Abstract
Background
Metabolic syndrome (MetS) is a cluster of multiple cardiometabolic risk factors that increase the risk of type 2 diabetes and cardiovascular diseases. Identifying novel biomarkers of MetS and their genetic associations could provide insights into the mechanisms of cardiometabolic diseases.
Methods
Potential MetS-associated metabolites were screened and internally validated by untargeted metabolomics analyses among 693 patients with MetS and 705 controls. External validation was conducted using two well-established targeted metabolomic methods among 149 patients with MetS and 253 controls. The genetic associations of metabolites were determined by linear regression using our previous genome-wide SNP data. Causal relationships were assessed using a one-sample Mendelian Randomization (MR) approach.
Findings
Nine metabolites were ultimately found to be associated with MetS or its components. Five metabolites, including LysoPC(14:0), LysoPC(15:0), propionyl carnitine, phenylalanine, and docosapentaenoic acid (DPA) were selected to construct a metabolite risk score (MRS), which was found to have a dose-response relationship with MetS and metabolic abnormalities. Moreover, MRS displayed a good ability to differentiate MetS and metabolic abnormalities. Three SNPs (rs11635491, rs7067822, and rs1952458) were associated with LysoPC(15:0). Two SNPs, rs1952458 and rs11635491 were found to be marginally correlated with several MetS components. MR analyses showed that a higher LysoPC(15:0) level was causally associated with the risk of overweight/obesity, dyslipidaemia, high uric acid, high insulin and high HOMA-IR.
Interpretation
We identified five metabolite biomarkers of MetS and three SNPs associated with LysoPC(15:0). MR analyses revealed that abnormal LysoPC metabolism may be causally linked the metabolic risk.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFC0907004).
Key Words: Metabolic syndrome, Metabolomics, mGWAS, Biomarkers, Zhejiang Metabolic Syndrome Cohort
Research in context.
Evidence before this study
Abnormal plasma metabolite levels have been reported to be associated with metabolic syndrome in previous human metabolomics studies. However, most of them lack of external replication, especially in the Chinese population. Furthermore, the genetic determinants for these metabolites remain to be uncovered. It has been demonstrated that identifying the genetic determinants of metabolites could improve our understanding of their role in diseases.
Added value of this study
Nine metabolites were ultimately found to be associated with metabolic syndrome or its components by multi-stage metabolomic analyses. Five metabolites, namely: LysoPC(14:0), LysoPC(15:0), propionyl carnitine, phenylalanine, and docosapentaenoic acid (DPA) were used to develop a metabolic risk score for MetS, and showed a good discriminating ability for MetS and metabolic abnormalities. Three SNPs including rs11635491, rs7067822, and rs1952458 were identified in association with the LysoPC(15:0) levels. Additionally, LysoPC(15:0) was found causally linked to multiple metabolic abnormalities.
Implications of all the available evidence
Our findings highlight that a higher leve of lysophosphatidylcholine is a marker of metabolic syndrome and metabolic abnormalities. Disturbed lysophosphatidylcholine metabolism may be a potential therapeutic target for metabolic syndrome.
Alt-text: Unlabelled box
1. Introduction
Metabolic syndrome (MetS) is a cluster of multiple cardiometabolic factors, including central obesity, hypertension, dyslipidaemia, and hyperglycaemia [1]. MetS increases the risk of type 2 diabetes, stroke, coronary heart diseases (CHD) and other disabilities [1]. Despite previous studies, the pathogenesis of MetS has not been fully elucidated. Identifying the altered metabolites in MetS is particularly important for the prevention and intervention of MetS and its associated complications.
MetS is a syndrome in which multiple metabolic abnormalities are simultaneously present within an organism. A systematic view is necessary for its mechanistic understanding [2]. As a part of system biology, advanced metabolomics could assist us in gaining comprehensive knowledge of the biological processes of MetS [3]. Considerable human metabolomics studies have identified multiple metabolites and lipids in relation to obesity, type 2 diabetes, dyslipidaemia, and cardiometabolic diseases [4], [5], [6], [7]. However, relevant research on MetS is scarce, and the sample size and the coverage of metabolites are relatively small [8], [9], [10], [11], [12]. In addition, the identified associations of metabolites often lacked replication, which prevented us from a systemic understanding of the pathogenesis of MetS [9,12].
However, although prior studies have detected several metabolites associated with MetS, their pathophysiological mechanisms remain poorly understood. It has been proposed that identifying the genetic determinants of metabolites would facilitate the mechanistic understanding of metabolites and their clinical endpoints [13], [14], [15]. Previous large-scale genome-wide association studies have reported a hundred gene loci regulating metabolite concentrations and found that some of these metabolic loci were located at gene encoding enzymes or transporters [14,16]. By integrating the genomic, metabolomics, and clinical trait data, Eugene Rhee et al. identified 31 genetic loci associated with 61 metabolites and demonstrated the role of AGXT2 in lipid metabolism [14]. Likewise, Jaana Hartiala et al. reported that two gene loci on 2q34 and 5q14.1 were associated with plasma betaine levels, and suggested that rs715 on 2q24 may contribute to a decreased risk of CAD in women, highlighting the potential of genome-wide association studies of metabolites in the exploration of disease mechanisms [17].
In this study, using untargeted and targeted metabolomics, we aimed to identify the novel metabolite biomarkers of MetS and its components in a relatively large population. In addition, a two-stage GWAS analysis was designed to determine the genetic associations for the replicated MetS-associated metabolite biomarkers, and the possible causal relationships between metabolites and MetS and its components will also be assessed.
2. Methods
2.1. Study design and subjects
Figure S1 presents the study design. First, using untargeted metabolomic analyses, we screened and internally validated the MetS-associated metabolites among 120 MetS patients and 120 controls matched by age and sex, and 575 MetS patients and 582 controls, respectively. Both populations were randomly selected from our previous cross-sectional survey on MetS in Hangzhou, Zhejiang, China [18]. External replication was performed using a targeted metabolomics approach in a sub-population of the Zhejiang Metabolic Syndrome Cohort, namely Xiazhi sub-cohort (N=1420). According to the same MetS criterion adopted in the untargeted metabolomics [19], 149 MetS patients and 253 controls were included into the external replication dataset. The detailed sample selection process has been described in the supplementary materials. A total of 1062 individuals from the internal validation dataset and 227 individuals from the discovery dataset that had good-quality genome-wide SNP data were included to discover and replicate the associations between SNPs and metabolites.
According to the Chinese Diabetes Society's criteria for MetS, the subjects with three or more metabolic abnormalities were defined as MetS [18], including body mass index (BMI)≥ 25·0 kg/m2 (overweight/obesity), systolic blood pressure (SBP) ≥140 mmHg or diastolic blood pressure (DBP) ≥ 90 mmHg (high BP), fasting plasma glucose (FPG) ≥ 6·1 mmol//L (high FPG), triglyceride (TG) ≥ 1·7 mmol//L or high-density lipoprotein cholesterol (HDL-C) <1·0 mmol//L in women or <0·9 mmol//L in men (dyslipidaemia). The healthy controls were free of metabolic disorders. The individuals who had metabolic-related interventions or had cancer or serious chronic liver, lung, heart, or kidney disorders were excluded.
The study protocol was approved by the research ethics committee at the School of Medicine, Zhejiang University. All participants provided informed consent.
2.2. Epidemiological investigation and clinical measurements
The epidemiological data was collected using a questionnaire-based interview by trained investigators using a standardised protocol. The information on age, sex, smoking, alcohol drinking behavior, drug use, and history of cardiovascular diseases, type 2 diabetes, hypertension, cancer, liver diseases, and kidney diseases was investigated.
The weight, height, SBP, and DBP were measured by experienced nurses using a standardised protocol. BMI was calculated as the body weight in kilogram divided by the square of the height in meters. The blood pressure values were measured in a sitting position using a mercury sphygmomanometer. The values of SBP and DBP were reported as the average of the three repeat measurements at 30-s intervals.
An overnight blood sample was collected for each subject and immediately frozen at -80 Celsius degrees. The serum TG, total cholesterol (TC), HDL-C, low-density lipoprotein cholesterol (LDL-C), uric acid (UA), FPG, and fasting insulin levels were measured using clinical methods. The homeostatic model assessment for insulin resistance (HOMA-IR) was calculated as fasting insulin (µU/L) × fasting glucose (nmol/L)/22·5.
2.3. Untargeted metabolic profiling
Untargeted metabolic profiling was performed with an Agilent 1290 infinity with 6545 Q/TOF-MS system (Agilent Technologies) under the positive and negative ion modes. The detailed methods for sample preparation, UPLC-Q/TOF-MS analyses, and data pre-processing have been published previously [20]. A total of 1793 ion features were detected in the discovery dataset. A total of 2238 ion features were detected in the internal validation dataset. The metabolites were identified by searching public metabolomics databases (The Human Metabolome Database and METLIN) and confirmed using available in-house reference compounds.
2.4. Targeted metabolomics analysis
A targeted metabolomics profiling was conducted at Shanghai Applied Protein Technology Co., Ltd, using high-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). The reagents and internal standards (IS) are described in Table S1. Dichloromethane was used to extract the serum metabolites. The detailed chromatographic conditions and MS parameters are provided in the supplementary materials and Table S2. The validation data for this method are shown in Table S3.
2.5. Genotyping, imputation, and quality control (QC)
Genotyping was conducted using Illumina Human-OmniExpress 760k chips (Illumina, San Diego, CA, USA) with standard quality-control procedures. The University of Michigan imputation server (http://imputationserver.sph.umich.edu/index.html) was used to conduct genotype imputation. Before imputation, all alleles were aligned to the forward strand of build 37 and converted to VCF files. The genotype was phased with EAGLE with 1000 Genomes Project Phase 3 Version 5 of EAS as the reference panel [21]. A detailed quality control procedure is provided in the supplementary materials. Consequently, 40,001,312 SNPs remained.
2.6. Statistical analysis
Normal distribution was visually checked by the Q-Q plots for all continuous variables. Variables with normal distribution were described as mean (standard deviation) and were compared using the Student's t-test, unless were described as median (interquartile range) and were compared using the Mann-Whitney U test. The categorical variables were described as numbers (percentages) and were compared using the chi‐square test.
In untargeted metabolomics analyses, the Mann-Whitney U test and orthogonal partial least-squared discriminant analysis (OPLS-DA) were used to select the differential features of MetS. The spearmen's correlation was used to analyse the correlations between the metabolites and MetS components. External validation was conducted on scaled data using logistic regression analysis after adjusting for age and sex. A detailed statistical analysis is shown in the supplementary materials.
Among the metabolites that were successfully replicated to be associated with MetS or its components, a stepwise logistic regression was utilised to select the metabolites to construct an additive metabolite risk score (MRS) weighted by the effect size of each metabolite. The associations between MRS and MetS and its components were analysed using a logistic regression analysis. The diagnostic value was evaluated using the receiver operating characteristic curve (ROC).
A GWAS analysis was performed with linear regression (for continuous variables) or logistic regression (for dichotomous variables) under an additive genetic model in PLINK2. A detailed data analysis is provided in the supplementary materials section. A meta-analysis was performed using inverse variance model implemented in the METAL software. Q-Q and Manhattan plots were drawn using the R package “qqmen”. Significant regions were visualised using the online tool Locustrack based on the EAS population.
Causal relationships were assessed using a one-sample MR approach with the additive weighted genetic risk score (wGRS) as the instrumental variable (IV). In addition, a two-sample MR analysis was conducted using summary statistics from Japan Biobank to replicate the causal associations between the metabolites and TG, HDL-C, SBP, DBP and BMI. The inverse variance weighted (IVW) method was used as the main analysis. The weighted median and MR-Egger methods were supplemented as sensitivity analyses. The MR approach must meet the following assumptions: 1) the IV is robustly associated with the interest exposure; 2) the IV is not associated with any potential confounders of the exposure-outcome relationship; and 3) the IV only affects the outcome through the exposure (no horizontal pleiotropy). The detailed study design is shown in the supplementary text and Figure S2. The power calculation for MR analyses was preformed using an online tool (https://shiny.cnsgenomics.com/mRnd/).
A two-tailed test was used for all statistical analyses in this study. All statistical analyses were conducted using R software 4.0.3.
2.7. Role of funding source
The funding sources had no role in the study design, data collection, data analyses, interpretation, or writing of the report of this manuscript.
3. Results
3.1. Subject characteristics
The demographic characteristics of the participants are summarised in Table 1. A total of 240, 1157, and 402 eligible subjects were recruited for the discovery, internal validation, and external validation datasets, respectively. Among them, 50%, 49·7% and 37·1% had MetS, respectively. A total of 60·8%, 50·5%, and 40·5% were men, respectively. The average age was 62·4, 56·8, and 58·6 years, respectively. The distributions of individual MetS components in the three datasets are displayed in Figure S3.
Table 1.
Baseline characteristics of the subjects
| Discovery (n=240) | Internal validation (n=1157) | External validation (n=402) | P | |
|---|---|---|---|---|
| MetS/Control (n) | 120/120 | 575/582 | 149/253 | <0.001 |
| Men (%) | 60.8 | 50.5 | 40.5 | <0.001 |
| Age, years † | 62.4 (0.8) | 56.8 (0.3) | 58.6 (0.6) | <0.001 |
| BMI (kg/m2) † | 23.92 (0.21) | 24.32 (0.11) | 23.19 (0.17) | <0.001 |
| WC (cm) † | 85.92 (0.69) | 82.2 (0.31) | 78.42 (0.54) | <0.001 |
| SBP (mmHg) † | 141.68 (1.64) | 139.5 (0.69) | 127.77 (1.14) | <0.001 |
| DBP (mmHg) † | 82.97 (0.88) | 82.94 (0.38) | 77.89 (0.56) | <0.001 |
| TG (mmol/L) * | 1.64 (1.15) | 1.61 (1.26) | 1.03 (0.84) | <0.001 |
| HDL-C (mmol/L) † | 1.51 (0.02) | 1.51 (0.01) | 1.28 (0.02) | <0.001 |
| FPG (mmol/L) * | 5.16 (1.14) | 5.12 (1.08) | 6.01 (0.13) | <0.001 |
| Insulin * | 3.6 (3.2) | 3.8 (3.3) | - | 0.23 |
| HOMA-IR * | 0.86 (0.98) | 0.90 (0.94) | - | 0.38 |
Data was presented as means (standard deviation) or median (interquartile range) or numbers (percentages). * denotes that data was presented as median (interquartile range). † denotes that data was presented as means (standard deviation). MetS, metabolic syndrome; BMI, body mass index; WC, waist circumference; SBP, systolic blood pressure; DBP, diastolic blood pressure; TG, triglyceride; HDL-C, high density lipoprotein cholesterol; FPG, fasting plasma glucose; HOMA-IR, homeostasis model assessment of insulin resistance.
3.2. Associations of metabolites with MetS in the discovery and internal validation datasets
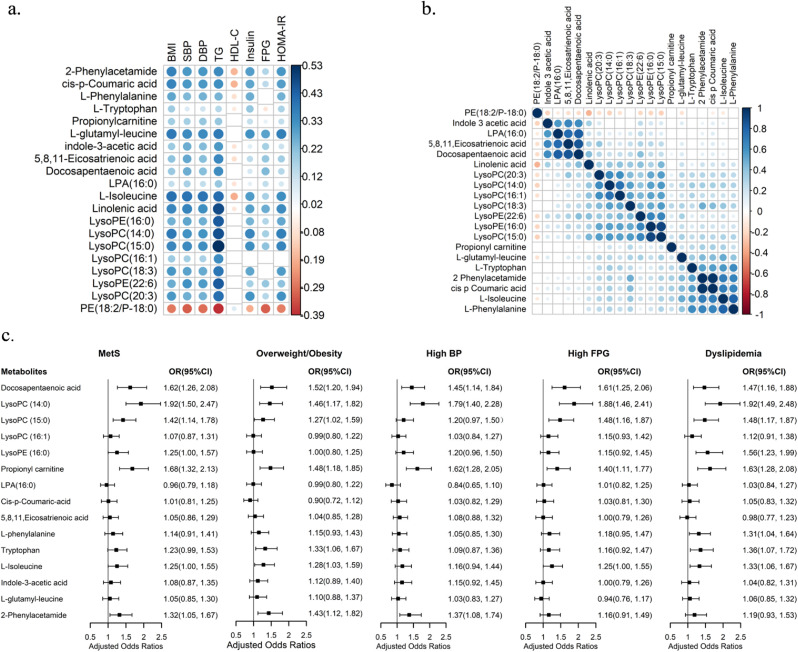
In the discovery dataset, nine amino acids, nine fatty acids or fatty acyls, six organic compounds and 10 glycerophospholipids were identified to be positively associated with MetS. It was observed that one amino acid and one glycerophospholipid were negatively associated with MetS (Table S4). In the internal validation dataset, three amino acids, four fatty acids or fatty acyls, four organic compounds and nine glycerophospholipids were ultimately confirmed as potential metabolite biomarkers of MetS (Table 2). Most of these metabolites had significant and modest correlations with MetS components, except for low HDL-C (Figure 1a). Two amino acids (L-isoleucine and L-phenylalanine) and two organic compounds (2-phenylacetamide and cis-p-coumaric acid) were found to be highly correlated with each other; additionally, LysoPCs and LysoPEs were also highly associated with each other (Figure 1b).
Table 2.
20 potential metabolite biomarkers of MetS in the internal validation dataset (N=1157)
| RT (min) | Mass to charge ratio | FC | VIP | P | Ion Mode | category | |
|---|---|---|---|---|---|---|---|
| L-Isoleucine* | 1.028 | 132.102 | 0.87 | 2.99 | 3.16E-47 | + | Amino acids |
| L-Phenylalanine* | 1.381 | 166.0864 | 0.90 | 2.11 | 3.67E-22 | + | Amino acids |
| L-Tryptophan* | 2.152 | 205.0971 | 0.93 | 1.19 | 2.81E-08 | + | Amino acids |
| 5,8,11-Eicosatrienoic acid* | 10.408 | 305.2492 | 0.80 | 1.94 | 3.79E-16 | - | Fatty acids |
| Docosapentaenoic acid* | 10.387 | 329.2487 | 0.78 | 2.01 | 2.01E-16 | - | Fatty acids |
| Linolenic acid* | 8.863 | 279.2317 | 0.67 | 2.99 | 4.44E-42 | + | Fatty acids |
| Propionyl carnitine | 1.031 | 218.1382 | 0.86 | 1.22 | 3.75E-10 | + | Fatty Acyls |
| LPA(16:0) | 10.189 | 409.2359 | 0.91 | 1.02 | 1.68E-06 | - | Glycerophospholipids |
| LysoPC(14:0) * | 7.373 | 468.3088 | 0.69 | 2.85 | 9.92E-43 | + | Glycerophospholipids |
| LysoPC(15:0) * | 8.448 | 482.3243 | 0.72 | 3.03 | 9.67E-43 | + | Glycerophospholipids |
| LysoPC(16:1) | 7.588 | 494.3247 | 0.82 | 1.72 | 2.10E-15 | + | Glycerophospholipids |
| LysoPC(18:3) | 7.937 | 518.3227 | 0.91 | 2.19 | 1.77E-29 | + | Glycerophospholipids |
| LysoPC(20:3) | 8.006 | 546.3542 | 0.81 | 2.26 | 2.92E-27 | + | Glycerophospholipids |
| LysoPE(16:0) * | 7.905 | 454.293 | 0.76 | 2.40 | 4.82E-28 | + | Glycerophospholipids |
| LysoPE(22:6) | 7.796 | 526.293 | 0.79 | 2.51 | 8.78E-27 | + | Glycerophospholipids |
| PE(18:2/P-18:0) | 11.27 | 728.5665 | 1.35 | 2.49 | 2.42E-31 | + | Glycerophospholipids |
| Indole-3-acetic acid | 4.839 | 174.054 | 0.78 | 1.54 | 1.90E-14 | - | Organic compounds |
| 2-Phenylacetamide* | 0.877 | 136.0757 | 0.87 | 2.65 | 1.57E-33 | + | Organic compounds |
| cis-p-Coumaric acid* | 0.878 | 165.0548 | 0.87 | 2.54 | 1.75E-32 | + | Organic compounds |
| L-glutamyl-leucine* | 2.694 | 261.1441 | 0.71 | 2.61 | 4.44E-42 | + | Organic compounds |
RT, retention time; FC, fold change (control/MetS). FC<1 means that the metabolite is upregulated in MetS; VIP, variable importance in the projection, which is calculated by orthogonal partial least-squared discriminant analysis and reveals the contribution of each variable for the discrimination between the MetS patients and controls. P values are corrected by the false discovery rate. The primary metabolite identification is based on its RT and mass to charge ratio. * means that metabolites were confirmed with available reference compounds.
Figure 1.
Associations between metabolites and between metabolites and MetS and its components. a: The associations between 20 MetS-associated metabolites and MetS components in the internal validation dataset (N=1157). The cells in blank signify that the correlation was non-significant (P>0.05); b: The associations between the 20 MetS-associated metabolites in the internal validation dataset (N=1157). The cells in blank signify that the correlation was non-significant (P>0.05); c: The associations of metabolites with MetS and its components in the external validation dataset (N=402). The OR was adjusted for age and sex. MetS, metabolic syndrome; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; TG, triglyceride; HDL-C, high density lipoprotein cholesterol; FPG, fasting plasma glucose; HOMA-IR, homeostasis model assessment of insulin resistance; OR, odds ratio; CI, confidence interval.
3.3. Associations of metabolites with MetS in the external validation dataset
A total of 15 metabolites with the available standard chemicals were quantified. After adjusting for age and sex, five metabolites were found to be positively associated with MetS, namely: DPA, LysoPC(14:0), LysoPC(15:0), propionyl carnitine and 2-phenylacetamide (Figure 1c). Three metabolites, namely: L-tryptophan, L-isoleucine, and LysoPE(16:0), were found marginally and positively associated with MetS (Figure 1c). Nine metabolites were associated with at least one MetS component, including eight metabolites associated with MetS and L-phenylalanine (Figure 1c).
DPA and LysoPC(14:0) were significantly associated with MetS in both sexes. LysoPC(15:0), propionyl carnitine, and tryptophan were significantly associated with MetS in women, but not in men. In addition, a positive relationship between indole-3-acetic acid and MetS was found in women. Only men displayed a positive association between L-isoleucine and MetS (Table S5). The sensitivity analyses showed similar association results (Table S6).
3.4. Associations of metabolite risk score (MRS) with MetS and its components
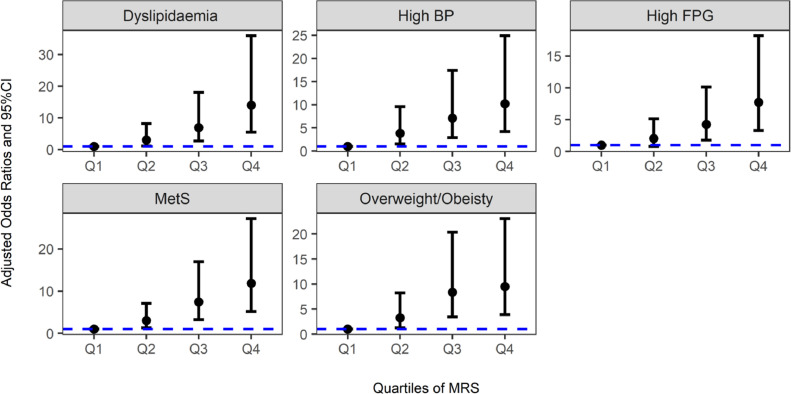
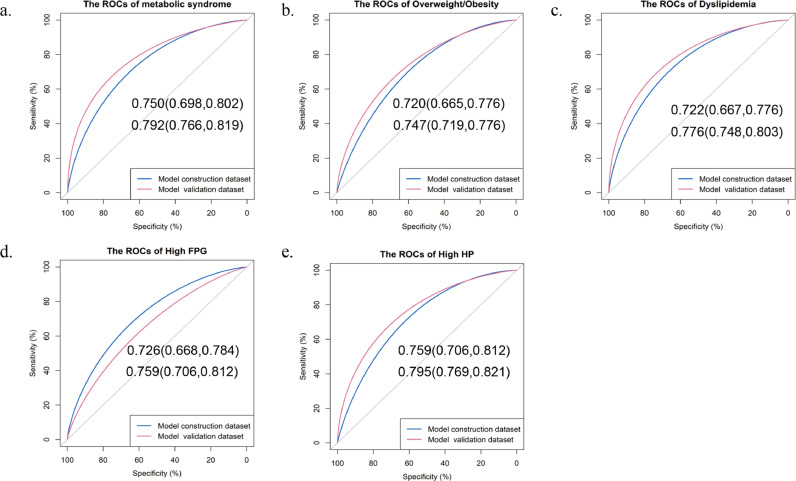
A panel of five metabolites was selected to construct a MRS, including DPA, LysoPC(14:0), LysoPC(15:0), propionyl carnitine, and L-phenylalanine. Figure 2 and Table S7 describe that with increasing quartiles of MRS, a higher presence of MetS and metabolic abnormalities was observed. Furthermore, the MRS displayed a good ability to differentiate MetS from the control, with an AUC of 0·750 (95%CI: 0·698–0·802) and 0·792 (95%CI: 0·766–0·819) in the model construction and validation datasets, respectively (Figure 3a). Similar results were observed for each MetS component (Figure 3b-e).
Figure 2.
Associations of quartiles of metabolite risk score (MRS) with MetS and its components in the external validation dataset (N=402). The OR was adjusted for age and sex. BP, blood pressure; FPG, fasting plasma glucose; OR, odds ratio; CI, confidence interval.
Figure 3.
ROCs of the metabolite risk score on MetS and its components in the model construction dataset (N=402) and model validation dataset (N=1157). a: ROC for metabolic syndrome; b: ROC for overweight/obesity; c: ROC for dyslipidaemia; d: ROC for high FPG; e: ROC for high HP. Data were shown as AUC (95%CI). BP, blood pressure; FPG, fasting plasma glucose; ROC, receiver operating characteristic curve; AUC, area under curve; CI, confidence interval.
3.5. Two-stage GWAS analyses for the five metabolite biomarkers of MetS
The characteristics of the subjects in the GWAS analyses are showed in Table S8. The genetic inflation factors for the five metabolites ranged from 0·997 to 1·006, suggesting little evidence of population stratification (Table S9). The Q-Q and Manhattan plots are drawn at Figure S4-S5. In the discovery stage, only two SNPs located in gene LIPC were identified in association with LysoPC(15:0); additionally, 131 suggestive SNPs were associated with at least one metabolite (Table S10). In the validation stage, three SNPs, rs11635491, rs1952458, and rs7067822 were reported to be associated with LysoPC(15:0), with P values <0·05 and consistent effect directions (Table 3, Table S11, Figure S6). The regional plots for the three SNPs are presented in Figure S7.
Table 3.
The SNP-metabolite associations identified in the two-stage GWAS
| SNP | CHR | POS | REF | ALT | MAF | Gene hit | Discovery Stage |
Validation Stage |
P combined | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BETA | SE | P | BETA | SE | P | |||||||||
| LysoPC(15:0) | rs11635491 | 15 | 58719741 | G | A | 0.37 | LIPC | 0.19 | 0.04 | 2.93E-09 | 0.25 | 0.1 | 0.015 | 1.47E-10 |
| rs7067822 | 10 | 110453691 | A | G | 0.61 | - | -0.19 | 0.04 | 5.46E-06 | -0.19 | 0.09 | 0.031 | 4.89E-07 | |
| rs1952458 | 6 | 51496168 | T | C | 0.54 | PKHD1 | 0.21 | 0.05 | 1.28E-05 | 0.29 | 0.11 | 0.044 | 1.53E-06 | |
SNP, single nucleotide polymorphism; CHR, chromosome; POS, position; REF. reference allele; ALT, alternative allele; MAF: minor allele frequency; SE, standard error.
Table S12 presented that out of the three SNPs, rs1952458 was also nominally associated with BMI, TG, uric acid, HOMA-IR, insulin, and MetS. rs11635491 was nominally associated with fasting insulin levels and HOMA-IR.
3.6. Causal relationships of LysoPC(15:0) with MetS and its components
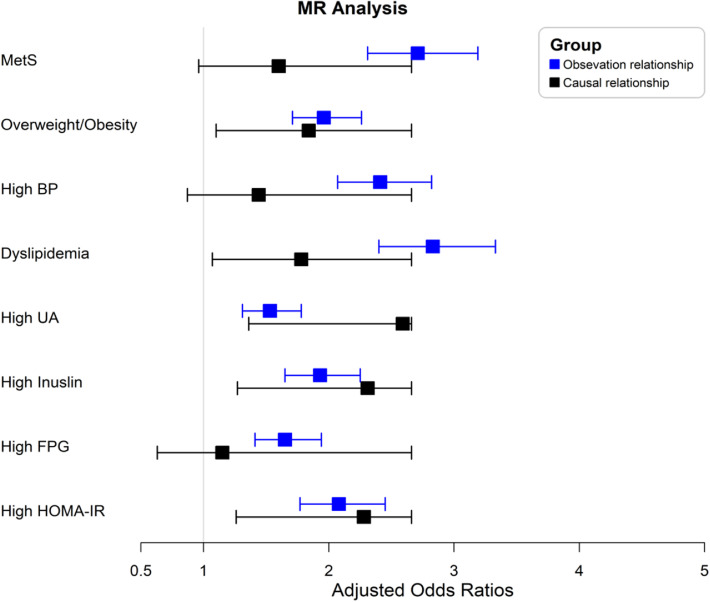
Three SNPs associated with LysoPC(15:0) were used to construct a gene risk score. This gene risk score then was used as the IV of LysoPC(15:0). The F statistic for the IV was 69·58. After adjusting for age and sex, observational associations revealed that higher LysoPC(15:0) levels were significantly associated with a higher risk of MetS and all listed metabolic abnormalities. For the causal estimates, the one-sample MR analysis showed that higher genetic predicted levels of LysoPC(15:0) were positively associated with the risk of overweight/obesity, dyslipidaemia (particularly due to the increased serum TG levels), high UA, high insulin, and high HOMA-IR, but not with MetS and high BP (P>0·05, Figure 4, Table S13-S14). In the two-sample MR analysis, the standard IVW analysis showed that genetically estimated higher LysoPC(15:0) levels were significantly associated with increased serum TG and HDL-C levels and no pleiotropy effect was indicated. No significant causal relationships between LysoPC(15:0) and BMI, SBP, DBP were found, and the power calculation indicates that there is limited statistical powers (<6%) to detect this moderate effect estimate (Table S15).
Figure 4.
MR analysis between LysoPC(15:0) and MetS and metabolic abnormalities. The OR was adjusted for age and sex. MetS, metabolic syndrome; BP, blood pressure; UA, uric acid; FBG, fasting blood glucose; HOMA-IR, homeostasis model assessment of insulin resistance; MR, mendelian randomization. The cutoff value for high UA, high HOMA-IR, and high insulin was based on their respective 75th percentile.
4. Discussion
Our study has two main findings. First, we screened and replicated nine metabolites associated with MetS or its components. An MRS based on five replicated metabolites showed a satisfactory discriminating ability for MetS and metabolic abnormalities, including DPA, LysoPC(14:0), LysoPC(15:0), propionyl carnitine, and L-phenylalanine. Second, we identified three SNPs associated with the LysoPC(15:0) levels. Two SNPs were found to be nominally correlated with MetS or its components. Moreover, genetically predicted higher LysoPC(15:0) was causally linked to a higher metabolic risk. Overall, our findings revealed the metabolic signature of MetS and suggested that disturbed lysophosphatidylcholine metabolism may be involved in the pathology of MetS.
Lysoglycerophospholipids (LPLs) is a class of bioactive phospholipids that are involved in multiple cellular activities, such as inflammatory response, cell proliferation, and cellular signal transductions [22,23]. Lysophosphatidylcholine (LysoPC) is the most abundant LPL in human plasma and can be generated by cleaving phosphatidylcholine (PC) via phospholipase A2 (PLA2) or by the transfer of fatty acids to free cholesterol via lecithin-cholesterol acyltransferase (LCAT) [23]. Previous studies have found that LysoPC participates in insulin resistance by disrupting hepatic fatty acid oxidation and insulin signal transduction [22,[24], [25], [26]]. LysoPC is also a component of ox-LDL, which can induce the formation of atherosclerotic plaques by stimulating the expression of inflammatory cytokines [22,27]. However, controversial results have been reported in epidemiological studies [24,28,29]. In line with this study, Kim et al. reported increased levels of 12 LysoPC species in metabolically unhealthy obese subjects, and found that the LysoPC levels were positively associated with several oxidative stress markers, including ox-LDL, 8-epi-PGF2α, and Lp-PLA2, suggesting that the altered LysoPC levels may be related to increased oxidative stress [24]. Additionally, Li et al. demonstrated that a high-fat diet induced an increased abundance of serum LysoPC(15:0) in NAFLD mice [30]. However, a recent lipidomic study on MetS identified converse relationships between LysoPCs and metabolic risk [28]. Although several population studies found that several LysoPCs (18:2, 18:1, 18:0) were associated with a reduced risk of T2D [31,32], no relevant studies have investigated the effect of LysoPCs (14:0, 15:0). Given that our one-sample MR analysis showed consistent causal relationships between LysoPC(15:0) and the cardiometabolic risk, such as obesity, dyslipideamia, insulin resistance, we speculated that this LysoPC species may be a promising target for metabolic diseases. Future functional studies are required to elucidate these mechanisms. Notably, in the two-sample MR analysis, only the causal association between LysoPC(15:0) and serum TG was well-replicated. A possible explanation was the relatively low statistical power and the variance explained by the IV of LysoPC(15:0). Genomic researches with larger sample size are required to discover more SNPs that associated with this metabolite.
Isoleucine has been demonstrated to be associated with obesity, MetS, and type 2 diabetes [25,33]. Newgard et al. found that a high-fat and branched-chain amino acid (BCAA)-rich diet induced insulin resistance and increased plasma C3 acylcarnitine levels, indicating that C3 acylcarnitine may be a direct product of BCAA metabolism [33]. Consistently, both previous studies and our study confirmed that higher C3 acylcarnitine levels were a risk factor for MetS [34], [35], [36]. Acylcarnitine plays a critical role in carbohydrate and lipid metabolism balance. Elevated acylcarnitine levels represent a disruption in fatty acid oxidation and mitochondrial function [37]. Altered plasma acylcarnitine levels have been previously indicated as promising biomarkers of obesity, insulin resistance, and type 2 diabetes, especially C3 carnitine. Plasma C3 acylcarnitine levels could reflect its levels in tissues in a better manner, whereas other species could not [36,[38], [39], [40]].
Previous studies on the relationship between DPA and MetS have been inconclusive. Only one study in children found that the levels of DPA in adipose tissue were positively associated with the MetS score [41]. In several Chinese epidemiological studies, DPA were found to be negatively associated with MetS [42], [43], [44]. A small number of interventional studies in rodents found that DPA lowered the serum triglyceride levels and pro-inflammatory cytokines and improved the insulin resistance [45]. However, genetic studies have reported that the C allele of rs174547 located at FADS1 and the T allele of rs780094 located at GCKR were linked to higher DPA levels [46]. Meanwhile, these risk alleles were also associated with a higher risk of MetS, indirectly supporting our study [47,48]. Therefore, whether DPA is beneficial to MetS needs to be further investigated.
Phenylalanine and tryptophan are aromatic amino acids (AAA) and have been frequently reported to be associated with multiple metabolic abnormalities [10,25,49,50]. Some researchers have proposed that an elevated phenylalanine level may be attributed to the competition of BCAA for the transportation of AAA [33,51]. Tryptophan is composed of a β carbon connected to an indole group, which can produce multiple signalling substances via the kynurenine, indole, and serotonin pathways [52]. Supporting our results, Ji Sun Oh et al. found that patients with MetS showed increased levels of tryptophan and its two downstream products (kynurenine and xanthurenic acid) and a higher tryptophan-to-kynurenine ratio [53]. The tryptophan-to-kynurenine ratio is an indicator of kynurenine pathway, and its upregulation was related to the low-grade inflammation state, oxidative stress, and immune response, which suggests that abnormal tryptophan levels may impact MetS by impairing the kynurenine pathway [54,55].
By using metabolites as the outcome measures of GWAS, Met-QTL has been a powerful tool in explaining the biological relevance of SNPs to a certain disease. In this study, three met-QTLs were identified to be associated with LysoPC(15:0). One of them, rs1163549, is located at the LIPC locus and displays strong eQTL signals of LIPC. Moreover, previous studies have reported that polymorphisms in LIPC are significantly associated with serum lipid profiles, MetS, T2D, and CAD risk [56], [57], [58], indicating that the disturbed serum LysoPC(15:0) levels may result from abnormal LIPC metabolic activity and LysoPC(15:0) may be a potential therapeutic target for MetS. rs1952458 is located at 6p12.3 of PKHD1. This gene mainly expressed in the kidney cortex and pancreas. The mutations in this gene cause autosomal recessive polycystic kidney disease. Patients with polycystic kidney disease have been reported to exhibit defective glucose metabolism and fatty acid oxidation [59]. In a previous GWAS, rs1952458 was found to be marginally associated with the prevalence of T2D [60]. Notably, our study showed nominal associations between rs1952458 and multiple metabolic abnormalities. Functional studies are required to determine whether this gene locus regulates metabolic processes.
Our study has several strengths. First, to the best of our knowledge, this study is the largest metabolomics study on MetS. Second, the use of untargeted metabolomics approach, targeted metabolomic approach, and the external validation provided precise and robust association results between metabolites and MetS. Third, by integrating with genomic data, we identified the genetic variants that associated with MetS-associated metabolites and MetS, providing insights into the mechanistic investigation of MetS.
This study has a few limitations. First, the biomarkers of MetS were screened among cross-sectional populations, precluding the causal relationship estimations. However, using previous SNP data, we assessed the possible causal relationships between metabolites and MetS. Second, the population of this study was from eastern China, thus all results of this study may not be generalised to other populations. A wider multi-center replication still required to verify the findings of this study in the future. Third, although our study was the largest metabolomics study on MetS, the sample size in the targeted metabolomics was relatively small, and the sample size in the GWAS analyses was also small, which may have contributed to some associations not being detected. Fourth, although we have applied optimal conditions to establish the quantitative methods in targeted metabolomics, some of the metabolites still could not achieve a good recovery rate. Fifth, information on dietary habits, nutrition, and physical activity was not collected in untargeted metabolomics, which hampered the investigation of its associations with metabolites, additionally, although using a standardised protocol and investigation techniques to reduce the possible self-reporting bias, we still cannot completely rule out the existence of self-reporting bias in the epidemiological data collection. Sixth, two instrumental SNPs used to construct the IV for LysoPC(15:0) did not arrive at the GWAS significance threshold, which may introduce a weak instrumental bias. However, the F statistic of the IV was larger than 10, indicating that the IV was sufficient for the MR analysis. Seventh, the gene pleiotropy cannot be completely avoided. Further functional genomic studies are required. Eighth, the statistical powers for the two-sample MR analysis for BMI, SBP, DBP were insufficient to detect these causal effects. Genetic association studies for this metabolite with larger sample size are urged.
5. Conclusions
We identified five metabolites that could be used as biomarkers of MetS. A metabolite risk score was constructed to assess the risk of MetS. Three SNPs were associated with LysoPC(15:0) level, additionally, LysoPC(15:0) was causally linked to the metabolic risk. In brief, MetS is associated with an altered metabolite profile, which may causally contribute to metabolic disorders.
Contributors
Conceptualisation: Maode Lai, Yimin Zhu. Data curation: Xiaohui Sun, Di He, Zongxue Cheng, Jun Li. Formal analysis: Jiankang Li. Funding acquisition:Yimin Zhu. Investigation: Qiong Wu, Xiaohui Sun, Di He, Zongxue Cheng, Jun Li. Methodology: Jiankang Li, Yongming Xie. Project administration: Maode Lai, Yimin Zhu. Resources: Yimin Zhu. Software: Qiong Wu, Xiaohui Sun. Supervision: Yimin Zhu. Validation: Qiong Wu. Visualisation: Qiong Wu. Writing – original draft: Qiong Wu. Writing– review & editing: Yimin Zhu, Xuhui Zhang.
All authors have read and approved the final version of the manuscript. Qiong Wu and Jiankang Li have verified the underlying data.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
We would like to thank all participants and investigators that took part in this study. We also would like to thank the grants from National Key Research and Development Program of China (2017YFC0907004).
Data sharing statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103707.
Contributor Information
Yimin Zhu, Email: zhuym@zju.edu.cn.
Maode Lai, Email: lmd@cpu.edu.cn.
Appendix. Supplementary materials
References
- 1.Saklayen MG. The Global Epidemic of the Metabolic Syndrome. Curr Hypertens Rep. 2018;20(2):12. doi: 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu N, Wang W, Yi M, Cheng S, Wang D. Study of the metabolomics characteristics of patients with metabolic syndrome based on liquid chromatography quadrupole time-of-flight mass spectrometry. Ann Endocrinol (Paris) 2018;79(1):37–44. doi: 10.1016/j.ando.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Lam SM, Wan Q, et al. High-Coverage Targeted Lipidomics Reveals Novel Serum Lipid Predictors and Lipid Pathway Dysregulation Antecedent to Type 2 Diabetes Onset in Normoglycemic Chinese Adults. Diabetes care. 2019;42(11):2117–2126. doi: 10.2337/dc19-0100. [DOI] [PubMed] [Google Scholar]
- 5.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126(9):1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floegel A, Kühn T, Sookthai D, et al. Serum metabolites and risk of myocardial infarction and ischemic stroke: a targeted metabolomic approach in two German prospective cohorts. European journal of epidemiology. 2018;33(1):55–66. doi: 10.1007/s10654-017-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rangel-Huerta OD, Pastor-Villaescusa B, Gil A. Are we close to defining a metabolomic signature of human obesity? A systematic review of metabolomics studies. Metabolomics. 2019;15(6):93. doi: 10.1007/s11306-019-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy P, Leong J, Jialal I. Amino acid levels in nascent metabolic syndrome: A contributor to the pro-inflammatory burden. J Diabetes Complications. 2018;32(5):465–469. doi: 10.1016/j.jdiacomp.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Shim K, Gulhar R, Jialal I. Exploratory metabolomics of nascent metabolic syndrome. J Diabetes Complications. 2019;33(3):212–216. doi: 10.1016/j.jdiacomp.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Surowiec I, Noordam R, Bennett K, et al. Metabolomic and lipidomic assessment of the metabolic syndrome in Dutch middle-aged individuals reveals novel biological signatures separating health and disease. Metabolomics. 2019;15(2):23. doi: 10.1007/s11306-019-1484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong LL, Yang S, Zhang W, et al. Discovery of metabolite profiles of metabolic syndrome using untargeted and targeted LC-MS based lipidomics approach. Journal of pharmaceutical and biomedical analysis. 2020;177 doi: 10.1016/j.jpba.2019.112848. [DOI] [PubMed] [Google Scholar]
- 12.Carioca AAF, Steluti J, Carvalho AM, et al. Plasma metabolomics are associated with metabolic syndrome: A targeted approach. Nutrition (Burbank, Los Angeles County, Calif) 2021;83 doi: 10.1016/j.nut.2020.111082. [DOI] [PubMed] [Google Scholar]
- 13.Rueedi R, Ledda M, Nicholls AW, et al. Genome-wide association study of metabolic traits reveals novel gene-metabolite-disease links. PLoS Genet. 2014;10(2) doi: 10.1371/journal.pgen.1004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee EP, Ho JE, Chen MH, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18(1):130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fall T, Salihovic S, Brandmaier S, et al. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia. 2016;59(10):2114–2124. doi: 10.1007/s00125-016-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long T, Hicks M, Yu HC, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49(4):568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 17.Hartiala JA, Tang WH, Wang Z, et al. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat Commun. 2016;7:10558. doi: 10.1038/ncomms10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Zhang D, Zhou D, et al. Susceptibility loci for metabolic syndrome and metabolic components identified in Han Chinese: a multi-stage genome-wide association study. J Cell Mol Med. 2017;21(6):1106–1116. doi: 10.1111/jcmm.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J, Zhang L, Wu Q, et al. Body roundness index is a superior indicator to associate with the cardio-metabolic risk: evidence from a cross-sectional study with 17,000 Eastern-China adults. BMC Cardiovasc Disord. 2021;21(1):97. doi: 10.1186/s12872-021-01905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Li J, Wang H, Qi LW, Zhu Y, Lai M. Tyrosine and Glutamine-Leucine Are Metabolic Markers of Early-Stage Colorectal Cancers. Gastroenterology. 2019;157(1):257–259. doi: 10.1053/j.gastro.2019.03.020. e5. [DOI] [PubMed] [Google Scholar]
- 21.Loh PR, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48(7):811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, Ke LY. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int J Mol Sci. 2019;20(5) doi: 10.3390/ijms20051149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grzelczyk A, Gendaszewska-Darmach E. Novel bioactive glycerol-based lysophospholipids: new data – new insight into their function. Biochimie. 2013;95(4):667–679. doi: 10.1016/j.biochi.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Kim M, Yoo HJ, Ko J, Lee JH. Metabolically unhealthy overweight individuals have high lysophosphatide levels, phospholipase activity, and oxidative stress. Clin Nutr. 2020;39(4):1137–1145. doi: 10.1016/j.clnu.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 25.Lent-Schochet D, McLaughlin M, Ramakrishnan N, et al. Exploratory metabolomics of metabolic syndrome: A status report. World J Diabetes. 2019;10(1):23–36. doi: 10.4239/wjd.v10.i1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soga T, Ohishi T, Matsui T, et al. Lysophosphatidylcholine enhances glucose-dependent insulin secretion via an orphan G-protein-coupled receptor. Biochem Biophys Res Commun. 2005;326(4):744–751. doi: 10.1016/j.bbrc.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 27.Kim M, Yoo HJ, Lee D, Lee JH. Oxidized LDL induces procoagulant profiles by increasing lysophosphatidylcholine levels, lysophosphatidylethanolamine levels, and Lp-PLA(2) activity in borderline hypercholesterolemia. Nutrition, metabolism, and cardiovascular diseases: NMCD. 2020;30(7):1137–1146. doi: 10.1016/j.numecd.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Yin X, Willinger CM, Keefe J, et al. Lipidomic profiling identifies signatures of metabolic risk. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantero I, Abete I, Del Bas JM, et al. Changes in lysophospholipids and liver status after weight loss: the RESMENA study. Nutrition & metabolism. 2018;15:51. doi: 10.1186/s12986-018-0288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad MI, Umair Ijaz M, Hussain M, et al. High fat diet incorporated with meat proteins changes biomarkers of lipid metabolism, antioxidant activities, and the serum metabolomic profile in Glrx1(-/-) mice. Food & function. 2020;11(1):236–252. doi: 10.1039/c9fo02207d. [DOI] [PubMed] [Google Scholar]
- 31.Wang-Sattler R, Yu Z, Herder C, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong H, Fang C, Fan Y, et al. Lipidomic profiling reveals distinct differences in plasma lipid composition in healthy, prediabetic, and type 2 diabetic individuals. GigaScience. 2017;6(7):1–12. doi: 10.1093/gigascience/gix036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Libert DM, Nowacki AS, Natowicz MR, et al. Metabolomic analysis of obesity, metabolic syndrome, and type 2 diabetes: amino acid and acylcarnitine levels change along a spectrum of metabolic wellness. PeerJ. 2018;6(1):e5410. doi: 10.7717/peerj.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JY, Park JY, Kim OY, et al. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS) J Proteome Res. 2010;9(9):4368–4375. doi: 10.1021/pr100101p. [DOI] [PubMed] [Google Scholar]
- 36.Huffman KM, Shah SH, Stevens RD, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes care. 2009;32(9):1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koves TR, Li P, An J, et al. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem. 2005;280(39):33588–33598. doi: 10.1074/jbc.M507621200. [DOI] [PubMed] [Google Scholar]
- 38.Schooneman MG, Achterkamp N, Argmann CA, Soeters MR, Houten SM. Plasma acylcarnitines inadequately reflect tissue acylcarnitine metabolism. Biochimica et biophysica acta. 2014;1841(7):987–994. doi: 10.1016/j.bbalip.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Franquesa A, Burkart AM, Isganaitis E, Patti ME. What Have Metabolomics Approaches Taught Us About Type 2 Diabetes? Current diabetes reports. 2016;16(8):74. doi: 10.1007/s11892-016-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mihalik SJ, Goodpaster BH, Kelley DE, et al. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity (Silver Spring, Md) 2010;18(9):1695–1700. doi: 10.1038/oby.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cespedes E, Baylin A, Campos H. Adipose tissue n-3 fatty acids and metabolic syndrome. Eur J Clin Nutr. 2015;69(1):114–120. doi: 10.1038/ejcn.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo XF, Li X, Shi M, Li D. n-3 Polyunsaturated Fatty Acids and Metabolic Syndrome Risk: A Meta-Analysis. Nutrients. 2017;9(7) doi: 10.3390/nu9070703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai XW, Chen YM, Zeng FF, Sun LL, Chen CG, Su YX. Association between n-3 polyunsaturated fatty acids in erythrocytes and metabolic syndrome in Chinese men and women. Eur J Nutr. 2016;55(3):981–989. doi: 10.1007/s00394-015-0912-3. [DOI] [PubMed] [Google Scholar]
- 44.Huang T, Bhulaidok S, Cai Z, et al. Plasma phospholipids n-3 polyunsaturated fatty acid is associated with metabolic syndrome. Molecular nutrition & food research. 2010;54(11):1628–1635. doi: 10.1002/mnfr.201000025. [DOI] [PubMed] [Google Scholar]
- 45.Huang JP, Cheng ML, Hung CY, et al. Docosapentaenoic acid and docosahexaenoic acid are positively associated with insulin sensitivity in rats fed high-fat and high-fructose diets. Journal of diabetes. 2017;9(10):936–946. doi: 10.1111/1753-0407.12505. [DOI] [PubMed] [Google Scholar]
- 46.Lemaitre RN, Tanaka T, Tang W, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7(7) doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lind L. Genome-Wide Association Study of the Metabolic Syndrome in UK Biobank. Metab Syndr Relat Disord. 2019;17(10):505–511. doi: 10.1089/met.2019.0070. [DOI] [PubMed] [Google Scholar]
- 48.Lind L. Genetic Determinants of Clustering of Cardiometabolic Risk Factors in U.K. Biobank. Metab Syndr Relat Disord. 2020;18(3):121–127. doi: 10.1089/met.2019.0096. [DOI] [PubMed] [Google Scholar]
- 49.Hernandez-Baixauli J, Quesada-Vázquez S, Mariné-Casadó R, et al. Detection of Early Disease Risk Factors Associated with Metabolic Syndrome: A New Era with the NMR Metabolomics Assessment. Nutrients. 2020;12(3) doi: 10.3390/nu12030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiklund PK, Pekkala S, Autio R, et al. Serum metabolic profiles in overweight and obese women with and without metabolic syndrome. Diabetol Metab Syndr. 2014;6(1):40. doi: 10.1186/1758-5996-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernstrom JD. Branched-chain amino acids and brain function. The Journal of nutrition. 2005;135(6 Suppl):1539s–1546s. doi: 10.1093/jn/135.6.1539S. [DOI] [PubMed] [Google Scholar]
- 52.Hosseinkhani F, Heinken A, Thiele I, Lindenburg PW, Harms AC, Hankemeier T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut microbes. 2021;13(1):1–22. doi: 10.1080/19490976.2021.1882927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esperanza MG, Wrobel K, Ojeda AG, et al. Liquid chromatography-mass spectrometry untargeted metabolomics reveals increased levels of tryptophan indole metabolites in urine of metabolic syndrome patients. European journal of mass spectrometry (Chichester, England) 2020;26(6):379–387. doi: 10.1177/1469066720964632. [DOI] [PubMed] [Google Scholar]
- 54.Mallmann NH, Lima ES, Lalwani P. Dysregulation of Tryptophan Catabolism in Metabolic Syndrome. Metab Syndr Relat Disord. 2018;16(3):135–142. doi: 10.1089/met.2017.0097. [DOI] [PubMed] [Google Scholar]
- 55.Rebnord EW, Strand E, Midttun Ø, et al. The kynurenine:tryptophan ratio as a predictor of incident type 2 diabetes mellitus in individuals with coronary artery disease. Diabetologia. 2017;60(9):1712–1721. doi: 10.1007/s00125-017-4329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reilly D, Hao K, Jensen MK, Girman CJ, Rimm EB. Use of systems biology approaches to analysis of genome-wide association studies of myocardial infarction and blood cholesterol in the nurses' health study and health professionals' follow-up study. PloS one. 2013;8(12):e85369. doi: 10.1371/journal.pone.0085369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edmondson AC, Braund PS, Stylianou IM, et al. Dense genotyping of candidate gene loci identifies variants associated with high-density lipoprotein cholesterol. Circ Cardiovasc Genet. 2011;4(2):145–155. doi: 10.1161/CIRCGENETICS.110.957563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh SW, Lee JE, Shin E, et al. Genome-wide association study of metabolic syndrome in Korean populations. PloS one. 2020;15(1) doi: 10.1371/journal.pone.0227357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Podrini C, Rowe I, Pagliarini R, et al. Dissection of metabolic reprogramming in polycystic kidney disease reveals coordinated rewiring of bioenergetic pathways. Communications biology. 2018;1:194. doi: 10.1038/s42003-018-0200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.