Single-cell RNA and ATAC sequencing identify transcriptomic and epigenomic features of acute- and memory-phase virus-responding CD4+ T cells and show that, unlike follicular helper T cells, memory CD4+ T cells develop despite lacking Bcl6 and Blimp1 transcription factors.
Abstract
During the immune response, CD4+ T cells differentiate into distinct effector subtypes, including follicular helper T (Tfh) cells that help B cells, and into memory cells. Tfh and memory cells are required for long-term immunity; both depend on the transcription factor Bcl6, raising the question whether they differentiate through similar mechanisms. Here, using single-cell RNA and ATAC sequencing, we show that virus-responding CD4+ T cells lacking both Bcl6 and Blimp1 can differentiate into cells with transcriptomic, chromatin accessibility, and functional attributes of memory cells but not of Tfh cells. Thus, Bcl6 promotes memory cell differentiation primarily through its repression of Blimp1. These findings demonstrate that distinct mechanisms underpin the differentiation of memory and Tfh CD4+ cells and define the Bcl6–Blimp1 axis as a potential target for promoting long-term memory T cell differentiation.
Introduction
CD4+ T cells are essential for durable immune responses. Upon antigen encounter, they differentiate into several effector subsets, including T follicular helper (Tfh) cells that provide help to B cells, or cytokine-producing cells, of which T helper type 1 (Th1) cells make IFNγ and contribute to defenses against intracellular pathogens (Laidlaw et al., 2016; MacLeod et al., 2009; Swain et al., 2012). In addition, antigen-specific memory CD4+ T cells persist after the resolution of infection. Among these, central memory (Tcm) cells, expressing the chemokine receptor CCR7, give rise to secondary effectors upon antigenic rechallenge (Pepper and Jenkins, 2011; Sallusto et al., 1999). One current perspective is that Tcm cells emerge from central memory precursors (Tcmp), a small subset of CD4+ T cells responding to infection that appear phenotypically distinct from effector cells (Marshall et al., 2011; Pepper et al., 2011).
However, whether memory CD4+ T cells develop along a trajectory separate from that of other CD4+ T cell subtypes is debated (Crotty, 2018). Unlike for precursors of memory CD8+ T cells (Crawford et al., 2014; Marshall et al., 2011), no simple combination of surface markers unambiguously distinguishes CD4+ Tcmp from effector T cells (Cho et al., 2017; Marshall et al., 2011; Vogelzang et al., 2014), and the same is true of transcription factors, including Tcf1 (encoded by Tcf7), expressed by both CD4+ Tcmp and Tfh cells (Choi et al., 2015; Nish et al., 2017; Wu et al., 2015; Xu et al., 2015). Additionally, both Tfh and memory cells depend on the zinc finger transcription factor Bcl6 (Ichii et al., 2007; Johnston et al., 2009; Nurieva et al., 2009; Pepper et al., 2011; Yu et al., 2009). Bcl6 and its antagonist Blimp1 (encoded by Prdm1, called Blimp1 here) promote the differentiation of Tfh and Th1 effector cells, respectively, and inhibit each other’s expression (Crotty et al., 2010). Accordingly, and mirroring Bcl6, Blimp1 inhibits the differentiation of memory T cells (Rutishauser et al., 2009; Shin et al., 2009). However, while Bcl6 is needed for CD4+ T cell memory, it has remained debated whether it promotes the survival of memory cells or the initial differentiation of memory precursors (Choi et al., 2013; Harrington et al., 2008; Ichii et al., 2007; Pepper et al., 2011; Tubo et al., 2016).
Recent studies using single-cell RNA sequencing (scRNAseq) have demonstrated that the Tcmp and Tcm transcriptomes are distinct from both Tfh and Th1 programs (Ciucci et al., 2019; Künzli et al., 2020). However, such studies could not address whether Tcmp cells derive separately or from Th1 or Tfh lineages. Here, we combined mouse genetics with single-cell chromatin accessibility and transcriptomic analyses to address this question. We demonstrate that CD4+ T cells deficient for both Bcl6 and Blimp1, unlike cells deficient for Bcl6 only, give rise to long-lived cells that develop gene expression and chromatin patterns associated with memory T cell differentiation and that mount antigen-driven cytokine and proliferative responses typical of memory cells. Thus, unlike for Tfh differentiation (Choi et al., 2020; Xie et al., 2017), the critical requirement for Bcl6 during CD4+ T cell memory differentiation is to repress the expression of Blimp1. These findings identify distinct mechanisms underpinning Tfh and Tcmp CD4+ T cell differentiation during the immune response.
Results and discussion
Blimp1-dependent and -independent function of Bcl6 during early CD4+ T cell differentiation
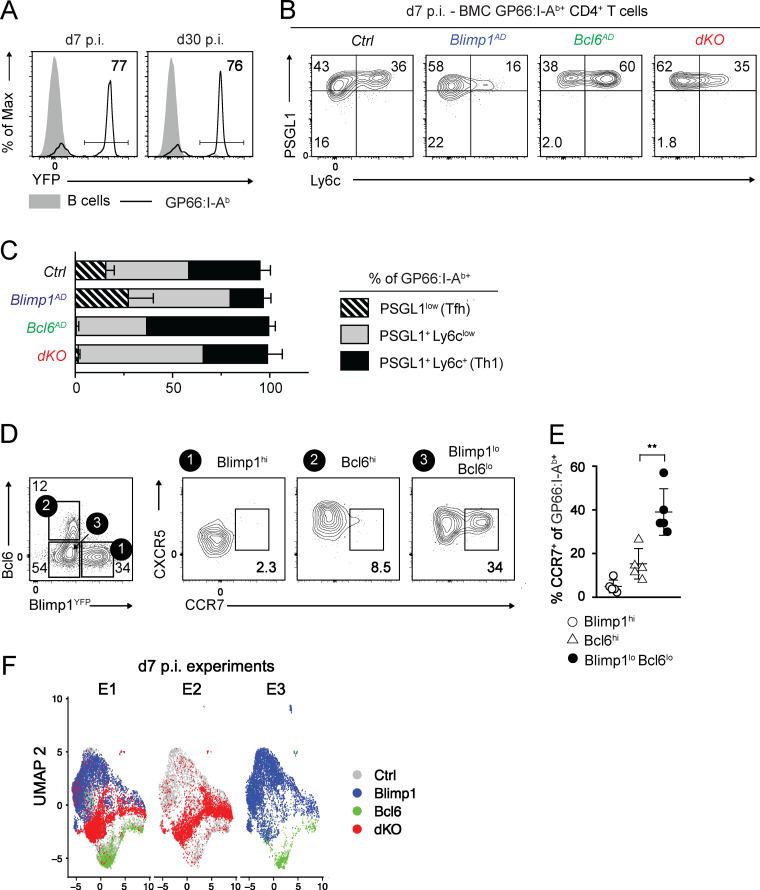
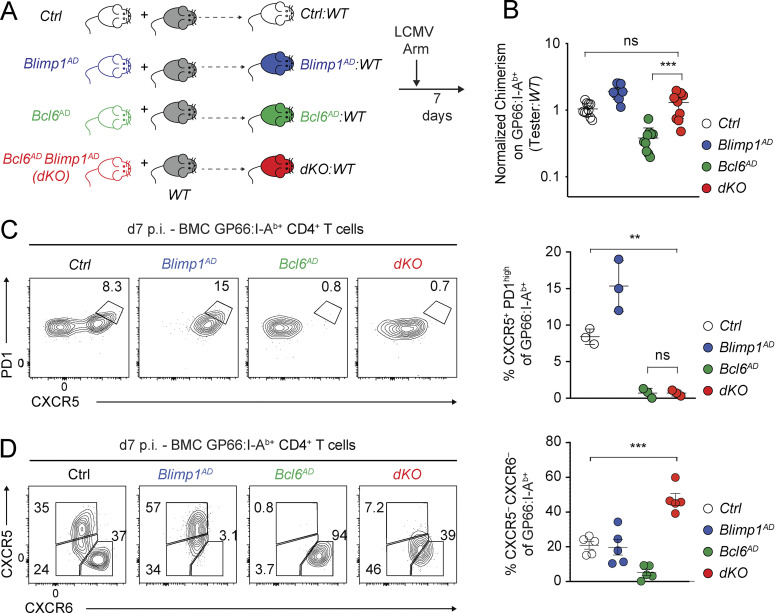
We investigated the functions of Bcl6 and Blimp1 in CD4+ T cells responding to the Armstrong strain of lymphocytic choriomeningitis virus (LCMV Arm), which WT mice clear within 7 d after inoculation (d7 p.i.; Oldstone, 2002). We tracked virus-specific cells binding MHC class II tetramers loaded with the LCMV GP66 peptide (GP66: I-Ab; Homann et al., 2001). We first inactivated Bcl6 or Blimp1 in mice expressing the Cre recombinase from the Tnfrsf4 gene, encoding Ox40 (called Ox40-Cre). After LCMV Arm infection, Ox40-Cre was expressed in >75% of antigen-specific CD4+ T cells but not in B cells or naive T cells (Ciucci et al., 2019; Fig. S1 A). Because of possible non–cell-intrinsic effects of Blimp1 and Bcl6 deletion on the immune response (Kallies et al., 2006; Martins et al., 2006), we performed experiments in mixed bone marrow chimeras (BMCs) generated by reconstituting lethally irradiated WT hosts with allelically marked WT “competitor” and Ox40-Cre+ “tester” bone marrow (Fig. 1 A). After immune reconstitution and viral infection, competitor cells support a normal virus response. Thus, we could investigate cell-intrinsic functions of Bcl6 and Blimp1 in the LCMV response using Ox40-Cre+ tester donors that deleted Blimp1 (Ox40-Cre+ Blimp1fl/fl, called Blimp1AD [for activation-induced deletion]), Bcl6 (Bcl6AD), or both (dKO). As a control (Ctrl), we used otherwise WT Ox40-Cre+ or Ox40-Cre– tester donors.
Figure S1.
(A) Ox40-Cre+ Rosa26YFP mice were infected with LCMV Arm, and spleen cells were analyzed at d7 or d30 p.i. Plots show Cre activity as reported by YFP expression on the indicated spleen T cell populations and is representative of more than three independent experiments. (B and C) Mixed BMCs made from Ctrl, Bcl6AD, Blimp1AD, or dKO tester and WT competitor were infected with LCMV Arm and analyzed at d7 p.i. (B) Ly6c versus PSGL1 expression on spleen GP66:I-Ab+ cells of the indicated genotype. (C) Bar graph shows the proportion of each subset as defined in B within the GP66:I-Ab+ populations in each tester genotype. (B and C) Data are from one experiment representative of three independent experiments with at least three animals per group. (D) Expression of Bcl6 and of YFP (indicative of Blimp1) on spleen GP66:I-Ab+ T cells (left plot) defines subsets analyzed for CCR7 versus CXCR5 expression (right three plots). (E) Graph shows the percentage of CCR7+ cells on the indicated GP66:I-Ab+ population. Each symbol represents a mouse. Data are from two pooled experiments representative of four independent experiments. **, P < 0.01; unpaired two-sided Welch’s t test. (F) UMAP plots from GP66:I-Ab+ T cells analyzed in Fig. 2. Each dot represents a cell and is color coded by genotype. Each plot includes cells of the experiment indicated at the top (defined in Table S1).
Figure 1.
Blimp1-independent impact of Bcl6 on CD4+ T cell responses. (A–D) Mixed BMC from Ctrl (Ox40-Cre+ Bcl6+/+ Blimp1+/+ or Ox40-Cre–), Ox40-Cre+ Bcl6fl/fl (Bcl6AD), Blimp1AD, or Ox40-Cre+ Bcl6fl/fl Blimp1fl/fl (dKO) tester, and WT competitor were infected with LCMV Arm and analyzed at d7 p.i. (A) Schematic representation of the experiment. (B) Tester/competitor ratios of GP66:I-Ab+ T cells normalized to that of naive CD4+ T cells in each animal. Data are from 3 independent transplantation experiments totaling 10 chimeras from each Ctrl, Blimp1AD, Bcl6AD, and dKO tester groups. Each symbol represents a mouse. (C) Flow cytometric analysis of CXCR5 versus PD1 expression on spleen GP66:I-Ab+ cells. Right graph shows the percentage of CXCR5+ PD1hi among I-Ab-GP66–specific cells for each genotype. Data are from one experiment representative (three mice per group, each symbol representing a mouse) of three independent experiments with at least three animals per group. (D) Flow cytometric expression of CXCR5 versus CXCR6 on spleen tester GP66:I-Ab+ cells of the indicated genotype. Graph on right summarizes data from more than three independent experiments. In B–D, **, P < 0.01; ***, P < 0.001; unpaired two-sided Welch’s t test.
We first assessed the tester/competitor ratios at d7 p.i. to evaluate how each factor affected the expansion of LCMV-responsive CD4+ T cells (Fig. 1 B). Whereas Blimp1AD tester cells had a small advantage over WT competitors, Bcl6AD tester cells failed to efficiently compete, suggesting a broad impact of Bcl6 on cell proliferation. In contrast, the tester/competitor ratio in dKO:WT chimeras reverted to that in Ctrl:WT chimeras, indicating that dKO and Ctrl CD4+ T cells competed as efficiently (Fig. 1 B). Thus, the initial expansion of LCMV-responding CD4+ T cells depended on Bcl6 largely through its repression of Blimp1. In agreement with previous reports (Choi et al., 2020; Xie et al., 2017), both Bcl6AD and dKO cells failed to become Tfh cells (defined as CXCR5hi PD1hi; Fig. 1 C). No subset of dKO cells expressed high levels of CXCR6, a Th1 cell marker (Fig. 1 D). Analyses of PSGL1 and Ly6c expression on dKO cells found little or no PSGL1lo Ly6c– (Tfh) cells and a preponderance of PSGL1+ Ly6clo cells, which, in WT mice, are enriched in memory precursors (Marshall et al., 2011; Fig. S1, B and C).
Distinct mechanisms underpin Bcl6 requirements for Tfh and Tcmp differentiation
These data suggested that memory precursors could develop from dKO cells; this would be in line with the observation that WT d7 p.i. GP66-I-Ab+ CD4+ T cells with little or no expression of Bcl6 and Blimp1 are enriched in CCR7+ CXCR5int cells, which include Tcmp cells (Pepper et al., 2011; Ciucci et al., 2019; Fig. S1, D and E). To address this and gain insight into the differentiation of LCMV-responding dKO cells, we performed scRNAseq, which provides single resolution over the whole transcriptome rather than on specific markers that could depend on either factor. We purified GP66:I-Ab+ cells of all genotypes from mixed BMC, processed them for scRNAseq using the 10x Genomics Chromium system (Zheng et al., 2017), and analyzed data with the Seurat pipeline (Stuart et al., 2019). Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction, performed on data integrated from three experiments (two biological replicates per genotype; Table S1), found that cells segregated by genotype rather than by experimental replicate (Fig. S1 F); this indicated little or no residual batch effect. Unsupervised clustering, performed separately on cells from each genotype, identified Ctrl clusters matching Th1, Tfh, and Tcmp groups, characterized by previously defined signatures (Ciucci et al., 2019; Fig. 2 A, left). Cells from each cluster segregated together on a UMAP plot displaying Ctrl cells only (Fig. 2 B, left). Blimp1AD cell clusters scored high for the Tfh or Tcmp signature but low for the Th1 signature (Fig. 2 A), contrasting with Bcl6AD clusters. Accordingly, the UMAP display positioned most Blimp1AD cells within outlines defined from Ctrl Tfh and Tcmp clusters and most Bcl6AD cells within Ctrl Th1 cluster outlines (Fig. 2 B). Thus, Bcl6- or Blimp1-deficient CD4+ T cells adopted transcriptomic programs similar to those existing in Ctrl cell subsets.
Figure 2.
Bcl6 promotes expression of the Tcmp transcriptome by repressing Blimp1. (A–C) Spleen GP66:I-Ab+ tester T cells from mixed BMC generated as in Fig. 1 A were analyzed by scRNAseq at d7 p.i. with LCMV Arm (Table S1 and Fig. S1 F). Data are from two replicates for each genotype, captured in experiments E1–E3. (A) Heatmap shows row-standardized signature scores (bottom rows) and expression of genes characteristic of Th1, Tfh, and Tcmp cells (top rows) among clusters (numbers below heatmap). Cells were clustered separately for each genotype. Top bar graph indicates the percentage of cells in each cluster within that genotype. (B) UMAP analysis of GP66:I-Ab+ T cells computed on cells of all genotypes from experiments E1–E3 and displayed separately for each genotype. Each dot represents a cell colored by cluster as in A. Outlines on the Ctrl plot circle clusters expressing genes characteristic of the Tcmp, Th1, and Tfh signatures and are projected on plots of mutant cells. (C) Violin plot shows relative gene expression across clusters and genotypes (key at bottom, color coded by cluster as in A). (D) Expression of CCR7 (top) and IL-7Rα (bottom) on spleen T cells from chimeric animals (left panels). Black lines show the expression on naive CD4+ T cells, and filled histograms show expression on GP66:I-Ab+ cells from the indicated genotype. Graphs on the right summarize the data; each symbol represents a mouse. Data are from one experiment representative of two independent ones, each with at least three mice per group. ***, P < 0.001; ****, P < 0.0001; unpaired two-sided Welch’s t test.
In contrast, most dKO cells segregated away from Ctrl and single mutant cells in the UMAP plot (Fig. 2 B, right). dKO clusters had low expression of genes associated with Tfh differentiation, including Cxcr5, Tox, Tox2, or Ascl2, unlike Blimp1AD cells (Fig. 2 A, right); this was consistent with the strict Bcl6 requirement for Tfh differentiation (Choi et al., 2020; Xie et al., 2017). However, contrary to Bcl6AD cells, most dKO clusters showed high Tcmp signature scores and expressed genes of the Tcmp program, including Tcf7, Bcl2, or Ccr7, as well as Il7r (Fig. 2, A and C). Unlike in Ctrl cells, the Tcmp transcriptome in dKO cells was coexpressed with Th1-related transcriptional regulators, including Id2, Runx3, Tbx21 (T-bet), and Ifng (IFNγ), although not with cytotoxic genes or Cxcr6 (Fig. 2 A). In agreement with the scRNAseq results, flow cytometry showed that most dKO cells expressed the Tcmp marker CCR7 (Fig. 2 D, top). Most also expressed IL-7Rα, which, unlike CCR7, is not normally expressed on Tcmp cells (Fig. 2 D); this indicated that Bcl6 and Blimp1 both contribute to Il7r repression.
A Bcl6- and Blimp1-independent Tcmp-like differentiation program
These findings suggested that the transcriptome of dKO cells was unique and distinct from that of Ctrl cells. However, it was also possible that it resulted from delayed disruption of Bcl6 or Blimp1. Indeed, differentiation programs diverge early after CD4+ T cell activation (Choi et al., 2013; DiToro et al., 2018), conceivably before Ox40-Cre–mediated gene deletion. To address this, we studied the responses of adoptively transferred naive Smarta TCR transgenic cells (GP66:I-Ab-specific) that had undergone gene deletion through activation of a tamoxifen-controlled Cre expressed from Cd4 regulatory elements (cd4-ERt2-Cre; Aghajani et al., 2012; Oxenius et al., 1998; Fig. S2 A). After verifying that this deletion strategy gave results similar to those with Ox40-Cre (compare Fig. S2, B–D, with Figs. 1 D and 2 D), we collected and processed d7 p.i. scRNAseq and Ctrl and dKO datasets from tamoxifen-induced transferred Smarta cells (experiment E5), which we combined and integrated with mixed BMC datasets (experiments E1 and E2; Ox40-Cre deletion) for further analyses (Table S1).
Figure S2.
Analyses of adoptively transferred Cd4-ERt2-Cre transgenic Smarta cells undergoing Bcl6 and Blimp1 deletion before antigen-induced activation, and scRNAseq and scATACseq analyses of Ctrl and dKO d7 p.i. LCMV-specific CD4+ T cells, from mixed BMC or adoptive transfer experiments, as in Fig. 3. (A) Schematic representation of Smarta T cell adoptive transfer experiments. (B) Graphs summarize numbers of (left) and Ki-67 expression in (right) Smarta cells from tamoxifen-treated donors that were either Bcl6fl/fl Blimp1fl/fl Cd4-ERt2-Cre (dKO, red-filled symbols) or Cre– littermates (Ctrl, open symbols), after transfer to LCMV Arm–infected WT recipients. (C) Expression of IL-7Rα (top) or CXCR5 (bottom) on Ctrl (gray filled) or dKO (red filled) adoptively transferred cells processed as in B. Dotted black lines represent the expression on naive CD4+ T cells from the same infected mice. Graph on right summarizes the data; symbol code as in B. MFI, mean fluorescence intensity. (D) Expression of CXCR5 versus CXCR6 on the same mice as in C. Graph on right summarizes the data with symbols as in B. (B–D) Data are from one experiment representative of two independent experiments with at least three animals per group. (E) UMAP plots display GP66:I-Ab+ T cells (tester cells from BMC; experiments E1, E2, and E4) or adoptively transferred Smarta cells (E5), captured at d7 p.i. LCMV Arm and processed for scRNAseq (top) or scATACseq (bottom) as in Fig. 3. Each dot represents a cell and is color coded by genotype. Each plot includes cells of a single experiment, as indicated at the top and defined in Table S1. (F) Bar plots indicate the distribution of scATACseq (top) or scRNAseq (bottom) clusters of d7 p.i. cells into Ctrl (gray) or dKO (red) genotypes. Data are from the three experiments shown in E. In B–D, **, P < 0.01; ***, P < 0.001; unpaired two-sided Welch’s t test.
UMAP analysis of the combined samples showed segregation of cells according to genotype, with minimal overlap between Ctrl and dKO cells (Fig. S2 E, top). We found good overlap across all three experiments (E1, E2, and E5), indicating comparable transcriptomic impact of both deletion approaches. Unsupervised clustering, performed on the combined samples to detect patterns common to both genotypes, identified clusters predominantly or exclusively comprising cells of either genotype (Fig. S2 F). Indeed, clusters identified in this combined analysis (numbered 10–19) matched those generated by separate clustering of Ctrl and dKO cells (Fig. 2 A). Clusters 11, 12, and 16, mostly made up of Ctrl cells, were respectively skewed toward Tcmp, Th1, and Tfh expression (Fig. 3 B), as were Ctrl clusters 0, 1, and 2 in Fig. 2. The dKO-enriched clusters, chiefly cluster 10, associated features of Th1 and Tcmp cells, similar to Fig. 2.
Figure 3.
Bcl6 and Blimp1 regulate the epigenomic landscape of CD4+ T cells. Integrated scRNAseq and scATACseq analyses of Ctrl and dKO LCMV-specific CD4+ T cells at d7 p.i. For each analysis platform, UMAP projection and clustering were performed on the full set of cells (both genotypes, all captures), and data are from three replicates for each genotype (Table S1 and Fig. S2 E). (A) UMAP plots of scATACseq data, displayed per genotype with cells color coded by scATACseq cluster (Roman numerals, top) or by projected transcriptomic cluster (Arabic numerals, bottom). Putative Tcmp, Th1, and Tfh clusters are outlined on the Ctrl UMAP plot (bottom). (B) Heatmaps show row-standardized gene expression (scRNAseq) or chromatin accessibility (scATACseq) among clusters of Ctrl and dKO cells. Row-standardized scores of transcriptomic signatures is shown at the bottom of the scRNAseq heatmap (color code underneath). The top bar graph indicates the percentage of cells in each individual cluster within the indicated genotype. Cluster numbers and color code are shown underneath heatmaps. For each genotype, clusters comprising <4% of cells were omitted from the heatmap. (C) Traces show scATACseq peaks at relevant loci in cells from Ctrl clusters II–IV.
Using the 10x Genomics Chromium system, we performed single-cell assay for transposase-accessible chromatin sequencing (scATACseq) on nuclei from d7 p.i. GP66-specific Ctrl and dKO spleen CD4+ T cells captured in two BMC experiments (E2, E4 with Ox40-Cre) and one Smarta adoptive transfer experiment (E5, Cd4-ERt2-Cre; Table S1); we combined all three datasets and used the Signac extension of Seurat for analysis (Stuart et al., 2019, 2020 Preprint). Cells were largely segregated by genotype on a UMAP plot of the combined samples (Fig. S2 E, bottom), and all clusters (labeled for scATACseq with roman numerals throughout this study) were dominated by either genotype (Fig. 3 A, top; and Fig. S2 F). Thus, scATACseq indicated distinct distributions of chromatin-accessible regions between dKO and Ctrl cells. Examination of the heatmap showed that clusters II, III, and IV, mostly comprising Ctrl cells, had chromatin accessibility at genes characteristic of Th1, Tcmp, and Tfh cells, respectively (Figs. 3 B, right; and Fig. 3 C). To examine if there was overall correspondence between gene expression and chromatin accessibility, we leveraged the Signac integration feature, which links each cell from the scATACseq dataset to the transcriptomic cluster that best matches that cell’s promoter accessibility status (Stuart et al., 2019). This linked the main three Ctrl RNA clusters 11, 12, and 16 to chromatin clusters III, II, and IV, respectively, and in dKO cells clusters 10 (RNA) and I (chromatin; Fig. 3 A, bottom). These analyses indicate a strong correspondence between gene expression and chromatin organization in virus-responding CD4+ T cells. They also show that combined disruption of Blimp1 and Bcl6 caused profound changes in both, generating a unique transcriptomic and epigenomic pattern associating features of both Th1 and Tcmp cells, but distinct from either.
Establishment of a memory program independently of Bcl6 and Blimp1
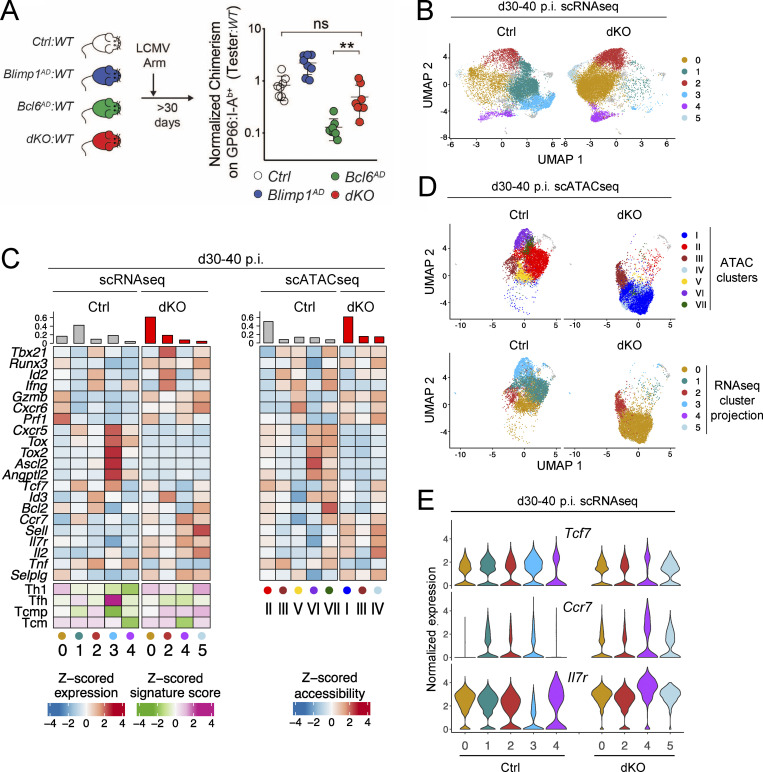
To study if dKO LCMV responders could generate memory cells, we first evaluated mixed BMC 30 d or more after LCMV Arm infection. We found that GP66-specific dKO CD4+ T cells persisted after d30 p.i., with only a modest reduction in tester/competitor ratios compared with Ctrl tester cells (Fig. 4 A). This contrasted with the reduced fitness of Bcl6-deficient tester cells, in line with previous results (Ichii et al., 2007).
Figure 4.
Bcl6 and Blimp1 regulate the transcriptome and epigenome of memory CD4+ T cells. Mixed BMCs made from WT competitor and tester of indicated genotypes were infected with LCMV Arm and analyzed ≥30 d later. (A) Right: Tester/competitor ratios of spleen GP66:I-Ab+ T cells normalized to that of naive CD4+ T cells in each animal 30 d p.i. Data are from three independent transplantation experiments, totaling eight mice in each group and performed as schematized on the left. Each symbol represents a mouse. **, P < 0.01. (B–E) Integrated scRNAseq and scATACseq analyses of tester I-Ab-GP66–specific Ctrl and dKO CD4+ T cells performed as reported in Table S1 and Fig. S3 A and displayed as in Fig. 3. Data are from two independent experiments for each genotype. (B) scRNAseq UMAP plot displayed per genotype and color coded by cluster. (C) Heatmaps show row-standardized gene expression or chromatin accessibility among clusters of Ctrl and dKO cells as in Fig. 3 B. (D) scATACseq UMAP plots shown separately for each genotype and color coded by scATACseq cluster (top) or by projected transcriptomic cluster (bottom). (E) Violin plot shows the relative expression of indicated genes across clusters and genotypes.
Thus, we examined the transcriptome and chromatin accessibility of Ctrl and dKO CD4+ T cells that persisted after LCMV infection. We captured and processed two samples of each genotype for both scRNAseq and scATACseq, all from BMCs processed as in Fig. 4 A, which we integrated for UMAP analysis and clustering (Table S1, experiments E6 and E7; Ctrls in Fig. S3, A and B). Similar to d7 p.i. data, we found a good match between scATACseq and scRNAseq clustering (Fig. 4, B–D). In line with our previous study (Ciucci et al., 2019), the predominant Ctrl scRNAseq clusters exhibited Tcm (Tcf7, Ccr7, cluster 1) and Tfh (Cxcr5, Ascl2, Tox2, cluster 3) attributes. They matched scATACseq clusters II and VI, respectively (Fig. 4 D, bottom), which showed corresponding chromatin accessibility at Sell and Ccr7 or Ascl2 (Fig. S3 C). These analyses suggested that Tfh and central memory functions are carried by separate cell subsets. Both cluster pairs 1:II and 3:VI were overwhelmingly composed of Ctrl cells and had a low Th1 score (Figs. S3 B and 4 C). In contrast, scRNAseq cluster 0, predominant among dKO cells but shared with Ctrl cells, associated expression of memory (Tcf7, Ccr7, Bcl2) and Th1 effector genes, but not of Tfh-related genes (Fig. 4, C and E). However, suggesting that Ctrl and dKO components were not strictly equivalent, each projected onto distinct scATACseq clusters: cluster V for Ctrl cells and predominantly cluster I for dKO cells. Ccr7 expression was similar in the main Ctrl and dKO clusters, whereas expression of Il7r trended higher in dKO than in Ctrl clusters (Fig. 4 E); flow cytometry analysis of CCR7 and IL-7Rα protein expression was consistent with such gene expression data (Fig. S3 D). We conclude from these analyses that antigen-specific CD4+ T cells can persist and acquire transcriptomic and chromatin attributes of memory independently of the transcription factors Bcl6 and Blimp1. Such cells had a unique differentiation pattern, combining retention of central memory features and expression of Th1- but not Tfh-related genes, unlike the more distinctive patterns of Ctrl Tcm and Tfh cells.
Figure S3.
LCMV-specific T cell responses in mixed BMCs made from WT competitor and either Ctrl or dKO tester cells, infected with LCMV Arm and analyzed ≥ 30 d later as in Figs. 4 and 5. (A–C) Ctrl and dKO GP66:I-Ab+ tester spleen T cells from mixed BMC were analyzed at d30-40 p.i. with LCMV Arm. (A) UMAP plots display tester GP66:I-Ab+ T cells captured at d35 (E6) or d40 (E7) p.i. and processed for scRNAseq (left) or scATACseq (right) as in Fig. 4 B. Each dot represents a cell and is color coded by genotype. Each plot includes cells from the indicated experiment. (B) Bar plots indicate the Ctrl (gray) versus dKO (red) genotype distribution of scATACseq (top) or scRNAseq (bottom) clusters of d30–40 p.i. cells referred to in A. Data are from the two experiments shown in A. (C) Traces show chromatin accessibility at selected loci across all cells from indicated Ctrl scATACseq clusters defined in Fig. 4, C and D. (D) Expression of CCR7 (left) or IL-7Rα (right) on Ctrl (gray filled) or dKO (red filled) tester I-Ab-GP66–specific CD4+ T cells at d30 p.i. with LCMV Arm of BMC mice as in Fig. 4 A. Dotted black lines represent the expression on CD44lo CD4+ T cells from the same infected mice. Graphs summarize the data, with symbol code on the right. Data are from one experiment representative of two independent experiments with at least three animals per group. Each symbol represents a mouse. ****, P < 0.0001; unpaired two-sided Welch’s t test. MFI, mean fluorescence intensity. (E) Graph summarizes the frequency of ex vivo Ctrl (black symbols) or dKO (red symbols) tester CD4+ CD44hi GP66:I-Ab+ T cells from mixed BMC, analyzed as in Fig. 4 A at d80 p.i. LCMV Arm. (F) Filled symbols show intracellular expression of TNFα (left) or IL-2 (right) by d80 p.i. tester CD44hi CD4+ T cells from BMC analyzed in E, as assessed by flow cytometry after 4-h in vitro culture with GP66 peptide. Open symbols summarize background staining as gated on CD44lo cells in the same cultures. Data in E and F are from a single experiment with three mice for each genotype and are not corrected for chimerism.
Bcl6- and Blimp1-independent functional memory responses
Last, we examined if persistent virus-specific dKO cells displayed functional attributes of memory. We first examined production of cytokines in response to antigen stimulation, which better evaluates cytokine expression than scRNAseq on ex vivo cells. We found similar production of IL-2 and TNFα by dKO versus Ctrl virus-specific cells that had been stimulated in vitro with GP66 30–40 d after primary Arm infection (Fig. 5 A). We made similar observations in a single set of chimeras analyzed 80 d after primary infection, suggesting long-lasting memory (Fig. S3, E and F). To measure in vivo responses to antigen rechallenge (recall), we inoculated the clone 13 strain of LCMV into chimeras infected with LCMV Arm ≥30 d earlier. Assessing tester/competitor ratios among GP66-specific cells at d5 after rechallenge, we found that both Ctrl and dKO tester cells responded to recall infection, a key property of memory T cells (Fig. 5 B). dKO cells were slightly outnumbered by the WT competitor, in line with cell distribution in the nonrechallenged d30 p.i. WT:dKO chimera (Fig. 4 A). Intracellular staining found similar expression of the cell cycle–associated antigen Ki67 in d5 postrecall Ctrl and dKO cells (Fig. 5 C). We conclude from these experiments that virus-responsive CD4+ T cells can provide long-lived antigenic responses, a defining attribute of memory T cells, despite lacking both Bcl6 and Blimp1.
Figure 5.
Bcl6 repression of Blimp1 mediates its impact on CD4+ T cell memory response. (A) Ctrl and dKO tester cells from mixed BMC generated and processed as in Fig. 4 A are analyzed by flow cytometry for (i) staining with anti-CD4 and I-Ab-GP66 tetramer (left column, ex vivo) and (ii) intracellular expression of TNFα and IL-2 after 4 h at 37°C with GP66 peptide (right columns). Bottom graphs summarize the percentage of GP66-specific cells (left, not corrected for chimerism to allow comparison with cytokine response; normalized tester/competitor ratio expressed as in Fig. 4 A was 0.5 in that experiment) and of cytokine-producing cells (right two plots). Data are from two pooled independent experiments (two mice per experiment) and are representative of a total of four experiments. (B) Mixed BMCs with Ctrl or dKO tester cells were infected with LCMV Arm and rechallenged 30 d later with LCMV clone 13 (left). Graph shows tester/competitor ratios of spleen GP66:I-Ab+ T cells in each animal 5 d after clone 13 rechallenge, normalized to the ratio of naive CD4+ T cells. Data are from three independent transplantation experiments, totaling eight Ctrl and seven dKO chimeras. (C) Histogram overlays (top) show, and bottom graph summarizes, Ki-67 expression on spleen GP66:I-Ab+ Ctrl and dKO tester cells from BMC analyzed 5 d after clone 13 rechallenge (as in B, filled histograms and symbols on graphs). Plain line histograms and open symbols on graphs show Ki-67 expression on memory cells from chimeras analyzed at d30 p.i. LCMV Arm with no rechallenge as in Fig. 4 A. Data are from one recall experiment with two or three animals per group (as shown on graph) and are representative of two other recall experiments. In A–C, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired two-sided Welch’s t test.
CD4+ memory T cells contribute to long-lived protection after infection or vaccination; their potential importance has recently been highlighted in severe acute respiratory syndrome coronavirus 2 infection (Lipsitch et al., 2020). Understanding the molecular mechanisms that control their development and maintenance is therefore necessary to expand immunization strategies. Here, we leveraged transcriptomic and epigenomic analyses to disentangle the differentiation of memory and effector CD4+ T cells. We demonstrate that the transcription factor Bcl6 uses distinct mechanisms to promote memory development and Tfh cell differentiation. Bcl6 repression of Blimp1 supports the development of long-lived antigen-specific responses with functional, transcriptomic, and epigenomic attributes of memory, contrasting with the strict requirement for Bcl6 in Tfh cell differentiation.
Given the involvement of Bcl6 in both Tfh and memory responses, it has been proposed that memory CD4+ T cells differentiate from early responding cells expressing a Tfh-like program (Choi et al., 2013; Crotty, 2018). Contrasting with this idea, we found that Tfh cell differentiation required Bcl6, regardless of Blimp1 status, as previously reported (Choi et al., 2020; Xie et al., 2017), whereas Tcmp and memory cells could develop in cells lacking both factors. These distinct genetic requirements indicate that these two subsets are mechanistically distinct.
Virus-responding Bcl6-Blimp1 dKO CD4+ T cells harbor hybrid transcriptomic and chromatin patterns, with features of Tcmp and Th1 cells, whether early in the response or at the memory phase. The expression of a memory program in dKO responder cells supports the idea that Bcl6 and Blimp1 normally divert undifferentiated cells toward Tfh and Th1 fates, respectively, and that they do so in part by repressing genes characteristic of a memory program. Indeed, previous chromatin immunoprecipitation followed by sequencing analyses have shown that both Bcl6 and Blimp1 bind genes associated with memory differentiation, including Ccr7 or Il7r (Hatzi et al., 2015; Liu et al., 2016; Mackay et al., 2016). These findings suggest that “effector” transcription factors, such as Bcl6 and Blimp1, serve in part to divert cells from a less differentiated program triggered by antigen receptor stimulation. Consistent with this idea, recent studies showed that epigenetic silencing of memory genes is necessary for the acquisition of effector functions in short-lived effector T cells (Gray et al., 2017; Pace et al., 2018).
Memory T cell potential is inversely correlated with T cell proliferation during the initial response to infection and immunization (Buchholz et al., 2013; Cho et al., 2017; Nish et al., 2017; Tubo et al., 2016). Unexpectedly, given the role of Bcl6 in memory cell survival, disrupting both Bcl6 and Blimp1 preserved both clonal expansion and long-lived responses. Thus, targeting Bcl6 and Blimp1 represents a new approach to amplify antigen-specific T cells while minimizing the loss of long-term potential or the accumulation of terminally differentiated effectors and could be explored to improve cancer adoptive T cell therapy.
Materials and methods
Mice
Mice carrying floxed alleles for Bcl6 (Hollister et al., 2013), Blimp1 (Shapiro-Shelef et al., 2003), and Rosa26YFP (Srinivas et al., 2001) were previously described, as were Ox40-Cre (Klinger et al., 2009; Zhu et al., 2004; obtained from N. Killeen, University of California at San Francisco, San Francisco, CA, via J. Zhu, National Institute of Allergy and Infectious Diseases, Bethesda, MD), Smarta (Oxenius et al., 1998), and Cd4ERT2Cre (Aghajani et al., 2012) animals. Blimp1YFP mice were from The Jackson Laboratory (008828). These mutant lines were backcrossed two or three times with C57BL/6 mice and then intercrossed to produce desired recombinant strains. CD45.1 or CD45.2 C57BL/6 mice were obtained from Charles River Laboratories. The Ox40-Cre allele was maintained heterozygous, and only female Ox40-Cre+ mice were used for breeding. For experiments with Ox40-Cre deletion, Ctrl mice included in experimental designs were Ox40-Cre+ Bcl6+/+ Blimp1+/+ or Ox40-Cre– animals from the same line as experimental mice. For experiments with Cd4ERT2Cre deletion, Ctrl mice were Cre-negative littermates from the same line. T cells from Ox40-Cre+ and Cd4ERT2Cre+ populations were gated on Rosa26YFP expression as an indicator of Cre recombinase activity, except where otherwise noted. Mice were housed in specific pathogen–free facilities. Animal procedures were approved by the National Cancer Institute Animal Care and Use Committee.
Mixed BMCs and infection
Mixed BMCs were made with T cell–depleted (Pan T Dynal kit; Invitrogen) bone marrow cells isolated from CD45 disparate mice, mixed together at various ratios (from 1:1 to 1:5), and injected into lethally irradiated (950 rad) CD45.1 recipients. Chimeric animals were infected 8–10 weeks after reconstitution.
Chimeric and nonchimeric 6–16-wk-old animals were infected by i.p. injection of 2 × 105 PFU of LCMV Arm. For rechallenge experiments, mice were infected i.v. with 2 × 106 PFU of LCMV clone 13. The chimerism of CD4+ GP66:I-Ab+ T cell populations was calculated from the ratio of the CD45.2+ tester population (Ctrl, Blimp1AD, Bcl6AD, or dKO) over the CD45.1+ CD45.2+ WT competitor relative to the tester/competitor ratio in naive (CD44lo)CD4+ T cells in the same animal.
Adoptive cell transfer
Gene disruption in CD4+ Smarta T cells was induced by tamoxifen gavage (5 mg/mouse/d in corn oil for 4 consecutive days) of donor Smarta Bcl6f/f Blimp1f/f Rosa26YFP Cd4-ERt2Cre+ or Cre-negative littermate Ctrls. After 3 d of rest, 30,000 spleen CD4+ T cells from tamoxifen-treated donors were bead purified as previously described (Ciucci et al., 2019) and i.v. injected into CD45 congenic recipients that were infected with LCMV Arm 24 h later.
Antibodies
Antibodies for the following specificities were purchased from BD Pharmingen, BioLegend, or Thermo Fisher eBioscience: CD4 (RM4.4 or GK1.5), CD8α (53-6-7), CD45.2 (104), CD45.1 (A20), TCRβ (H57-597), CD5 (53-7.3), B220 (RA3-6B2), CD44 (IM7), IL-7Rα (A7R34), CCR7 (4B12), CXCR5 (SPRCL5), CXCR6 (SA051D1), IL-2 (Jes6-5H4), IFNγ (XMG1.2), TNFα (MP6-XT22), Ki-67 (SolA15), PSGL1 (2PH1), Ly6C (HK1.4), CD27 (LG.7F9), and BCL6 (K112-91). MHC tetramers loaded with the LCMV GP66 peptide were obtained from the National Institutes of Health Tetramer Core Facility.
Cell preparation and staining
Spleen cells were prepared and stained as previously described (Ciucci et al., 2017). Surface staining with GP66:I-Ab+ tetramer and for CCR7 or CXCR5 was performed at 37°C for 1 h before staining with antibodies for other cell surface markers. Intracellular staining for Bcl6 or CCR7 was performed as previously described with a two-step fixation to preserve YFP and GFP fluorescence (Heinen et al., 2014). Cytokine staining was performed after 4 h on splenocytes incubated in the presence of 2 µg/ml GP66 (GLKGPDIYKGVYQFKSVEFD) peptide (AnaSpec) and GolgiStop. Flow cytometry data were acquired on LSR II and FACSymphony devices (BD Biosciences) and analyzed with FlowJo software (BD Biosciences). Dead cells and doublets were excluded by Live/Dead staining (Invitrogen) and forward scatter height by width gating. Except if otherwise mentioned, the numbers in the cytometry plot indicate the percentages of cells in the gates. Purification of lymphocytes by cell sorting was performed on a FACS Fusion device (BD Biosciences).
scRNAseq
Allelically marked GP66:I-Ab+ Rosa26YFP+ spleen T cells from LCMV infected chimera or adoptive transfer hosts (experiment E5) were sorted, then loaded onto a 10x Genomics Chromium platform to generate cDNAs carrying cell- and transcript-specific barcodes that were used to construct sequencing libraries using Chromium Single Cell 3′ (version 2 or version 3) or 5′ (version 1.1) Library & Gel Bead Kits according to the manufacturer’s instructions. Cells of each genotype were captured separately, except for experiment E2, in which Ctrl and dKO populations were sorted and barcoded separately with TotalSeq antibodies (BioLegend) before mixing and cell captures. Libraries were sequenced on multiple runs of an Illumina NextSeq system using paired-end 26 × 98 bp or 26 × 57 bp in order to reach a sequencing saturation >70%, resulting in at least 50,000 reads/cell. Single-cell sequencing files were processed, and count matrixes were extracted using the Cell Ranger Single Cell Software Suite (version 1.3.1). Further analyses were performed in R using the Seurat package (Stuart et al., 2019).
Data were preprocessed by removing genes expressed in fewer than two cells and excluding cells that were potential doublets or empty droplets or cells expressing at least 10% mitochondrial genes. Linear dimension reduction (principal component analysis) was performed based on the highly variable genes for each preprocessed dataset. For the data captured in experiments E1–E3, they were integrated using the Seurat integration method with the FindIntegrationAnchors and IntegrateData R functions. Afterward, clustering was performed separately for each genotype. Clusters representing not >2% of each population were grayed out in the UMAP plot. For other experiments, datasets were integrated through the Seurat integration method, and UMAP projection along with clustering was performed on the full set of cells. Clusters representing not >2% of all cells were grayed out in the UMAP plots, whereas clusters with >4% cells in each genotype were shown in the heatmaps. Gene signature scores from published signatures (Ciucci et al., 2019) were calculated using the AddModuleScore function.
scATACseq
GP66:I-Ab+ Rosa26YFP+ spleen T cells from LCMV-infected chimera (all experiments except E5) or Smarta cells from LCMV-infected WT recipients (experiment E5) were sorted, and nuclei were isolated before transposition at 37°C for 1 h and captured onto the 10x Genomics Chromium platform. Libraries were constructed using the V1 Chromium Single Cell ATAC Solution according to the manufacturer’s instructions (10x Genomics). Libraries were sequenced on multiple runs of the Illumina NextSeq system using paired-end 50 × 50 bp in order to reach at least 9,000 unique fragments/cell. Single-cell sequencing files were processed, and count matrixes were extracted using the Cell Ranger ATACseq Software (version 1.0.1). Further analyses were performed in R using the Seurat and Signac packages (Stuart et al., 2019).
Only cells with 3,000–40,000 fragments, nucleosome signal <1, transcription start site enrichment >2, ratio of reads in genomic blacklist region <0.05, and percentage of reads in peaks >50% were analyzed. Reduction of data dimensionality was performed using the latent semantic indexing algorithm. Data were integrated using the Signac integration method with FindIntegrationAnchors and IntegrateEmbeddings R functions. The integration anchors were identified using the reciprocal latent semantic indexing projection, and the low-dimensional cell embeddings were integrated across replicates. UMAP projection and clustering were performed on the full set of cells. Clusters representing not >2% of all cells were grayed out in the UMAP plots, whereas clusters representing with >4% of cells in each genotype were shown in the heatmaps. Gene activity was calculated based on fragments matching gene genomic coordinates from 2 kb pairs upstream of the transcription start site to 2 kb pairs downstream of the transcription termination site.
Statistical analyses
Except for deep-sequencing data, statistical significance was calculated with Prism software. Except where otherwise indicated in figure legends, error bars in graphs indicate the SD, and statistical comparisons were done by unpaired two-sided Welch’s t test. Statistical significance indicated in figure panels uses the following convention: *, P < 0.05, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows (i) deletion efficiency as assessed from Rosa26YFP expression, (ii) subsets of antigen-responding CD4+ T cells defined by expression of Ly6C and PSGL1, (iii) correspondence between expression of Bcl6 or Blimp1 and that of CXCR5 and CCR7, and (iv) UMAP replicate information on scRNAseq experiments E1-E3. Fig. S2 shows (i) schematic and results of Tamoxifen-induced deletion of Bcl6 and Blimp1, and (ii) replicate information on and cluster distribution of d7 p.i. scATACseq and scRNAseq experiments shown in Fig. 3. Fig. S3 shows (i) replicate information on and cluster distribution of memory scATACseq and scRNAseq experiments shown in Fig. 4, (ii) memory ATAC peaks on select genes, (iii) expression of CCR7 and IL-7Rα on memory cells, and (iv) frequency of and cytokine production by d80 p.i. memory cells. Table S1 summarizes metrics for individual scRNAseq and scATACseq experiments.
Supplementary Material
summarizes metrics for individual scRNAseq and scATACseq experiments.
Acknowledgments
We thank H. Kwak and Q. Xiao for technical assistance; E. Conner, J. Shetty, B. Tran, and Y. Zhao for assistance with sequencing; J. Zhu and J. O’Shea for mice; the National Institutes of Health Tetramer Core Facility for reagents; the Center for Cancer Research Flow Cytometry Core, the National Institutes of Health High-Performance Computing cluster, and the University of Rochester Flow Cytometry Core and Genomics Research Center for assistance; J. Ashwell, V. Lazarevic, and J. Zhu for reading the manuscript; and D. Goldstein, M. Malik, and the National Cancer Institute Office of Science and Technology Resources for their support. Sequencing was performed with the Center for Cancer Research Genomics Core, the Center for Cancer Research Sequencing Facility, and the University of Rochester Genomics Research Center.
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. The Center for Cancer Research Single Cell Analysis Facility is funded by the Frederick National Laboratory for Cancer Research under contract HHSN261200800001E.
Author contributions: T. Ciucci designed and performed research, analyzed data, and wrote the original manuscript. M.S. Vacchio, J. Nie, and L.B. Chopp performed research and analyzed data. M.C. Kelly contributed to single-cell captures and provided guidance. T. Cheng performed bioinformatics analyses. D.B. McGavern provided viral stocks and guidance with LCMV experiments. R. Bosselut supervised the research and wrote the final manuscript with contributions from all authors.
Data availability
Sequence data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession nos. GSE149912, GSE149913, and GSE184848. All other data and code are available upon request.
References
- Aghajani, K., Keerthivasan S., Yu Y., and Gounari F.. 2012. Generation of CD4CreER(T2) transgenic mice to study development of peripheral CD4-T-cells. Genesis. 50:908–913. 10.1002/dvg.22052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz, V.R., Flossdorf M., Hensel I., Kretschmer L., Weissbrich B., Gräf P., Verschoor A., Schiemann M., Höfer T., and Busch D.H.. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science. 340:630–635. 10.1126/science.1235454 [DOI] [PubMed] [Google Scholar]
- Cho, Y.L., Flossdorf M., Kretschmer L., Höfer T., Busch D.H., and Buchholz V.R.. 2017. TCR signal quality modulates fate decisions of single CD4+ T cells in a probabilistic manner. Cell Rep. 20:806–818. 10.1016/j.celrep.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Choi, Y.S., Yang J.A., Yusuf I., Johnston R.J., Greenbaum J., Peters B., and Crotty S.. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190:4014–4026. 10.4049/jimmunol.1202963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.S., Gullicksrud J.A., Xing S., Zeng Z., Shan Q., Li F., Love P.E., Peng W., Xue H.H., and Crotty S.. 2015. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16:980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., Diao H., Faliti C.E., Truong J., Rossi M., Bélanger S., Yu B., Goldrath A.W., Pipkin M.E., and Crotty S.. 2020. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat. Immunol. 21:777–789. 10.1038/s41590-020-0706-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci, T., Vacchio M.S., and Bosselut R.. 2017. A STAT3-dependent transcriptional circuitry inhibits cytotoxic gene expression in T cells. Proc. Natl. Acad. Sci. USA. 114:13236–13241. 10.1073/pnas.1711160114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci, T., Vacchio M.S., Gao Y., Tomassoni Ardori F., Candia J., Mehta M., Zhao Y., Tran B., Pepper M., Tessarollo L., et al. 2019. The emergence and functional fitness of memory CD4+ T cells require the transcription factor Thpok. Immunity. 50:91–105.e4. 10.1016/j.immuni.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., and Wherry E.J.. 2014. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 40:289–302. 10.1016/j.immuni.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S. 2018. Do memory CD4 T cells keep their cell-type programming: plasticity versus fate commitment? Complexities of interpretation due to the heterogeneity of memory CD4 T cells, including T follicular helper cells. Cold Spring Harb. Perspect. Biol. 10:a032102. 10.1101/cshperspect.a032102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S., Johnston R.J., and Schoenberger S.P.. 2010. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11:114–120. 10.1038/ni.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiToro, D., Winstead C.J., Pham D., Witte S., Andargachew R., Singer J.R., Wilson C.G., Zindl C.L., Luther R.J., Silberger D.J., et al. 2018. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science. 361:eaao2933. 10.1126/science.aao2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, S.M., Amezquita R.A., Guan T., Kleinstein S.H., and Kaech S.M.. 2017. Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity. 46:596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.E., Janowski K.M., Oliver J.R., Zajac A.J., and Weaver C.T.. 2008. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 452:356–360. 10.1038/nature06672 [DOI] [PubMed] [Google Scholar]
- Hatzi, K., Nance J.P., Kroenke M.A., Bothwell M., Haddad E.K., Melnick A., and Crotty S.. 2015. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212:539–553. 10.1084/jem.20141380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen, A.P., Wanke F., Moos S., Attig S., Luche H., Pal P.P., Budisa N., Fehling H.J., Waisman A., and Kurschus F.C.. 2014. Improved method to retain cytosolic reporter protein fluorescence while staining for nuclear proteins. Cytometry A. 85:621–627. 10.1002/cyto.a.22451 [DOI] [PubMed] [Google Scholar]
- Hollister, K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., and Dent A.L.. 2013. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 191:3705–3711. 10.4049/jimmunol.1300378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann, D., Teyton L., and Oldstone M.B.. 2001. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat. Med. 7:913–919. 10.1038/90950 [DOI] [PubMed] [Google Scholar]
- Ichii, H., Sakamoto A., Arima M., Hatano M., Kuroda Y., and Tokuhisa T.. 2007. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int. Immunol. 19:427–433. 10.1093/intimm/dxm007 [DOI] [PubMed] [Google Scholar]
- Johnston, R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Hawkins E.D., Belz G.T., Metcalf D., Hommel M., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2006. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 7:466–474. 10.1038/ni1321 [DOI] [PubMed] [Google Scholar]
- Klinger, M., Kim J.K., Chmura S.A., Barczak A., Erle D.J., and Killeen N.. 2009. Thymic OX40 expression discriminates cells undergoing strong responses to selection ligands. J. Immunol. 182:4581–4589. 10.4049/jimmunol.0900010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzli, M., Schreiner D., Pereboom T.C., Swarnalekha N., Litzler L.C., Lötscher J., Ertuna Y.I., Roux J., Geier F., Jakob R.P., et al. 2020. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci. Immunol. 5:eaay5552. 10.1126/sciimmunol.aay5552 [DOI] [PubMed] [Google Scholar]
- Laidlaw, B.J., Craft J.E., and Kaech S.M.. 2016. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 16:102–111. 10.1038/nri.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch, M., Grad Y.H., Sette A., and Crotty S.. 2020. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 20:709–713. 10.1038/s41577-020-00460-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Lu H., Chen T., Nallaparaju K.C., Yan X., Tanaka S., Ichiyama K., Zhang X., Zhang L., Wen X., et al. 2016. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 14:1735–1747. 10.1016/j.celrep.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, L.K., Minnich M., Kragten N.A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- MacLeod, M.K., Clambey E.T., Kappler J.W., and Marrack P.. 2009. CD4 memory T cells: what are they and what can they do? Semin. Immunol. 21:53–61. 10.1016/j.smim.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, H.D., Chandele A., Jung Y.W., Meng H., Poholek A.C., Parish I.A., Rutishauser R., Cui W., Kleinstein S.H., Craft J., and Kaech S.M.. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 35:633–646. 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, G.A., Cimmino L., Shapiro-Shelef M., Szabolcs M., Herron A., Magnusdottir E., and Calame K.. 2006. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol. 7:457–465. 10.1038/ni1320 [DOI] [PubMed] [Google Scholar]
- Nish, S.A., Zens K.D., Kratchmarov R., Lin W.W., Adams W.C., Chen Y.H., Yen B., Rothman N.J., Bhandoola A., Xue H.H., et al. 2017. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J. Exp. Med. 214:39–47. 10.1084/jem.20161046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone, M.B. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr. Top. Microbiol. Immunol. 263:83–117. 10.1007/978-3-642-56055-2_6 [DOI] [PubMed] [Google Scholar]
- Oxenius, A., Bachmann M.F., Zinkernagel R.M., and Hengartner H.. 1998. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400. [DOI] [PubMed] [Google Scholar]
- Pace, L., Goudot C., Zueva E., Gueguen P., Burgdorf N., Waterfall J.J., Quivy J.P., Almouzni G., and Amigorena S.. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science. 359:177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Pepper, M., and Jenkins M.K.. 2011. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 12:467–471. 10.1038/ni.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser, R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef, M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., and Calame K.. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620. 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Shin, H., Blackburn S.D., Intlekofer A.M., Kao C., Angelosanto J.M., Reiner S.L., and Wherry E.J.. 2009. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 31:309–320. 10.1016/j.immuni.2009.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas, S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., and Costantini F.. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4. 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Srivastava A., Lareau C., and Satija R.. 2020. Multimodal single-cell chromatin analysis with Signac. bioRxiv. (Preprint posted November 10, 2020) 10.1101/2020.11.09.373613 [DOI]
- Swain, S.L., McKinstry K.K., and Strutt T.M.. 2012. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 12:136–148. 10.1038/nri3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo, N.J., Fife B.T., Pagan A.J., Kotov D.I., Goldberg M.F., and Jenkins M.K.. 2016. Most microbe-specific naïve CD4+ T cells produce memory cells during infection. Science. 351:511–514. 10.1126/science.aad0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang, A., Perdomo C., Zedler U., Kuhlmann S., Hurwitz R., Gengenbacher M., and Kaufmann S.H.. 2014. Central memory CD4+ T cells are responsible for the recombinant Bacillus Calmette-Guérin ΔureC:hly vaccine’s superior protection against tuberculosis. J. Infect. Dis. 210:1928–1937. 10.1093/infdis/jiu347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Shin H.M., Moseman E.A., Ji Y., Huang B., Harly C., Sen J.M., Berg L.J., Gattinoni L., McGavern D.B., and Schwartzberg P.L.. 2015. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 12:2099–2110. 10.1016/j.celrep.2015.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, M.M., Koh B.H., Hollister K., Wu H., Sun J., Kaplan M.H., and Dent A.L.. 2017. Bcl6 promotes follicular helper T-cell differentiation and PD-1 expression in a Blimp1-independent manner in mice. Eur. J. Immunol. 47:1136–1141. 10.1002/eji.201747034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Cao Y., Xie Z., Huang Q., Bai Q., Yang X., He R., Hao Y., Wang H., Zhao T., et al. 2015. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat. Immunol. 16:991–999. 10.1038/ni.3229 [DOI] [PubMed] [Google Scholar]
- Yu, D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Zheng, G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Min B., Hu-Li J., Watson C.J., Grinberg A., Wang Q., Killeen N., Urban J.F. Jr., Guo L., and Paul W.E.. 2004. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5:1157–1165. 10.1038/ni1128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes metrics for individual scRNAseq and scATACseq experiments.
Data Availability Statement
Sequence data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession nos. GSE149912, GSE149913, and GSE184848. All other data and code are available upon request.