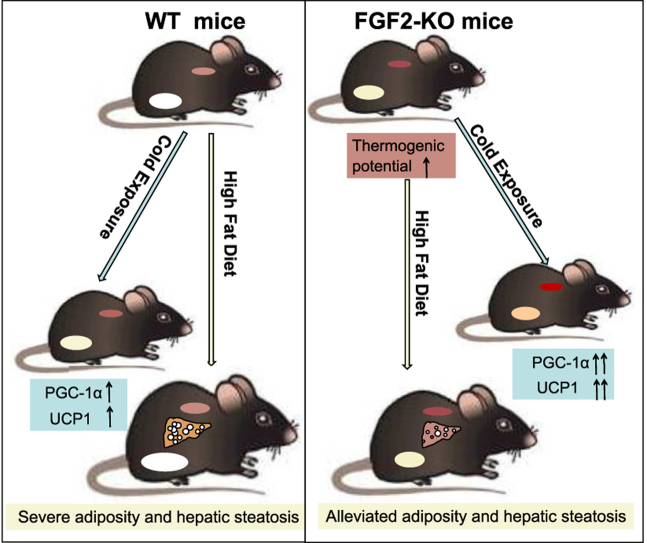
Abstract
Objective
Fibroblast growth factor 2 (FGF2) has been reported to play divergent roles in white adipogenic differentiation, however, whether it regulates thermogenesis of fat tissues remains largely unknown. We therefore aimed to investigate the effect of FGF2 on fat thermogenesis and elucidate the underlying mechanisms.
Methods
FGF2-KO and wild-type (WT) mice were fed with chow diet and high-fat diet (HFD) for 14 weeks. The brown and white fat mass, thermogenic capability, respiratory exchange ratio, and hepatic fat deposition were determined. In vitro experiments were conducted to compare the thermogenic ability of FGF2-KO- with WT-derived brown and white adipocytes. Exogenous FGF2 was supplemented to in vitro-cultured WT brown and ISO-induced beige adipocytes. The FGFR inhibitor, PPARγ agonist, and PGC-1α expression lentivirus were used with the aid of technologies including Co-IP, ChIP, and luciferase reporter assay to elucidate the mechanisms underlying the FGF2 regulation of thermogenesis.
Results
FGF2 gene disruption results in increased thermogenic capability in both brown and beige fat, supporting by increased UCP1 expression, enhanced respiratory exchange ratio, and elevated thermogenic potential in response to cold exposure. Thus, the deletion of FGF2 protects mice from high fat-induced adiposity and hepatic steatosis. Mechanistically, in vitro investigations indicated FGF2 acts in autocrine/paracrine fashions. Exogenous FGF2 supplementation inhibits both PGC-1α and PPARγ expression, leading to suppression of UCP1 expression in brown and beige adipocytes.
Conclusions
These findings demonstrate that FGF2 is a novel thermogenic regulator, suggesting a viable potential strategy for using FGF2-selective inhibitors in combat adiposity and associated hepatic steatosis.
Keywords: Fibroblast growth factor 2 (FGF2), Thermogenesis, Brown and beige fat, PPARγ, PGC-1α
Graphical abstract
Highlights
-
•
FGF2-KO mice show potentiated stimulation on thermogenic capability under both basal and cold challenge stimulation.
-
•
FGF2 disruption protected mice against HFD-induced adiposity and hepatic steatosis.
-
•
FGF2 acts in autocrine/paracrine fashions in vitro.
-
•
Both PPARγ and PGC-1α play roles in FGF2 suppression of thermogenesis.
1. Introduction
In mammals, there are three types of adipose tissue: white, brown, and beige [1]. While white adipose tissue (WAT) serves as a repository for fatty acids, brown and beige adipocytes burn fatty acids and glucose to generate heat, leading to increased energy expenditure [[1], [2], [3]]. Brown adipocytes derive from a myf5-positive cell lineage, characterized by the presence of small, dense lipid droplets enriched with mitochondria and high expression of uncoupling protein 1 (UCP1), which enables the uncoupling of oxidative phosphorylation from ATP production [2,4]. The expression of UCP1 is driven by peroxisome proliferator-activated receptor gamma (PPARγ) in cooperation with other transcriptional components, including PPARγ coactivator-1α (PGC-1α) [5,6]. In contrast, beige adipocytes emerge from white fat through a process called browning or beiging [3]. The PGC-1α transcriptional cofactor is also critical to trigger white-to-beige fat conversion [6,7]. Like brown adipocytes, beige adipocytes possess, albeit to a lesser degree, several features indispensable to thermogenesis, such as multilocular lipid droplet morphology, high UCP1 expression, and densely packed mitochondria [1,3]. Previous studies have provided evidence that metabolically active brown and beige fat are present in adult humans [8,9], and their abundance is inversely correlated with fat mass and obesity-associated disorders [8,10].
Large-scale studies have demonstrated the inducibility of both brown and beige fat [[11], [12], [13], [14]]. β-Adrenergic signaling induced by cold exposure and/or β-adrenergic agonists serve as the primary physiological signal pathway for activation of brown fat thermogenesis and stimulation of beige adipocyte development [11,13]. In addition, several other secreted factors and hormones, such as BMP8 [15], FGF21 [16], Irisin [17], and apelin [18], have been shown to participate in regulating thermogenic activity in brown and/or beige adipocytes, the activation of which may profoundly decrease fat accumulation while improving lipid metabolism and glucose homeostasis. Identifying key regulators of the thermogenic functions of brown and beige adipocytes represents a major goal for development of potential therapeutic avenues for obesity and metabolic diseases, such as fatty liver [14,19].
Fibroblast growth factor 2 (FGF2), also known as basic FGF (bFGF), is among the first recognized members of the FGF family [20]. Through loss-of-function studies, FGF2 has been reported to play essential or predominant roles in the development of vessels [21], nerves [22], and bone [23,24]. The regulation of white adipogenic differentiation by FGF2 has been rigorously demonstrated [[25], [26], [27], [28]]. Kawaguchi et al. [25] showed the induction of de novo adipogenesis in reconstituted basement membrane supplemented with FGF2. FGF2 significantly enhances the adipogenic differentiation of human adipose-derived stem cells (hASCs) and the expression of PPARγ [26]. Hao et al. [27] described a positive correlation between plasma FGF2 levels and fat mass, as well as an increased risk of obesity. However, Xiao et al. [28] reported that bone marrow stem cells of FGF2-deficient mice showed enhanced lipogenic ability with up-regulation of key adipogenic signaling molecules.
In our preliminary studies, we found that the plasma FGF2 levels were significantly elevated in obese mice as well as at thermoneutrality (Figure S1). Because animals with obesity or at thermoneutrality tend to be accompanied with reduced thermogenesis [29,30], we speculated that FGF2 might play a negative role in regulating the thermogenic function of brown and/or beige fat. By using a FGF2 gene knockout (KO) mouse model, we discovered that FGF2 disruption strongly enhances the thermogenic action of both brown and beige fat, which lead to an increase in energy expenditure and improvement of lipid homeostasis. Consequently, FGF2 deletion alleviates HFD-induced adiposity and hepatic steatosis. Further in vitro investigations indicated that FGF2 suppresses PGC-1α and PPARγ expression, which leads to attenuated UCP1 expression and thermogenic activity in brown and beige adipocytes. These findings establish that FGF2 negatively regulates thermogenesis in both brown and beige fat, suggesting a strong potential therapeutic approach for the treatment of adiposity via FGF2-specific signaling inhibitors.
2. Materials and methods
2.1. Animals
All experiments were conducted in FGF2-KO (FGF2−/−) mice and WT (FGF2+/+) littermates with the same genetic background (C57BL/6J). KO was conducted using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 methods. Two single-guide RNAs (sgRNA1: GGAGACAGAGGCCTGCAATG and sgRNA2: TCTCGCGGACGCCATCCAC G) were designed to target promoter and exon 1. Successful deletion was confirmed by PCR genotyping using tail genomic DNA with primers 5′-TCTAACAACTGAGGCAGGGCAA-3′ and 5′-GAAGTGGCAACTCAC CGTG TG-3’. FGF2 heterozygous (+/−) mice were bred to obtain FGF2-KO mice and their WT littermates.
Mice were housed in a temperature- and humidity-controlled, pathogen-free facility with 12-h dark–light cycles. For HFD studies, animals were fed a diet of 45% kcal from fat (Ref. D12451, Research Diets Inc., USA). The body weight and food and water intake of mice were recorded weekly. The body composition of mice was determined by dual energy X-ray absorptiometry (DXA) (MEDIKORS, Korea). At the end of the experiment, the animals were kept fasting for 12 h and sacrificed by isoflurane inhalation followed by cervical dislocation. Interscapular BAT (iBAT), inguinal WAT (iWAT), and liver tissues were harvested and weighed. All animal experiments were performed in accordance with the guidelines for the Care and Use of Laboratory Animals, and animal maintenance and experimental procedures were approved by the Animal Care and Use Committee of Shandong Agricultural University, China.
2.2. Histology, H&E staining, ORO staining, and immuno-histochemistry/immuno-fluorescence analysis
iBAT, iWAT, and liver tissues were fixed in 4% paraformaldehyde for more than 24 h and embedded in paraffin. Paraffin samples were sectioned (5 μm) and stained with hematoxylin and eosin for histochemical examination. Area of adipocytes in adipose tissues were measured by using an image software (Nikon, Japan). For ORO staining, liver samples were frozen in liquid nitrogen and sectioned at 8 μm in thickness using a cryostat. The sections were stained with ORO solution for 10 min. After washing with water, sections were stained for 1 min in hematoxylin. For immuno-histochemistry/immune-fluorescence, de-paraffinized iBAT and iWAT sections were blocked with FBS, incubated with specific UCP1 primary and HRP-/Alexa Fluor 555-conjugated secondary antibodies, and detected accordingly.
2.3. Scanning electron microscopy (SEM)
iBAT and iWAT samples were fixed with 2% glutaraldehyde and post-fixed in 1% osmium tetroxide for 1 h, dehydrated in graded concentrations of ethanol and 100% acetone. The specimens were dried at the critical point. Subsequently, the specimens were stuck on a colloidal silver and sputtered with gold by a MED010 coater (Balzers) and analyzed with a scanning electron microscope (JEOL, Japan).
2.4. Quantification of mtDNA copy number
Equal amounts of WT and KO iBAT and iWAT were used to extract total DNA after digestion with proteinase K, respectively. The isolated DNA was used to amplify mtDNA using primers for the mitochondrial cytochrome c oxidase subunit 2 (COX2) gene, with the Rsp18 nuclear gene as an internal control of genomic DNA, as described previously [31].
2.5. Hepatic triglyceride (TG) and total cholesterol (TCH) content determination
Liver tissue (500 mg) was homogenized in 300 μL RIPA lysis buffer in a Polytron disrupter. The homogenate was centrifuged at 12,000 g for 5 min, and the supernatant was collected. TG and TCH content in the tissue was quantified with commercial assay kits (Dongou, China), which was normalized to total protein and expressed as mmol/g total protein.
2.6. Metabolic studies
Mice were housed in individual CLAMS-HC Comprehensive Lab Animal Monitoring System (Columbus, USA) with free access to water and food, and allowed to acclimate for a 24 h period. Data were collected every 9 min for another 24 h. O2 consumption (VO2), CO2 production (VCO2), and heat production were measured by the TSE system, and respiratory exchange ratio (RER) were calculated using the manufacturer's system software.
2.7. Infrared thermography
The temperature of WT and KO mice was recorded with an infrared camera (FOTRIC 225) and analyzed with a specific software (FOTRIC Tools Software). At least five pictures of each mouse were taken and analyzed.
2.8. Cold challenge and β3-AR agonist treatment
For cold exposure experiment, WT and FGF2−/− mice were kept at a 5 °C room for 24 h. Core body temperature was monitored using a rectal probe every 3 or 6 h for the 24 h-duration of the study. Twenty-four hours later, mice at RT and cold room were sacrificed to collect iBAT and iWAT for further thermogenic-capability determinations. For β3-AR agonist treatment, CL was intraperitoneally injected into mice at 0.5 mg kg−1 body weight/day. Three days later, iBAT and iWAT were collected to determine the mRNA expression of thermogenic genes and make paraffin slices.
2.9. Isolation of brown and white SVFs, in vitro differentiation and treatments
Brown and white SVFs were obtained and induced to differentiate into mature adipocytes, respectively, as previously described with minor modification [12,32]. Briefly, BAT in the interscapular and WAT in the inguen were isolated from 3-week old mice by plastic and reconstructive surgery. BAT was digested with 1.0 mg/mL collagenase I for 30 min at 37 °C. WAT was digested with 1.5 mg/mL collagenase I for 45 min at 37 °C. The cells were cultured to subconfluence in basal medium containing 15% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Subsequently, the BAT SVFs were exposed to induction medium containing 5 μM dexamethasone, 5 μg/mL insulin, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 nM T3, and 15% FBS for 2 days. The cells were then maintained in media containing 5 μg/mL insulin, 1 nM T3, and 15% FBS for another 6 days. The WAT SVFs were induced to differentiate into white adipocytes using cocktail adipogenic stimuli, consisting of 0.1 μM dexamethasone, 5 μg/mL insulin, and 0.5 mM IBMX for the first 2 days, and 0.1 μM dexamethasone and 5 μg/mL insulin for further 8 days.
To induce the beiging of white adipocytes, ISO (0.1 μM) was added to the WAT SVF differentiating medium for the latter 8 days. For FGF2 treatments, 10 ng/mL FGF2 and 10 ng/mL HP were added during the whole induction period. For SSR and Rosi treatments, 0.5 μM SSR or 1 μM Rosi were also supplemented in the entire induction process. The media were replaced every 2 or 4 days.
2.10. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues or cells using RNAiso Plus Reagent and converted to cDNA using the HiScript II Q RT SuperMix for qPCR Kit (Vazyme, China). qRT-PCR was performed with SYBR green, fluorescent dye (Takara, Japan) using a Real-Time PCR System (Applied Biosystems). Transcript levels were quantified using the 2−ΔΔCt method values to that of GAPDH. Primers used were shown in Table 1.
Table 1.
Primers used in this study.
| Genes | Sequence 5′ to 3′ |
|---|---|
| GAPDH | Forward-AGAGTGTTTCCTCGTCCCG Reverse-CCGTTGAATTTGCCGTGA |
| FGF2 | Forward-TCCCACCAGGCCACTT Reverse-CGTCCATCTTCCTTCATA |
| Primer-GT | Forward-TCTAACAACTGAGGCAGGGCAA Reverse-GAAGTGGCAACTCACCGTGTG |
| PPARγ | Forward-GACCACTCGCATTCCTTT Reverse-ACAGACTCGGCACTCAAT |
| aP2 | Forward-CCTTTGTGGGAACCTGGAA Reverse-TGTCGTCTGCGGTGATTT |
| C/EBPα | Forward-TCGGTGCGTCTAAGATGAGG Reverse-TGAGTATCCAAGGCACAAGGT |
| FAS | Forward-GGGTCTATGCCACGATTC Reverse-GTGTCCCATGTTGGATTTG |
| LPL | Forward-GAGGATGGCAAGCAACAC Reverse-AGCAGTTCTCCGATGTCC |
| SREBP-1c | Forward- GACTACATCCGCTTCTTGC Reverse- CACCACTTCGGGTTTCAT |
| ACC | Forward- AGGCTATGTGAAGGATGTG Reverse- CTGAAGAGGTTAGGGAAG |
| PPARα | Forward- CAAGTGCCTGTCTGTCGG Reverse- GCGGGTTGTTGCTGGTCT |
| HSL | Forward-AGCACTACAAACGCAACGA Reverse-CGACAGCACCTCAATCTCA |
| CPT1 | Forward-CGTGACGTTGGACGAATC Reverse-TCTGCGTTTATGCCTATC |
| PGC-1α | Forward-AGAAGCGGGAGTCTGAAA Reverse-CAGGTGTAACGGTAGGTG |
| PRDM16 | Forward-GCGGTCAGCAATAGCAGC Reverse-CCCGTGGTAGTGTCCAAGTC |
| UCP1 | Forward-GCTTAATGACTGGAGGTGTG Reverse-GCTTTCTGTGGTGGCTAT |
| Cidea | Forward-CTTCCTCGGCTGTCTCAA Reverse-TGGCTGCTCTTCTGTATCG |
| C/EBPβ | Forward-TGTCCACGTCGTCGTCGTCC Reverse-CCGTCAGCTCCAGCACCTTGT |
| TMEM26 | Forward-GACTCCACCAAACACTCC Reverse-GCATACTCCACGTCCACA |
| Tbx1 | Forward-ACCGAGATGATCGTCACCAAG Reverse-ACCAGCCAGGAGGAGCTATG |
| mtTF | Forward-CCTCGTCTATCAGTCTTGT Reverse-GCTTCTGGTAGCTCCCTC |
| CytC | Forward-ATCTCCACGGTCTGTTCG Reverse-GCCCTTTCTCCCTTCTTC |
| Cox7a | Forward-GGCTCTGGTCCGGTCTT Reverse-CTGGGAGGTCATTGTCG |
| Cox8b | Forward-TGCGAAGTTCACAGTGGTT Reverse-GGCGGAAGTGGGAGTTTT |
| Aco2 | Forward-ATCGAGCGGGGAAAGACATAC Reverse-TGATGGTACAGCCACCTTAGG |
| Uqcrc2 | Forward-AAAGTTGCCCCGAAGGTTAAA Reverse-GAGCATAGTTTTCCAGAGAAGCA |
| COX2 (mtDNA) | Forward-ATAACCGAGTCGTTCTGCCAAT Reverse-TTTCAGAGCATTGGCCATAGAA |
| Rsp18 (Genomic DNA) | Forward-CATCACCCACTTACCCCCAAAA Reverse-TGTGTTAGGGGACTGGTGGACA |
| UCP1 promoter | Forward- AGCTTGCTGTCACTCCTCTACA Reverse- TGAGGAAAGGGTTGACCTTG |
2.11. Western blotting
Protein lysates were obtained from tissues or cells using RIPA lysis buffer containing protease and phosphatase inhibitor cocktails (Roche, USA). Western blotting was conducted as described previously [31]. The following primary antibodies were used: GAPDH (AB0038, Abways, China), β-actin (P60709, Abways, China), PPARγ (sc-7273, Santa Cruz), PGC-1α (sc-13067, Santa Cruz), and UCP1 (ab15517, Abcam). The band density was quantified by Image J software.
2.12. Lentivirus infection
For experiments with lentivirus, the recombinant Lenti-control and Lenti-PGC-1α were individually supplemented into 50% confluency SVFs at 5 MOI with 6 μg/mL polybrene, and the medium was refreshed 24 h after infection. After recovering for another 72 h, the infected cells were induced for differentiation together with other treatments.
2.13. MitoTracker staining
Different-treated brown and beige adipocytes were stained with MitoTracker Red CMXRos (200 nM) (CST, #9082) in DMEM containing 15% FBS at 37 °C for 30 min. Following washing twice with DMEM containing 15% FBS, the cells were incubated with DAPI (1 μg/mL) for 5 min at RT. The intracellular MitoTracker-stained mitochondria were detected using a confocal laser scanning microscopy (CLSM) (Zeiss, Germany). Images were acquired and processed with the same setting for different treatments. The fluorescence intensity was quantified using Image J software.
2.14. Immuno-fluorescence staining of UCP1
For immune-fluorescence staining, formalin-fixed and Triton X 100- permeabilized brown and beige adipocytes were pre-incubated with a blocking buffer (PBS containing 5% FBS) for 60 min and incubated with UCP1 antibody (1:100 dilution) in blocking buffer at 4 °C overnight. The slides were washed and incubated with Alexa Flour 555-conjugated secondary antibody (1:500 dilution). After staining with DAPI (1 μg/mL) for 5 min, images were acquired by using a CLSM (Zeiss, Germany) and processed with the same setting for different treatments. The fluorescence intensity was quantified using Image J software.
2.15. Co-immunoprecipitation (Co-IP)
To determine the influence of FGF2 and/or FGFR1 inhibitor SSR on interaction of PPARγ with PGC-1α in differentiating brown and beige adipocytes, Co-IP was performed as previously described [33] with some minor modifications. In brief, the white and brown SVFs were induced to differentiate with the induction medium for 4 days in the absence or presence of FGF2 and/or SSR. Co-IP was conducted using anti-PPARγ antibody as indicated, and western blotting was performed using anti-PPARγ and anti-PGC-1α antibodies, respectively. IgG was used as a negative control.
2.16. Chromatin immunoprecipitation (ChIP) assay
To determine the interaction of PPARγ with the promoter of UCP1, ChIP assay was performed using a ChIP Assay Kit, as described previously [31]. The brown and white SVFs were induced using individual differentiation media, which were simultaneously treated with Vehicle, FGF2, Rosi, Rosi + FGF2, Lenti-control, Lenti-control + FGF2, Lenti-PGC-1α, Lenti-PGC-1α+FGF2, Lenti-PGC-1α+Rosi, or Lenti-PGC-1α+Rosi + FGF2 for 6 days. After immunoprecipitation with the anti-PPARγ antibody (1 μg, Santa Cruz, #SC-7273) at 4 °C overnight, DNA-protein complexes were collected and treated according to the instruction of the manufacturer. The extracted DNA was used to test the promoter regions of UCP1 by qPCR. The primers for UCP1-promoter were listed in Table 1.
2.17. Dual luciferase reporter assay
HEK293 cells cultured in 96-well plates were co-transfected with 1000 ng/mL of pGL3-basic, pGL3-UCP1p, or pGL3-control, along with 50 ng/mL of pRL-TK, respectively. Six hours later, the cells were infected with Lenti-control or Lenti-PGC-1α recombinant viruses for 24 h in some treatments. The cells were treated with Vehicle, FGF2, Rosi, Rosi + FGF2, Lenti-control, Lenti-control + FGF2, Lenti-PGC-1α, Lenti-PGC-1α+FGF2, Lenti-PGC-1α+Rosi, or Lenti-PGC-1α+Rosi + FGF2 for 2 days. Finally, the treated cells were washed and lysed in 20 μL of lysis buffer. The firefly luciferase activity was examined according to the protocol (dual reporter assay system, Promega), and efficiency was normalized to renilla luciferase activity directed by a co-transfected control plasmid pRL-TK.
2.18. Statistical analysis
Statistical analysis was performed on data from at least 3 repeated experiments. All data were presented as means ± SEM. Significant difference between treatments was tested by one-way ANOVA, two-tailed Student's t-test, or ANCOVA analysis (SPSS statistics 25.0). p < 0.05 was regarded as significant.
3. Results
3.1. FGF2 gene disruption is associated with enhanced brown fat thermogenesis and beiging of white fat
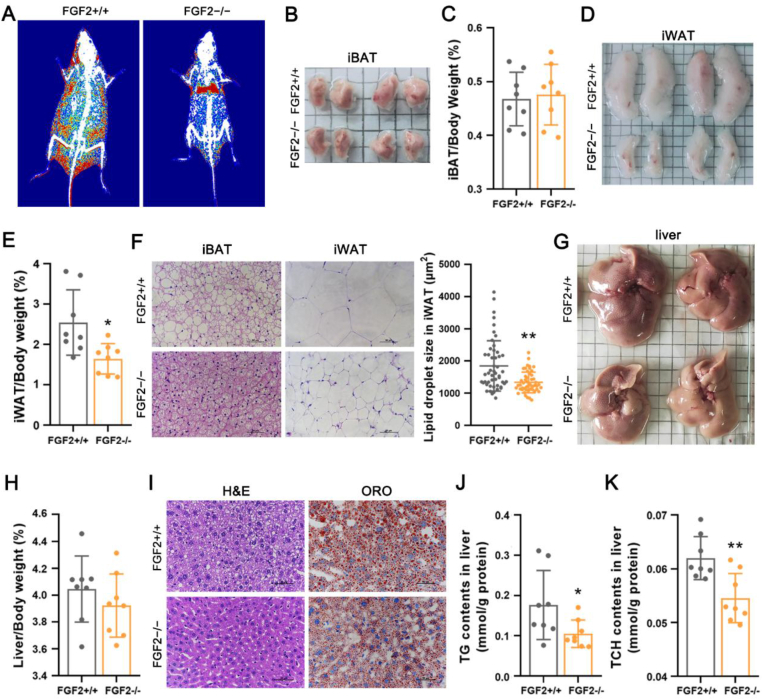
To evaluate the role of FGF2 in the thermogenic potential of adipose tissues, we generated FGF2-KO mice (genetic background C57BL/6J) (Figure S2). To initially characterize the KO phenotype, we fed 3-week-old male FGF2+/+ and FGF2−/− mice with chow diet for 14 weeks and found that, although the dynamic changes in body weight and average body weight were indistinguishable between groups at 17 weeks old (Figure S3, A and B), FGF2−/− mice consumed more food than WT littermates during the course of the experiment (Figure S3 C). FGF2−/− mice had a significantly higher whole feed/gain ratio (Figure S3 D). The fat mass and the lean mass of FGF2−/− mice were significantly decreased and increased, respectively (Figure S3 F). Although the iBAT index of FGF2−/− mice was comparable with that of FGF2+/+ mice (Figure 1A,B, Figure S3 E), FGF2−/− mice had markedly less subcutaneous white fat (Figure 1C,D, Figure S3 E).
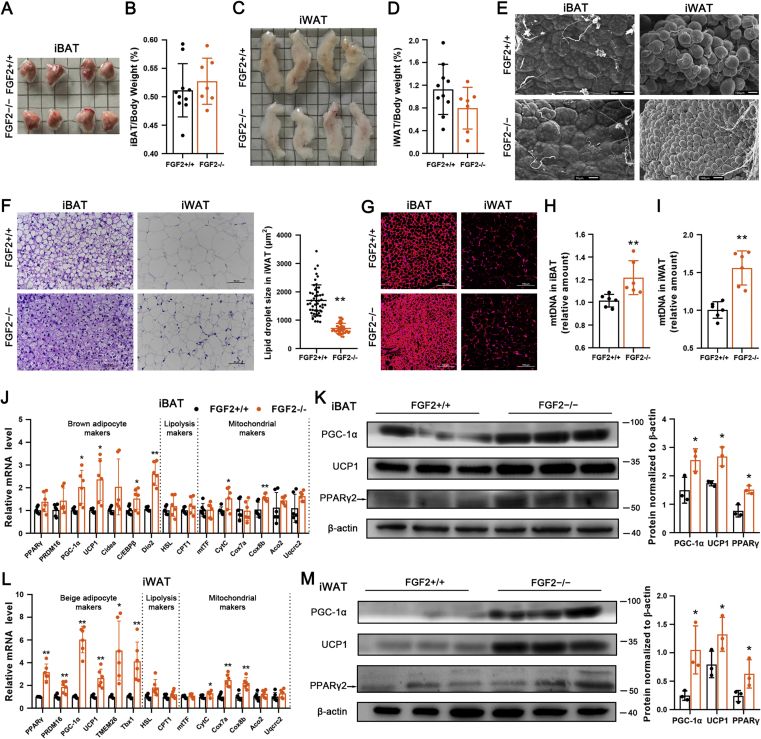
Figure 1.
FGF2 gene disruption leads to enhanced brown fat thermogenesis and beiging of white fat. Tissue samples were collected from 17-week-old FGF2+/+ and FGF2−/− mice. (A, B) Representative iBAT images (A) and ratios of iBAT/body weight (B) of FGF2+/+ and FGF2−/− mice (n = 7–10). (C, D) Representative iWAT images (C) and ratios of iWAT/body weight (D) of FGF2+/+ and FGF2−/− mice (n = 7–10). (E) Representative scanning electron microscopy images of iBAT (Scale bar = 10 μm) and iWAT (Scale bar = 100 μm) sections (n = 3). (F) Representative images of H&E staining of iBAT and iWAT sections (n = 5), and the lipid droplet size in iWAT. Scale bar = 50 μm. (G) Immuno-fluorescence staining of UCP1 (red) in FGF2+/+ and FGF2−/− iBAT and iWAT sections (n = 5). Scale bar = 100 μm. (H, I) Quantification of relative mtDNA levels in iBAT (H) and iWAT (I) in FGF2+/+ and FGF2−/− mice (n = 10–12). (J) The relative mRNA levels of brown adipocyte, lipolysis, and mitochondrial markers in iBAT of FGF2+/+ and FGF2−/− mice, determined by qRT-PCR (n = 6). (K, M) Western blot analysis of PPARγ, PGC-1α, and UCP1 protein contents in iBAT (K) and iWAT (M) of FGF2+/+ and FGF2−/− mice (n = 3). The blotting bands were quantified with β-actin as a loading control. (L) The relative mRNA levels of beige adipocyte, lipolysis, and mitochondrial markers in iWAT of FGF2+/+ and FGF2−/− mice, determined by qRT-PCR (n = 6). Values represent means ± SEM. Statistical analysis was performed using two-tailed Student's t-test. ∗p < 0.05, ∗∗p < 0.01 compared with FGF2+/+ samples.
Histologically, iBAT in KO mice was comparable with that in WT tissue (Figure 1E,F). However, the UCP1 immuno-reactivity and mtDNA levels in KO iBAT were evidently elevated (Figure 1G,H). The transcription levels of thermogenic-associated genes (PGC-1α, UCP1, C/EBPβ, and Dio2) and mitochondrial markers (CytC, and Cox8b) were drastically increased in FGF2−/− iBAT comparing with those in the WT iBAT (Figure 1 J). In addition, the protein levels of PGC-1α, UCP1, and PPARγ were all elevated in iBAT of FGF2−/− mice (Figure 1 K). The adipogenic potential of KO iBAT was enhanced, as the mRNA levels of aP2, FAS and LPL were highly augmented, and the protein expression of aP2 was also marginally elevated (Figure S4, A and C). These data indicate that FGF2-KO mice recruit and expend more fat in BAT via elevated thermogenesis.
Moreover, FGF2 disruption led to a marked decrease in lipid droplet size in iWAT (Figure 1E,F), characteristic of reduced triglyceride accumulation or accelerated triglyceride release. Immune-fluorescence analysis revealed greater intensity of UCP1 immuno-reactivity in the iWAT of FGF2-KO mice (Figure 1 G). The mtDNA copy number in KO iWAT was increased by ∼50% compared with that in the WT tissue (p < 0.01) (Figure 1 I). Furthermore, iWAT from FGF2−/− mice displayed elevated transcription of thermogenic markers (PRDM16, PGC-1α and UCP1), beige adipocyte-specific genes (TMEM26 and Tbx1), and mitochondrial markers (CytC, Cox7a, and Cox8b) (Figure 1 L). Likewise, KO iWAT also showed increased protein levels of PPARγ, PGC-1α and UCP1 (Figure 1 M). The transcription of adipogenic-related genes (PPARγ, C/EBPα, FAS, LPL, and aP2) was significantly induced, and the protein expression of aP2 was marginally up-regulated in KO iWAT (Figure 1L, Figure S4, B and D). These results suggest that the decrease in adipocyte size in FGF2−/− iWAT depends greatly on the degree of induced beiging and the thermogenic potential rather than on reduced triglyceride synthesis.
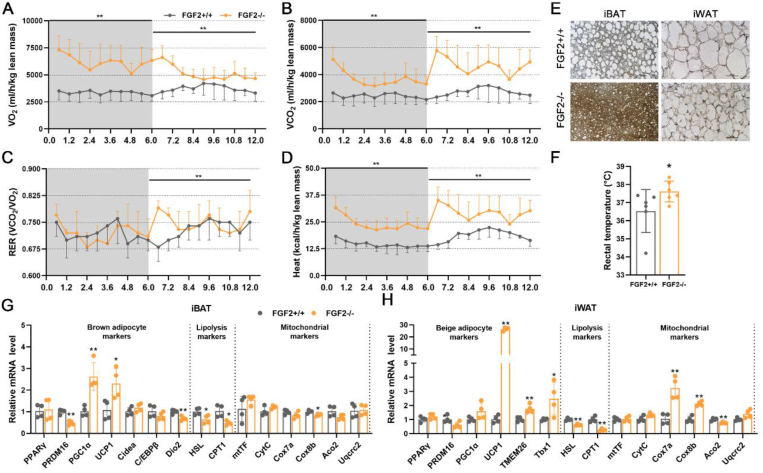
3.2. FGF2-KO mice show increased RER and body temperature, as well as activated responses to cold challenge
In view of the role of brown and beige fat in non-shivering thermogenesis, we compared the respiratory metabolic parameters of the FGF2+/+ and FGF2−/− mice at ambient temperature (25 °C). In contrast to WT mice, carbon dioxide production, oxygen consumption, and RER in FGF2−/− mice both increased substantially in the dark (p < 0.05) (Figure 2A–C). In the light, oxygen consumption, carbon dioxide production, and RER were also higher in KO mice, although not significantly (Figure 2A–C).
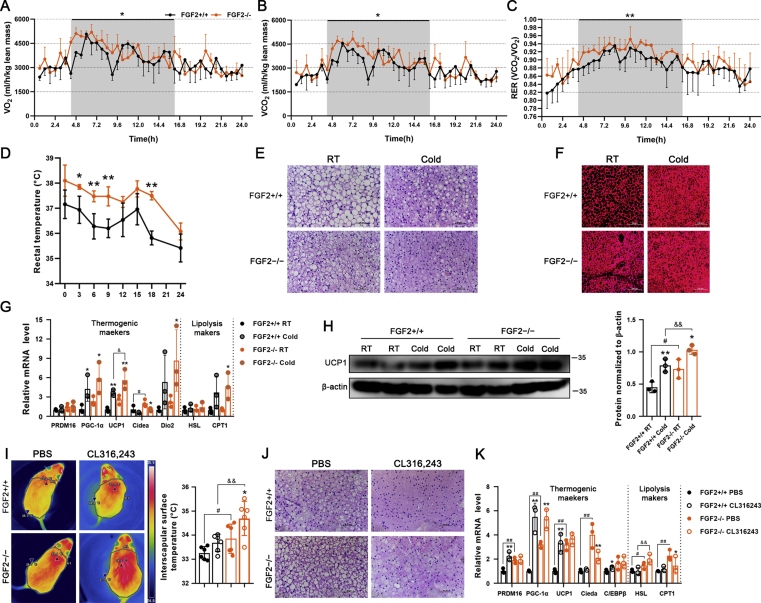
Figure 2.
FGF2-KO mice show increased whole-body energy expenditure and activated thermogenic capability of brown fat. (A, B) O2 consumption (A) and CO2 production (B) (expressed as ml/h/kg lean mass) measured in 12-week-old FGF2+/+ and FGF2−/− mice during a 24-h period measured using a CLAMS apparatus (n = 3). (C) RER dynamics calculated by VCO2/VO2 (n = 3). (D) Core body temperature changes in FGF2+/+ and FGF2−/− mice following cold challenge, determined by rectal probe every 3 or 6 h for a 24-h duration (n = 4–5). (E, F) Representative images of H&E staining (Scale bar = 50 μm) (E) and immune-fluorescence staining of UCP1 (red) (Scale bar = 100 μm) (F) of iBAT sections, under room temperature (RT) or cold challenge for 24 h (n = 3). (G) qRT-PCR analysis of thermogenic- and lipolysis-related gene expression in FGF2+/+ and FGF2−/− iBAT under normal temperature or cold challenge for 24 h (n = 3). (H) Western blot analysis of UCP1 protein levels in FGF2+/+ and FGF2−/− iBAT under RT or cold challenge for 24 h (n = 3). GAPDH was used as a loading control. (I) Representative thermal images and dorsal interscapular surface temperatures of FGF2+/+ and FGF2−/− mice after injection with CL316,243 or PBS control (n = 6). (J) Representative images of H&E staining of iBAT sections upon PBS or CL316,243 injection (n = 3). Scale bar = 50 μm. (K) qRT-PCR analysis of thermogenic- and lipolysis-related gene expression in FGF2+/+ and FGF2−/− iBAT under PBS or CL316,243 treatments (n = 3). Data represent means ± SEM. Statistical analysis was performed using ANCOVA (A–C), two-tailed Student's t-test (D), and one-way ANOVA after Tukey's multiple comparison test (G, H, I and K). ∗p < 0.05, ∗∗p < 0.01 vs. FGF2+/+ mice in A-D. ∗p < 0.05, ∗∗p < 0.01 vs. Vehicle in the same littermates; #p < 0.05, ##p < 0.01 vs. FGF2+/+ mice upon Vehicle treatment; &p < 0.05, &&p < 0.01 vs. FGF2+/+ mice upon cold challenge or CL316,243 treatment in G, H, I and K.
Given that increased metabolic rate is often accompanied by higher thermogenic capacity, we examined the animals’ body temperature under cold challenge conditions. As expected, FGF2−/− mice showed elevated rectal temperature both prior to and during the 24-hour cold challenge, which was approximately 1 °C higher on average than that of WT littermates at corresponding time points (Figure 2 D). By using an infrared camera, we found that the surface temperature of KO iBAT was significantly higher than that of WT in both untreated and CL treatments (Figure 2 I).
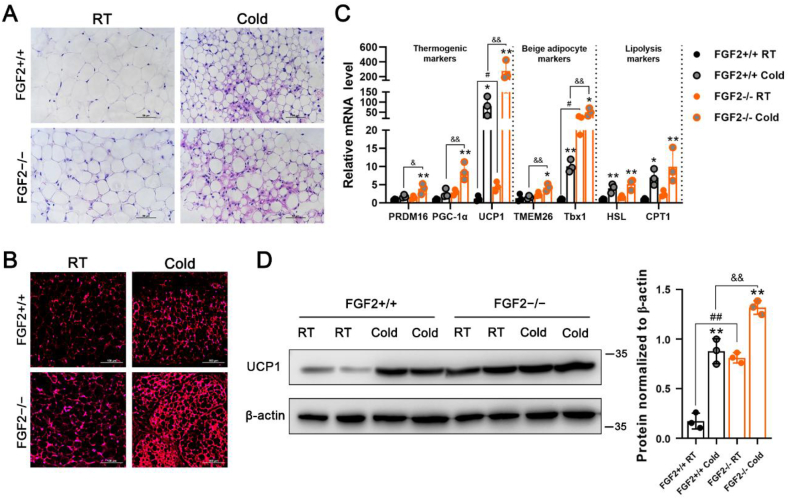
In light of our findings of potentiated activation of body temperature in KO mice under both cold and β3-AR stimulation conditions, we examined the thermogenic markers in treated adipose tissues. In iBAT and iWAT, both cold challenge and β3-AR stimulation led to the occurrence of smaller lipid droplets and higher expression of thermogenic genes (Figure 2E–H, J and K, Figure 3, Figure S5 A). In response to cold challenge, FGF2−/− iBAT exhibited a strongly potentiated induction of UCP1 mRNA and protein expression relative to WT controls (Figure 2G,H). However, following CL injection, only the mRNA level of HSL was significantly higher in FGF2−/− iBAT than that in FGF2+/+ iBAT (Figure 2 K). KO iWAT showed a more pronounced increase of PRDM16, PGC-1α, UCP1, TMEM26, and Tbx1 transcription levels, and a higher UCP1 protein content upon cold challenge (Figure 3C,D), and only an elevated transcription of Tbx1 following CL treatment (Figure S5 B) compared with WT iWAT. These data reflect a stronger cold-response by both KO iBAT and iWAT than by WT tissues.
Figure 3.
The thermogenic capability of iWAT exhibits higher potentiation in FGF2-KO mice than in WT under cold challenge conditions. (A, B) Representative images of H&E staining (Scale bar = 50 μm) (A) and immune-fluorescence staining of UCP1 (red) (Scale bar = 100 μm) (B) of iWAT sections under RT or cold challenge for 24 h (n = 3). (C) qRT-PCR analysis of thermogenic-, beige adipocyte-, and lipolysis-related gene expression in FGF2+/+ and FGF2−/− iWAT under normal temperature or cold challenge for 24 h (n = 3). (D) Western blot analysis of UCP1 protein levels in FGF2+/+ and FGF2−/− iWAT under RT or cold challenge for 24 h (n = 3). GAPDH was used as a loading control. Data represent means ± SEM. Statistical analysis was performed using one-way ANOVA after Tukey's multiple comparison test. ∗p < 0.05, ∗∗p < 0.01 vs. Vehicle in the same littermates; #p < 0.05, ##p < 0.01 vs. FGF2+/+ mice upon Vehicle treatment; &p < 0.05, &&p < 0.01 vs. FGF2+/+ mice upon cold challenge.
3.3. FGF2-KO mice show higher stability in lipid homeostasis and amelioration of HFD-induced adiposity and hepatic steatosis
In view of the enhanced function of brown and beige fat resulting from FGF2-KO, its effects on lipid homeostasis were next investigated. Upon chow feeding, significantly reduced plasma TG but not of TCH content was observed in the FGF2-disrupted mice (Figure S6, A and B). The AST but not the ALT activity was substantially decreased by FGF2 deficiency (Figure S6, C and D). In response to HFD feeding, the plasma TG content was significantly lower in FGF2-KO mice than in WT animals (p < 0.05), while TCH content, ALT activities, and AST activities were modestly downregulated (p > 0.05) (Fig S6).
HFD is prone to induce adiposity and ectopic fat deposition in livers. We determined the influence of FGF2 disruption on fat accumulation and hepatic steatosis following 14 weeks of HFD feeding. Compared to WT, HFD-fed FGF2-KO mice exhibited significantly lower body weight with no significant change of feed/gain ratio (Figure S7, A-C). Specifically, DXA and anatomical imaging revealed vastly lower subcutaneous fat mass in FGF2-KO mice, compared with that in WT (Figure 4A, Figure S7 D). Moreover, the iWAT index was greatly down-regulated, while the iBAT index was not significantly altered in HFD-fed KO mice (Figure 4B–E). In addition, the adipocyte size in iWAT and lipid droplet size in iBAT were generally smaller among FGF2-KO mice than among WT (Figure 4 F).
Figure 4.
FGF2-KO protects mice against HFD-induced adiposity and hepatic steatosis. (A) Representative DXA images of 17-week-old FGF2+/+ and FGF2−/− mice fed with HFD (n = 4). Red represents areas with more than 50% fat. (B, C) Representative iBAT images (B) and ratios of iBAT/body weight (C) of HFD-fed FGF2+/+ and FGF2−/− mice at age of 17 weeks old (n = 8). (D, E) Representative iWAT images (D) and ratios of iWAT/body weight (E) of HFD-fed FGF2+/+ and FGF2−/− mice at age of 17 weeks old (n = 8). (F) Representative images of H&E staining of iBAT and iWAT sections (n = 3), and the lipid droplet size in iWAT. Scale bar = 50 μm. (G, H) Representative liver images (G) and the liver/body weight ratios (H) of FGF2+/+ and FGF2−/− HFD-fed mice (n = 8). (I) Representative images of H&E and ORO staining of liver sections (n = 3). Scale bar = 50 μm. (J, K) TG (J) and TCH (K) contents in the livers of HFD-fed FGF2+/+ and FGF2−/− mice (n = 8). Values represent means ± SEM. Statistical analysis was performed using two-tailed Student's t-test. ∗p < 0.05, ∗∗p < 0.01 vs. HFD-fed FGF2+/+ mice.
The livers of FGF2−/− mice were visibly smaller and the liver index was not significantly lower than WT fed with HFD (Figure 4G,H), thus indicating that the HFD-induced fatty liver phenotype may be ameliorated by FGF2 disruption. As expected, H&E and ORO staining of liver sections (Figure 4 I), as well as lower hepatic levels of TG and TCH (Figure 4J,K), provided further evidence that FGF2 KO led to alleviation of HFD-induced hepatic steatosis. Furthermore, liver-specific expression of fat synthesis- or fatty acid oxidation-associated genes were not significantly different between HFD-fed KO mice and WT, except for a decrease in ACC (Figure S8), suggesting that the amelioration of hepatic fat deposition was not attributable to the direct alteration of in situ fat metabolism.
To ascertain whether the amelioration of HFD-induced adiposity and hepatic steatosis mediated by FGF2 deficiency was due to the elevation of thermogenic activity, we compared the thermogenic capability between HFD-fed WT and KO mice. Oxygen consumption, carbon dioxide production, heat production, and the RER at day were all vastly elevated (p < 0.01) in the KO HFD-fed mice compared to that in WT (Figure 5A–D). HFD-fed KO mice displayed substantially greater UCP1 immunostaining in both iBAT and iWAT sections (Figure 5 E). The rectal temperature was significantly higher in HFD-fed KO mice (Figure 5 F). In addition, the transcriptional levels of PGC-1α and UCP1 in iBAT, as well as those of UCP1, TMEM26, Tbx1, Cox7a, and Cox8b in iWAT, were all profoundly elevated in HFD-fed KO mice (Figure 5G,H). These results suggest that the alleviated HFD-induced adiposity and hepatic steatosis phenotype associated with FGF2 deficiency is due in large measure to the enhanced thermogenic ability of brown and beige fats.
Figure 5.
FGF2-KO HFD-fed mice exhibit increased whole-body energy expenditure and activated thermogenic function in both brown and white fats. (A, B) O2 consumption (A) and CO2 production (B) (expressed as ml/h/kg lean mass) measured in HFD-fed FGF2+/+ and FGF2−/− mice during a 12-hour period measured using a CLAMS apparatus (n = 3). (C) RER dynamics calculated by VCO2/VO2 (n = 3). (D) The whole-body energy expenditure (heat production) in HFD-fed FGF2+/+ and FGF2−/− mice, which was expressed as kcal/h/kg lean mass (n = 3). (E) Immuno-histochemistry staining of UCP1 (brown) in HFD-fed FGF2+/+ and FGF2−/− iBAT and iWAT sections (n = 3). Scale bar = 50 μm. (F) The core body temperature of HFD-fed FGF2+/+ and FGF2−/− mice (n = 6). (G, H) The relative mRNA levels of brown/beige adipocyte-, lipolysis-, and mitochondrial-related markers in iBAT (G) and iWAT (H) of HFD-fed FGF2+/+ and FGF2−/− mice, determined by qRT-PCR (n = 4). Data represent means ± SEM. Statistical analysis was performed using ANCOVA (A–D) or two-tailed Student's t-test (F–H). ∗p < 0.05, ∗∗p < 0.01 vs. HFD-fed FGF2+/+ mice.
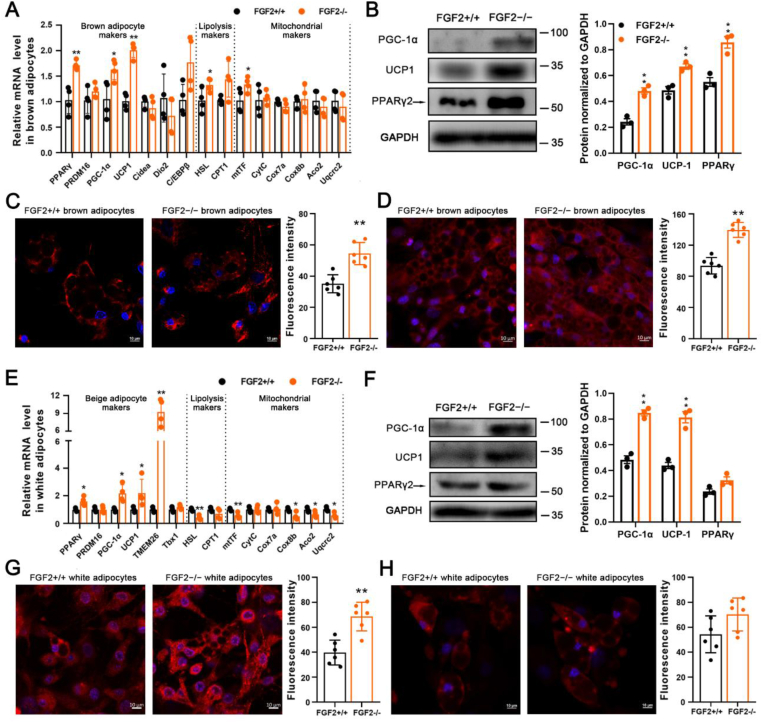
3.4. FGF2-KO-derived brown and beige adipocytes exhibit higher adipogenic and thermogenic gene expression in vitro
Given the enhancement of in vivo thermogenesis resulting from FGF2 disruption, in vitro experiments were conducted to compare the adipogenic and thermogenic gene expression levels between WT- and KO-derived adipocytes. We found that the FGF2−/− brown SVF-differentiated adipocytes exhibited higher transcription of thermogenic genes, including PGC-1α, and UCP1, as well as the adipogenic gene of PPARγ, the lipolysis marker HSL and the mitochondrion marker mtTF than did controls (Figure 6 A). Similarly, the protein expression of PPARγ, PGC-1α, and UCP1 was enhanced in FGF2−/− brown adipocytes, compared to that in WT cells (Figure 6 B). The mitochondria density and the UCP1 immuno-reactivity were also significantly elevated in KO brown adipocytes (Figure 6C,D). We used ISO to induce the beiging of in vitro cultured WT- and KO-derived white adipocytes, and identified that the beiging-associated genes, including PPARγ, PGC-1α, UCP1, and TMEM26 were all activated in KO cells (Figure 6 E). In contrast to WT, the KO beiged white adipocytes displayed enhanced PGC-1α and UCP1 protein levels, mitochondria density, and a marginal higher UCP1 immuno-reactivity (Figure 6F–H). These results indicated that FGF2-KO brown and beige adipocytes also show higher adipogenic and thermogenic gene expression in vitro, hinting the autocrine regulation of FGF2 on thermogenesis.
Figure 6.
FGF2–/–-derived brown and beige adipocytes exhibit higher adipogenic and thermogenic gene expression in vitro. (A) Relative mRNA levels of brown adipocyte, lipolysis, and mitochondrial genes in differentiated FGF2+/+ and FGF2−/− brown adipocytes (n = 4). (B) Western blot analysis of PPARγ, PGC-1α, and UCP1 protein contents in FGF2+/+ and FGF2−/− brown adipocytes (n = 3). GAPDH was used as a loading control. (C, D) MitoTracker staining (red) (C) and immuno-fluorescence staining of UCP1 (red) (D) in FGF2+/+ and FGF2−/− brown adipocytes (n = 6). The nuclei (blue) were stained with DAPI. Scale bar = 10 μm. (E) Relative mRNA expression beige adipocyte, lipolysis, and mitochondrial genes in differentiated FGF2+/+ and FGF2−/− beige adipocytes (n = 4). (F) Western blot analysis of PPARγ, PGC-1α, and UCP-1 protein contents in FGF2+/+ and FGF2−/− beige adipocytes (n = 3). GAPDH was used as a loading control. (G, H) MitoTracker staining (red) (G), immune-fluorescence staining of UCP1 (red) (H), and fluorescence quantification of FGF2+/+ and FGF2−/− beiging white adipocytes (n = 6). The nuclei (blue) were stained with DAPI. Scale bar = 10 μm. Values represent means ± SEM. Statistical analysis was performed using two-tailed Student's t-test. ∗p < 0.05, ∗∗p < 0.01 compared with that of FGF2+/+.
3.5. Exogenous FGF2 application inhibits expression of adipogenic and thermogenic genes in cultured brown and beige adipocytes
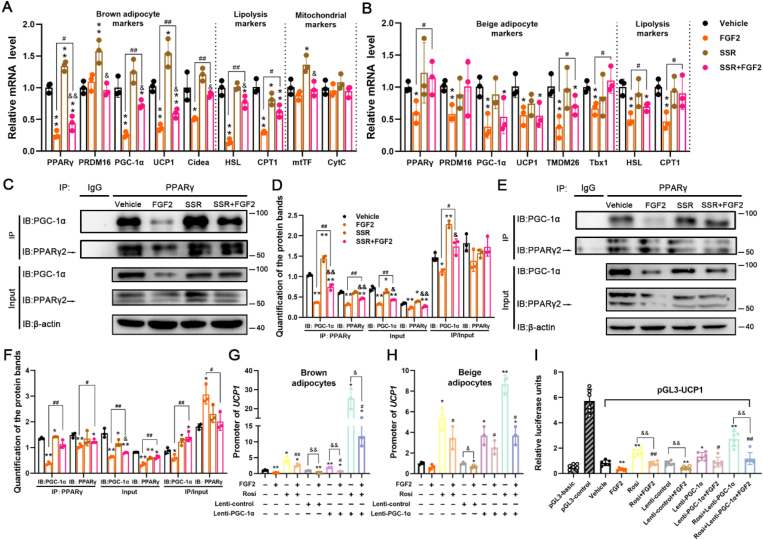
Previous reports have established that FGF2 acts in either an autocrine or a paracrine fashion via FGFR1 binding that requires concurrent interaction with heparin (HP) [21]. Thus, we supplemented exogenous FGF2 together with HP to the cell culture medium to test the alteration of thermogenic genes. In the SVF-differentiated brown adipocytes, FGF2 plus HP treatment strongly suppressed the mRNA levels of PGC-1α, UCP1, and CytC, as well as the protein expression of PGC-1α (Figure S9). We used FGF2 in conjunction with HP in all the following experiments.
In in vitro cultured SVF-differentiated brown adipocytes, exogenous FGF2 supplementation led to large suppression of PPARγ, PGC-1α, UCP1, and Cidea, as well as HSL and CPT1, whereas SSR alone showed elevation of PPARγ and UCP1 (Figure 7 A). SSR was able to antagonize the inhibition by FGF2 on thermogenic gene expression, including PGC-1α, UCP1, and Cidea, as well as on the adipogenic marker PPARγ (Figure 7 A). Moreover, when comparing the genes expression in the SSR alone group, FGF2 and SSR co-treatment significantly down-regulated transcription of PPARγ, PGC-1α, UCP1, Cidea, HSL, and mtTF (Figure 7 A). For the protein expression of PPARγ and PGC-1α, FGF2 showed marked inhibition, while SSR only displayed modest stimulation of PPARγ, and SSR + FGF2 co-treatment just rescued the FGF2 suppression of PGC-1α (Figure 7C,D). However, the protein contents of PPARγ and PGC-1α were both lower in the SSR + FGF2 group than the SSR alone (Figure 7C,D). These results indicate that FGF2 inhibits expression of thermogenic and adipogenic genes via a paracrine manner in brown adipocytes.
Figure 7.
Exogenous FGF2 application suppresses thermogenesis in both brown and beige adipocytes by inhibiting PGC-1α and PPARγ expression. (A) The inhibition of FGF2 on brown adipocyte-, lipolysis-, and mitochondrial-associated genes could be partially rescued by SSR, as indicated by qRT-PCR analysis (n = 3). GAPDH serves as an internal control. (B) The inhibition of FGF2 on beige adipocyte-, and lipolysis-associated genes could be partially rescued by SSR, as indicated by qRT-PCR analysis (n = 3). GAPDH serves as an internal control. (C and D) Co-IP analysis and quantification of association between PGC-1α and PPARγ in differentiating brown adipocytes treated with Vehicle, FGF2, SSR, or SSR + FGF2 (n = 3). (E and F) Co-IP analysis and quantification of association between PGC-1α and PPARγ in differentiating beige adipocytes treated with Vehicle, FGF2, SSR, or SSR + FGF2 (n = 3). (G) ChIP assay of the association between PPARγ and the UCP1 promoter in brown adipocytes treated with Vehicle, FGF2, Rosi, Rosi + FGF2, Lenti-control, Lenti-control + FGF2, Lenti-PGC-1α, Lenti-PGC-1α+FGF2, Rosi + Lenti-PGC-1α, or Rosi + Lenti-PGC-1α+FGF2 (n = 3). (H) ChIP assay of the interaction between PPARγ and the UCP1 promoter in beige adipocytes treated with Vehicle, FGF2, Rosi, Rosi + FGF2, Lenti-control, Lenti-control + FGF2, Lenti-PGC-1α, Lenti-PGC-1α+FGF2, Rosi + Lenti-PGC-1α, or Rosi + Lenti-PGC-1α+FGF2 (n = 3). (I) Dual luciferase activity driven by UCP1 transcription in HEK293 cells treated with Vehicle, FGF2, Rosi, Rosi + FGF2, Lenti-control, Lenti-control + FGF2, Lenti-PGC-1α, Lenti-PGC-1α+FGF2, Rosi + Lenti-PGC-1α, or Rosi + Lenti-PGC-1α+FGF2 (n = 6). Values represent means ± SEM. Statistical analysis was performed using one-way ANOVA after Tukey's multiple comparison test. ∗p < 0.05, ∗∗p < 0.01 vs. Vehicle; #p < 0.05, ##p < 0.01 vs. FGF2 treatment; &p < 0.05, &&p < 0.01 vs. individual control treatments without FGF2.
In SVF-differentiated beige adipocytes, FGF2 greatly inhibited the mRNA expression levels of all the tested beige adipocyte markers (e.g. PPARγ, PRDM16, PGC-1α, UCP1, TMEM26, and Tbx1), as well as the lipolysis markers (HSL and CPT1). However, only the FGF2-suppressed transcription of PPARγ, TMEM26, Tbx1, HSL and CPT1 was efficiently recovered by SSR (Figure 7 B). We found that SSR alone could not significantly affect the genes transcription compared to the Vehicle, and there was no evident difference of the genes expression between SSR + FGF2 and SSR treatments (Figure 7 B). Strikingly, FGF2 decreased the protein contents of PPARγ and PGC-1α, which could be modestly rescued by SSR (Figure 7E,F). However, SSR alone even slightly suppressed the protein expression of PPARγ and PGC-1α, and SSR + FGF2 treatment only exhibited a significant down-regulation of PGC-1α compared to SSR treatment (Figure 7E,F). These findings suggest that FGF2 strongly suppresses the beiging process of white adipocytes, but the FGFR inhibitor SSR just modestly negates the FGF2-mediated blockade of some beiging markers.
3.6. Both PPARγ and PGC-1α play roles in FGF2 suppression of UCP1 expression in brown and beige adipocytes
The expression levels of UCP1 determine thermogenic capability for both brown and beige adipocytes, which could be induced by the PPARγ transcription factor in conjunction with other regulatory co-factors, such as PGC-1α [2,6,35]. In light of the FGF2 suppression of PPARγ and PGC-1α accumulation, we further examined whether FGF2 interfered with interactions between PPARγ/PGC-1α and the UCP1 promoter region.
In brown adipocytes, while exogenous FGF2 application led to decreased interaction between PGC-1α with PPARγ, the addition of SSR alleviated the inhibitory impact of FGF2 (Figure 7C,D). The PPARγ-interacted PGC-1α was significantly increased in the SSR group than Vehicle, and also than the SSR + FGF2 treatment. This was partially but not fully dependent on the altered protein contents of PPARγ and PGC-1α in different treatments, as there was also significant difference when comparing the IP's PGC-1α protein to the individual Input protein (Figure 7C,D). Results of ChIP assay indicated that PPARγ interaction with the UCP1 promoter was significantly decreased in the presence of FGF2, which was rescued largely by Rosi treatment but was more efficiently recovered by Rosi and Lenti-PGC-1α co-treatment (Figure 7 G). It was shown significant variation on the PPARγ-interacted UCP1 promoter levels when comparing the Lenti-PGC-1α+FGF2 with Lenti-PGC-1α treatment, and comparing the Rosi + Lenti-PGC-1α+FGF2 with Rosi + Lenti-PGC-1α treatment (Figure 7 G).
In beige adipocytes, FGF2 significantly inhibited the interaction of PGC-1α with PPARγ, and SSR negated the blockade of FGF2 (Figure 7E,F). However, the alteration was primarily resulted from the changed protein content of PGC-1α to PPARγ, because the PPARγ level was just slightly varied among different treatments (Figure 7E,F). When comparing the IP proteins between SSR with Vehicle, or between SSR + FGF2 with SSR, there were few differences (Figure 7E,F). ChIP assay showed that PPARγ interaction with the UCP1 promoter was markedly inhibited by FGF2, which was strongly recovered by Rosi, Lenti-PGC-1α, and Rosi + Lenti-PGC-1α, respectively (Figure 7H). However, there was no significant difference when comparing the Rosi + FGF2 with Rosi treatment, comparing the Lenti-PGC-1α+FGF2 with Lenti-PGC-1α treatment, or comparing the Rosi + Lenti-PGC-1α+FGF2 with Rosi + Lenti-PGC-1α treatment (Figure 7H), which suggested the relative lower suppression degree of FGF2 on UCP1 expression in beige adipocytes.
In HEK293 cells, the luciferase reporter activity driven by the UCP1 promoter showed that Rosi, Lenti-PGC-1α, and the two together could highly rescue the UCP1 expression that was decreased by FGF2 (Figure 7 I). Although Rosi, Lenti-PGC-1α, and Rosi + Lenti-PGC-1α each showed significant elevation of UCP1 expression, suppression was observed when comparing the Rosi + FGF2 with Rosi or comparing the Rosi + Lenti-PGC-1α+FGF2 with Rosi + Lenti-PGC-1α (Figure 7 I).
Taken together, while FGF2 suppression of UCP1 expression in brown adipocytes is dependent on both PPARγ and PGC-1α, its action in beige adipocytes is primarily attributable to PGC-1α and slightly to PPARγ.
4. Discussion
Here, we show that FGF2 disruption stimulates the thermogenic potential of both brown and beige fat, revealing a previously unrecognized role of FGF2 in adipocyte function. The major findings of this study are as follows: a) FGF2-KO mice exhibit an elevated capacity for thermogenesis in both brown and beige fats, thus showing higher energy expenditure under both basal and cold conditions; b) FGF2 gene deletion protects against HFD-induced adiposity and hepatic steatosis; c) FGF2 leads to inhibition of the expression of PPARγ and PGC-1α, thereby suppressing thermogenic gene expression.
Although several FGF family members, including FGF2, have been established to play unique roles in fat metabolism and/or function, white fat has received considerably more attention [[36], [37], [38], [39]]. For example, Jonker et al. [36] reported on the role of the PPARγ-FGF1 signaling axis in adaptive adipose remodeling, in which FGF1-KO mice show impaired adipose expansion under high fat conditions. FGF21 functions in a feed-forward loop to regulate PPARγ activity via prevention of sumoylation, and FGF21-KO mice show attenuated expression of PPARγ-dependent genes and decreased body fat [38]. Fisher et al. [16] showed that FGF21 enhances PGC-1α protein expression as well as the browning of WAT in adaptive thermogenesis in an autocrine/paracrine manner. Contrary to the role of FGF21, we find that FGF2 negatively affects PGC-1α mRNA and protein abundance, subsequently blocking the white-to-brown fat switch. In addition, we observe an increase in mtDNA copy number and thermogenic gene expression in FGF2-KO BAT. FGF2 secretion is up-regulated by thermoneutrality. These findings collectively provide the first description of FGF2 function in thermogenic potentials in both brown and beige fat cells. The negative FGF2 thermogenic regulation seems to contradict other recently reported FGFs, in which FGF6 and FGF9 induce a high level of UCP1 expression in brown and white preadipocytes independent of adipogenic differentiation [40], while FGF8b triggers UCP1 expression in epididymal white preadipocytes and interfere in adipogenesis [41]. In our study, we reveal the inhibition of FGF2 on UCP1 expression as well as on adipogenesis (Figure 7, Figure S10). It can be concluded that different FGF family members function variably with different mechanisms.
As expected, oxygen consumption, RER, and body temperature are all increased due to FGF2 disruption, which is consistent with the increase in whole-body metabolic rate associated with activated brown and/or beige fat function [31,33]. Accordingly, FGF2 disruption amplifies the enhancement of cold-induced thermogenic activity for both brown and white fat cells. Thermogenic activity of activated brown and/or beige fat is often accompanied by improved metabolic homeostasis [2,10]. Our results indicate the potential contribution to improved lipid homeostasis made by increasing thermogenic capability of fat tissues, which is evidenced by the resistance to diet-induced adiposity and hepatic steatosis in FGF2−/− mice. The elimination of FGF2 KO protection on white adipose tissue expansion at thermoneutrality further support that the benefits stem from changes in thermogenic activity of adipose tissue (Figure S11). These correlations are similar to findings from other studies in which the abundance and/or the thermogenic ability of brown or beige cells are increased in KO strains (such as retinoblastoma protein, tumor necrosis factor-α receptor 1, liver X receptor, and histone deacetylase 11) that also exhibit improved lipid homeostasis and resistance to HFD-induced obesity and/or fatty liver [32,33,42,43]. These findings suggest a fascinating possibility that priming brown and beige fat function to tackle adiposity and associated hepatic steatosis via inhibition of FGF2 signaling.
In fact, we also examined the glucose level and insulin sensitivity of the WT and FGF2-KO mice as enhanced energy expenditure is often accompanied with improved glucose homeostasis [4,36]. However, neither the glucose tolerance nor the insulin sensitivity is significantly altered by FGF2 deficiency, both under chow- and HFD-feeding conditions (Figure S12). It has been established that besides adipose tissue, FGF2 is expressed in other tissues during development and in adult mice and has been shown to play critical roles in cellular proliferation, angiogenesis, and differentiation to several lineages [21,22,24,44,45]. Since we used a whole body FGF2-KO mouse model, it is conceivable that the influence of FGF2 gene disruption on other tissue(s) might counteract the anticipated improvement of glucose metabolism. This needs to be elucidated in future studies.
We reveal the cell-autonomous regulation of FGF2 on thermogenesis through FGFR, which is similar with that of FGF6, FGF8b, FGF9, and FGF21, although in contrast, those FGFs enhance the thermogenic potentials [16,40,41]. It has been demonstrated that FGFs activate a series of downstream signaling pathways, such as PI3K-AKT, Ras-PKC, MAPK, and JAK/STAT3 [[46], [47], [48], [49], [50]]. However, the RNA sequencing data showed that only small proportion of genes in the above pathways are significantly regulated by FGF2 disruption (Table S1 and S2). Because the ERK signaling pathway has been reported to be involved in the FGF2 inhibition of hASC adipogenesis [34], we further determined whether this signaling also plays a role in FGF2 regulation of thermogenesis. In in vitro-cultured brown and beige adipocytes, FGF2 application indeed led to enhanced phosphorylation of ERK, whereas the ERK inhibitor PD only slightly antagonized the FGF2 inhibition of thermogenic genes (Figure S13 and S14). The modest repression of ERK phosphorylation in FGF2 KO-derived brown and white adipocytes (Figure S15), as well as the unaltered phosphorylation level of ERK in KO iWAT and iBAT (Figure S16) further supported the small contribution of this signaling to the FGF2 regulation of thermogenesis. We postulate that the FGF2 regulation of thermogenesis might be dependent on several signaling pathways rather certain one, which should be clarified in future investigations.
Given the negative correlation of PPARγ and PGC-1α with FGF2, we further discovered the essential regulatory contributions of PPARγ and PGC-1α in modulating FGF2 activity by using Rosi (a PPARγ-specific agonist) and Lenti-PGC-1α (a recombinant PGC-1α expression lentivirus construct) in the in vitro-cultured brown and beige adipocytes (Figure S13 and S14, and Figure 7). This finding is in agreement with previous studies which showed PGC-1α is a critical transcription co-factor of PPARγ for UCP1 expression [5,7,35]. Collectively, these results indicate that FGF2 application inhibits PPARγ and PGC-1α expression to suppress thermogenic gene expression.
In conclusion, we describe an unreported role for FGF2 in the negative regulation of thermogenesis of brown and beige fat in a cell-autonomous manner. FGF2-KO mice show elevated thermogenic ability of brown and beige fat, with higher energy expenditure and improved lipid homeostasis, as well as protection against HFD-induced adiposity and hepatic steatosis. Both PPARγ and PGC-1α play roles in the negative regulation of thermogenic gene expression by FGF2. Future studies will further elucidate the contribution of autocrine FGF2 to the inhibition of brown fat function and white-to-beige fat conversion by using mice with adipocyte-specific conditional deletion of FGF2. Further investigation is needed to determine the role of FGF2-specific signaling inhibitors in the thermogenic activities of adipose tissues in vivo.
Author contributions
Haifang Li, Xinzhi Zhang, Cheng Huang, Huan Liu, and Qiang Zhang performed in vitro and in vivo experiments, wrote the manuscript, organized the literature and figures and performed statistical analysis. Haifang Li, and Hai Lin conceived the project, led and supervised the study, and reviewed/edited the manuscript. Qianying Sun, Yanxin Jia, Shuang Liu, Mei Dong, Mengjie Hou, and Yiming Liu performed experiments, contributed to discussion and revision.
Acknowledgments
The authors would like to thank Zizhang Zhou (Shandong Agricultural University), Qingxin Liu (Shandong Agricultural University), Xuguo Zhou (University of Kentucky), Yingli Shang (Shandong Agricultural University), Yi Tang (Shandong Agricultural University), and Jinfang Zhang (Guangzhou University of Chinese Medicine) for their assistance in preparation of this manuscript. We thank Wencheng Zhang (Shandong University) for assistance with metabolic studies, and to Yanqin Wang (Hebei Normal University) for help in lentivirus production. This work was supported by the National Key Research and Development Program of China (No. 2016YFD0500510, 2017YFE0129800), the Taishan Scholars Program [No. 201511023, 201712022], Funds of Shandong “Double Tops” Program, the Natural Science Foundation of Shandong Province, China [No. ZR2019MC016], and the Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province [2019KJE009].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101358.
Contributor Information
Haifang Li, Email: haifangli@sdau.edu.cn.
Hai Lin, Email: hailin@sdau.edu.cn.
Conflict of interest
The authors declare that they have no conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poekes L., Lanthier N., Leclercq I.A. Brown adipose tissue: a potential target in the fight against obesity and the metabolic syndrome. Clinical Science. 2015;129:933–949. doi: 10.1042/CS20150339. [DOI] [PubMed] [Google Scholar]
- 3.Wu J., Boström P., Sparks L.M., Ye L., Choi J.H., Giang A.H., et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanford K.I., Middelbeek R.J., Townsend K.L., An D., Nygaard E.B., Hitchcox K.M., et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. Journal of Clinical Investigation. 2013;123:215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 6.Puigserver P., Spiegelman B.M. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocrine Reviews. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 7.Xue B., Coulter A., Rim J.S., Koza R.A., Kozak L.P. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Molecular and Cellular Biology. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T., et al. Functional brown adipose tissue in healthy adults. New England Journal of Medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 9.Rogers N.H. Brown adipose tissue during puberty and with aging. Annals of Medicine. 2015;47:142–149. doi: 10.3109/07853890.2014.914807. [DOI] [PubMed] [Google Scholar]
- 10.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J., et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D., et al. Cold-activated brown adipose tissue in healthy men. New England Journal of Medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 12.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J., et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. Journal of Clinical Investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachman E.S., Dhillon H., Zhang C.Y., Cinti S., Bianco A.C., Kobilka B.K., et al. BetaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 14.Villarroya F., Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metabolism. 2013;17:638–643. doi: 10.1016/j.cmet.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Whittle A.J., Carobbio S., Martins L., Slawik M., Hondares E., Vázquez M.J., et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell. 2012;149:871–885. doi: 10.1016/j.cell.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher F.M., Kleiner S., Douris N., Fox E.C., Mepani R.J., Verdeguer F., et al. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes & Development. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boström P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C., et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Than A., He H.L., Chua S.H., Xu D., Sun L., Leow M.K., et al. Apelin enhances brown adipogenesis and browning of white adipocytes. Journal of Biological Chemistry. 2015;290:14679–14691. doi: 10.1074/jbc.M115.643817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeve D., Tang W., Graff J. Fighting fat with fat: the expanding field of adipose stem cells. Cell Stem Cell. 2009;5:472–481. doi: 10.1016/j.stem.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers C.J., McLeskey S.W., Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocrine-Related Cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 21.Zhou M., Sutliff R.L., Paul R.J., Lorenz J.N., Hoying J.B., Haudenschild C.C., et al. Fibroblast growth factor 2 control of vascular tone. Nature Medicine. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raballo R., Rhee J., Lyn-Cook R., Leckman J.F., Schwartz M.L., Vaccarino F.M. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. Journal of Neuroscience. 2000;20:5012–5023. doi: 10.1523/JNEUROSCI.20-13-05012.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hurley M.M., Lee S.K., Raisz L.G., Bernecker P., Lorenzo J. Basic fibroblast growth factor induces osteoclast formation in murine bone marrow cultures. Bone. 1998;22:309–316. doi: 10.1016/s8756-3282(97)00292-5. [DOI] [PubMed] [Google Scholar]
- 24.Montero A., Okada Y., Tomita M., Ito M., Tsurukami H., Nakamura T., et al. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. Journal of Clinical Investigation. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi N., Toriyama K., Nicodemou-Lena E., Inou K., Torii S., Kitagawa Y. De novo adipogenesis in mice at the site of injection of basement membrane and basic fibroblast growth factor. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1062–1066. doi: 10.1073/pnas.95.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakudo N., Shimotsuma A., Kusumoto K. Fibroblast growth factor-2 stimulates adipogenic differentiation of human adipose-derived stem cells. Biochemical and Biophysical Research Communications. 2007;359:239–244. doi: 10.1016/j.bbrc.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 27.Hao R.H., Guo Y., Dong S.S., Weng G.Z., Yan H., Zhu D.L., et al. Associations of plasma FGF2 levels and polymorphisms in the FGF2 gene with obesity phenotypes in Han Chinese population. Scientific Reports. 2016;6:19868. doi: 10.1038/srep19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao L., Sobue T., Esliger A., Kronenberg M.S., Coffin J.D., Doetschman T., et al. Disruption of the Fgf2 gene activates the adipogenic and suppresses the osteogenic program in mesenchymal marrow stromal stem cells. Bone. 2010;47:360–370. doi: 10.1016/j.bone.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suchacki K.J., Stimson R.H. Nutritional regulation of human brown adipose tissue. Nutrients. 2021;13(6):1748. doi: 10.3390/nu13061748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui X., Nguyen N.L., Zarebidaki E., Cao Q., Li F., Zha L., et al. Thermoneutrality decreases thermogenic program and promotes adiposity in high-fat diet-fed mice. Physiological Reports. 2016;4(10) doi: 10.14814/phy2.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao L., Cui X., Chen Q., Yang X., Fang F., Zhang J., et al. Cold-inducible SIRT6 regulates thermogenesis of brown and beige fat. Cell Reports. 2017;20:641–654. doi: 10.1016/j.celrep.2017.06.069. [DOI] [PubMed] [Google Scholar]
- 32.Hansen J.B., Jørgensen C., Petersen R.K., Hallenborg P., De Matteis R., Bøye H.A., et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagchi R.A., Ferguson B.S., Stratton M.S., Hu T., Cavasin M.A., Sun L., et al. HDAC11 suppresses the thermogenic program of adipose tissue via BRD2. Journal of Clinical Investigation. 2018;3 doi: 10.1172/jci.insight.120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S., Ahn C., Bong N., Choe S., Lee D.K. Biphasic effects of FGF2 on adipogenesis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmadian M., Suh J.M., Hah N., Liddle C., Atkins A.R., Downes M., et al. PPARγ signaling and metabolism: the good, the bad and the future. Nature Medicine. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonker J.W., Suh J.M., Atkins A.R., Ahmadian M., Li P., Whyte J., et al. A PPARγ-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature. 2012;2485:391–394. doi: 10.1038/nature10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Badman M.K., Pissios P., Kennedy A.R., Koukos G., Flier J.S., Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metabolism. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Dutchak P.A., Katafuchi T., Bookout A.L., Choi J.H., Yu R.T., Mangelsdorf D.J., et al. Fibroblast growth factor-21 regulates PPARγ activity and the antidiabetic actions of thiazolidinediones. Cell. 2012;148:556–567. doi: 10.1016/j.cell.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakaue H., Konishi M., Ogawa W., Asaki T., Mori T., Yamasaki M., et al. Requirement of fibroblast growth factor 10 in development of white adipose tissue. Genes & Development. 2002;16:908–912. doi: 10.1101/gad.983202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shamsi F., Xue R., Huang T.L., Lundh M., Liu Y., Leiria L.O., et al. FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nature Communications. 2020;11(1):1421. doi: 10.1038/s41467-020-15055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westphal S., Gantert T., Kless C., Hüttinger K., Klingenspor M., Fromme T. Fibroblast growth factor 8b induces uncoupling protein 1 expression in epididymal white preadipocytes. Scientific Reports. 2019;9(1):8470. doi: 10.1038/s41598-019-44878-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romanatto T., Roman E.A., Arruda A.P., Denis R.G., Solon C., Milanski M., et al. Deletion of tumor necrosis factor-alpha receptor 1 (TNFR1) protects against diet-induced obesity by means of increased thermogenesis. Journal of Biological Chemistry. 2016;291:26934. doi: 10.1074/jbc.A109.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Zhang Y., Yehuda-Shnaidman E., Medvedev A.V., Kumar N., Daniel K.W., et al. Liver X receptor alpha is a transcriptional repressor of the uncoupling protein 1 gene and the brown fat phenotype. Cellular and Molecular Biology. 2008;28:2187–2200. doi: 10.1128/MCB.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefaucheur J.P., Sebille A. Basic fibroblast growth factor promotes in vivo muscle regeneration in murine muscular dystrophy. Neuroscience Letters. 1995;202(1–2):121–124. doi: 10.1016/0304-3940(95)12223-0. [DOI] [PubMed] [Google Scholar]
- 45.Doukas J., Blease K., Craig D., Ma C., Chandler L.A., Sosnowski B.A., et al. Delivery of FGF genes to wound repair cells enhances arteriogenesis and myogenesis in skeletal muscle. Molecular Therapy - Nucleic Acids. 2002;5(5 Pt 1):517–527. doi: 10.1006/mthe.2002.0579. [DOI] [PubMed] [Google Scholar]
- 46.Okada T., Enkhjargal B., Travis Z.D., Ocak U., Tang J., Suzuki H., et al. FGF-2 attenuates neuronal apoptosis via FGFR3/PI3k/akt signaling pathway after subarachnoid hemorrhage. Molecular Neurobiology. 2019;56(12):8203–8219. doi: 10.1007/s12035-019-01668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geary L., LaBonne C. FGF mediated MAPK and PI3K/Akt Signals make distinct contributions to pluripotency and the establishment of Neural Crest. Elife. 2018;7 doi: 10.7554/eLife.33845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathew G., Hannan A., Hertzler-Schaefer K., Wang F., Feng G.S., Zhong J., et al. Targeting of Ras-mediated FGF signaling suppresses Pten-deficient skin tumor. Proceedings of the National Academy of Sciences of the U S A. 2016;113(46):13156–13161. doi: 10.1073/pnas.1604450113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ranieri D., Nanni M., Persechino F., Torrisi M.R., Belleudi F. Role of PKCepsilon in the epithelial-mesenchymal transition induced by FGFR2 isoform switch. Cell Communication and Signaling. 2020;18(1):76. doi: 10.1186/s12964-020-00582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong Z., Santeford A., Ban N., Lee T.J., Smith C., Ornitz D.M., et al. FGF2-induced STAT3 activation regulates pathologic neovascularization. Experimental Eye Research. 2019;187:107775. doi: 10.1016/j.exer.2019.107775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.