Summary
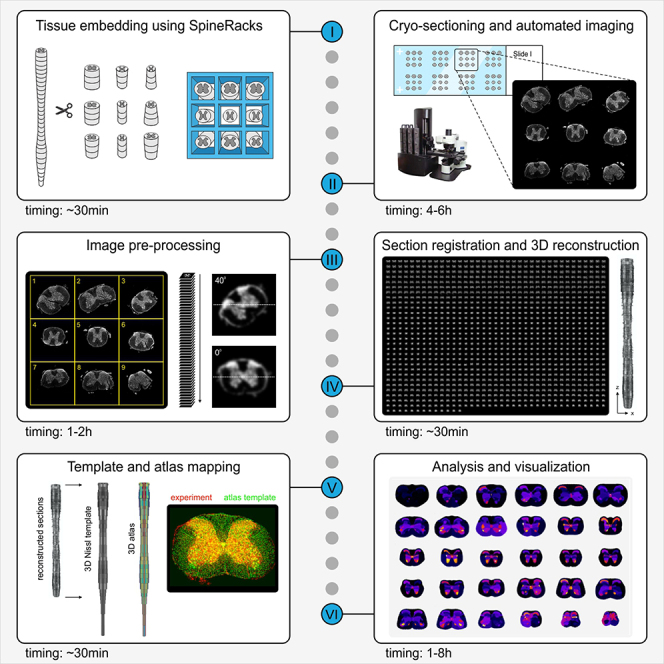
Spatial analysis of spinal neurons is currently limited by a lack of tools for efficient preparation and imaging of the whole spinal cord (SC) and the absence of a 3D reference atlas. Here, we describe protocols for efficient sectioning of whole SC using SpineRacks and subsequent image registration, atlas mapping, and 3D analysis of cells and projections, using SpinalJ. Together, these tools enable high-throughput analyses of adult mouse SC and direct comparison of spatial information of neurons between animals and studies.
For complete details on the use and execution of this protocol, please refer to Fiederling et al. (2021).
Subject areas: Developmental biology, Microscopy, Model Organisms, Neuroscience
Graphical abstract
Highlights
-
•
SpineRacks facilitate efficient sectioning and imaging of whole mouse spinal cord
-
•
SpinalJ allows registration, mapping and analysis of spinal sections
-
•
A 3D atlas provides a common framework for comparative analyses of neurons
Spatial analysis of spinal neurons is currently limited by a lack of tools for efficient preparation and imaging of the whole spinal cord (SC) and the lack of a 3D reference atlas. Here, we describe protocols for efficient sectioning of whole SC using SpineRacks and subsequent image registration, atlas mapping, and 3D analysis of cells and projections, using SpinalJ. Together, these tools enable high-throughput analyses of adult mouse SC and direct comparison of spatial information of neurons between animals and studies.
Before you begin
Given the technical limitations of traditional histological methods and more recent clearing approaches, SC analyses in mice have typically focused on sparse sampling through the entire SC or on subsets of spinal segments, resulting in incomplete characterization of the distributions and diversity of spinal neurons and their projections along the rostro-caudal (r-c) axis. Moreover, data generated by sparse sampling of whole SC is difficult to map accurately to defined r-c positions without the context of neighboring sections or vertebral landmarks. To overcome these problems, we have developed tools to improve sectioning efficiency of whole SC and to facilitate section registration, atlas mapping and 3D analysis of SC sections (Fiederling et al., 2021). These tools fill the prevailing gap in methodology for SC analysis that limits detailed characterization of spinal cell types and circuitry. Moreover, mapping of section data to a common coordinate framework using SpinalJ permits SC wide analysis with high spatial precision and comparison of results from different samples and across research groups.
In the protocol below, we describe in detail the use of SpineRacks and SpinalJ for efficient analysis of whole mouse SC. To follow along with this protocol and for easy testing and troubleshooting of SpinalJ, we provide an example dataset that comprises raw image data of a full spinal cord sample, all intermediate processing files generated during analysis, and our resulting output files. A video tutorial that guides the user through all steps is also included. Links to these resources are provided in the key resources table.
Print SpineRacks
Timing: ∼3 h print time for 50 SpineRacks on the Ultimaker3 printer (may vary using other printers)
-
1.Download SpineRack.stl print files (https://github.com/felixfiederling/SpinalJ/blob/main/SpineRack.stl).
-
a.Slice file for printing.We used the following settings in Cura v4.8 for printing on the Ultimaker3 printer: Material: natural PVA; print core: BB 0.4; layer height: 0.15 mm; print temp: 220°C, bed: 60°C; infill: 20%; build plate adhesion: 3 mm brim.
-
a.
-
2.
Print using PVA filament.
-
3.
Store SpineRacks in a sealed container (such as a 50 mL screw top tube) at room temperature (20°C–22°C) in the dark with a desiccant bag to prevent absorption of ambient moisture.
SpinalJ installation
Timing: 15–30 min
We have successfully tested SpinalJ on systems running Microsoft Windows 10 using Fiji version 1.53c. Memory requirements depend strongly on the size of the input images, but at least 16 GB RAM are recommended. SpinalJ is generally compatible with MacOS, but does not support machine learning detection of cells and projections with Ilastik (step 20 c).
-
4.
Download and install Fiji software (http://fiji.sc/Fiji).
-
5.Download and install SpinalJ for Fiji (https://github.com/felixfiederling/SpinalJ).
-
a.Copy the SpinalJ.jar file into your Fiji plugins folder and restart Fiji to install the plugin (Alternatively, use Plugins > Install).
-
b.Store the segmentation masks “Split_mask.roi” and “Split_mask_scaled.roi” on your computer.
-
c.Download the SpinalJ atlas files (https://data.mendeley.com/datasets/4rrggzv5d5/1) and store on your computer.
-
a.
-
6.
Install Attenuation Correction (Biot et al., 2008). Download and copy into your Fiji plugins folder and restart Fiji.
Attenuation Correction can be downloaded at: https://imagejdocu.tudor.lu/plugin/stacks/attenuation_correction/start
-
7.
Install TurboReg and MultiStackReg (Thévenaz et al., 1998). Download and copy into your Fiji plugins folder and restart Fiji.
TurboReg can be downloaded at: http://bigwww.epfl.ch/thevenaz/turboreg/
MultiStackReg can be downloaded at: http://bradbusse.net/sciencedownloads.html
-
8.
Install Elastix 5.0.0 (Klein et al., 2010).
Elastix can be downloaded at: https://elastix.lumc.nl/download.php
-
9.
Install Ilastik 1.3.3post3 (Berg et al., 2019). Only required for machine learning detection of cells and projections (step 20 c).
Ilastik can be downloaded at: https://www.ilastik.org/download.html
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde 16% aqueous solution | Electron Microscopy Sciences | Cat#15700 |
| PBS 10x | Corning | Cat#46-013-CM |
| Sucrose | Fisher Scientific | Cat#AC177140050 |
| Tissue-Tek OCT Compound | Sakura Finetek | Cat#4583 |
| Fluoro-Gel mounting medium | Electron Microscopy Sciences | Cat#17985-11 |
| Neurotrace 500/525 (1:200) | Thermo Fisher Scientific | Cat#N21480 |
| Neurotrace 640/660 (1:200) | Thermo Fisher Scientific | Cat#N21483 |
| DAPI (1:1000) | Thermo Fisher Scientific | Cat#D1306 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice, adult (25–90 weeks old), males and females | The Jackson Laboratory | Stock#000664 |
| Deposited data | ||
| 3D SC reference atlas p56 | Fiederling et al. (2021) | https://data.mendeley.com/datasets/4rrggzv5d5/1 |
| SpinalJ example dataset: processed files |
This protocol | https://data.mendeley.com/datasets/mkc37w4rrv/1 |
| SpinalJ example dataset: raw files (1/6) |
This protocol | https://data.mendeley.com/datasets/dsn9kggdpx/1 |
| SpinalJ example dataset: raw files (2/6) |
This protocol | https://data.mendeley.com/datasets/mrbjz83xf6/1 |
| SpinalJ example dataset: raw files (3/6) |
This protocol | https://data.mendeley.com/datasets/dfyvhrh4cd/1 |
| SpinalJ example dataset: raw files (4/6) |
This protocol | https://data.mendeley.com/datasets/43ky5by55m/1 |
| SpinalJ example dataset: raw files (5/6) |
This protocol | https://data.mendeley.com/datasets/xdj4kxkvnp/1 |
| SpinalJ example dataset: raw files (6/6) |
This protocol | https://data.mendeley.com/datasets/4hx2cj4j67/1 |
| SpinalJ video tutorial | This protocol | https://data.mendeley.com/datasets/63m34hg3pt/1 |
| SpineRack .stl print file | Fiederling et al. (2021) |
https://doi.org/10.5281/zenodo.5177185 Also available through: https://github.com/felixfiederling/SpinalJ/blob/main/SpineRack.stl |
| Software and algorithms | ||
| Cura v4.8 | Ultimaker | https://ultimaker.com/software/ultimaker-cura |
| Fiji (ImageJ) | Schindelin et al. (2012) | https://fiji.sc/ |
| SpinalJ | Fiederling et al. (2021) |
https://doi.org/10.5281/zenodo.5177185 Also available through: https://github.com/felixfiederling/SpinalJ |
| Attenuation Correction | Biot et al. (2008) | https://imagejdocu.tudor.lu/plugin/stacks/attenuation_correction/start#attenuation_correction |
| TurboReg | Thévenaz et al. (1998) | http://bigwww.epfl.ch/thevenaz/turboreg/ |
| MultiStackReg | Thévenaz et al. (1998) | http://bradbusse.net/sciencedownloads.html |
| 3D Viewer | Schmid et al. (2010) | https://imagej.nih.gov/ij/plugins/3d-viewer/ |
| Elastix | Klein et al. (2010) | https://elastix.lumc.nl/download.php |
| Ilastik | Berg et al. (2019) | https://www.ilastik.org/ |
| NIS-Elements JOBS | Nikon Instruments | https://www.microscope.healthcare.nikon.com/en_EU/products/software/nis-elements/nis-elements-jobs |
| JOBS settings for imaging | Fiederling et al. (2021) | https://doi.org/10.5281/zenodo.5177185 |
| Other | ||
| Ultimaker3 3D printer (or equivalent) | Ultimaker | https://ultimaker.com/de/3d-printers/ultimaker-3 |
| PVA filament | Ultimaker | Cat#9731 |
| Truncated embedding mold Peel-A-Way(R) T12 | Polysciences | Cat#189861 |
| Cryostat | Leica Biosystems | CM3050S or equivalent |
| Disposable microtome blades | Sakura Finetec | Cat#4685 |
| Microscope slides, Fisherbrand Superfrost Plus | Fisher Scientific | Cat#12-550-15 |
| Wheaton dish | DWK Life Sciences | Cat#900200 |
| Slide Scanning Microscope e.g., Nikon AZ100 Multizoom | Nikon Instruments | Model AZ100, discontinued |
| Automated slide feeder | Prior Scientific Inc. | Model PL-200 |
| Andor Zyla camera | Oxford Instruments | Model Zyla 4.2 sCMOS |
Step-by-step method details
Tissue preparation
Timing: ∼1 h hands on + 2 days unattended
This protocol has been established on perfusion-fixed SC tissue of adult C57BL/6J mice.
CRITICAL: Make sure to acquire approval for anesthesia and perfusion of mice and to comply with institutional guidelines on the care and use of animals in research.
-
1.
Perfuse the deeply anesthetized animal with PBS followed by 4% PFA in PBS and expose the SC by standard laminectomy or by hydraulic extrusion (Kennedy et al., 2013). Postfix spinal tissue in 4% PFA in PBS on an orbital shaker for 2–12 h at 4°C.
-
2.
Wash SC 3x (10 min each) in PBS on ice, remove dura and meninges, then transfer into a 15 mL tube filled with PBS containing 30% sucrose and incubate at 4°C for 1–2 days or until the tissue has sunk, for cryo-protection.
Spinal cord embedding using SpineRacks
Timing: 10–15 min hands on + 20–30 min unattended
After fixation and isolation of the SC, the tissue is divided into nine short pieces that are embedded with precise upright orientation and in rostrocaudal (r-c) order within the SpineRack support scaffold. Placing tissue pieces in the same block, in order, ensures parallel cryosectioning that reduces whole SC sectioning time and preserves section identity.
-
3.
Prepare a ∼5 cm × 5 cm piece of Parafilm, and pour ∼5 mL OCT onto it. This pool of OCT will be used to wash and orient all tissue pieces prior to embedding.
-
4.
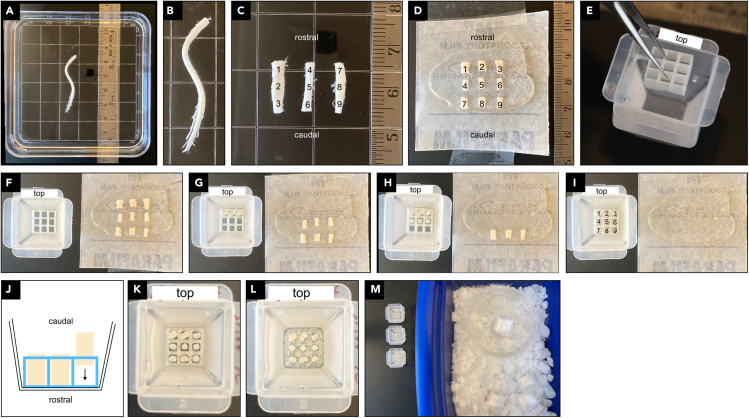
Transfer the SC together with the sucrose solution to a square petri dish (Figure 1A). Cut the SC into three equal parts (Figures 1B and 1C) using sharp spring scissors while viewing with a dissecting microscope.
CRITICAL: Make sure to cut perpendicular to the long axis of the SC (angled cuts produce incomplete cross sections that cannot be used for registration). Keep track of the identity and r-c orientation of the tissue pieces.
-
5.
Cut each piece from step 4 into three equal sized pieces of 3–4 mm (Figures 1C and 1D). Starting with the most rostral segment, after each cut gently transfer the resulting tissue pieces 1–3 into the OCT puddle with their rostral end facing the top side of the Parafilm square, using forceps. Repeat with the other pieces and collect resulting tissue pieces 4–6 and 7–9 in OCT.
CRITICAL: Keeping track of the identity and orientation of each tissue piece is essential. For maximal efficiency of cryo-sectioning and subsequent imaging, all tissue pieces should be of equal length.
-
6.
Fill a truncated embedding mold with OCT and mark one side for orientation (here marked “top”). Submerge a SpineRack into OCT and push all the way to the bottom of the mold using forceps (Figure 1E). Release any trapped air bubbles.
CRITICAL: After immersing the SpineRack into OCT, the PVA material will start to dissolve and will soften and expand in the process, and this may prevent positioning or placement of tissue pieces. Thus, tissue must be embedded immediately, within the first 15 min after immersing the SpineRack in OCT.
-
7.
Transfer one tissue piece at a time into the SpineRack, taking care to load the pieces into wells from top-left to bottom-right in r-c order (Figures 1F–1I). Using forceps, carefully push each tissue piece within a well to the bottom of the mold with its rostral end facing down and the dorsal side facing the marked (top) side of the mold (Figure 1J). Wide cervical segments 1–3 are rotated by ∼45° to fit the well.
Optional: If interested only in a smaller sub-region of SC (e.g., L3-L6), SpineRacks can also be used to embed and section multiple samples in parallel.
Optional: SpineRacks (with same or adjusted geometry) can also be used to embed other tissues or organisms that have a high length-to-width ratio (muscles, fish embryos) or that require parallel sectioning in a single orientation (eg. mouse eyes).
-
8.
After embedding all tissue pieces, leave mold containing SpineRack and tissue at room temperature for 20–30 min. This allows the PVA material to soften for ease of sectioning (Figures 1K and 1L).
-
9.
Freeze the block on dry ice in a slush of ethanol and crushed dry ice until the block is solid (Figure 1M).
Pause point: Use immediately OR, if the histology protocol in use for the tissue permits, store blocks at −80°C until use. For NeuroTrace or DAPI labelling, blocks can be stored for at least 6 months (longer has not been tested).
Figure 1.
Embedding of SC tissue using SpineRacks
(A and B) Isolated SC in sucrose solution.
(C) SC cut into 3 equal tissue pieces. Numbers 1–9 indicate r-c orientation and placement of next cuts. Ruler scale in mm.
(D) SC cut into 9 equal pieces arranged according to position in SpineRack in OCT.
(E) Submerging SpineRack in embedding mold filled with OCT.
(F–I) Placement of tissue pieces 1–9 into SpineRack.
(J) Vertical orientation of tissue pieces in the mold.
(K and L) Incubation of SpineRack in OCT expands and softens the material for sectioning (K: before, L: after 20 min incubation).
(M) Freezing the block.
Sectioning and staining
Timing: 2 h hands on + incubation time for immunohistochemistry (optional)
Using SpineRacks, >1000 25μm sections comprising spinal segments C1-L6 can be produced within 1h and collected on as few as 16 slides. Cutting thicker sections can further reduce sectioning and processing time at the expense of r-c resolution.
-
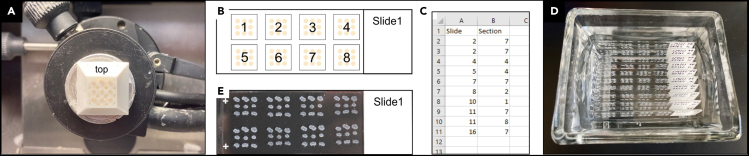
10.
Place the block into the cryostat chamber for 20–30 min to allow temperatures to equilibrate. We section at approximately −21°C using a Leica Cryostat CM3050S and Sakura Finetec microtome blade. For sectioning, remove the mold, keeping track of orientation, and mount the block on the block holder with the marked side up and set sectioning thickness to 25μm (Figure 2A).
-
11.
Section the block and collect all sections continuously on numbered slides. To permit automated imaging, collect eight block sections in two rows of four with equal spacing on each slide (Figure 2B). Keep note of the order and orientation of sections on each slide, as this information is crucial to maintain r-c order of images. Make notes of lost sections (slide # and position on slide; lost sections can be replaced with neighboring sections during image processing; Figure 2C).
Pause point: Unless user-specific antibody or histology procedure of choice does not support it, slides can now be stored at −80°C. For subsequent NeuroTrace or DAPI labelling slides can be stored for at least 3 months (longer has not been tested).
-
12.
Wash slides in a glass Wheaton dish filled with PBS for 5–10 min (gentle shaking at room temperature) to wash away SpineRack material and OCT (Figure 2D). Proceed with immunohistochemistry, or other staining methods, following standard protocols (Evilsizor et al., 2015) and mount slides under coverslips (Figure 2E).
CRITICAL: For subsequent image registration and atlas mapping, co-staining with NeuroTrace (preferred) or DAPI is required.
Pause point: Mounted slides can be stored at 4°C in the dark. Slides can be kept for at least 3 months after NeuroTrace or DAPI labelling. User specific antibodies or stains will dictate other storage times.
Figure 2.
Sectioning, collecting and staining of block sections
(A) Tissue block mounted in cryostat.
(B) Schematic arrangement of 8 block sections on a slide in a left-and-up order. See also Figure 4A.
(C) Example csv. list of lost sections.
(D) Slides washing in PBS.
(E) Slide after staining and mounting.
Imaging
Timing: 10–15 min hands on + 3–5 h unattended
-
13.Image slides on a slide scanning microscope.
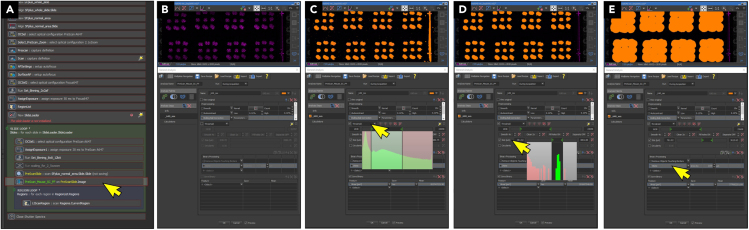
The workflow for automatic scanning of block sections using the Nikon system, running NIS-Elements JOBS software is described below (Figure 3; JOBS file available for download):Note: Processing in SpinalJ requires either images of individual tissue sections or images of block sections (array of 9 tissue sections), and alphanumeric file naming that reflects section order. In this study, we used a motorized Nikon AZ100 Multizoom microscope equipped with an automated slide feeder and Andor Zyla sCMOS camera to acquire images of block sections. Any other automated or manual systems can be used to collect images, but may require additional manual intervention, depending on the capabilities of the individual system to identify, scan and save block sections on the slide automatically. For example, images taken on slide scanners that scan the whole slide have to be cropped to individual block section images. Block section images should be saved as .nd2 or .tif files with file naming “SlideXX_SectionYY” (e.g., Slide12_Section04.nd2). For non-motorized systems, images of individual sections have to be acquired and saved manually. Single section images should be saved as .tif files with file naming “SegmentXX_SlideYY_ImageZZ” (e.g., Segment06_Slide12_Image01.tif) if pre-processing in SpinalJ is desired.Setting up Nikon slide scanner for scanning block sections with NIS-Elements JOBS:-
a.Load all slides in order into a slide magazine (1–16, bottom to top) and initialize the slide scanner using 4x objective, 2.1x zoom.
-
b.Open JOBS scan file for SC sections and pre-scan 1–2 slides using the autofocusing channel (DAPI or NT). To save the pre-scan, edit settings before scanning as follows (Figure 3A):
-
i.Under PreScanSlide set Save Image → always.
-
ii.Disable Region Loop.
-
i.
-
c.After pre-scanning, open the pre-scan image.
-
d.Open JOBS scan file and open scan parameters (Figure 3B).
-
i.Set lower and upper intensity threshold to filter out background and unspecific signals (Figure 3C).
-
ii.Set size cutoff to exclude objects that are not tissue sections (Figure 3D).
-
iii.Set dilation factor to merge all sections (1-9) of a block section into a single object (Figure 3E). Avoid merging multiple block sections.
-
i.
-
e.Save settings, disable save pre-scan, enable Region Loop and start the scanning process.
-
a.
Figure 3.
Scan settings for imaging block sections using NIS-Elements JOBS
(A) Load a pre-scan image of a slide and open pre-scan settings in JOBS file.
(B) Check ‘rolling ball correction’.
(C) Set lower and upper threshold to isolate pixel intensities within SC sections.
(D) Apply size cutoff to filter out objects smaller and larger than SC sections.
(E) Apply dilation factor to create a mask that connects all sections of a block section, but does not touch sections from neighboring block sections. Save settings and start scan.
Image pre-processing in SpinalJ
Timing: 0.5–1 h hands on + 1–2 h unattended
SpinalJ provides a series of tools to pre-process SC section images for registration and analysis. These include segmentation of block section images, compensation for lost sections, r-c ordering of sections, cleaning out damaged sections, as well as re-orientation and horizontal alignment of sections. If preprocessing has already been performed without using SpinalJ, skip to step 19.
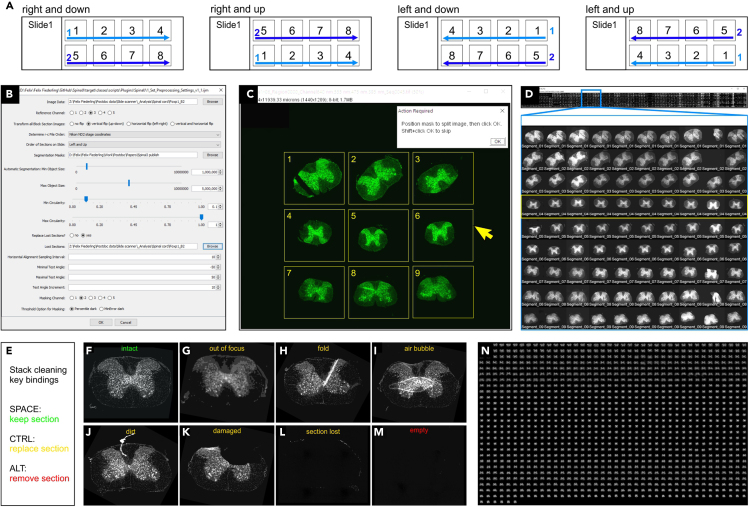
Achieving r-c order of images critically depends on how sections were collected on the slide. SpinalJ supports a series of collection orders, as defined in Figure 4A.
-
14.
Setting Pre-Processing Settings:
To prepare image pre-processing, create a parameter file that contains information on how the images of a sample are processed (Figure 4B). Run Plugins > SpinalJ > 1 Set Preprocessing Settings. In the GUI that appears, complete the fields below.-
a.Image Data - Specify location of the folder containing the raw sections.Note: Files in .nd2 format cannot exceed 2GB. If image resolution does not match 1micron/pixel, scale images using Fiji>Image>Scale or create custom segmentation masks (see troubleshooting 4).
-
b.Reference Channel - Select channel containing NeuroTrace or DAPI.
-
c.Transform all Block Section Images - Select transformation option to flip images as necessary so that sections are oriented with their dorsal side up and array order as follows: Top row, left to right: 1, 2, 3. Middle row: 4, 5, 6. Bottom row: 7, 8, 9 (see Figure 4C).
-
d.Determine r-c File Order - Select method used to bring image files into r-c order (using stage coordinates or alphanumerical file names).
-
e.Order of Sections on Slide - Indicate order of sections on slide, see Figure 4A. Required only if choosing stage coordinate sorting.
-
f.Segmentation Masks - Specify the location of the folder containing the segmentation masks .roi files. If using images with resolution other than 1micron\pixel, see troubleshooting 4.
-
g.Automatic Segmentation:
-
i.Min. Object Size – Specify lower area cutoff for auto-detection of tissue sections. Default: 1e6.
-
ii.Max. Object Size – Specify upper area cutoff for auto-detection of tissue sections. Default: 5e6.
-
iii.Min. Circularity – Specify lower circularity cutoff for auto-detection of tissue sections. Default: 0.05.
-
iv.Max. Circularity – Specify upper circularity cutoff for auto-detection of tissue sections. Default: 1.
-
i.
-
h.Replace Lost Sections - Check ‘yes’ to compensate for lost sections during cryo-sectioning by duplicating neighboring sections. Requires a .csv list of lost sections (see below).
-
i.Lost Sections - Specify the location of the folder containing a .csv list of lost sections. Required only if choosing to replace lost sections. To create the list, enter pairs of values that specify the slide (column A) and the position (column B) on which the section was lost e.g., if the seventh section on the second slide was lost, enter 2, 7 (see Figure 2C). If several consecutive sections are lost, repeat the list entry. The first row contains the titles ‘Slide’ and ‘Section’, respectively. Save list as ‘_Lost_Sections.csv’.
-
j.Horizontal Alignment Sampling Interval - Specify n. Total number of sections / n sections are randomly selected to determine the horizontal orientation for each tissue piece. Default: 10.
-
k.Minimal Test Angle - Specify negative test angle extreme. Must be > −90°. Default: −50°.
-
l.Maximal Test Angle - Specify positive test angle extreme. Must be < 90°. Default: 50°.
-
m.Test Angle Increment - Specify interval between test angles. Default: 10.
-
n.Masking Channel - Select channel for section segmentation. NeuroTrace or DAPI.
-
o.Thresholding Option for Masking - Choose automatic thresholding option. Empirically, we found ‘Percentile dark’ to work best for NT and ‘Min error’ for DAPI.
-
a.
-
15.
Segmentation of block section images:
Run Plugins > SpinalJ > 2 Block Section Segmentation to split block section images into individual tissue section images. Confirm path to the folder containing raw image data. Input files need to contain slide information as first part of the filename, e.g., “Slide4_Section3.nd2”. Images in.nd2 format will be automatically sorted rostrocaudally using stage coordinates when selecting the corresponding option in step 14. Images in other file formats have to be renamed manually to reflect r-c order. After ordering, block section images are automatically segmented. Images that contain less than 9 tissue sections cannot be automatically segmented and are saved to folder \_Temp\_0_Scaled\ as a down-scaled version and displayed at the end of the script for manual segmentation. For these files, position the segmentation mask displayed on top of each image so that each tissue section falls within the appropriate outline of the section array mask, reflecting segment identity (Figure 4C). Click OK to proceed to the next section. If identification of segment identity is impossible because of missing sections, hold SHIFT + click OK to skip segmentation of the current image. Output files are saved to folder \_Temp\_I_Split\ with names indicating tissue piece (segment) identity, e.g., “Segment05_Slide12_Image02.tif”.
-
16.
Segment Rotation:
Run Plugins > SpinalJ > 3 Batch Rotation to re-orient section images from tissue pieces embedded upside down or rotated by 90° (e.g., piece 4 in Figure 4C). Confirm path to the folder containing raw image data. A preview montage of all sections will show the reference channel of all section images ordered in rows representing tissue pieces 1–9. Identify rows in which all sections are oriented at an angle ≥ 90° or ≤ −90° and select those rows for rotation (Figure 4D). Set rotation value and click OK. Confirm proper orientation in the updated preview montage saved at \_Temp\_II_Preview_Split\.
-
17.
Section Clean up:
Run Plugins > SpinalJ > 4 Clean Stack to clean out compromised and empty sections and to obtain a continuous stack of intact tissue sections. Confirm path to the folder containing raw image data. A reference channel preview of the image stack of individual tissue sections will be displayed. Navigate through the stack and decide for each section whether to ‘keep’, ‘replace’, or ‘delete’ the current image by pressing the SPACE, CTRL or ALT key, respectively (Figures 4E–4M). To the left and right of the stack window, additional windows show the previous (left) and next (right) image in the stack. Use ‘replace’ only after at least one previous image has been selected for ‘keep’. Processed files are saved to folder \_Temp\_III_Clean\ and a preview montage of the reference channel is saved to \_Temp\_IV_Preview_Clean\ for validation.
-
18.
Horizontal alignment of section images:
Run Plugins > SpinalJ > 5_Horizontal Alignment to align tissue sections horizontally in order to improve image registration performance. Confirm path to the folder containing raw image data. After automatic alignment is completed, a preview montage of the cleaned and aligned sections is created for validation (Figure 4N). In addition, pre-processed images are scaled to match the resolution of the atlas and saved to a new folder /_Output/ 1_Reformatted_Sections/ with subfolders for each image channel.Note: Data generated during image pre-processing will be saved to subfolder \_Temp\ and can be deleted after reformatted images have been created (step 18).
Figure 4.
Image pre-processing
(A) Schematic of commonly used arrangements of sections on the slide.
(B) Pre-processing settings (for details see descriptions in step 14).
(C) For manual segmentation of block section images, position the segmentation mask (yellow) to include all tissue sections 1-9.
(D) Batch rotation of images from tissue pieces embedded upside down. Zoom in (blue box) reveals that tissue piece 4 (yellow box) needs 180° rotation.
(E) Cleaning out of compromised and empty section images using SPACE, CTRL and ALT keys.
(F–M) Example NT images of sections to keep (green), replace (orange), or delete (red).
(N) Montage of all horizontally aligned sections from a sample after pre-processing.
Image registration, atlas mapping, and analysis in SpinalJ
Timing: 5–30 min hands on + 1–8 h unattended
All subsequent steps assume a single folder containing intact, consecutive single section images of a sample with an alphanumerical name format that reflects r-c order.
-
19.
Setting Registration and Analysis Parameters for SpinalJ:
SpinalJ is designed to batch process multiple samples, even when the experimental conditions or analysis settings are different. This step creates a parameter file containing analysis information for individual samples. Run Plugins > SpinalJ > 6 Set Analysis Settings. In the GUI that appears, complete the fields below:-
a.Select folder - Specify the location of the folder containing the ordered tissue sections (/_Output/).
-
b.Final resolution of image output (μm/px) - Specify the resolution used by the rest of the analysis, default: 2μm/pixel.
-
c.Counterstain channel (DAPI or NeuroTrace) – Select channel containing NeuroTrace, or DAPI.
-
d.Background intensity of counterstain channel - this value should be slightly higher than the intensity of the background surrounding the tissue. To determine the background intensity: open a section image in Fiji and hover the cursor over the tissue and background to find a suitable intensity level for this setting. If set too low, the sections will not be adequately isolated, and if set too high, the sections will be patchy and over-cropped.
-
e.Background removal prior to segmentation (rolling ball radius in px, 0 if none) - Specify the radius of a rolling ball filter used to clean up images before cell detection and projection analysis. A value of 7 works well for the recommend image pixel size of 2μm/px.
-
f.Section cut thickness - Indicate section cut thickness. If using a sub-series, multiply accordingly. E.g., every second section, cut at 20μm = 40μm.
-
g.Spinal cord range (start segment, end segment) - Indicate the range of segments included in the dataset (e.g., C2-L5). To determine range, open preview stack of sections and determine the segment identity of the first and last section based on morphological landmarks and by comparing to neighboring sections.
-
h.Reference section - Specify the starting section used for initial section-to-section registration. This should be roughly in the middle of the dataset. e.g., section 500 in a 1000 section dataset.
-
i.Generate full resolution registered image of reference channel – Check only if attempting to perform cell or projection analysis on the reference channel.
-
j.Perform a second pass section registration - This feature can be used to register badly damaged sections with the help of fiducial markers but is not required for normal use.
-
k.Method for cell detection:
-
i.Manual cell detection takes a list of cell locations created by manual cell counting – see step 20a.
-
ii.Find Maxima will locate the center of cells and is fast and effective for cells with clearly labeled soma – see step 20b.
-
iii.Machine learning segmentation with Ilastik can yield better results, especially when cells are difficult to discern from brightly labeled axons and dendrites. This will require training of an Ilastik Project for each channel – see step 20c.
-
i.
-
l.Channel selection - Select up to three channel/s for cell analysis. Set the first selector to the channel to analyze. If no remaining channels will be analyzed, set the remaining selectors to 0.
-
m.Minimum intensity threshold: Specify the minimum threshold used for detecting cells when using Find Maxima – see step 20b.
-
n.Minimum cell area: Specify the area used for cell detection (when using Ilastik for segmentation).
-
o.Method for projection detection:
-
i.Binary Threshold: fluorescence above this threshold will be included in the density analysis. This is a fast and effective method, useful when it’s not necessary to remove cell bodies from the analysis.
-
ii.Machine learning segmentation with Ilastik can yield better results than binary thresholding, especially when it is necessary to differentiate between cells and projections. This will require training of an Ilastik Project for each channel – see 20c.
-
i.
-
p.Channel selection: Select up to three channel/s for projection analysis. Set the first selector to the channel to analyze. If no remaining channels will be analyzed, set the remaining selectors to 0.
-
q.Minimum intensity threshold: Choose the minimum threshold used for detecting projections when using Binary Threshold – see 20b.
-
r.Ilastik location: Specify the directory where Ilastik is installed.
-
s.Elastix location: Specify the directory where Elastix is installed.
-
a.
-
20.After setting up the Registration and Analysis Parameter file, follow the steps below according to the selected option for cell detection (skip if interested only in intensity mapping, see step 21 i):
-
a.Manual cell detection (19 k.i.):
-
i.Run through steps 1–19 of this guide. In step 19 k, select ‘Manual Cell Count’.
-
ii.Run step 21 a-d. The experiment folder will now contain a sub-folder \3_Registered_Sections\. Inside this folder, you will find subfolders for each channel e.g., Folder \2\ corresponds to channel 2.
-
iii.Drag the entire channel folder on Fiji. An ‘Open Folder’ window will appear. Click ‘Yes’.
-
iv.Under Edit > Options > Input/Output… turn off rows and columns headers.
-
v.Under Analyze > Set Measurements deselect everything except centroid. Set decimal places to 0.
-
vi.Under Image > Properties… set unit of length to pixel. Pixel width, pixel height, voxel depth = 1.
-
vii.Select the Multipoint Tool.
-
viii.Go through slice by slice and click on every cell. If you miss a cell, it is ok to go back through the stack and add new cells. CTRL+ click removes markers.
-
ix.After counting, you can save this image if you wish to keep a record of how the cells were counted.
-
x.Press CTRL+M to get the centroid and slice position of each cell.
-
xi.Save this list as ‘Cell_Points_Ch#.csv’ into the subfolder \4_Processed_Sections\Detected_Cells\ (if you are counting cells on channel 2 this file would be called Cell_Points_Ch2.csv).
-
xii.Run step 21. Uncheck c and d - already performed. Check e to perform cell detection and analysis, and any other desired analysis steps.
-
i.
-
b.Determining thresholds for Find Maxima cell detection and binary thresholds for projection analysis (19 k.ii):
-
i.Run through steps 1–19 of this guide. Run steps 21 a-e, as described in step 21.
-
ii.The experiment folder will now contain a sub-folder \3_Registered_Sections\. Inside this folder, you will find subfolders for each channel e.g., Folder \2\ corresponds to channel 2.
-
iii.Apply a rolling ball filter with the same diameter used within SpinalJ, then confirm the typical intensities of the cells you wish to detect using Find Maxima, and use this value in the analysis settings. For binary thresholding of axons/dendrites use the threshold tool (CTRL+SHIFT+T) to determine the minimum threshold value to be used.
-
i.
-
c.Training Ilastik for machine learning based mesoscale mapping / cell detection (19 k.iii):We recommend using Ilastik for best results, especially when analyzing densely labeled neurons, although other cell detection methods are provided. Be aware that oversaturation of axons/dendrites may result in false positive cell detections when using automated analysis. If analyzing both cells and neuronal projections, it is important to ensure your images are free from oversaturated pixels. For best results in quantifying cell numbers, use a probe with nuclear localization, such as GFP fused to histone 2B.
-
i.Prepare representative images for training the Ilastik project.Training Ilastik’s pixel classifier requires several representative images. A minimum of 5 (better 10–20) sections from different r-c positions of the SC is recommended. If training a project on multiple animals or if the label is highly variable between sections, additional training images may be required.Training images can come from the \1_Reformated_Sections\ or \3_Registered_Sections\ folders. During the SpinalJ pipeline, background subtraction is applied to all sections prior to running Ilastik. After providing a list of sections, images suitable for training will be created and saved to folder \Ilastik_Training_Images\.
 CRITICAL: When adding images to the Ilastik project for training, ensure they also have this same background subtraction applied. You can do this by running Plugins > SpinalJ > Other Tools > Create Ilastik training images in Fiji.
CRITICAL: When adding images to the Ilastik project for training, ensure they also have this same background subtraction applied. You can do this by running Plugins > SpinalJ > Other Tools > Create Ilastik training images in Fiji. -
ii.Open the provided Ilastik project file.An example project is provided as a template to build the projects for each of your channels containing all settings required to export the necessary probability images for use in SpinalJ.
-
iii.Select the existing images in the project file. Right-click and choose ‘Reset’ - this clears out the existing training data, but keeps project file settings, allowing you to add new images for training.
-
iv.Copy in the images selected for training.We recommend saving these images into the project file for auditing and to avoid problems if further training is required. To do so, Right-click and select ‘Dataset Properties…’, in the window that appears, change ‘Storage’ to ‘Copy into project file’. Then save the project.
-
v.Train the project file.The Ilastik Project comes preconfigured with three labels: Cells (for soma/cell bodies), Projections (for axons/dendrites), and Background.If you are not detecting cells, you do not have to train the cells label (likewise for projections).For best results when training on 16-bit images, it can be helpful to adjust how the image is displayed in Ilastik. You can do this by right-clicking on ‘Raw Input’ in Group Visibility list (lower left-hand corner), then selecting ‘Adjust thresholds’. Uncheck the ‘automatic range’ option and reduce the ‘maximum value’ so that you can clearly visualize low intensity structures.For further advice on training an Ilastik project:
-
vi.Save the project file as: ‘Ilastik_Project_Channel_#’ (e.g.,‘Ilastik_Project_Channel_1’ for channel 1) in the \Ilastik_Projects\ folder.
-
vii.Repeat steps 20 c.i-vi to create an Ilastik project for each channel to be analyzed. These projects can be copied to other animals/datasets to be analyzed.
-
i.
-
a.
-
21.
Atlas registration analysis
This step will automatically process each folder and generate three subfolders that contain 1) the registered sections at the resolution set for analysis (\3_Registered_Sections\), 2) the processed images used for analysis, such as the output from Ilastik, and validation images for confirming the accuracy of cell segmentation (\4_Processed_Sections\; Figure 5A) and 3) the final analysis tables and images (\5_Analysis_Output\; Figures 5B and 5C). Run Plugins > SpinalJ > 7 Registration and Atlas Analysis. In the GUI that appears, complete the fields below:Note: For efficient processing, we recommend confirming good section registration prior to performing further analyses steps. The registered sections are stored in folder /_Output/3_Registered_Sections/ after performing step 21 a-c and can be visualized in 3D using the ImageJ 3D Viewer plugin (Schmid et al., 2010), see Methods video S1.-
a.Select experiment/spinal cord folder(s) - Specify the location(s) of the folder(s) for analysis (/_Output/).Methods video S1. Registered sections, related to step 21Animation through a stack of 1302 25μm registered sections (Neurotrace channel) covering segments C1-S3 and the same data re-sliced in the horizontal and sagittal plane. Data is provided in the example dataset.Download video file (5.1MB, mp4)
-
b.Select spinal cord atlas to be used - Specify the directory where the atlas files are located.
-
c.1. Perform section registration? - Choose to perform initial rigid body registration of all tissue sections.
-
d.2. Perform atlas registration? - Choose to perform affine and elastic registration for atlas mapping to experimental data.
-
e.3. Perform cell detection and analysis? - Choose to perform cell detection using the method selected in the previous step. If performing manual cell counting, run both registration steps first, manually count cells, then rerun this step with this and the following options enabled. The cell analysis generates two tables for each channel used for cell detection. The Detected Cells table provides information on the coordinates of each cell, the region and segment it is contained in, and the measured mean intensity in each channel. The Detected Cells Summary table provides a total cell count for each region and segment.
-
f.Generate cell analysis images and heatmaps? - Choose to generate images showing cell locations in the atlas colored according to annotation ID.
-
g.4. Perform mesoscale mapping projection analysis? - Choose to perform projection analysis. The mesoscale mapping analysis uses reverse mapping to generate output tables for each channel containing information on the density of axons/dendrites present in each atlas region, and on the relative density according to the volume of the entire labeled volume. Importantly, all density measurements are calculated using the region volume as represented in the dataset, allowing for accurate measurement even when a region is only partially represented in the experimental data.
-
h.Generate mesoscale mapping visualization images and heatmaps? - Choose to generate images showing the projections mapped to the atlas and density heatmaps.
-
i.5. Measure mean intensities of annotated regions? - Choose to perform region specific intensity measurements. Mean intensities for each channel, atlas region and segment are saved to output tables.
-
j.Generate intensity based atlas images? - Choose to generate an atlas image with regions colored with their measured mean intensity.
-
k.6. Extract specific regions at full resolution (requires >128GB RAM)
-
l.Provide annotation IDs for extraction (e.g., 4,10) - Specify location of a list (comma separated) with annotation regions for extraction.
-
a.
Note: After the analysis is complete, all output tables and graphs (e.g., cell density heatmaps, as shown in Figure 5C) can be found in folder \5_Analysis_Output\. In order to re-run analysis with different settings, delete this folder and repeat step 21.
Figure 5.
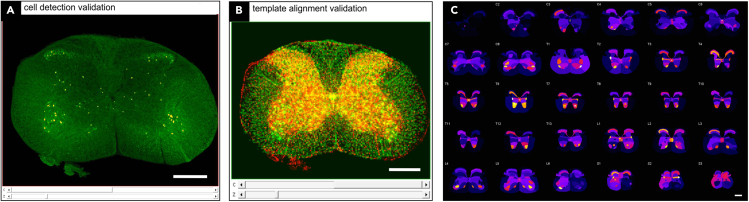
Quality control and output
(A) Overlay of automatically detected cells and corresponding image channel for visual validation of object detection. Shown is data of the example dataset (ChatCre;Ai14tdTomato); green: channel 1 (555nm), red: objects detected using Ilastik.
(B) Overlay of the atlas template (green) and the experimental example dataset (red, Neurotrace channel) after atlas alignment.
(C) SpinalJ output graphs like this heatmap montage of cell densities within channel 1 of the example dataset provide a quick visual summary of the data. Scale: 500μm.
Expected outcomes
We provide a toolbox for the analysis of neuronal number and position of cells and processes within the full r-c extent of mouse SC. We have developed SpineRacks for oriented embedding and efficient sectioning, SpinalJ for user-friendly image processing and analysis, and a 3D SC atlas that provides a standardized reference for analysis of spinal neurons and projections. The availability of a common coordinate framework and 3D anatomical annotations permits comparative mapping of spinal neurons across samples and labs.
We have validated the accuracy and reproducibility of SpinalJ mapping with reference to published manual studies, attesting to its usefulness for a variety of experiments (Fiederling et al., 2021).
Analysis output data including intensity values, cell coordinates as well as cell/projection density is provided in table (csv.) format and summarized in heatmap graphs for each spinal segment (Figure 5C).
An example dataset including the raw image data, parameter settings files and output images are provided and available for download (see key resources table). Table 1 lists all folders and important files within this dataset. To reproduce this experiment and re-run all pre-processing and analysis steps (14–21), copy the raw .nd2 files (provided in a total of 6 subfolders, 120 GB) to a new location and start with step 14. Alternatively, to skip pre-processing and start directly with section registration (19–21), copy folder “example_data/_Output/1_Reformatted_Sections/” (21.3 GB) and start with step 19. The settings we used for processing and analysis are provided in files “_PreProcessing_Parameters.csv” and “Analysis_Settings.csv”. To repeat parts of the analysis with adjusted settings make sure to delete folder “…/5_Analysis_Output/” before re-running step 21.
Table 1.
List of files included in example dataset
| Folder | File | Description | Size | Step |
|---|---|---|---|---|
| SpinalJ_example_data_processed/ | Example dataset | 299 GB | n/a | |
| _Sample_Info.csv | Mouse genetics, staining and imaging info | 1 KB | n/a | |
| _Lost_Sections.csv | Sections lost during cryosectioning | 1 KB | 11 | |
| _PreProcessing_Parameters.csv | Parameters used for pre-processing | 1 KB | 14 | |
| _Cleaning_log.csv | User input during section clean up | 10 KB | 17 | |
| _II_Preview_Montage.tif | Down scaled single section images (reference channel montage) | 48.9 MB | 15 | |
| _IV_Preview_Montage_Clean.tif | Down scaled cleaned single section images (reference channel montage) | 43.5 MB | 17 | |
| _V_Preview_Montage_Aligned.tif | Down scaled, cleaned and aligned single section images (reference channel montage) | 341 MB | 18 | |
| SpinalJ_example_data_processed/_Output/ | Pre-processed image data, temporary files created during analysis, as well as resulting files | 38.5 GB | 19, 21 | |
| Analysis_Settings.csv | Parameters used for registration and analysis | 2 KB | 19 | |
| …/_Output/1_Reformatted_Sections/ | Single section images in rostro-caudal order, split by channel, and reference channel montage | 21.3 GB | 18 | |
| …/_Output/3_Registered_Sections/ | Registered single section images, split by channel | 8.7 GB | 21 | |
| …/_Output/4_Processed_Sections/ | Temporary processing data | 6.3 GB | 21 | |
| …/4_Processed_Sections/Detected_Cells/ | Cell detection data | 2 MB | 21 | |
| …4_Processed_Sections/Enhanced/ | Images enhanced for Ilastik pixel classification; split by channel | 4.1 GB | 20, 21 | |
| …/4_Processed_Sections/Measured_Intensities/ | Intensity data | 3 MB | 21 | |
| …/4_Processed_Sections/Object_Detection_Validation/ | Overlay of section images and detected cells; split by channel | 2.1 GB | 21 | |
| …/4_Processed_Sections/Probability_Masks/ | Probability masks created by Ilastik | 66 MB | 20, 21 | |
| …/4_Processed_Sections/Resampled_Cells/ | Cells mapped to atlas | 2 MB | 21 | |
| …/_Output/5_Analysis_Output/ | Analysis output | 2 GB | 21 | |
| Annotated_Volumes_XY_10_Z_25micron.csv | Volumes of atlas regions | 34 KB | 21 | |
| Transformed_Segment_Annotations.csv | Segment annotations transformed to match template length | 9 KB | 21 | |
| Template_Brain_Aligned.tif | Overlay of experimental SC data and SC template | 341 MB | 21 | |
| Transformed_Annotations.tif | Transformed atlas annotations | 341 MB | 21 | |
| Transformed_Hemisphere_Annotations.tif | Transformed hemisegment boundaries | 86 MB | 21 | |
| Transformed_Segments.tif | Transformed segment boundaries | 86 MB | 21 | |
| …/5_Analysis_Output/Cell_Analysis/ | Cell analysis output lists and graphs | 1.1 GB | 21 | |
| …/5_Analysis_Output/ Region_Mean_Intensity_Measurements_XY_10_Z_25micron/ | Intensity analysis output lists and graphs | 51 MB | 21 | |
| …./5_Analysis_Output/ Transform_Parameters/ | Transformation data | 9 MB | 21 | |
| …/_Output/Ilastik_Projects/ | Ilastik training file(s) | 255 MB | 20 c | |
| SpinalJ_example_data_raw 1-6 | Raw .nd2 images of block sections | 120 GB | 13 |
Limitations
The quality of section registration relies critically on intact spinal sections. Images of damaged and incomplete sections, as well as out-of-focus images, cannot be used and must be replaced with neighboring sections. In the samples analyzed, we had to replace up to 15% of sections distributed throughout the length of any given SC. Comparative analysis of multiple samples suggests that this intervention did not confound the real distribution of cells (Fiederling et al., 2021). Replacing larger numbers of sections may, however, substantially reduce axial resolution locally and introduce artifacts in cell distributions, especially when analyzing small, local cell clusters. Thus, good section quality is critical and tissue segment break points must be avoided in areas of particular interest.
To minimize damage and loss of information when cutting SC tissue for parallel embedding, careful placement of cuts orthogonal to the long axis of the SC is essential to reduce incomplete transverse sections. To generate a complete dataset, cuts can be placed at slightly staggered intervals along the r-c axis of SC in different samples, allowing smooth analysis through breakpoints.
SpinalJ provides the first 3D atlas and a common coordinate framework for mouse SC. The creation of this 3D atlas from 2D annotations that comprise only one annotated section per spinal segment (Allen Institute for Brain Science, 2008) does not provide additional r-c resolution. Rather, the new atlas brings the existing annotations into r-c context and into a format that allows 3D registration and atlas mapping. Thus, the 3D atlas does not account for anatomical differences within the length of individual segments. Moreover, it is important to note that this atlas and the Allen Spinal Cord Atlas annotations are based on sections of a single animal and therefore cannot account for inter-animal variability in SC anatomy.
We propose, however, that iterative mapping of defined populations of neurons to the standard reference in SpinalJ can be used to create and improve 3D annotations and segment boundaries. Providing our tools as an open resource, we hope progressively to improve the quality of the atlas with increasing numbers of datasets and markers mapped by individual labs and/or shared within the community.
Atlas mapping in SpinalJ is achieved by the registration of the DAPI or NT experimental volume to the 3D Nissl template of the atlas. We use an affine 3D transformation to prevent deformation and warping of the experimental data during registration. With this, the width-to-length ratio of the experimental dataset is fixed and must match the template. However, loss of sections can shorten the experimental data along the r-c axis (the average shortening in the samples processed in Fiederling et al., 2021 was 39 sections = 975μm), resulting in mismapping. To compensate for this, SpinalJ matches the total r-c length of the experimental dataset to the length of the corresponding segment range in the template by interpolating additional sections. The current compensation approach assumes random/equal spacing of missing sections and does not account for locally concentrated losses, which can, however, be manually accommodated.
Troubleshooting
Problem 1
Embedding - Tissue pieces do not fit into the wells of the SpineRack.
Related to step 7. Notable resistance when placing tissue pieces into the wells of the SpineRack.
Potential solution
Make sure that the dimensions of the SpineRack match the print file. If the printed product is smaller than expected, calibration of the 3D printer may solve this issue. The SpineRack should also be checked for rough surfaces and print irregularities (especially in the first print layer) protruding into the wells. In such cases, affected print layers can be peeled off.
Alternatively, the print file can be sliced and printed slightly enlarged (e.g., at 110%).
Problem 2
Sectioning - Difficulties in sectioning through the block.
Related to step 11. Notable resistance when cutting through the block including SpineRack and tissue; sections deform or split frequently.
Potential solution
After embedding tissue pieces, increase SpineRack softening wait time by a few minutes before freezing the block (step 8). Make sure to remove air bubbles that may prevent contact of OCT with the SpineRack. Use a fresh, sharp blade on the cryostat.
Problem 3
SpinalJ - Automatic segmentation of block section images fails.
Related to step 15. Automatic segmentation of block section images fails on a high proportion of images, even if image contains nine tissue sections. Segmented images in folder “\Temp\_I_Split\” show empty images or partial sections.
Potential solution
Adjust segmentation settings in step 14. The lower circularity cutoff value is critical to distinguish tissue sections from debris/auto-fluorescence and may be adjusted upwards (0.1–0.2) if too many objects are detected or downwards (0.0–0.1) if tissue sections are not reliably detected. Optimal settings can be determined manually by running the following sequence of operations on the reference channel of a representative block section image:
run("Enhance Contrast", "saturated=0.35");
run("8-bit");
setAutoThreshold("Mean dark");
run("Convert to Mask");
run("Fill Holes");
roiManager("reset");
run("Analyze Particles...", "size=1000000-5000000 pixel circularity=0.1–1.00 show=Nothing include add");
If manual segmentation is preferred, setting extremely stringent auto-segmentation values (e.g., minimal circularity = 0.9) will essentially negate any automatic segmentation.
Problem 4
SpinalJ – Mask for manual segmentation of block section images does not fit.
Related to step 15. Mask for manual segmentation does not fit the section array.
Potential solution
Check if arrays of tissue sections appear regularly spaced. If not, sections may have been compressed during sectioning → see problem 2. If this is the case for all sections, a custom segmentation mask can be created, see below.
Check if the input image resolution matches 1um/px. If not, scale images accordingly, using Fiji>Image>Scale and re-run the analysis. Alternatively, a custom segmentation mask can be created based on an example input image using SpinalJ > Other Tools > Create Segmentation Mask. As input, browse a single channel tiff image of a block section that shows nine tissue sections.
Problem 5
SpinalJ – Bad atlas alignment.
Related to steps 19 & 21. Experimental dataset does not align well to reference template.
Potential solution
Make sure to select the correct segment range for the experimental dataset. To identify start and end segments, refer to the Allen Spinal Cord Atlas (https://mousespinal.brain-map.org/imageseries/showref.html). When re-adjusting segment range, make sure to delete “DAPI_25.tif” and “DAPI_25_Mask.tif” in folder /3_Registered_Sections/ and re-run section registration and atlas mapping.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Jane Dodd (jd18@columbia.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank Samaher Fageiry for technical advice on SC tissue preparation and histology, Marcela Carmona and Anna Kim for help with cortical virus injections, Crystal K. Colón Ortiz for providing mouse eye samples, and Amy Norovich and Young Mi Kwon for providing betta fish brains. We thank Susan B. Morton and the Jessell Lab for gifts of custom antibodies. We are grateful to Carl E. Schoonover and Sarah Ohashi for help with 3D printing and thank Isobel Jessell for the term “SpineRack”. Thank you to the Mason-Dodd and Bendesky labs for discussion and Christoph Gebhardt for critical comments on the manuscript. Imaging was performed with support from the Zuckerman Institute’s Cellular Imaging platform. This work was funded by NIH grants 1R21NS120665-01, 5U19NS104649, and DFG grant FI 2367/1-1.
Author contributions
The study was conceptualized and directed by F.F., J.D., and C.M. F.F. developed SpineRacks and performed histology studies, imaging, and data interpretation. L.A.H. and F.F. developed SpinalJ software. D.N. provided VAChT/Foxp1 mice for validation. F.F., L.A.H., and J.D. wrote the manuscript with input from all authors.
Declaration of interests
The authors declare that they have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100897.
Contributor Information
Felix Fiederling, Email: ff2397@columbia.edu.
Jane Dodd, Email: jd18@columbia.edu.
Data and code availability
The SpinalJ package created in Fiederling et al. (2021) is freely accessible through Github: https://github.com/felixfiederling/SpinalJ and Zenodo: https://doi.org/10.5281/zenodo.5177185. Example raw and processed image data has been deposited at Mendeley Data: https://data.mendeley.com/datasets/mkc37w4rrv/1.
References
- Allen Institute for Brain Science . Allen Institute for Brain Science; 2008. Allen Spinal Cord Atlas.https://mousespinal.brain-map.org/imageseries/showref.html Available at: [Google Scholar]
- Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. Ilastik: Interactive machine learning for (Bio)Image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- Biot E., Crowell E., Höfte H., Maurin Y., Vernhettes S., Andrey P. 2008 5th IEEE Int. Symp. Biomed. Imaging from Nano to Macro, Proceedings, ISBI. 2008. A new filter for spot extraction in N-dimensional biological imaging; pp. 975–978. [Google Scholar]
- Evilsizor M.N., Ray-Jones H.F., Lifshitz J., Ziebell J. Primer for immunohistochemistry on cryosectioned rat brain tissue: example staining for microglia and neurons. J. Vis. Exp. 2015;2015:1–9. doi: 10.3791/52293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiederling F., Hammond L.A., Ng D., Mason C., Dodd J. Tools for efficient analysis of neurons in a 3D reference atlas of whole mouse spinal cord. Cell Rep. Methods. 2021;1:100074. doi: 10.1016/j.crmeth.2021.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy H.S., Jones C., Caplazi P. Comparison of standard laminectomy with an optimized ejection method for the removal of spinal cords from rats and mice. J. Histotechnol. 2013;36:86–91. doi: 10.1179/014788813X13756994210382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B., Schindelin J., Cardona A., Longair M., Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Animation through a stack of 1302 25μm registered sections (Neurotrace channel) covering segments C1-S3 and the same data re-sliced in the horizontal and sagittal plane. Data is provided in the example dataset.
Data Availability Statement
The SpinalJ package created in Fiederling et al. (2021) is freely accessible through Github: https://github.com/felixfiederling/SpinalJ and Zenodo: https://doi.org/10.5281/zenodo.5177185. Example raw and processed image data has been deposited at Mendeley Data: https://data.mendeley.com/datasets/mkc37w4rrv/1.