene sets using Enrichr (https://maayanlab.cloud/Enrichr/) and ToppGene (https://toppgene.cchmc.org/). For the trajectory, Monocle package version 2.18.0 was used.
Data availability and correspondence
The raw single cell RNA sequencing data along with their associated metadata were deposited in Sequence Read Archive (SRA) under the accession number PRJNA773987. The metadata of each dataset corresponds to information of the Table 1 in the manuscript. The codes generated during this study are available at Github (https://github.com/mbgld/SINGLE). Please contact Y.S.C for further correspondence regarding requests for materials and data.
Statistical analysis
The differentially expressed genes between the two clusters of interest were calculated using the Wilcoxon rank sum test, which is the default option of Seurat V 3.2.2, and the adjusted p-value was obtained using Bonferroni correction. The difference in distribution between the normal and tumor tissue of the cluster of interest was calculated by dividing the number of cells belonging to individual subclusters by the total number of cells belonging to the subcluster of the corresponding case and then compared using the unpaired Wilcoxon rank sum test.
Results
Single-cell sequencing of tumors and paired normal lung tissues
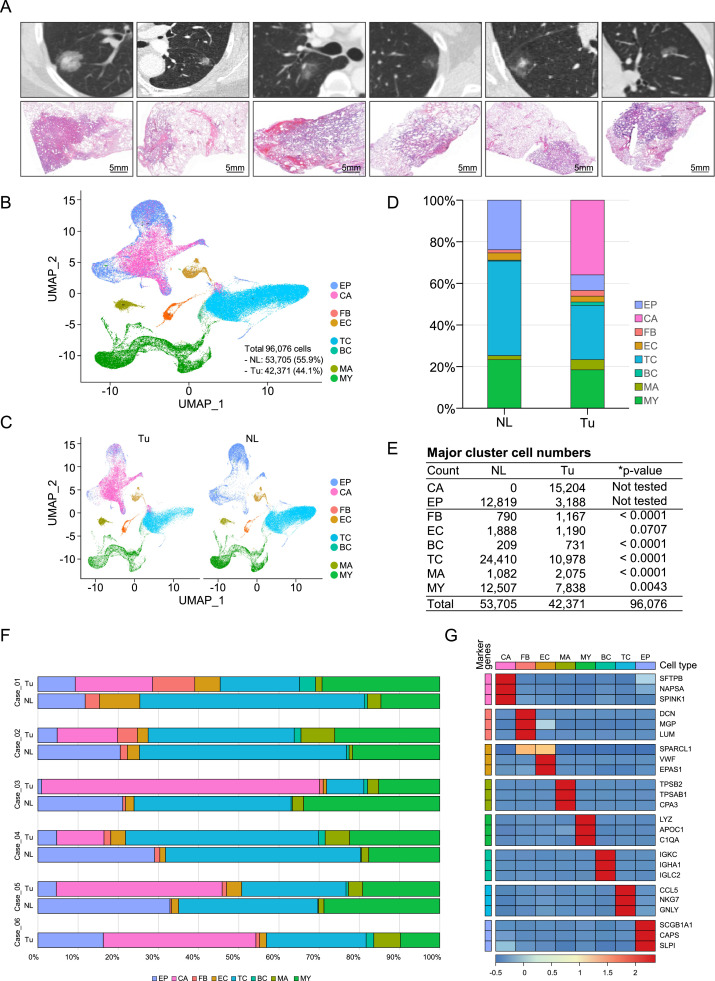
Single-cell sequencing was conducted using 11 specimens obtained from a total of 6 subjects. The inclusion criteria and exclusion criteria for the recruited patients are described in the Materials and Methods, and the clinical characteristics, chest CT and low-magnification slide images of individual patients are shown in Table 1 and Fig. 1A, respectively. The QC parameters for single-cell sequencing are shown in Supplementary Table 1. After the filtering process, the number of cells obtained from normal lung tissue was 53,705 (55.9%) and that from tumor tissue was 42,371 (44.1%). A total of 96,076 cells were initially divided into 7 major cell groups through dimensional reduction and classification, and each cluster was annotated by comparing markers representing each cluster obtained from Seurat's FindConservedMarkers function and known canonical markers of lung cells [7], [8], [9] (Fig. 1B,C, Supplementary Fig. 1). In the obtained epithelial cell cluster, barcodes of lung cancer cells were secured using the InferCNV package, and then the remaining cells were annotated as epithelial cells. For refinement of lung cancer cells, clusters with epithelial features were obtained from each tumor tissue, and then the cells that had distinct genetic aberrations were defined as lung cancer cells by InferCNV using cells obtained from normal lung tissue and nonmalignant cells obtained from tumor tissue of the same patient as references (Supplementary Figs. 2, 3) [10,11]. T cells occupied the largest proportion among the obtained cell clusters, followed by epithelial cells, including cancer cells and myeloid cells (Fig. 1D–G).
Fig. 1.
Overview of the dataset used in this study, single-cell RNA sequencing analysis, and clustering of cells from Tu and NL. (A) Representative lung HRCT image (top) of each patient included in the study and the corresponding low-magnification micrographs (bottom). (B) UMAP of 96,076 cells clustered and colored by major lung cell type and (C) by tissue origin. (D) Stacked column chart and (E) table showing the number of cells belonging to the major cell cluster. The p-value was obtained by prop.test dividing the number of cells belonging to each cell cluster in NL and Tu by the total number of cells in NL and Tu, respectively. *Note that the number of EP and CA was excluded from statistical analysis. (F) Horizontal bar plots showing the number of cells belonging to each cluster split by tissue origin. (G) Heatmap showing the top three marker genes representing each major cell cluster. NL; normal lung tissue, Tu; tumor, EP; epithelial cell, CA; cancer cell, FB; fibroblast, EC; endothelial cell, TC; T cell, BC; B cell, MA; mast cell, MY; myeloid cell.
Lung cancer cells are located close to terminally differentiated lung epithelial cell clusters
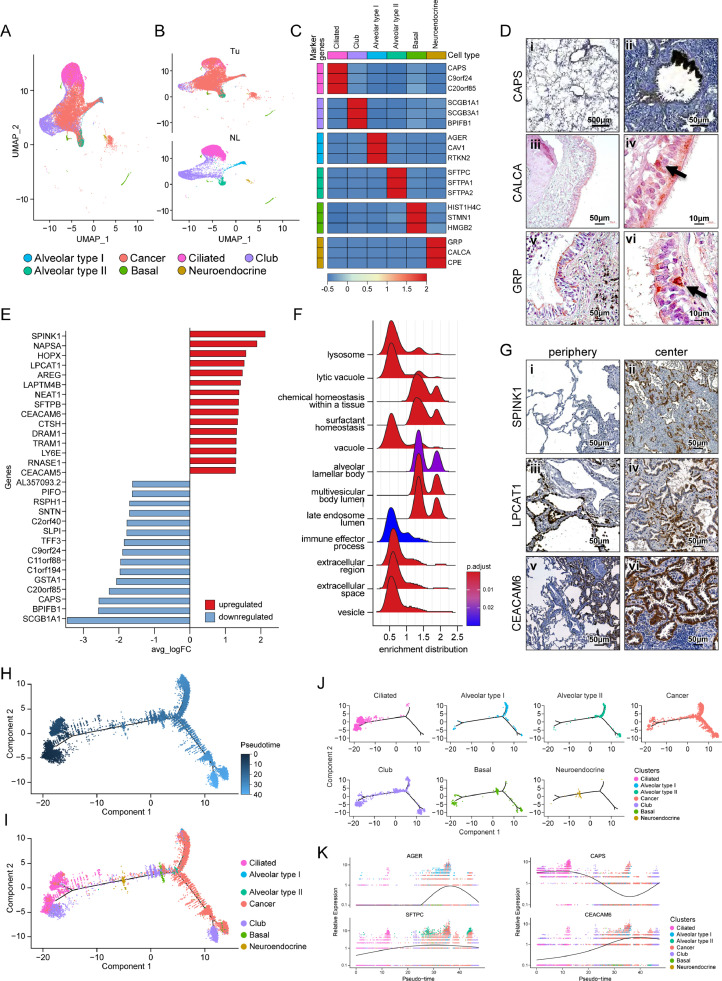
Lung epithelial cells were further characterized after subclustering after removal of the cancer cells from all epithelial cells of merged tumor tissues and normal lung tissues (Fig. 2A–C). Among the epithelial cells, the prominent cell type was ciliated bronchial epithelial cells, characterized by overexpression of CAPS, C9orf24, and C20orf85 (Fig. 2D i,ii), followed by secretory club cells, characterized by expression of SCGB1A1, SCGB3A1, and BPIFB1. The respiratory bronchioles and alveoli are mainly composed of terminally differentiated cell groups: type I alveolar cells and type II alveolar cells. Type I alveolar cells showed overexpression of unique genes such as AGER, CAV1, and RTKN2, whereas type II alveolar cells showed cell fractions that were similar to those in type I alveolar cells and characterized by overexpression of surfactant family proteins such as SFTPC, SFTPA1, and SFTPA2. Interestingly, a few cells with neuroendocrine features overexpressing unique genes, such as GRP, CALCA, and CPE were observed and were presumed to be pulmonary neuroendocrine cells. When it was further evaluated by immunohistochemical (IHC) staining, a few flask-shaped cells strongly stained with GRP and CALCA adjacent to the basement membrane of the bronchial epithelium could be observed (Fig. 2D iii–vi).
Fig. 2.
Lung cancer cells were located amid the lung epithelial cell subclusters. (A) UMAP of lung cancer cells and lung epithelial cell subclusters (B) colored according to tissue origin. (C) Heatmap showing the representative genes of each lung epithelial cell subcluster. (D) Expression of CAPS in pulmonary alveoli (i) and bronchial epithelia (ii). Expression of CALCA (iii, iv) and GRP (v, vi) in normal lung bronchial epithelium (black arrows). (E) Horizontal bar plot of the 15 top and bottom differentially expressed genes between lung cancer cells and the other lung epithelial cell populations. (F) A ridge plot showing the enriched pathways of differentially expressed genes overexpressed in lung cancer cells compared to lung epithelial cells. (G) Expression of SPINK1 (i, ii), LCAT1 (iii, iv), and CEACAM6 (v, vi) in the periphery (left panels) and center (right panels) of GGNs. Periphery indicates a border between NL and Tu, and center indicates inside tumor. (H) Unsupervised trajectory plot of lung cancer cells and lung epithelial cell subclusters according to pseudotime and (I) the same plot including cell subclusters colored by cell type. (J) Unsupervised trajectory plot of lung cancer cells and epithelial cell subclusters split by cell type. (K) Plot showing changes in AGER, CAPS, SFTPC, and CEACAM6 gene expression according to pseudotime.
In our UMAP model, lung cancer cells clustered in the center of ciliated cells, club cells, alveolar type I cells and alveolar type II cells (Fig. 2A,B). The top 15 most DEGs between lung cancer cells and normal lung epithelial cells are shown in Fig. 2E, and the entire gene set is shown in Supplementary Table 2. These DEGs were enriched for secretory vesicle and surfactant pathways, suggesting that the origin of early lung carcinogenesis is closely related to type II alveolar cells (Fig. 2F). Lung cancer cells share many markers with type II alveolar cells and fewer markers with type I and ciliated bronchial cells. Among the top DEGs, IHC staining was performed by selecting those that did not overlap with the common marker of type II alveolar cells (Fig. 2G). SPINK1 prevents trypsin-catalyzed premature activation of zymogens, and LPCAT1 converts lysophosphatidylcholine to phosphatidylcholine in the presence of acyl-CoA. Both were specifically stained in the cancer cells at the border between normal and cancer tissues and inside the tumor. Lung cancer cells also overexpress the CEACAM6 and CEACAM5 surface glycoproteins, which play a role in intercellular adhesion in a calcium- and fibronectin-independent manner [12]. CEACAM6 staining was not present on the components of normal lung tissues, but it was strongly positive in lung cancer cells regardless of histologic subtype, suggesting its possible involvement in early tumorigenesis. When DEGs obtained by comparing lung cancer cells with normal lung epithelial cells were analyzed using KEGG (https://www.genome.jp/kegg/pathway.html), significant enrichment of ERBB signaling pathways and apoptotic pathways was observed (Supplementary Fig. 4).
When the lung cancer cells were projected onto the trajectories of the normal lung epithelial cell group, they traced either on type I alveolar cells or type II alveolar cells. Club cells were observed in all processes according to pseudotime of differentiation between bronchial cells and alveolar cells, whereas neuroendocrine cells were observed at the branching time point from ciliated cells to alveolar cells (Fig. 2H–J). When representative genes of major lung epithelial cell clusters were aligned according to pseudotime, concurrency was observed between the genes related to surfactant homeostasis, such as SFTPC, and cancer-specific genes, such as CEACAM6 (Fig. 2K), suggesting that the surfactant-related pathway is strongly involved in early lung carcinogenesis.
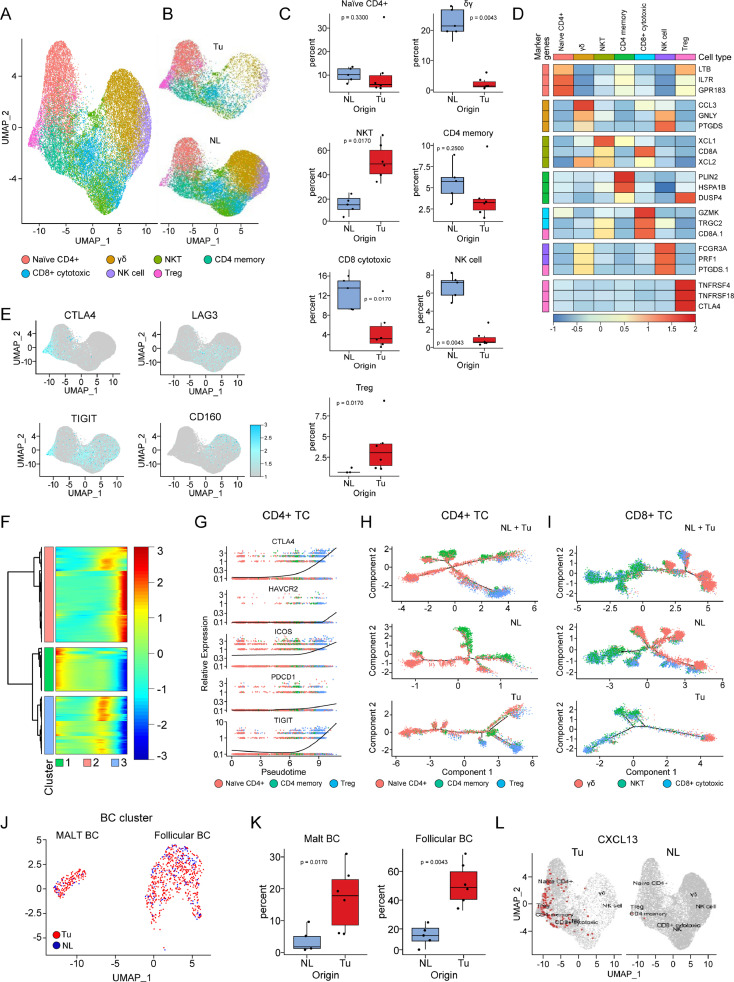
Depletion of CD8+ TCs and γδ TCs and enrichment of Tregs and BCs in the early TME
TCs are a group of cells with a very heterogeneous distribution between tumor and normal lung tissues. All TC subtypes, except Tregs characterized by TNFRSF4+/TNFRSF18+/CTLA4+, were less frequently detected in tumor tissues than in normal lung tissues (Fig. 3A-D). Immune fatigue markers showed different expression patterns according to TC type and associated cluster; CTLA4 expression was commonly observed in CD4+ TCs and Tregs, whereas LAG3 expression was commonly observed in CD8+ TCs, and HAVCR2, CD244, and CD160 expression was commonly observed in γδ TCs and natural killer (NK) cells (Fig. 3E). Compared to normal lung tissues, tumor tissues had an overwhelming number of Tregs; these Tregs had distinct expression of CTLA4 and TIGIT, showing an exhausted phenotype (Fig. 3E). In the trajectory analysis with CD4+ TCs, we identified 3 pseudotime-dependent subclusters (Fig. 3F). When the CD4+ TC clusters were aligned according to the pseudotime designating the naïve CD4+ TCs as the root state, Tregs were located in the last stage of differentiation and possessed immune fatigue phenotypes (Fig. 3G). Within tumor tissue, Tregs differentiated from naïve CD4+ TCs and fully developed at the end of the trajectory as an independent cluster, whereas those inside normal tissue showed no significant flow with scant numbers (Fig. 3H). The trajectory analysis of cytotoxic TC clusters within normal tissue, which included cytotoxic CD8+ TCs, γδ TCs, and NKT cells, and setting CD8+ TCs as the root state, showed gradual and even distribution of the cellular composition throughout the pseudotime (Fig. 3I). Whereas trajectory of these cytotoxic TC clusters inside tumor tissue showed small number of terminally differentiated γδ TCs.
Fig. 3.
Disturbance of the distribution of lymphocytes residing in lung adenocarcinoma begins as early as the appearance of GGNs. (A) UMAP of TC and related cell clusters divided by color and (B) by tissue origin. (C) Box plots showing the distribution of individual TC subtypes in NL and Tu. Each dot represents the fraction obtained by dividing the number of cells belonging to individual TC subclusters from a case by the total number of TCs obtained from the corresponding case. The p-value was obtained using a two-sided unpaired Wilcoxon rank sum test. (D) Heatmap showing the representative genes of TC clusters. (E) DimPlots of TC clusters projected by individual exhaustion makers. (F) Heatmap showing the change in total gene expression in CD4+ TCs according to pseudotime. Note that CD4+ TCs are classified into three groups. (G) Plot showing the change in gene expression related to immune cell exhaustion in CD4+ TCs according to pseudotime. (H) Unsupervised trajectory plot of all CD4+ TCs (top), those from NL tissues (middle), and those from Tu tissues (bottom) colored by cell type. (I) Unsupervised trajectory plot of all CD8+ TCs (top), those from NL tissues (middle), and those from Tu tissues (bottom) colored by cell type. (J) UMAP showing subclusters of BC colored by tissue origin. (K) Box plots showing the distribution of individual BC subtypes in NL and Tu. Each dot represents the fraction obtained by dividing the number of cells belonging to individual BC subclusters from a case by the total number of BCs obtained from the corresponding case. The p-value was obtained using a two-sided unpaired Wilcoxon rank sum test. (L) A dimension plot projecting CXCL13-expressing cells on the TC clusters divided by tissue origin. Tu; tumor, NL; normal lung tissue, TC; T cell, BC; B cell.
BCs were more strongly enriched in tumor tissues than in normal lung tissue. In this study, the 940 BCs analyzed comprised 2 subtypes of BCs (follicular and mucosa-associated lymphoid tissue (MALT) BCs). Of these BCs, 77.8% were found in tumor tissues, making them the most prominent cells enriched in tumor tissues among the immune cells found in the lung tissues (Fig. 3J). Although follicular BCs showed a relatively high proportion compared to the MALT BCs, their degree of elevation in the proportion of follicular BCs in tumors compared to normal lung tissues was similar to that of MALT BCs (Fig. 3K). The BC chemoattractant CXCL13, known to be secreted from cancer cells, follicular dendritic cells (DCs), and T follicular helper cells, may be related to BC influx in tumor tissues [13,14]. When the cell population overexpressing CXCL13 were examined in our dataset, majority of them were detected in CD4+ TC cells located inside tumor tissue, especially in Treg and CD4+ memory TCs. (Fig. 3L, Supplementary Fig. 5A).
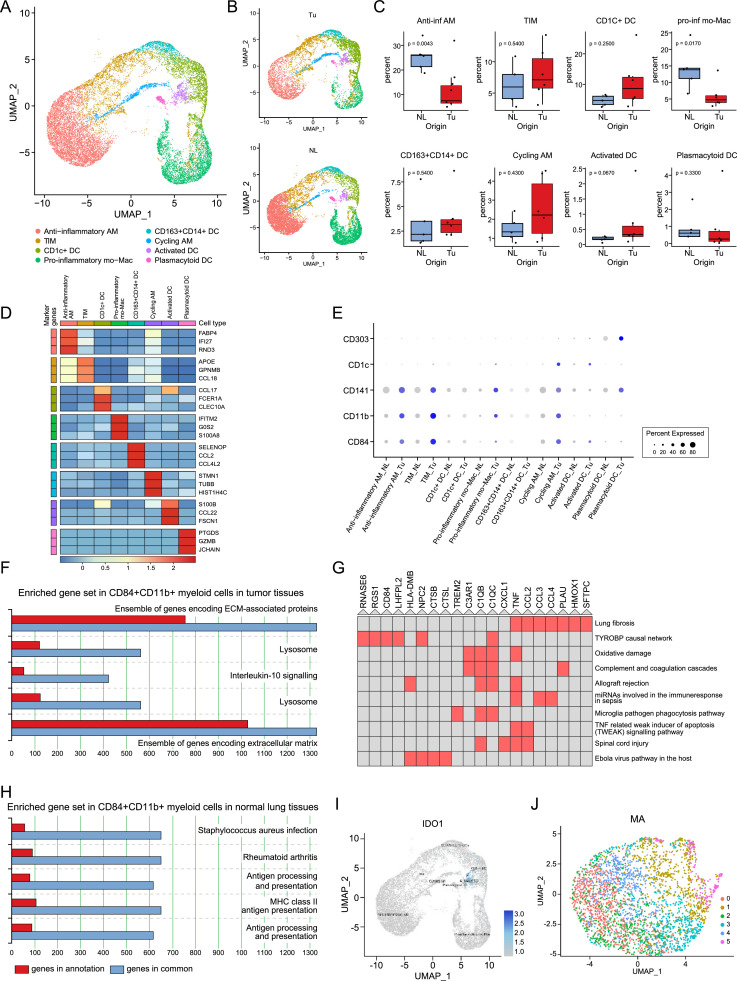
Myeloid cells show an immunosuppressive immature phenotype in the TME
In this analysis, 20,345 myeloid cells were recovered and grouped into eight clusters (Fig. 4A–D). Anti-inflammatory alveolar macrophages were significantly decreased in the tumor tissue compared with the normal lung tissue. In UMAP, tissue-infiltrating macrophages (TIMs) were located adjacent to anti-inflammatory alveolar macrophages and on the opposite side of pro-inflammatory monocyte-derived macrophages. When TIMs inside tumors, tumor-associated macrophages (TAMs), in other words, were compared to macrophages in normal lung tissues, they were enriched in genes related to leukocyte chemotaxis (Supplementary Fig. 5B).
Fig. 4.
Immunosuppressive immature myeloid cells make the tumor microenvironment prone to tumor progression. (A) UMAP of MY clusters divided by color and (B) by tissue origin. (C) Box plots showing the distribution of individual MY subtypes in NL and Tu. Each dot represents the fraction obtained by dividing the number of cells belonging to individual MY subclusters from a case by the total number of MY obtained from the corresponding case. The p-value was obtained using a two-sided unpaired Wilcoxon rank sum test. (D) Heatmap showing representative genes of the MY subcluster. (E) Dot plot of representative markers for myeloid-derived suppressor cells (CD84 and CD11b) and dendritic cells (CD141, CD1c, and CD303). (F, H) Pathway enrichment analysis results visualized by ToppGene. The top 5 pathways with P-value < 0.001 are shown and numbers mean total genes in term. Genes overexpressed in CD84+CD11b+ myeloid cells in Tu (F) and NL (H). (G) Cluster grams of gene pathways enriched in CD84+CD11b+ myeloid cells in Tu identified using Enrichr's WikiPathways 2019 Human. The red colored box denotes that p-value < 0.001. (I) A plot showing IDO1 overexpression in myeloid cells. (J) UMAP of mast cells. MY; myeloid cell, AM; alveolar macrophage, TIM; tissue-infiltrating macrophages, mo-Mac; monocyte-derived macrophage, DC; dendritic cell, Tu; tumor, NL; normal lung tissue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Myeloid-derived suppressor cells (MDSCs) are of myeloid origin, harbor an immunosuppressive function, and reside in a cancer-related context [15]. Along with CD11b+, we adopted CD84+ as an additional marker, which has been shown to improve the detection of MDSCs through single-cell RNA sequencing analysis of human tissue [16]. CD84+CD11b+ myeloid cells were a rare heterogeneous population scattered in macrophage and DC clusters (Supplementary Fig. 5C). Macrophages overexpressing the CD84 and CD11b genes were more scattered in tumor tissues than in normal tissues. (Fig. 4E). When the enriched genes were subjected to GO analysis with ToppGene (https://toppgene.cchmc.org/) and Enrichr (https://maayanlab.cloud/Enrichr/), the genes enriched in the CD84+CD11b+ myeloid cells from the tumor tissues were related to the extracellular matrix, pulmonary fibrosis and IL-10 signaling (Fig. 4F,G, Supplementary Table 3), indicating that CD84+CD11b+ myeloid cells inside the tumor produce tumor-promoting cytokines (IL-10) and make the TME more permissive for tumor progression through alteration of extracellular matrix composition. On the other hand, the CD84+CD11b+ myeloid cells in normal lung tissue showed enrichment of genes related to host protection from infection and inflammation (Fig. 4H, Supplementary Table 4). DCs clustered into four groups, and among them, IDO1, which inhibits T-cell proliferation by degrading tryptophan, was more enriched in activated DCs in tumor tissues than in normal lung tissues (Fig. 4I). Mast cells were separated from myeloid cells and formed a unique and very homogenous population, so there were no significant DEGs between individual subclusters (Fig. 4J).
CAFs are associated with the disruption of normal vascular structures
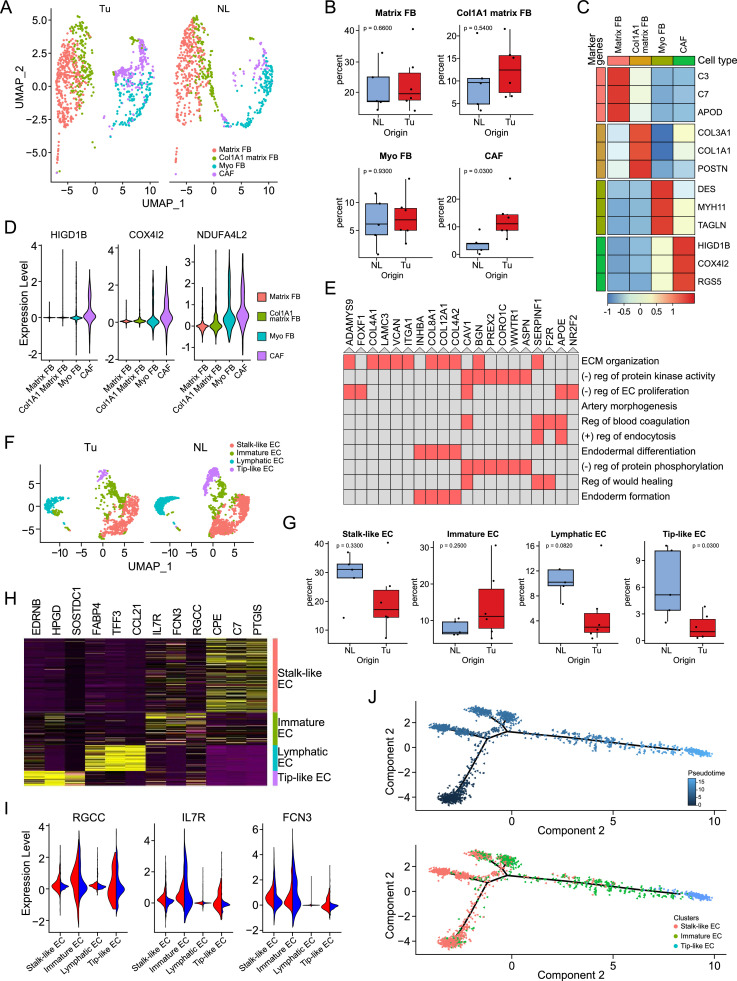
Fibroblasts (FBs) were the second most enriched cell type in tumor tissues than in normal lung tissue in this GGN-type early lung cancer. FBs were classified into four subtypes: matrix FBs, COL1A1+ FBs, myofibroblasts (myo FBs) and CAFs (Fig. 5A–C). Among them, the unique fibroblast group called CAFs showed higher expression of HIGD1B, COX4I2, and RGS5 than other FB clusters. The CAF clusters showed significant overexpression of hypoxia-related genes compared to other FB clusters (Fig. 5D). When the genes enriched in these clusters were subjected to GO analysis with Enrichr (https://maayanlab.cloud/Enrichr/), gene sets related to negative regulation of protein kinase activity and negative regulation of endothelial cell proliferation were enriched, and those associated with extracellular matrix organization were enriched (Fig. 5E, Supplementary Table 5).
Fig. 5.
Cancer-associated fibroblasts (CAFs) interfere with the differentiation of vascular structures inside tumors. (A) UMAP of FB split by tissue origin. (B) Box plots showing the distribution of individual FB subtypes in NL and Tu. Each dot represents the fraction obtained by dividing the number of cells belonging to individual FB subclusters from a case by the total number of FBs obtained from the corresponding case. The p-value was obtained using a two-sided unpaired Wilcoxon rank sum test. (C) Heatmap showing the representative genes of FB subclusters. (D) Feature plot comparing the expression of CAF signature genes related to hypoxia among the fibroblast subclusters. (E) Cluster grams of gene pathways enriched in CAFs identified using Enrichr's GO Biological Process 2018. The red colored box denotes that p-value < 0.001. (F) UMAP of EC clusters divided by tissue origin. (G) Box plots showing the distribution of individual EC subtypes in NL and Tu. Each dot represents the fraction obtained by dividing the number of cells belonging to individual EC subclusters from a case by the total number of ECs obtained from the corresponding case. The p-value was obtained using a two-sided unpaired Wilcoxon rank sum test. (H) Heatmap showing representative genes of each subcluster of EC. (I) Feature plots of RGCC, IL7R, and FCN3 in the EC clusters split by tissue origin. (J) Unsupervised trajectory plot of ECs according to pseudotime (top) and the same plot including cell subclusters colored by cell type (bottom). FB; fibroblast, Myo FB; myofibroblast, CAF; cancer-associated fibroblast, EC; endothelial cell, Tu; tumor, NL; normal lung tissue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Not irrelevant to this finding, the EC cluster was adjacent to the FB cluster, and fewer cells were found in EC clusters than in FB clusters in tumor tissue (Fig. 5F–H). Interestingly, in tumor tissues, an increased number of undifferentiated EC clusters located in the middle of stalk-like and tip-like ECs were observed in UMAP, with a strikingly decreased number of tip-like ECs. These cells were characterized by RGCC, IL7R, and FCN3 expression (Fig. 5I), and when compared with the other EC clusters, this cluster showed enrichment of gene sets related to cellular locomotion and motility as well as gene sets related to vascular development and tube morphogenesis (Supplementary Table 6). When trajectory analysis was conducted to further confirm their characteristics, it was shown that their location between the stalk-like EC and the tip-like EC clusters was closer to the stalk-like EC cluster (Fig. 5J). Taken together, these findings suggest that there is an undifferentiated vascular cell cluster between stalk-like ECs and tip-like ECs and that CAFs inhibit EC differentiation into tip-like ECs.
Discussions
The natural course of GGN has been studied from a radiology point of view, but investigations on the molecular and biological aspects are very limited. GGN-type lung cancer is a perfect model for studying the initiation of lung carcinogenesis, and in this study, paired samples were obtained from never smokers and those who had quit smoking long before the study to observe the subtle changes that occur in early lung cancer and its microenvironment.
Discriminating normal lung cells and lung cancer cells was a very important part of this study. Traditionally, lung cancer is diagnosed by identifying the characteristic morphology of the cells. Non-small cell lung cancer (NSCLC) cells have ample cytoplasm and several inconspicuous nucleoli, whereas early lung cancer cells have less cellular atypia, leading to diagnostic difficulties. The definition of lung cancer cells by single-cell transcriptomic analyses is slightly different between studies. In the earlier studies of Lambrechts et al., EPCAM-overexpressing epithelial cells obtained from tumor tissue were defined as lung cancer cells [8,17]. Kim et al. defined epithelial cells obtained from tumor tissue as lung cancer cells that showed perturbations in their CNV signal > 0.02 mean squares or > 0.2 CNV correlation [9]. Comparing the earlier methods, the strategy we described in the materials and methods made it easier to interpret tumor biomarkers, trajectories and DEG results by accurately discriminating cancer cells from other nonmalignant epithelial components within the tumor tissue. Interestingly, cancer cells obtained from never smokers with early lung cancer not only showed heterogeneity between samples but also clearly showed CNV heterogeneity within individual samples (Supplementary Fig. 3). The clinical implications of CNV heterogeneity within an individual sample are currently unknown, but significant findings could be obtained in the near future.
Among the top DEGs obtained by comparing lung cancer cells with other lung epithelial clusters, there were SPINK1, CEACAM6, and CEACAM5 as biomarkers distinguishing them from type II alveolar cells. SPINK1 is a trypsin inhibitor secreted from pancreatic acinar cells and protects the pancreas by inhibiting trypsin-mediated premature activation of zymogens in the pancreas [18]. Previous studies on SPINK1 mainly focused on the diseases of the pancreas [19], but recently it has been found that its expression is increased in the tissues or blood of many carcinomas including ovary [20], prostate [21], liver [22], and breast [23]. SPINK1, which acts as a growth factor or inhibitor of apoptosis, draws attention as a diagnostic or therapeutic target for various carcinomas, despite of concerns that inhibition may promote pancreatitis (Reviewed by Rasanen [24])
Among the proteins belonging to the 12 CEACAM family located on Chromosome 19q133.1-13.2 [25], CEACAM6 along with CEACEM5, also known as CEA, were top DEGs which is specifically overexpressed in lung cancer cells. The mechanism by which CEACAM6, which does not have an intracellular domain, inhibits anoikis is still not well known, but overexpression of CEACAM6 is presumed to activate the EGFR pathway [26] or the Src-FAK pathway [27]. Researches and clinical trials targeting the CEACAM family protein are being actively conducted in several carcinomas including colorectal cancer, and additional evidence and in-depth consideration are needed to properly apply them to lung cancer.
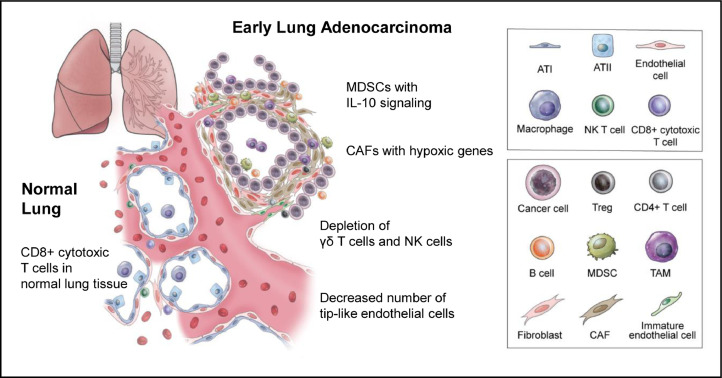
Among the cells that make up the TME, cell clusters with high heterogeneity are in the order of TC, BC, FB and EC. CD84+CD11b+ myeloid cells play a role as a propagator of the heterogeneity of the TME, promoting the formation of CAFs, and CAFs inhibit normal vascular formation and further affect lymphoid infiltration. BCs and TCs showed the most remarkably different distribution between tumor and normal lung tissues (Fig. 6).
Fig. 6.
A schematic view of the tumor microenvironment components of early lung adenocarcinoma. Even in very early lung adenocarcinoma, tumors are surrounded by infiltrating immune cells and a wide range of stromal cells, such as MDSCs, TAMs, Tregs, B cells, CAFs, and immature endothelial cells, which are prone to immune evasion. ATI; alveolar type I cell, ATII; alveolar type II cell, Treg; regulatory T cell, MDSC; myeloid-derived suppressor cell, TAM; tumor-associated macrophage, CAF; cancer-associated fibroblast.
BCs were most strongly enriched inside tumor tissues compared to normal lung tissues. It has been consistently reported that an increase in BC population in lung cancer tissue has a favorable effect on long-term prognosis ([28], [29], [30], [31], Reviewed by Wang et al. [13]). A strong BC attractant CXCL13, is mainly secreted from follicular helper TC, that locates in the periphery of the BC follicle of secondary lymphoid organs and involved in the proliferation of BC and the development of antigen-specific BC immunity [32]. In this study, overexpression of the CXCL13 gene was observed in Treg and CD4+ memory TCs inside tumor tissue, that make it interesting to further investigate the relationship between follicular helper TC and Treg and CD4+ memory cells in lung cancer tissue. By doing, not only the origin of Treg, which is the only TC cluster increased in the lung cancer, but also the therapeutic targets that is related to BC infiltration and to the regulation of other cytotoxic TC component would be discovered.
Recently, two relevant articles were published by Chinese researchers [33,34]. Their studies differ from ours in the point that they did not perform paired sequencing of normal-tumor tissue obtained from the same individual. Lu et al. compared the difference in TME between minimally invasive adenocarcinoma and solid adenocarcinoma of stage I without data from normal appearing adjacent lung tissue [33]. In order to overcome the defect caused by the absence of results obtained from paired normal appearing lung tissues, Xing et al. used the opened data from Lambrecht's et al. as normal lung tissue control [8,34]. However, the majority of the tumor tissues used by Xing et al. were obtained from never smokers, whereas the normal lung tissue data from opened source were all obtained from Ex or active smokers. We believe that these factors have been put together to provide evidence that can more accurately examine changes in the TME of never smoker lung adenocarcinoma.
In conclusion, even in GGN-type early lung cancer, there was both intertumor and intratumor heterogeneity, which were estimated by InferCNV. In addition, we also found changes in the microenvironment in tumor tissues, such as the formation of tumor-specific subclusters, that were not observed in normal lung tissues and differences in the proportions of cellular components between tumor and normal lung tissues. Coordination of components of the TME towards immune evasion is governed by TNFRSF4+/TNFRSF18+/CTLA4+ Tregs from the onset of lung cancer, requiring unremitting efforts to target and overcome them. The provision of information on changes in cancer cell-specific biomarkers and TME using early lung cancer from never smokers will provide new insight into early lung carcinogenesis and useful targets for treatment.
Funding
This study was supported by the National Research Foundation of Republic of Korea (Grant No. NRF-2020R1A2B5B01001883) awarded to Y. S. C.
CRediT authorship contribution statement
Eun Young Kim: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Yoon Jin Cha: Data curation, Formal analysis, Investigation, Resources, Software, Validation, Visualization, Writing – original draft. Sang Hoon Lee: Data curation, Formal analysis, Software, Validation, Visualization. Sukin Jeong: Investigation. Young Jun Choi: Data curation, Formal analysis, Software, Validation, Visualization. Duk Hwan Moon: Resources. Sungsoo Lee: Resources. Yoon Soo Chang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
CRediT authorship contribution statement
Eun Young Kim: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Yoon Jin Cha: Data curation, Formal analysis, Investigation, Resources, Software, Validation, Visualization, Writing – original draft. Sang Hoon Lee: Data curation, Formal analysis, Software, Validation, Visualization. Sukin Jeong: Investigation. Yong Jun Choi: Data curation, Formal analysis, Software, Validation, Visualization. Duk Hwan Moon: Resources. Sungsoo Lee: Resources. Yoon Soo Chang: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101277.
Appendix. Supplementary materials
References
- 1.Islami F., Goding Sauer A., Miller K.D., Siegel R.L., Fedewa S.A., Jacobs E.J., McCullough M.L., Patel A.V., Ma J., Soerjomataram I., Flanders W.D., Brawley O.W., Gapstur S.M., Jemal A. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J. Clin. 2018;68:31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Sun S., Schiller J.H., Gazdar A.F. Lung cancer in never smokers–a different disease. Nat. Rev. Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 4.Austin J.H., Müller N.L., Friedman P.J., Hansell D.M., Naidich D.P., Remy-Jardin M., Webb W.R., Zerhouni E.A. Glossary of terms for CT of the lungs: recommendations of the nomenclature committee of the Fleischner society. Radiology. 1996;200:327–331. doi: 10.1148/radiology.200.2.8685321. [DOI] [PubMed] [Google Scholar]
- 5.Gao J.W., Rizzo S., Ma L.H., Qiu X.Y., Warth A., Seki N., Hasegawa M., Zou J.W., Li Q., Femia M., Lv T.F., Song Y. Pulmonary ground-glass opacity: computed tomography features, histopathology and molecular pathology. Transl. Lung Cancer Res. 2017;6:68–75. doi: 10.21037/tlcr.2017.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu W.S., Hong S.R., Lee J.G., Lee J.S., Jung H.S., Kim D.J., Chung K.Y., Lee C.Y. Three-dimensional ground glass opacity ratio in CT images can predict tumor invasiveness of stage IA lung cancer. Yonsei Med. J. 2016;57:1131–1138. doi: 10.3349/ymj.2016.57.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travaglini K.J., Nabhan A.N., Penland L., Sinha R., Gillich A., Sit R.V., Chang S., Conley S.D., Mori Y., Seita J., Berry G.J., Shrager J.B., Metzger R.J., Kuo C.S., Neff N., Weissman I.L., Quake S.R., Krasnow M.A. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwé H., Pircher A., Van den Eynde K., Weynand B., Verbeken E., De Leyn P., Liston A., Vansteenkiste J., Carmeliet P., Aerts S., Thienpont B. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 9.Kim N., Kim H.K., Lee K., Hong Y., Cho J.H., Choi J.W., Lee J.I., Suh Y.L., Ku B.M., Eum H.H., Choi S., Choi Y.L., Joung J.G., Park W.Y., Jung H.A., Sun J.M., Lee S.H., Ahn J.S., Park K., Ahn M.J., Lee H.O. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., Deschler D.G., Varvares M.A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J.W., Faquin W.C., Lin D.T., Regev A., Bernstein B.E. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624. doi: 10.1016/j.cell.2017.10.044. e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., Louis D.N., Rozenblatt-Rosen O., Suvà M.L., Regev A., Bernstein B.E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumenthal R.D., Hansen H.J., Goldenberg D.M. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen) Cancer Res. 2005;65:8809–8817. doi: 10.1158/0008-5472.CAN-05-0420. [DOI] [PubMed] [Google Scholar]
- 13.Wang S.S., Liu W., Ly D., Xu H., Qu L., Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2019;16:6–18. doi: 10.1038/s41423-018-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campa M.J., Moody M.A., Zhang R., Liao H.X., Gottlin E.B., Patz E.F. Interrogation of individual intratumoral B lymphocytes from lung cancer patients for molecular target discovery. Cancer Immunol. Immunother. 2016;65:171–180. doi: 10.1007/s00262-015-1787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronte V., Brandau S., Chen S.-.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alshetaiwi H., Pervolarakis N., McIntyre L.L., Ma D., Nguyen Q., Rath J.A., Nee K., Hernandez G., Evans K., Torosian L., Silva A., Walsh C., Kessenbrock K. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 2020;5:eaay6017. doi: 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu T., Yang X., Shi Y., Zhao M., Bi G., Liang J., Chen Z., Huang Y., Jiang W., Lin Z., Xi J., Wang S., Yang Y., Zhan C., Wang Q., Tan L. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov. 2020;6:69. doi: 10.1038/s41421-020-00200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin-Tóth M. Human mesotrypsin defies natural trypsin inhibitors: from passive resistance to active destruction. Protein Pept. Lett. 2005;12:457–464. doi: 10.2174/0929866054395356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haglund C., Huhtala M.L., Halila H., Nordling S., Roberts P.J., Scheinin T.M., Stenman U.H. Tumour-associated trypsin inhibitor, TATI, in patients with pancreatic cancer, pancreatitis and benign biliary diseases. Br. J. Cancer. 1986;54:297–303. doi: 10.1038/bjc.1986.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venesmaa P., Stenman U.H., Forss M., Leminen A., Lehtovirta P., Vartiainen J., Paavonen J. Pre-operative serum level of tumour-associated trypsin inhibitor and residual tumour size as prognostic indicators in stage III epithelial ovarian cancer. Br. J. Obstet. Gynaecol. 1998;105:508–511. doi: 10.1111/j.1471-0528.1998.tb10150.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang C., Wang L., Su B., Lu N., Song J., Yang X., Fu W., Tan W., Han B. Serine protease inhibitor Kazal type 1 promotes epithelial-mesenchymal transition through EGFR signaling pathway in prostate cancer. Prostate. 2014;74:689–701. doi: 10.1002/pros.22787. [DOI] [PubMed] [Google Scholar]
- 22.Lu F., Lamontagne J., Sun A., Pinkerton M., Block T., Lu X. Role of the inflammatory protein serine protease inhibitor Kazal in preventing cytolytic granule granzyme A-mediated apoptosis. Immunology. 2011;134:398–408. doi: 10.1111/j.1365-2567.2011.03498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-mezayen H.A., Metwally F.M., Darwish H. A novel discriminant score based on tumor-associated trypsin inhibitor for accurate diagnosis of metastasis in patients with breast cancer. Tumour Biol. 2014;35:2759–2767. doi: 10.1007/s13277-013-1366-y. [DOI] [PubMed] [Google Scholar]
- 24.Räsänen K., Itkonen O., Koistinen H., Stenman U.H. Emerging roles of SPINK1 in cancer. Clin. Chem. 2016;62:449–457. doi: 10.1373/clinchem.2015.241513. [DOI] [PubMed] [Google Scholar]
- 25.Johnson B., Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin. Cancer Drugs. 2015;2:100–111. doi: 10.2174/2212697X02666150602215823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang W.F., Cheng T.M., Chang C.C., Pan S.H., Changou C.A., Chang T.H., Lee K.H., Wu S.Y., Chen Y.F., Chuang K.H., Shieh D.B., Chen Y.L., Tu C.C., Tsui W.L., Wu M.H. Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation. Oncogene. 2018;37:116–127. doi: 10.1038/onc.2017.303. [DOI] [PubMed] [Google Scholar]
- 27.Duxbury M.S., Ito H., Ashley S.W., Whang E.E. CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J. Biol. Chem. 2004;279:23176–23182. doi: 10.1074/jbc.M402051200. [DOI] [PubMed] [Google Scholar]
- 28.Schalper K.A., Brown J., Carvajal-Hausdorf D., McLaughlin J., Velcheti V., Syrigos K.N., Herbst R.S., Rimm D.L. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J. Natl. Cancer Inst. 2015;107:dju435. doi: 10.1093/jnci/dju435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinoshita T., Muramatsu R., Fujita T., Nagumo H., Sakurai T., Noji S., Takahata E., Yaguchi T., Tsukamoto N., Kudo-Saito C., Hayashi Y., Kamiyama I., Ohtsuka T., Asamura H., Kawakami Y. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann. Oncol. 2016;27:2117–2123. doi: 10.1093/annonc/mdw319. [DOI] [PubMed] [Google Scholar]
- 30.Eerola A.K., Soini Y., Pääkkö P. Tumour infiltrating lymphocytes in relation to tumour angiogenesis, apoptosis and prognosis in patients with large cell lung carcinoma. Lung Cancer. 1999;26:73–83. doi: 10.1016/s0169-5002(99)00072-0. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier M.P., Edwardes M.D., Michel R.P., Halwani F., Morin J.E. Prognostic markers in resectable non-small cell lung cancer: a multivariate analysis. Can. J. Surg. 2001;44:180–188. [PMC free article] [PubMed] [Google Scholar]
- 32.Cosgrove J., Novkovic M., Albrecht S., Pikor N.B., Zhou Z., Onder L., Mörbe U., Cupovic J., Miller H., Alden K., Thuery A., O'Toole P., Pinter R., Jarrett S., Taylor E., Venetz D., Heller M., Uguccioni M., Legler D.F., Lacey C.J., Coatesworth A., Polak W.G., Cupedo T., Manoury B., Thelen M., Stein J.V., Wolf M., Leake M.C., Timmis J., Ludewig B., Coles M.C. B cell zone reticular cell microenvironments shape CXCL13 gradient formation. Nat. Commun. 2020;11:3677. doi: 10.1038/s41467-020-17135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu T., Yang X., Shi Y., Zhao M., Bi G., Liang J., Chen Z., Huang Y., Jiang W., Lin Z., Xi J., Wang S., Yang Y., Zhan C., Wang Q., Tan L. Single-cell transcriptome atlas of lung adenocarcinoma featured with ground glass nodules. Cell Discov. 2020;6:69. doi: 10.1038/s41421-020-00200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing X., Yang F., Huang Q., Guo H., Li J., Qiu M., Bai F., Wang J. Decoding the multicellular ecosystem of lung adenocarcinoma manifested as pulmonary subsolid nodules by single-cell RNA sequencing. Sci. Adv. 2021;7:eabd9738. doi: 10.1126/sciadv.abd9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw single cell RNA sequencing data along with their associated metadata were deposited in Sequence Read Archive (SRA) under the accession number PRJNA773987. The metadata of each dataset corresponds to information of the Table 1 in the manuscript. The codes generated during this study are available at Github (https://github.com/mbgld/SINGLE). Please contact Y.S.C for further correspondence regarding requests for materials and data.