Mutual antagonism between the transcriptional repressors Bcl-6 and Blimp-1 has been appreciated as a key mechanistic determinant of lymphoid differentiation programs. Now, in this issue of JEM, Ciucci et al. demonstrate that this relationship is central to the generation of T cell memory.
Abstract
For over a decade, mutual antagonism between the transcriptional repressors Bcl-6 and Blimp-1 has been appreciated as a key mechanistic determinant of lymphoid differentiation programs. Now, in this issue of JEM, Ciucci et al. (2021. J. Exp. Med. https://doi.org/10.1084/jem.20202343) demonstrate that this relationship is "central" to the generation of T cell memory.
The establishment of effective immunological memory is key to the efficacy of one of the most successful therapies of modern medicine: the vaccine. The importance of understanding the mechanisms underlying this process has been highlighted most recently by the dire need for successful vaccination strategies against SARS-CoV-2. Two types of CD4+ T helper cell populations that are critical for this process are T follicular helper (Tfh) cells, which facilitate humoral immune responses by assisting B lymphocytes with the production of pathogen-neutralizing antibodies, and long-lived memory T cells capable of generating effector responses more rapidly and robustly to repeat pathogenic encounters (Crotty, 2019; Krueger et al., 2021). Of the latter, central memory T (Tcm) cells reside in the T cell zone of secondary lymphoid organs to enable long-term antigen surveillance. Despite their distinct functions, it has been established that Tfh and Tcm cell populations share certain regulatory requirements, including dependence on a partially conserved set of transcriptional regulators that includes the repressor Bcl-6 (Ichii et al., 2007, Johnston et al., 2009, Nurieva et al., 2009, Pepper et al., 2011, Yu et al., 2009). The differentiation of both cell types is also inhibited by IL-2/STAT5 signaling and the STAT5-induced transcriptional repressor Blimp-1, which is a direct target of Bcl-6–mediated repression (Crotty et al., 2010). These overlapping regulatory mechanisms have led to speculation that Tfh and Tcm cells may arise from a shared cell lineage, yet the phenotypic and functional differences between these populations argue that a divergence must occur at some point. Exactly how this divergence occurs has been unclear. In this issue of JEM, Ciucci et al. (2021) begin to provide needed clarity to this subject by identifying distinct roles for Bcl-6 in regulating Tcm versus Tfh cell differentiation, notably that the obligate role for Bcl-6 in the former process is to repress Blimp-1 expression.

Insights from Kaitlin A. Read and Kenneth J. Oestreich.
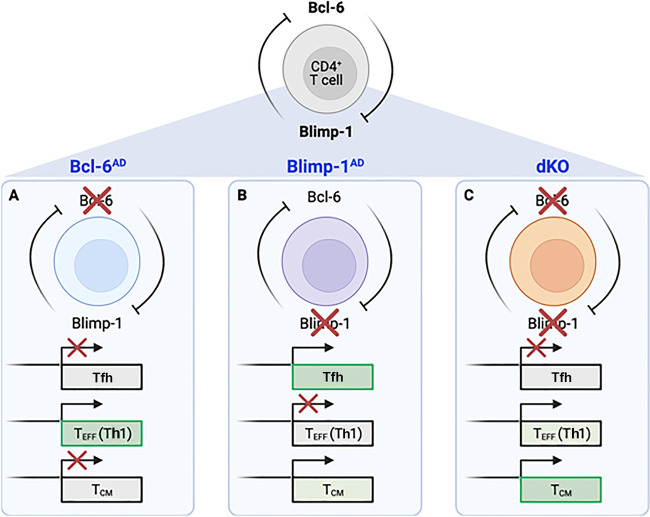
The authors began by using a lymphocytic choriomeningitis virus (LCMV) infection model to examine the effects of conditional deletion of Bcl-6 (Bcl-6AD), Blimp-1 (Blimp-1AD), or both (dKO) on responding antigen-specific CD4+ T cell populations in a mixed bone marrow chimera setting. Consistent with previous findings, loss of Bcl-6 compromised both Tfh and Tcm differentiation relative to WT controls, while Blimp-1–deficient animals displayed the opposite phenotype. Intriguingly, while dKO animals displayed a decrease in Tfh populations, there was no defect in the percentage of cells expressing Tcm surface markers, suggesting that the essential role for Bcl-6 in memory cell differentiation is to repress Blimp-1 (see figure).
Model depicting the phenotypic outcomes of selective deletion of Bcl-6 (A), Blimp-1 (B), or both (C) in CD4+ T cell populations during LCMV infection. Ciucci et al. (2021) find that differential requirements for these mutually antagonistic factors ultimately shape the transcriptional landscape underlying Tfh and Tcm cell generation. While Bcl-6 has been shown to be important for the generation of both cell types, CD4+ T cells lacking both Bcl-6 and Blimp-1 are able to generate Tcm—but not Tfh—cell populations, indicating that the obligate role for Bcl-6 in Tcm cells is to repress expression of Blimp-1.
The authors further characterized WT, Bcl-6AD, Blimp-1AD, and dKO CD4+ T cells by performing single cell RNA-sequencing (scRNAseq) at a peak effector time point (7 d after infection). Supporting the idea that Bcl-6 is not required for Tcm formation in the absence of Blimp-1, the authors observed a “rescue” of cells expressing a T central memory precursor (Tcmp) cell gene signature in dKO cells relative to Bcl-6AD. Curiously, dKO cells also expressed genes encoding transcriptional regulators (Tbx21, Id2, and Runx3) and an effector cytokine (Ifng) associated with T helper 1 (Th1) cells. These findings were in contrast to the expected, and observed, increases in cells with Th1 and Tfh signatures in Bcl-6–deficient and Blimp-1–deficient cells, respectively. Collectively, these data suggested that despite loss of Bcl-6, Tcmp cell–like populations were present in dKO cells at the effector stage of the immune response. To determine whether such differences were maintained following the resolution of infection, the authors also performed scRNAseq analysis 30 d after infection. As with the effector time point, a population of cells expressing a Tcm cell signature was absent in the Bcl-6AD setting, yet present in the dKO setting. These data were consistent with increased chromatin accessibility at Tcm cell gene regulatory elements, observed via concurrent single-cell assay for transposase accessible chromatin sequencing analysis, suggesting a profound impact on the Tcm cell chromatin landscape in the absence of both Bcl-6 and Blimp-1.
Finally, the authors probed the functional nature of the Tcm cell–like populations in dKO cells by assessing both their ability to produce cytokines and proliferate in response to antigenic rechallenge. Consistent with bona fide memory cell function, dKO cells produced IL-2 and TNF-α at similar levels to WT cells when stimulated with LCMV-specific peptides ex vivo ≥30 d after infection. Experiments to assess recall responses also revealed dKO memory cell proliferation via Ki67 expression following rechallenge with LCMV. Based on these findings, the authors concluded that dKO cells provide long-lasting, antigen-specific memory responses.
The findings by Ciucci et al. (2021) are exciting and impactful for several reasons. First, their work helps to define the specific role of Bcl-6 and, in a broader context, the role of the Bcl-6–Blimp-1 regulatory axis, more precisely in CD4+ Tcm cell differentiation. Specifically, that Bcl-6 is not required for Tcm cell generation when Blimp-1 is absent. Further, this work complements two studies demonstrating that the role of Bcl-6 extends beyond Blimp-1 repression during Tfh cell differentiation, and thus helps to provide much-needed clarity regarding the conserved and divergent roles of Bcl-6 in Tcm versus Tfh cell populations (Choi et al., 2020; Xie et al., 2017). From a mechanistic standpoint, this study also opens the door to exciting new questions. First, how is the repertoire of target genes repressed by Bcl-6 specified between Tcm and Tfh cell populations? The findings by Ciucci et al. suggest that Prdm1 (Blimp-1) is the critical Bcl-6 target gene in Tcm cell differentiation, while additional gene targets have been identified in Tfh cells (Choi et al., 2020). It is possible that differential activities for Bcl-6 are related to its relative expression, as Bcl-6 protein is elevated in Tfh compared with Tcm cell populations (Pepper et al., 2011). However, a number of studies have shown that interactions between Bcl-6 and other transcriptional regulators or chromatin remodeling complexes, such as T-bet, AP1, and switch/sucrose non-fermentable, can alter the identity of specific Bcl-6 gene targets (Choi et al., 2015, Oestreich et al., 2011; Hatzi et al., 2015). Thus, it is interesting to speculate that cell type–specific interactions with transcription factors or chromatin modifiers may direct the repressive capabilities of Bcl-6, and ultimately explain its varied roles between Tcm and Tfh cell populations.
This work also provides a springboard from which to continue to examine developmental relationships between effector programs and long-lived memory populations, as well as temporal roles for Blimp-1 and Bcl-6 in these processes. The present study demonstrates that Tcmp cells arise at the effector stage following the early deletion of both Blimp-1 and Bcl-6. It is also intriguing to consider whether this relationship is conserved at later, post-effector stages. For example, it has been proposed that polyfunctional Tcm-like cells can arise from the effector Tfh cell subset (Krueger et al., 2021). In support of this, cells expressing both Tfh (Cxcr5) and Tcm (Ccr7) markers have been shown to give rise to diverse progeny during recall responses, including both Th1 and Tfh effector populations (Pepper et al., 2011; Lüthje et al., 2012). Thus, does the obligate role of Bcl-6 repressing Blimp-1 remain the key mechanism in these polyfunctional Tcm cell populations, or are there additional, temporal roles for Bcl-6 at the post-effector stage? It is interesting to note that the authors do observe phenotypic differences in Tcm cells generated in WT versus dKO animals. Specifically, Tcm cells from the latter appear to be biased toward a Th1 cell gene signature and away from a Tfh cell signature, indeed suggesting additional roles for Bcl-6 in maintaining Tcm cell polyfunctionality. Finally, as Tcmp/Tcm cell generation is not limited to type 1 immune responses, it will be of interest to determine whether the mechanism explored by Ciucci et al. in the current work is conserved during additional types of immune challenges. Ultimately, this study provides a strong foundation from which to answer these intriguing and therapeutically valuable questions.
The field of immunology has moved well beyond a cell differentiation model in which the presence of a single “master regulator” transcription factor equals the differentiation of a singular immune cell type. As with Bcl-6 in the present study, it has become increasingly clear that the functions of transcription factors are altered to regulate gene expression signatures as part of cell-specific differentiation programs. The findings by Ciucci et al. have provided much-needed insight into discrete roles for Bcl-6 in regulating Tcm and Tfh cell differentiation, and these findings in turn may be leveraged for therapeutic benefit. Tfh cells are clinically important to vaccine responses and the treatment of autoimmunity (Crotty, 2019). Tcm cells are robust mediators of anti-tumor immunity, including cellular therapy approaches such as chimeric antigen receptor T cells (Busch et al., 2016). Thus, distinguishing the mechanisms that regulate Tfh and Tcm cell differentiation will continue to be significant for human health, as such insights may allow for more refined strategies for the improvement of vaccination, the treatment of autoimmunity, and cancer immunotherapy approaches.
References
- Busch, D.H., et al. 2016. Semin. Immunol. 10.1016/j.smim.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., et al. 2015. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1418592112 [DOI] [Google Scholar]
- Choi, J., et al. 2020. Nat. Immunol. 10.1038/s41590-020-0706-5 [DOI] [Google Scholar]
- Ciucci, T., et al. 2021. J. Exp. Med. 10.1084/jem.20202343 [DOI] [Google Scholar]
- Crotty, S. 2019. Immunity. 10.1016/j.immuni.2019.04.011 [DOI] [Google Scholar]
- Crotty, S., et al. 2010. Nat. Immunol. 10.1038/ni.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi, K., et al. 2015. J. Exp. Med. 10.1084/jem.20141380 [DOI] [Google Scholar]
- Ichii, H., et al. 2007. Int. Immunol. 10.1093/intimm/dxm007 [DOI] [PubMed] [Google Scholar]
- Johnston, R.J., et al. 2009. Science. 10.1126/science.1175870 [DOI] [Google Scholar]
- Krueger, P.D., et al. 2021. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a038141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje, K., et al. 2012. Nat. Immunol. 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- Nurieva, R.I., et al. 2009. Science. 10.1126/science.1176676 [DOI] [Google Scholar]
- Oestreich, K.J., et al. 2011. J. Exp. Med. 10.1084/jem.20102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, M., et al. 2011. Immunity. 10.1016/j.immuni.2011.09.009 [DOI] [Google Scholar]
- Xie, M.M., et al. 2017. Eur. J. Immunol. 10.1002/eji.201747034 [DOI] [Google Scholar]
- Yu, D., et al. 2009. Immunity. 10.1016/j.immuni.2009.07.002 [DOI] [Google Scholar]