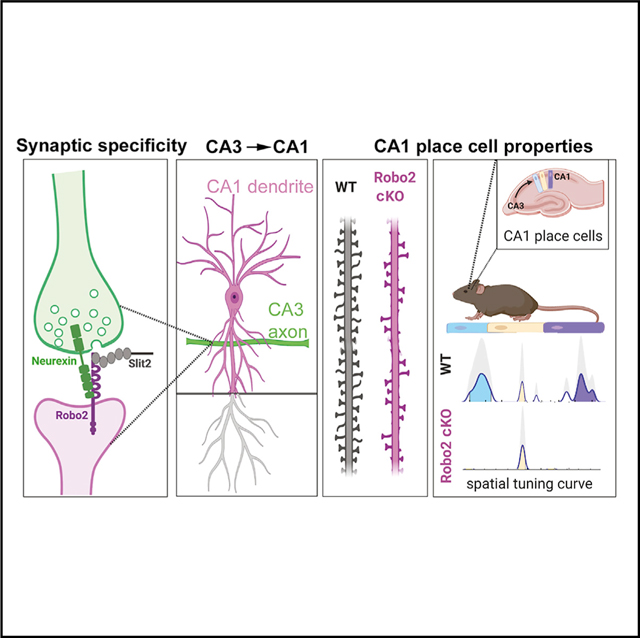
SUMMARY
Synaptic connectivity within adult circuits exhibits a remarkable degree of cellular and subcellular specificity. We report that the axon guidance receptor Robo2 plays a role in establishing synaptic specificity in hippocampal CA1. In vivo, Robo2 is present and required postsynaptically in CA1 pyramidal neurons (PNs) for the formation of excitatory (E) but not inhibitory (I) synapses, specifically in proximal but not distal dendritic compartments. In vitro approaches show that the synaptogenic activity of Robo2 involves a trans-synaptic interaction with presynaptic Neurexins, as well as binding to its canonical extracellular ligand Slit. In vivo 2-photon Ca2+ imaging of CA1 PNs during spatial navigation in awake behaving mice shows that preventing Robo2-dependent excitatory synapse formation cell autonomously during development alters place cell properties of adult CA1 PNs. Our results identify a trans-synaptic complex linking the establishment of synaptic specificity to circuit function.
In brief
Blockus et al. demonstrate that the axon guidance receptors Robo1/2 are synaptogenic cues required postsynaptically for the establishment of synaptic specificity between CA3 axons and dendrites of CA1 pyramidal neurons (PNs). Developmental deletion of Robo2 in CA1 PNs disrupts the emergence of place cell properties in adult hippocampus.
Graphical Abstract
INTRODUCTION
Proper circuit function relies on the establishment of synaptic connections characterized by a high degree of cellular and subcellular specificity. How this striking degree of synaptic specificity is achieved during development remains poorly understood, especially in the complex brain of mammals. Many cell surface molecules mediating molecular recognition between axons and dendrites of specific neuronal cell types have been identified (de Wit and Ghosh, 2016; Schroeder and de Wit, 2018; Südhof, 2018; Zipursky and Sanes, 2010). These trans-synaptic adhesion molecules are often expressed in a cell-type specific manner (Schreiner et al., 2017) thereby determining the subset of a neuron’s possible synaptic partners. Some classes of these molecules have synaptic organizing properties evidenced by their direct or indirect ability to recruit key pre- and postsynaptic proteins. When axons reach their target areas, a cellular switch from a phase of growth and branching to synaptogenesis must occur. This switch is characterized by dynamic instability of adhesion progressively leading to more stable patterns of synaptic connectivity characterizing adult circuits. One of the foremost outstanding questions in neuroscience is how synapse formation is mechanistically integrated with earlier developmental steps such as axon guidance and branching. One potential mechanism for coordinating this transition relies on the possibility that axon guidance cues acquire synaptogenic functions during circuit wiring. Indeed, evidence for such dual use of developmental molecules has accumulated over the years suggesting this might be a common theme to overcome constraints imposed by a limited genetic repertoire that has to orchestrate a much larger variety of functional processes and behaviors (Blockus and Polleux, 2021; Dorskind and Kolodkin, 2021; Hunyara and Kolodkin, 2020; Koropouli and Kolodkin, 2014; Shen and Cowan, 2010). Several axon guidance molecules have been studied for their function in subsequent developmental processes, including synapse formation, as well as synaptic plasticity, among them EphB (for a recent review see Henderson and Dalva, 2018), Netrin-1/DCC (Glasgow et al., 2020; Glasgow et al., 2021) and Semaphorins (Tran et al., 2009; Wang et al., 2017). However, very few studies have tested the impact of disruption of these synaptogenic cues and their role in establishing synaptic specificity on circuit function in vivo.
Here, we report that the well-studied axon guidance ligand-receptor pair Slit/Robo plays a role in synaptogenesis that depends on an interaction between postsynaptic Robo, their extracellular Slit ligands and a trans-synaptic interaction with the presynaptic organizing proteins Neurexins at nascent synapses. Slit-Robo signaling has been extensively studied for its role in axon guidance (Blockus and Chédotal, 2016) and branching (Campbell et al., 2007) for over three decades across many model organisms, but a role for Slit-Robo signaling in synaptogenesis and circuit function remains largely unexplored. Among all axon guidance receptor/ligand pairs, we focused on Robo/Slit to test their potential role in synaptogenesis and the establishment of synaptic specificity for three main reasons: (1) We found that their expression is maintained following completion of axon guidance; (2) Robo2 protein distribution in CA1 PNs is strikingly layer-specific; and (3) recent work demonstrated that a large class of transmembrane proteins regulating synaptic specificity contain Leucine-rich repeat (LRR) and Ig domains (de Wit and Ghosh, 2014), which are present in Slits and Robo receptors, respectively.
Our results identify a function for Slit-Robo signaling, beyond axon guidance, in synaptic specificity through formation of a trans-synaptic complex with presynaptic Neurexins. We show that Robo2 is critical for the establishment of synaptic connectivity characterizing hippocampal CA1 PNs in vivo, which play a key role in navigation, and episodic learning and memory. We demonstrate that cell-autonomous deletion of Robo2 impacts excitatory synapse formation in CA1 PNs in an input-specific way, with significant consequences for the emergence of place cell properties. Our results provide insights into the molecular mechanisms linking synaptic specificity and the emergence of circuit properties during mammalian brain development.
RESULTS
Robo2 is expressed in the developing and mature hippocampus and localizes at excitatory synapses
To determine whether Slit/Robo could be involved in stages of neuronal development after completion of axon guidance, we first characterized its region-specific and subcellular localization in the postnatal mouse hippocampus. Using publicly accessible resources (Allen Brain Atlas), we found that Robo2 mRNA is expressed throughout all Cornu Ammonis (CA) regions of the hippocampus at P14, and its expression is maintained into adulthood (P28, Figure 1A). Expression of Robo1 and the Robo ligand Slit2 within the hippocampus is confined mostly to the CA3 region, which provides presynaptic input to CA1 (Figure 1A). Since Robo2 but not Robo1 is expressed in CA1, this allowed us to analyze functions of Robo2 without redundancy with Robo1. We next determined Robo2 protein expression at P35 in CA1. Strikingly, Robo2 protein is expressed specifically in CA1 stratum oriens (SO) and stratum radiatum (SR), but is not detected in stratum lacunosum moleculare (SLM) (Figures 1B and 1E). SO and SR correspond to layers where axons from CA3 and CA2 PNs synapse onto dendrites CA1 PNs, while the apical tuft of CA1 PNs in SLM receives long-range inputs from the entorhinal cortex (EC). Co-immunostaining with the presynaptic marker Bassoon and the postsynaptic marker PSD-95 shows a punctate staining pattern for Robo2 (Figure S1A) at low magnification. At higher magnification of the synaptic profile, characterized by direct apposition of Bassoon and PSD95, Robo2 is found significantly overlapping with PSD95 strongly suggesting a postsynaptic enrichment (Figure S1B).
Figure 1. Robo2 expression pattern in the hippocampus and its subcellular localization at synapses.
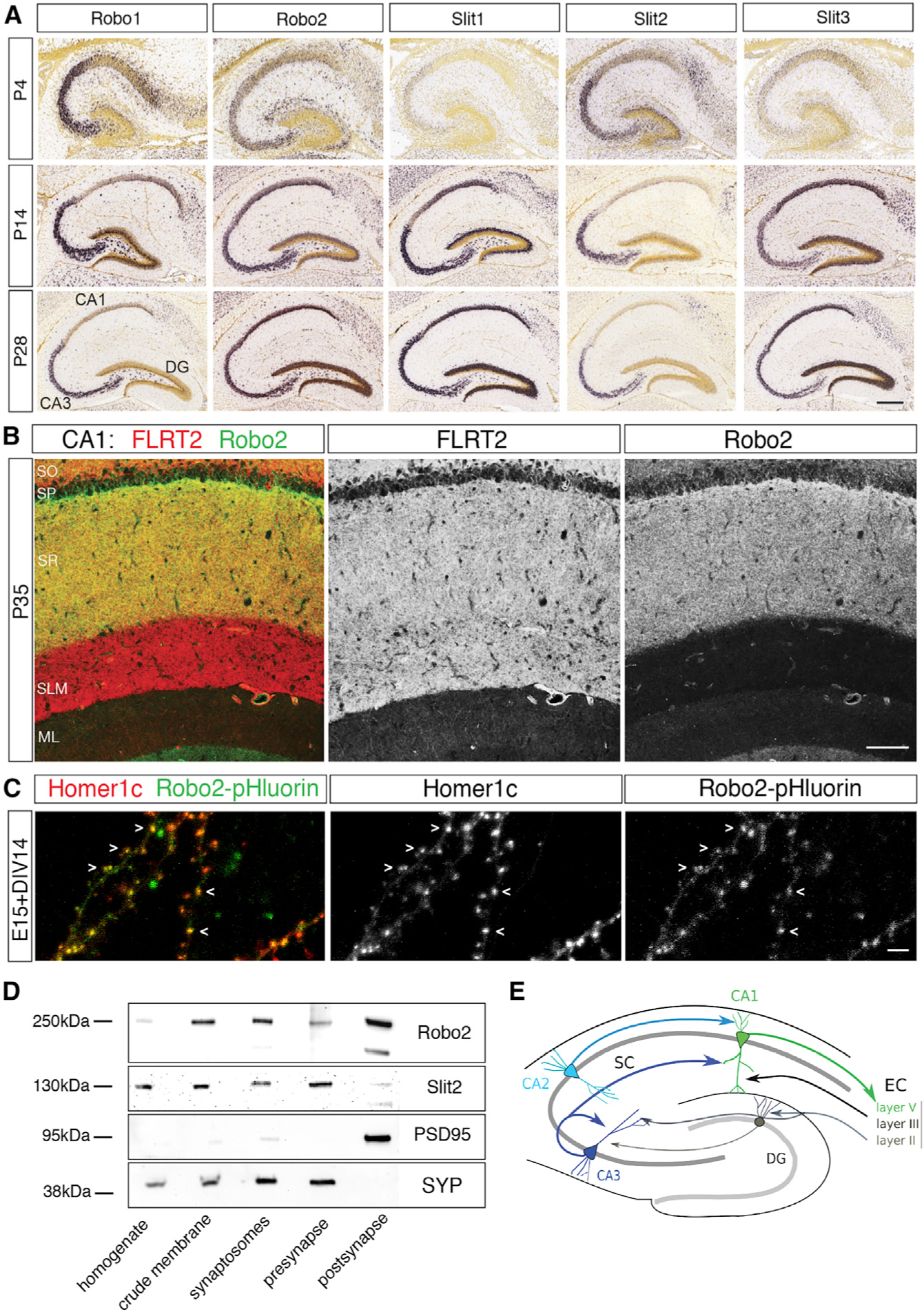
(A) In situ mRNA hybridization showing the expression pattern for Robo1, Robo2 and Slit1,2,3 in the early postnatal and mature mouse hippocampus. Robo2, Slit1,3 are expressed in all CA regions in the hippocampus, Robo1 and Slit2 expression is confined to CA3. Robo2 expression increases between P4 and P14. Scale bar: 50μm.
(B) Immunohistochemistry for FLRT2 (red) and Robo2 (green) proteins in the rat hippocampus at P35. While FLRT2 is expressed throughout all layers in CA1, expression of Robo2 is confined to SO and SR. Scale bar: 100μm. SR, stratum radiatum; SLM, stratum lacunosum-moleculare; ML, molecular layer of DG; SO, stratum oriens; SP, stratum pyramidale.
(C) Subcellular localizationofRobo2 in vitro. Ex utero electroporation of Robo2-pHluorin and Homer1-c-tdtomato in pyramidal neurons in vitro. Robo2-pHluorin colocalizes with the excitatory post-synapticmarker Homer1c at the majority of dendritic spines at 14 DIV. Scale bar: 15μm.
(D) Synaptic fractionation and immunoblot for Robo2, Slit2 and synaptic markers. Successful isolation of pre-and postsynaptic compartments is validated by presence of synaptophysin and PSD95 respectively. Robo2 localizes to pre-and post-synaptic compartments, its ligand Slit2 is enriched presynaptically.
(E) Schematic of hippocampal circuit connectivity.
To provide further evidence that Robo2 is presented at the plasma membrane postsynaptically at excitatory synapses, we used ex utero electroporation to express pHluorin-tagged Robo2 (Pignata et al., 2019) in cortical pyramidal neurons in dissociated cultures (E15+14DIV) corresponding to the peak of synaptogenesis in vitro. A pHluorin tag (Miesenböck et al., 1998) fused to the extracellular domain of Robo2 allows specific visualization of the plasma membrane targeted form of Robo2, but not the pool of the protein contained in intracellular vesicles (Pignata et al., 2019; Figures S1C–S1E). Live imaging shows that Robo2-pHluorin co-localizes with the excitatory postsynaptic scaffolding protein Homer1c in dendritic spines of cortical pyramidal neurons (Figure 1C). To further delineate which synaptic compartment Robo and Slit proteins localize to, we also performed biochemical synaptic membrane fractionation. By western blotting, we found that endogenous Robo2 and Slit2 localize to both, pre-and postsynaptic membranes, with Robo2 enriched in postsynaptic membranes and its ligand Slit2 in presynaptic membranes (Figure 1D).
Robo2 is required postsynaptically for excitatory synapse formation in CA1 pyramidal neurons
Having established that Robo2 localizes to excitatory postsynaptic compartments, we next tested if Robo2 is required cell-autonomously for excitatory synaptic development in CA1 PNs in vivo. To do this, we used targeted hippocampal in utero electroporation (HIUE) to cell-autonomously delete Robo2 from a subset of CA1 PNs using a Robo2 conditional knockout mouse (Robo2F/F). We performed HIUE using a limiting amount of plasmid encoding Cre-recombinase together with a non-limiting amount of a Cre-dependent reporter (FLEX-tdTomato) into CA1 progenitors of control (wild-type, WT) and Robo2F/F embryos at embryonic day E15.5 (Figures 2A and S2E; see STAR Methods for details). This approach is not only cell-autonomous with regard to CA1 PNs, but is essentially postsynaptic-autonomous. This is due to the fact that axons of CA1 PNs form almost no recurrent excitatory connections with other CA1 PNs (Sun et al., 2014) Thus, all presynaptic inputs received by the sparse population of Robo2-deficient CA1 PNs are wild-type. Hence, our approach enables us to specifically assess a postsynaptic function of Robo2 in CA1 PNs in vivo. Using this approach, we found that conditional deletion of Robo2 from CA1 PNs throughout development leads to a significant reduction of spine density in proximal dendritic compartments (SO: basal, 39.6%, SR: apical oblique 41.9%), but does not affect spine density in distal apical tuft dendrites (SLM: tuft) (Figure 2B). This decrease in spine density occurs in all CA1 PNs, irrespective of their position across the radial axis of the pyramidal layer (Figures S2A–S2C). Overall dendritic growth of Robo2-deficient CA1 PNs was not affected compared to wild-type controls (Figure S2D). Furthermore, Robo2 protein is not detected in vGlut1+ sublayers of CA1 at P4, suggesting low abundance of the receptor in incoming growing axons from CA2/3 invading SR and SO, as well as growing CA1 PN dendrites at this stage (Figure S3). Taken together, these data argue against the spine density reduction being a secondary effect of an axon guidance deficit. The Robo2 conditional knockout (Robo2F/F) was validated using a genetic NexCre mouse line which induces recombination in all glutamatergic pyramidal neurons throughout the hippocampus but not in GABAergic interneurons, or any non-neuronal cell types (Goebbels et al., 2006). The corresponding qRT-PCR amplification was used to measure Robo2 mRNA abundance (but also Hprt mRNA as a control) in hippocampi isolated from control (Robo2F/F) versus NexCre;Robo2F/F. Our analysis demonstrates a significant (~70%–80%) reduction of Robo2 mRNA in NexCre;Robo2F/F hippocampi compared to control demonstrating successful excision of the loxP-flanked exons by Cre recombinase (Figure S4).
Figure 2. Robo2 is required for synapse formation in hippocampal CA1 PNs.
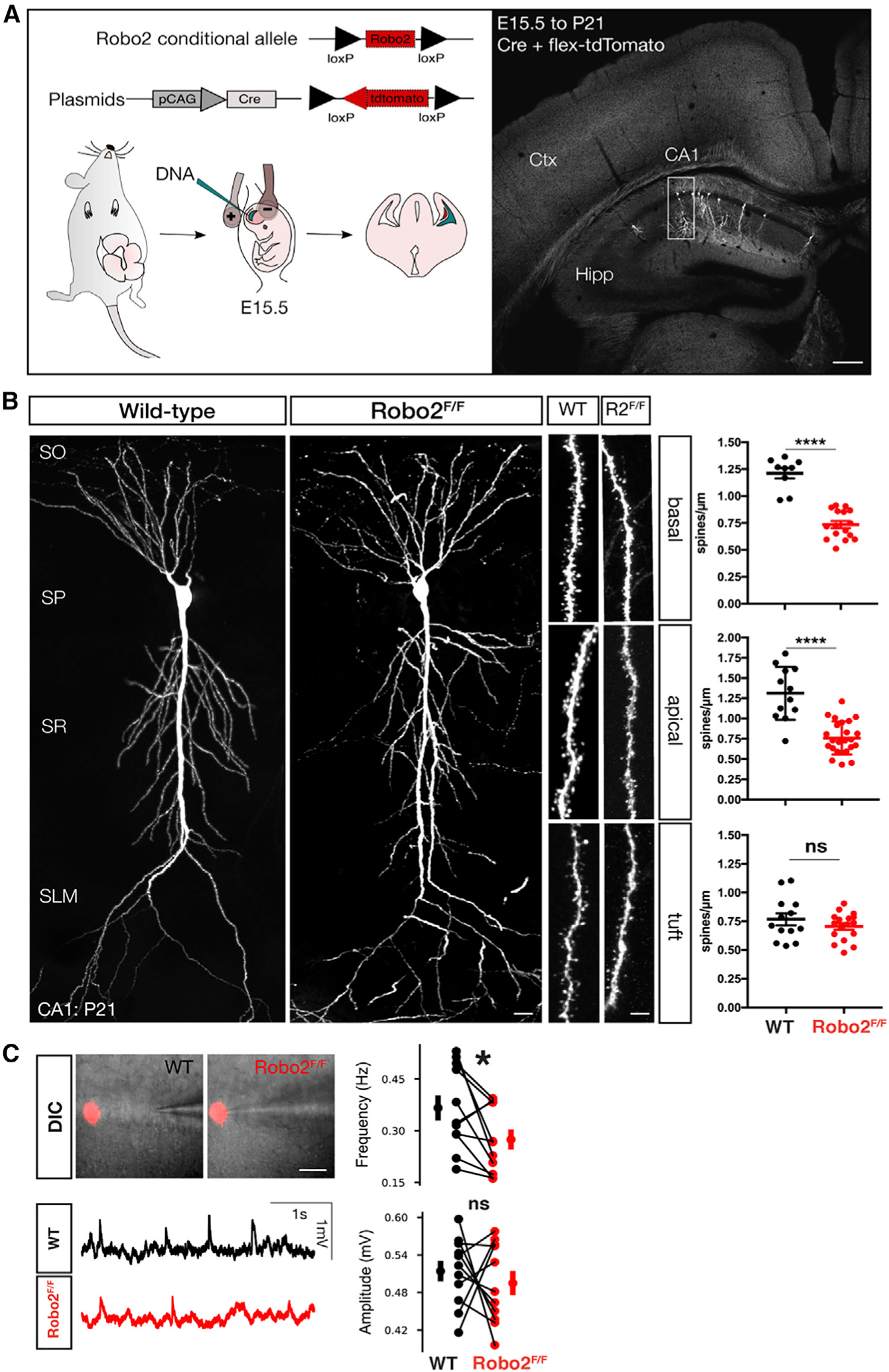
(A) Schematic of in utero electroporation to sparsely target CA1 PN progenitors in order to introduce plasmids expressing Cre-recombinase and flex-tdtomato. Scale bar: 150μm.
(B) Optically isolated CA1 PNs in either WT or Robo2F/F mice. Analysis of spine distribution reveals a significant decrease in spine density in proximal dendritic compartments of Robo2-deficient CA1 PNs (basal: WT: n = 9 dendritic segments, mean = 1.21 spines/μm ± 0.049 (SEM), KO: n = 16 dendritic segments, mean = 0.73 spines/μm ± 0.033 (SEM), reduction = 39.67%, ****p < 0.0001, Mann-Whitney; apical: WT: n = 12 dendritic segments, mean = 1.31 spines/μm ± 0.095 (SEM), KO: n = 24 dendritic segments, mean = 0.76 spines/μm ± 0.041 (SEM), reduction = 41.98%, ****p < 0.0001, Mann-Whitney), but not distal tuft dendrites (tuft:WT: n = 13 dendritic segments, mean = 0.76 spines/μm ± 0.052 (SEM), KO: n = 17 dendritic segments, mean = 0.71 spines/μm ± 0.029 (SEM), p = 0.51, ns, Mann-Whitney). Scale bar: 25μm.
(C) Whole-cell patch clamp and mEPSP measurements of WT and Robo2-deficient CA1 PNs. DIC images and example traces of patched Robo2 KO neuron as indicated by presence of flex-mcherry and neighboring WT neuron (n = 11 pairs). Quantification of amplitude (mV, WT avg: 0.514; SEM: 0.016 mV, KO avg: 0.495; SEM: 0.019 mV, p = 0.54592, paired t test, ns) and frequency (Hz, WT avg: 0.366; SEM: 0.037 Hz, KO avg: 0.275; SEM: 0.029 Hz, *p = 0.0329, Wilcoxon-signed rank test).
See also Figures S1 and S2.
Next, to corroborate our findings that conditional deletion of Robo2 from CA1 PNs interferes specifically with synaptic development, we used a postnatal CA1-PN specific-Cre-mediated deletion of Robo2 (Lypd1Cre;Robo2F/F, see Figure S5B). We find that postnatal deletion of Robo2 phenocopies the compartment-specific reduction in spine density observed in the sparse HIUE approach. We confirmed that in this in Lypd1Cre mouse line (Tg(Lypd1-cre)NR151Gsat), the onset of Cre recombination ramps up around P7 in CA1 PNs (Figure S5A) (i.e., after axon guidance is completed for principal cells in the hippocampus).
Finally, this reduction in spine density was already apparent at postnatal day 12 (P12; Figure S6A), strongly arguing for a requirement of Robo2 in early excitatory synapse formation, rather than for synaptic maintenance during hippocampal circuit maturation. Taken together with its laminar and subcellular protein localization pattern (Figures 1B and 1D), our results demonstrate that Robo2 is required postsynaptically in CA1 PNs for the formation of ~40% of excitatory synapses in an input-specific manner (required for CA2/CA3 inputs but not for EC inputs). Using the same HIUE approach, we also analyzed the density of inhibitory synapses by quantifying Gephyrin puncta (Chen et al., 2012) in sparsely electroporated Robo2-KO and WT CA1 PNs (Figure S6C). We did not find any significant difference in the density of inhibitory synapses between Robo2 null and control CA1 PNs in any of the three main dendritic subdomains.
To analyze any effects of our sparse, conditional deletion of Robo2 on excitatory synaptic physiology in CA1 PNs, we performed patch-clamp recordings in acute adult hippocampal slices. Using the same HIUE approach in Robo2F/F mice to achieve a sparse deletion of Robo2 from CA1 PNs, we performed whole-cell current-clamp recordings of miniature excitatory postsynaptic potentials (mEPSP) in Robo2 KO (mCherry-expressing through Cre-dependent FLEX::mCherry recombination) and neighboring WT CA1 PNs within the same hippocampal slices (Figures 2C and S6B). As expected from the reduction in spine density, the frequency of sEPSPs was significantly reduced in Robo2 null compared to adjacent WT CA1 PNs, whereas the amplitude of the remaining events was not significantly different (Figure 2C). These data demonstrate that postsynaptic Robo2 expression is required for excitatory synapse physiology in CA1 PNs. Using the same approach, we also recorded mIPSPs, and in line with our mapping of Gephyrin+ inhibitory synapses, we did not find any significant changes in amplitude or frequency of inhibitory synaptic events in Robo2 null compared to control CA1 PNs (Figures S6C–S6E).
Robo2 induces excitatory synapse formation in a Slit-dependent manner
Our data thus far show that Robo2 localizes to, and is required postsynaptically for excitatory (but not inhibitory) synapse formation in CA1 PNs in a compartment-specific manner. Next, we used a reductionist approach to determine whether Robo1/2 are directly involved in synapse formation. Specifically, we used an in vitro hemi-synapse assay (Scheiffele et al., 2000) to determine whether Robo proteins expressed on the surface of HEK293 cells can induce formation of presynaptic boutons from axons of co-cultured primary cortical neurons. Using Neuroligin1 (NLG1) as a positive control since it can induce both excitatory and inhibitory synapses in vitro (Scheiffele et al., 2000) and CD8 as negative control, we tested whether Robo receptors were able to induce the formation of presynaptic boutons from axons of co-cultured cortical neurons. Indeed, expression of either Robo1 or Robo2 in HEK293 cells led to a strong clustering of axonal Vglut1 around the cell perimeter (Figure 3A). Interestingly, Robo3, did not induce accumulation of Vglut1+ presynaptic boutons. Robo3 is a divergent member of the Robo family and has lost the ability to bind Slit ligands during mammalian evolution (Zelina et al., 2014). The finding that Robo3 is not able to induce Vglut1+ presynaptic bouton clustering prompted us to determine if the synaptogenic activity of Robos was Slit-dependent. To do so, we expressed a form of Robo2 harboring a deletion of the Slit binding domain (Robo2−ΔIg1,2; Liu et al., 2004) in the HEK293 cells and found that Robo2ΔIg1,2 did not induce Vglut1+ presynaptic bouton clustering. We verified that the deletion of these domains does not influence trafficking of the receptor to the membrane (Figure S7). Finally, we co-expressed a secreted, Robo2-Fc construct in the HEK293 cells, which efficiently sequesters extracellular Slit away from Robo receptors (Brose et al., 1999). This approach likewise abolishes presynaptic Vglut1 clustering, supporting a critical role for Slit binding in the synaptogenic activity of Robo2 in vitro. Some synaptogenic transmembrane proteins such as NLG1 indiscriminately induce the formation of excitatory and inhibitory presynaptic boutons in the hemi-synapse assay (Scheiffele et al., 2000). By contrast, HEK293 cells expressing Robo2 did not induce clustering of the presynaptic inhibitory Vgat1 from surrounding axons (Figure 3B). Altogether, our data show that Robo1 and Robo2 specifically induces the formation of excitatory, but not inhibitory, synapses in a Slit-dependent manner.
Figure 3. Robo2 induces excitatory synapse formation in a Slit-dependent manner.
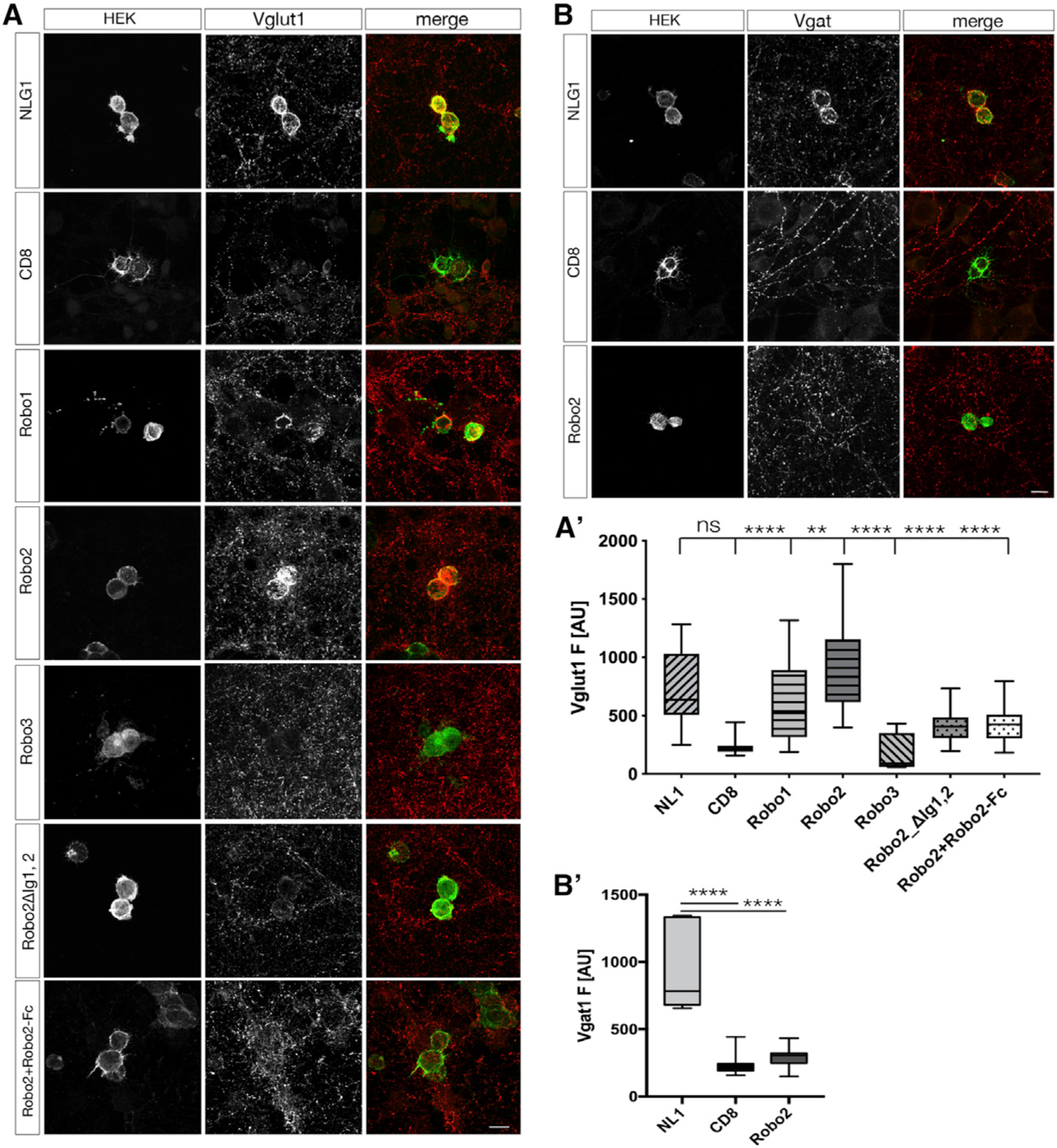
(A) In vitro hemi-synapse assay (24h coculture of primary neurons at DIV12 with HEK293 cells expressing cDNAs as indicated in text boxes on the left for each row of images). Immunostaining for Vglut1 indicates presence of excitatory presynaptic boutons, which cluster around the perimeter of the HEK293 cell in case the cDNA is synaptogenic. NLG1: positive control, CD8: negative control. Robo1 and Robo2 expression in HEK293 cells leads to Vglut1 clustering around the cell perimeter. Slit-binding deficient receptors, Robo3 and Robo2ΔIg1,2, Robo2[L93P], are unable to induce Vglut1 clustering. Scale bar: 7μm.
(B) Robo2 does not induce inhibitory synapse formation as evidence by the absence of clustering the inhibitory presynaptic marker Vgat1 in immunohistochemistry. Scale bar: 7 μm.
(C and D) Quantification of hemi-synapse assay. F: Fluorescence intensity in AU (arbitrary units). Statistics: one-way ANOVA, ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, whiskers show min/max, n = 4 independent experiments with at least 20 cells/coverslip (triplicates of one condition per independent experiment).
Presynaptic Neurexins bind directly to Robo2 and are required for Robo2-dependent synaptogenesis
Our in vitro and in vivo results show that Robo2 is both necessary and sufficient to induce excitatory synapses, prompting us to determine the identity of the trans-synaptic binding partners in the presynaptic membrane required for Robo2′s synaptogenic activity. We first sought to test whether the synaptogenic activity of Robo1/2 requires a potential homophilic trans interaction with Robo1/2 expressed in axons presynaptically. This reasoning is based on the following: (1) Homophilic trans-interactions of Robos can occur in vivo in the context of axon guidance in Drosophila (Evans et al., 2015); (2) axonal function of Robo as an axon guidance Slit receptor is well known; and (3) we detected a small fraction of the Robo2 pool on presynaptic membranes (Figure 1D). To test if presynaptic Robo1/2 could be involved in ‘postsynaptic’ Robo2-mediated synaptic induction, we repeated the hemi-synapse assay using cortical neurons deficient for Robo1 or Robo1/2 double knockout embryos. Since a homozygous constitutive deletion of Robo2 (but not Robo1) is perinatally lethal (Grieshammer et al., 2004), we isolated cortical neurons from a combination of Robo1 constitutive knockout and a Robo2 conditional allele and infected the cultures with Cre-expressing lentivirus at DIV0 (Figure 4A). Interestingly, Robo1/2-deficient axons were still able to cluster Vglut1 around HEK293 cells expressing Robo2. Our results show that presynaptic Robo1/2 expression is not required to support the synaptogenic function of postsynaptic Robo2.
Figure 4. Robo2 is part of a heterophilic complex with presynaptic Neurexins in trans.
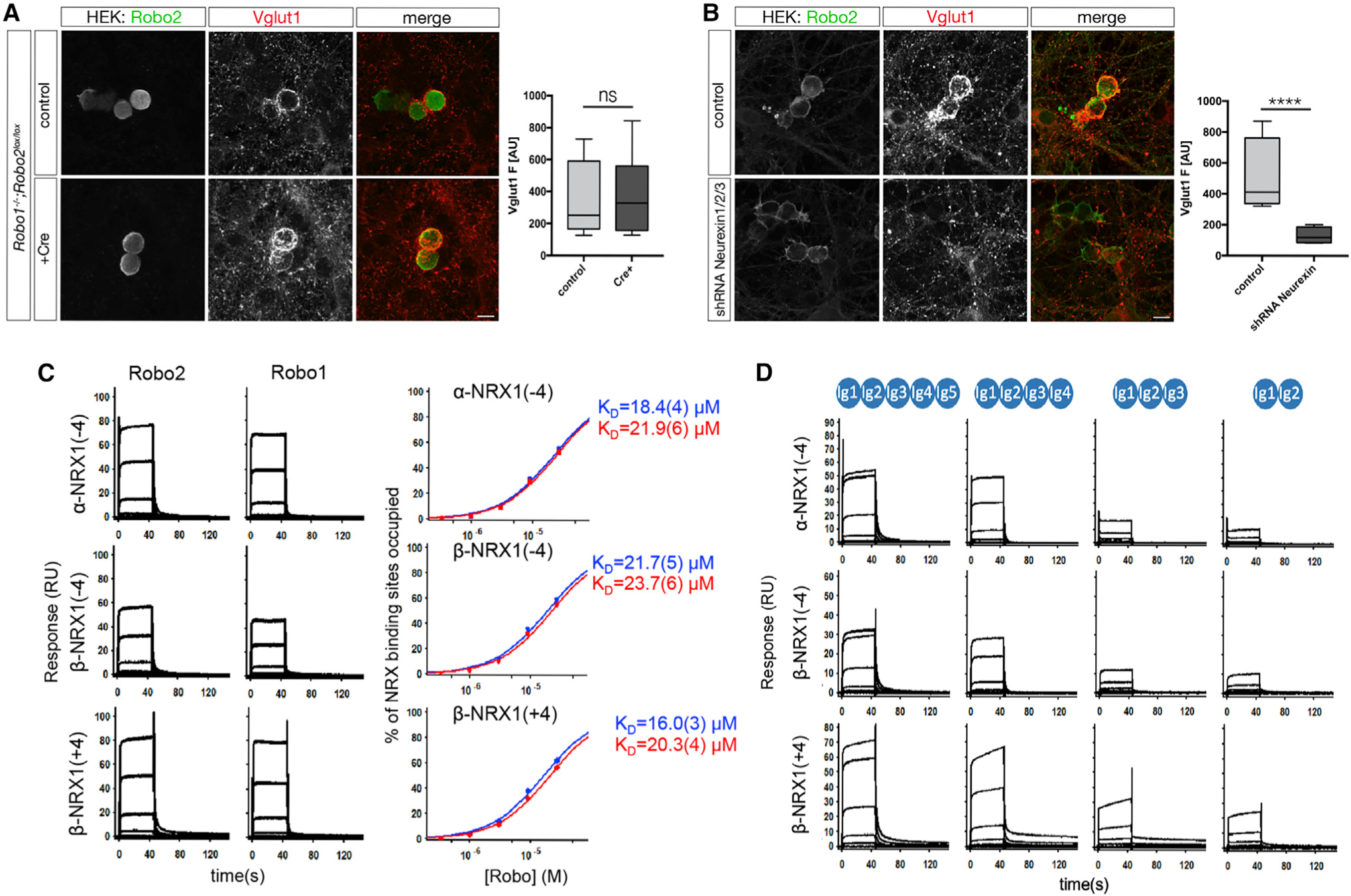
(A) Presynaptic Robo1/2 are not required for Vglut1 clustering around Robo2-expressing HEK293 cells. In vitro hemi-synapse assay was performed using primary neurons from either Robo1−/− or Robo1−/−;Robo2F/F (infected with Lentivirus expressing Cre-recombinase at DIV0, one-way ANOVA, ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, whiskers show min/max, n = 3 independent experiments). Scale bar: 7μm.
(B) Presynaptic Neurexins are essential for Robo2-dependent Vglut1-clustering. In vitro hemi-synapse assay was performed in the presence (DIV3) of a Lentivirus expressing a pan-Neurexin shRNA. Neurexin knockdown completely abolishes Robo2-dependent Vglut1-clustering. Scale bar: 7μm.
(C) SPR binding experiments of Robo1 and Robo2 ectodomains over NRX surfaces: Binding of Robo2 and Robo1 (Ig1–5) over surfaces immobilized with α-NRX1Δ4, β-NRX1Δ4, and β-NRX1+4 ectodomains. Robo 2 and Robo1-binding was detected with all three NRX-immobilized surfaces. Binding isotherms of the percentage of NRX binding sites occupied versus Robo concentration (right panels) yield the KDs for Robo2 (Ig1–5) and Robo1 (Ig1–5), shown in blue and red, respectively. The number in brackets represents the error of the fit.
(D) Binding of Robo2 Ig deletion fragments over α-NRX1Δ4, β-NRX1Δ4, and β-NRX1+4 ectodomains. Robo2 fragments encompassing Ig domains 1–5, 1–4, 1–3 and 1–2 respectively were tested for binding. A sharp decrease in signal is observed between the Ig1–4 and Ig1–3 fragments suggesting that Nrxn binds mostly with the Ig4–5 domains of Robo2.
See also Figure S3.
We therefore hypothesized that the synaptogenic function of Robo and its secreted ligand Slit depends on an interaction with an as of yet unknown presynaptic transmembrane protein. Hence, we took an unbiased proteomic approach based on an experimental pipeline developed to identify synaptic interactomes (Savas et al., 2014). We purified synaptosome fractions from P21 rat brains and performed pulldown with recombinant Slit2-Fc protein (Figure S8A). Shotgun mass-spectrometry (MS) analysis of synaptic proteins binding to recombinant Slit2-Fc identified Neurexin1/2/3 as one of the transmembrane proteins pulled down by Slit2. We also identified Robo2, Glypican1, and PlexinA1—membrane-associated proteins previously identified as Slit-interacting proteins (Bülow et al., 2008; Delloye-Bourgeois et al., 2015; Ronca et al., 2001)—validating our approach. We focused on Neurexins as potential presynaptic Slit-Robo interacting partners mediating their synaptogenic activity because of the well-characterized function for Neurexins as a presynaptic organizing protein family (Südhof, 2018).
In order to determine whether presynaptic Neurexins are required for the synaptogenic function of Robo2, we repeated the hemi-synapse assay after knockdown of presynaptic Neurexins (Gokce and Südhof, 2013). Neurexins are key presynaptic organizing proteins capable of forming multiple trans-synaptic complexes with postsynaptic proteins in a synapse-specific way. Three main Neurexin genes (Nrx1–3) can each generate two main isoforms (α [long] and β [(short]) that display a considerable degree of alternative splicing. As a first approach, we targeted all isoforms of Neurexin1/2/3(α and β) with a previously validated shRNA-expressing lentivirus (Gokce and Südhof, 2013) to infect primary neurons 7 days before adding the HEK293 cells expressing Robo2. Strikingly, Neurexin1/2/3(α/β)-knockdown completely abolished Robo2-synaptogenic activity (Figure 4B), establishing presynaptic Neurexins as an essential component of the trans-synaptic adhesion complex mediating Slit-dependent Robo2 synaptogenic activity.
To further characterize the potential interaction between Neurexins and Robo2 at synapses, we turned to surface plasmon resonance (SPR), a biophysical method to analyze direct interactions between macromolecules allowing quantitative measurement of binding affinities. Given the high structural diversity of Neurexin isoforms, we chose to immobilize three Neurexin1 isoforms: α-Neurexin1 without insertion of the major splice-site (SS) 4 (α-NRX1(−4)), β-Neurexin without or with SS4 (β-NRX1(−4), and β-NRX1(+4), respectively) on a dextran-chip via amide coupling and flowed purified recombinant Robo1 and Robo2 (Ig domains 1–5; Figure S8F) over the chip as analyte (Figure 4C). Robo1 and Robo2 bound directly to all Neurexin isoforms tested with KDs of ~16–23μM, an affinity in the same order of magnitude as β-Neurexin1/2/3-Neuroligin1/2/3 interactions (ranges between ~0.8–56μM) (Koehnke et al., 2010). Interestingly, we did not observe any direct interaction of Slits and Neurexins in SPR (data not shown). However, the fact that we identified Robo in our Slit2-Fc pull-down/MS experiments from synaptosomes (Figure S8A) may suggest that Neurexin identification in the same experiment may be due to its interaction with Robo as part of a tripartite complex.
To identify the binding interface between Neurexins and Robo1/2, we performed similar SPR experiments with Ig domain deletions of Robo2. We found a drastic reduction in binding efficiency of Robo2 to Neurexins when Ig4–5 were deleted (Figure 4D). This reveals that binding sites for Slit (Ig1–2) and Neurexins (Ig4–5) on Robo2 probably do not overlap. Notably, Robo-Neurexin interactions were dependent on the presence of Heparin as well as Ca2+ (Figures S8B–S8E). Together with our findings that the Slit-binding domain of Robo2 is important for its synaptogenetic function (Figure 3), our results strongly suggest the existence of a tripartite Robo-Slit-Neurexin trans-synaptic complex promoting excitatory (but not inhibitory) synapse formation.
Sparse developmental deletion of Robo2 alters place cell properties of CA1 PNs in vivo
We then sought to determine whether interfering with Robo2-dependent development of excitatory synapses in CA1 PNs affects their functional activity patterns in vivo. CA1 PNs represent an ideal model to study the impact of interfering with a specific set of synapses on circuit function. A subset of CA1 PNs exhibit spatially tuned activity when the animal is exploring an environment (O’Keefe and Dostrovsky, 1971). However, how different input streams from CA2/3 and EC contribute to CA1 PN place coding is still a topic of intense investigation with previous studies yielding mixed results (Brun et al., 2002; Davoudi and Foster, 2019; Mizumori et al., 1989; Nakashiba et al., 2008; Nakazawa et al., 2002) (see Discussion). Manipulation of Robo2 in CA1 PNs presents a unique tool to address how a reduction specific to excitatory drive from CA2/3 affects place cell properties. Hence, we assessed CA1 PN place cell properties at the population level using in vivo two-photon (2p) microscopy-based calcium (Ca2+) imaging in head-fixed, awake-behaving mice (Danielson et al., 2016; Kaifosh et al., 2013; Lovett-Barron et al., 2014).
We used the same HIUE approach to conditionally delete Robo2 via Cre expression alongside expression of FLEX::mCherry from a sparse subpopulation of CA1 PNs in Robo2F/F animals. We then stereotactically injected dorsal CA1 with a recombinant adeno-associated virus (rAAV) expressing the genetically encoded Ca2+ indicator GCaMP6f. This approach allows us to image the activity of hundreds of GCaMP6f-expressing CA1 PNs simultaneously within stratum pyramidale. About 10% of recorded CA1 PNs are deleted for Robo2, as identified by the presence of Cre-dependent mCherry expression while all other CA1 PNs are wild-type control (Figure 5B–D). Mice were trained to run for randomly delivered water rewards (random foraging) on a linear treadmill belt decorated with spatial cues as navigational landmarks, as previously described (Danielson et al., 2016).
Figure 5. Robo2 knockout alters place cell properties of CA1 PNs in vivo.
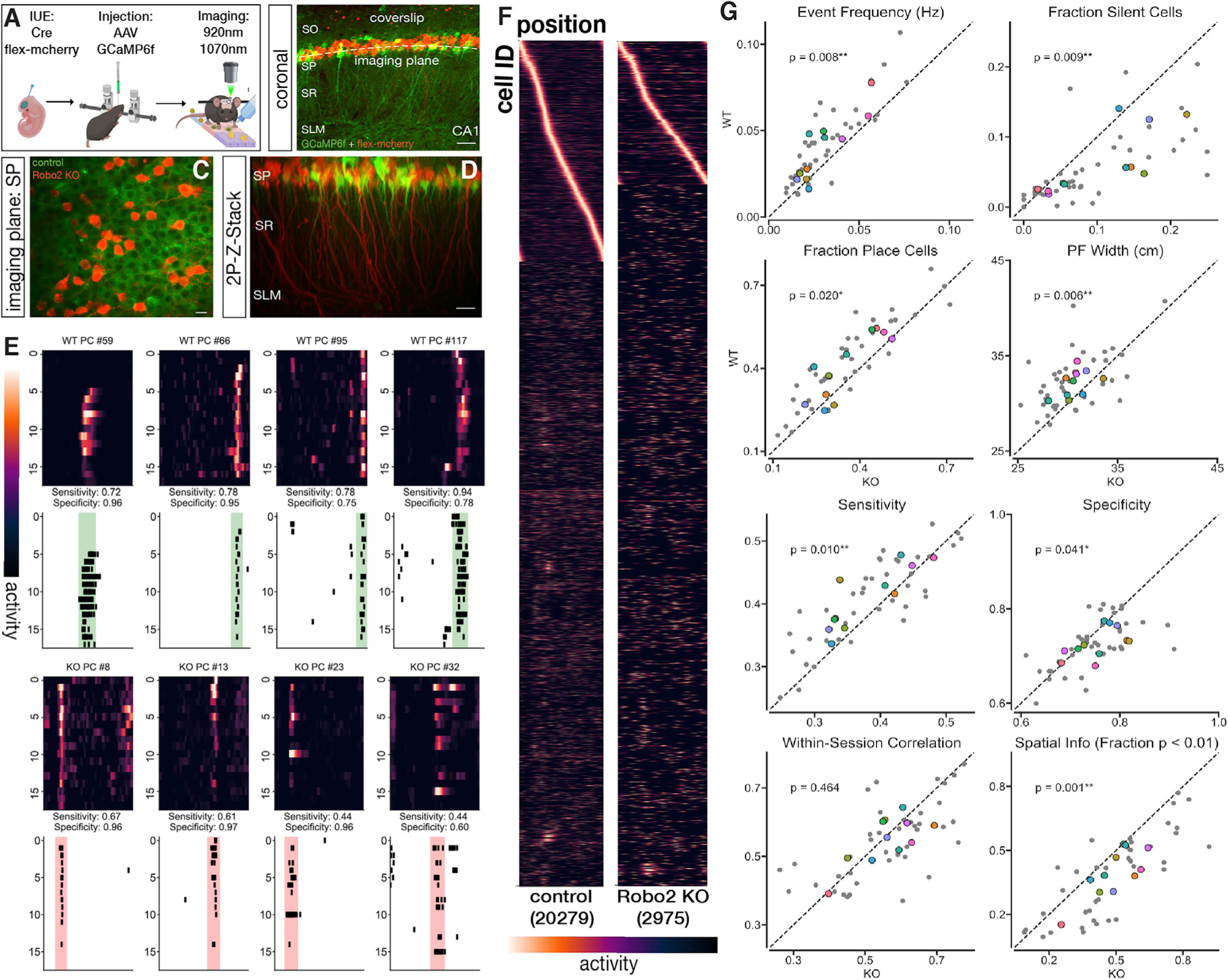
(A) Overview of the imaging paradigm. Once adult, mice were injected with AAV-hSyn-GCaMP6f and implanted with a metal headpost to allow for in vivo 2-photon imaging in awake-mice. Mice explore a treadmill belt with spatial cues in a 1D environment.
(B) Representative image of field of view (post hoc). HIUE was used to create a mosaic Robo2 knockout of CA1 PN subpopulations. Scale bar top left: 35μm.
(C) Imaging field of view acquired during a session. Scale bar 15μm.
(D) In vivo Z stack volume of imaged area. Scale bar bottom right: 20μm.
(E) Example WT (green) and Robo2 KO (red) place cells from a single session. Lap by lap z-scored dF/F heatmap (above) and corresponding deconvolved event raster (below) with place field highlighted.
(F) Normalized tuning of all imaged cells (total cell counts in parentheses; place cells sorted by peak) across sessions for WT and Robo2 KO neurons. G. Differences in spike frequency and place cell properties between WT and Robo2 KO neurons. Robo2 KO neurons exhibit a decrease in spike frequency and an increase in the fraction of silent (i.e., no detected events in a session) cells. Robo2 KO neurons also exhibit a decrease in place cell fraction, place field width, and sensitivity, but a modest increase in specificity. Details see text (paired t test; event frequency (Hz): **p = 0.008, fraction silent cells: **p = 0.009, sensitivity: **p = 0.01, specificity: *p = 0.041, fraction place cells: *p = 0.02, place field width (cm): p = 0.006**, significant spatial info: **p = 0.001, within-session correlation: p = 0.464, ns; n = 11 FOVs). Grey dots indicate individual sessions, colored dots indicate FOV means across sessions. Dashed line indicates equal values for WT and Robo2 KO subpopulations.
We then analyzed inferred events from deconvolved Ca2+ activity and compared place cell properties of Robo2 KO cells and their WT counterparts in adult mice (> 60 days). We found that Robo2 KO CA1 PCs have significantly reduced event frequency during running (WT: 0.075 ± 0.022 Hz, KO: 0.064 ± 0.017 Hz; mean ± sd) (Figure 5G). This reduction corresponded to an increase in the fraction of silent cells per session (i.e., the fraction for which no significant events were recorded; WT: 0.063 ± 0.047, KO: 0.106 ± 0.069; mean ± sd). Overall, we observed a significant (~13%) reduction in the fraction of place cells within Robo2 KO compared to WT CA1 PNs (WT: 0.404 ± 0.118, KO: 0.352 ± 0.105) (Figures 5F–5G). Furthermore, the remaining Robo2 KO place cells showed significant alterations in their response properties. While within-session correlation is not significantly different between the two cell populations, relative to WT CA1 PCs, Robo2 KO CA1 PCs showed significantly reduced sensitivity (WT: 0.440 ± 0.048, KO: 0.412 ± 0.059), defined as the reliability of spiking during place field traversals. Interestingly, we observed a significant increase in specificity of spiking activity in the Robo2 KO compared to WT CA1 PCs (WT: 0.676 ± 0.032, KO: 0.708 ± 0.041), along with an increase in the fraction of PNs with significant spatial information for Robo2 KO (Figure 5G). In sum, our data suggest an important role for CA2/CA3 in driving ongoing place-specific firing and location encoding in CA1 PNs, and shows specifically that Robo2-dependent alteration in excitatory synapse development has a significant impact on in vivo properties of hippocampal CA1 PNs in awake-behaving mice.
DISCUSSION
We have uncovered a role for the axon guidance molecules Slit and Robo in excitatory synapse formation and synaptic specificity in CA1 pyramidal neurons of the hippocampus. Our data demonstrate that postsynaptic Robo2 promotes excitatory (but not inhibitory) synapse formation by forming a trans-synaptic complex with its LRR domain-containing Slit ligand and presynaptic Neurexins. Interestingly, Robo2 protein localization is restricted to specific dendritic domains of CA1 PNs (apical oblique and basal dendrites but not apical tufts), and conditional deletion of Robo2 leads to a selective ~40% decrease in excitatory synapse formation within these two domains corresponding to dendritic domains receiving excitatory inputs from CA2/CA3 PNs. Finally, we found that cell-autonomous deletion of Robo2 from CA1 PNs alters their place cell properties in vivo. Altogether, our data show that Robo2 functions outside of axon guidance in excitatory synapse formation. Our results unravel a function for Robo signaling in synapse formation and link the establishment of synapse specificity to the emergence of place cell properties in CA1 PNs with implications for hippocampal circuit function.
Slit-Robo signaling in axon guidance and synapse formation
In the context of axon guidance, overwhelming evidence has demonstrated that Slit binding to Robo elicits signaling leading to growth cone repulsion (Nguyen Ba-Charvet et al., 1999). Even though we did not observe any change in dendritic growth comparing Robo2 null and wild-type in CA1 PNs, several studies previously suggested that Robo/slit signaling plays a role in dendritic branching and tiling through either chemorepulsive or chemoattractive functions (Gibson et al., 2014; Hocking et al., 2010; Whitford et al., 2002). Furthermore, Slit2 was previously shown in culture to be sufficient to induce presynaptic clustering of axons emerging from spinal cord explants (Wu et al., 2015). Interestingly, Robo1 and Robo2 both exhibit synaptogenic activity, but not the divergent Robo3, which lost the ability to bind Slits during mammalian evolution (Zelina et al., 2014). Our results suggest that in the context of synapse formation, postsynaptic Robo promotes excitatory synapse formation in a Slit- and Neurexin-dependent manner. However, our results are based on the in vitro synaptogenesis assay and future work will be required to test the in vivo relevance of Slit/Neurexin binding to postsynaptic Robo for its synaptogenic activity. Slit1/2/3 as well as many isoforms of Neurexins 1/2/3 (Fuccillo et al., 2015) are expressed in complex spatial and temporal dynamics in multiple cell-types in the developing and mature hippocampus, making the analysis of their requirement for Robo-dependent synaptogenesis in vivo challenging.
Together with our biochemical data, we propose a model whereby, during the switch between axon guidance and synapse formation, Slit binding to axonal Robo receptors elicits chemorepulsion whereas during synapse formation, dendritic Robo receptors elicit excitatory synapse formation by forming a trans-synaptic complex with Slit and presynaptic Neurexins. Future experiments will need to identify the downstream signalling pathways mediating such divergent cell biological outcomes of Robo/Slit signaling in axons and dendrites which has been previously documented for other axon guidance cues such as Semaphorins (Chen et al., 2008; Polleux et al., 1998; Polleux et al., 2000).
Slit binds through its LRR domains to the first two Ig domains (Ig1–2) of Robo (Liu et al., 2004) and our results show that Neurexins bind most efficiently to Ig4–5 of Robo. Furthermore, recent structural work shows that the Robo trans-dimerization domain is contained within Ig4–5 (Barak et al., 2019). We therefore propose a model whereby Slit-binding to Robo releases the inhibitory Robo trans-interaction inducing a conformational change (Barak et al., 2019) which might allow postsynaptic Robo to bind to presynaptic Neurexins. However, using recombinant proteins in our SPR assay, α- and β-Neurexin1 can bind to Robo in the absence of Slit, therefore it is possible that, in these conditions, Robo is rather unconstrained conformationally and does not require Slit to enable Neurexin-Robo interaction.
Furthermore, the cellular and spatial source of Slit in Robo2-dependent synaptogenesis remains an open question. In the hippocampus, Slit2 expression is restricted CA3, presynaptic to CA1 PNs, while Slit1 and Slit3 are more broadly expressed in CA regions (Figure 1A). In future experiments, it will be important to further our understanding of the structural mechanisms underlying the Robo-Slit-Neurexin trans-synaptic complex to dissect how these proteins interact in a context-specific manner.
Trans-synaptic coincidence detection as a molecular mechanism to increase synaptic specificity
Our results uncover a trans-synaptic molecular complex constituted of Robo, Slit, and Neurexin. The finding that Slit as well as Neurexin are both important to support Robo2-dependent synaptogenesis in vitro, posits the existence of a tripartite trans-synaptic protein complex. This suggests a model whereby synaptic specificity during recognition of a presynaptic axon and the corresponding postsynaptic dendrites requires coincidence detection at the molecular level between at least three components as recently exemplified (Sando et al., 2019). Another example of this coincidence detection are ligand-induced adhesion mechanisms at synapses (Ledda et al., 2007). We identified other transmembrane components in our synaptic Slit pull-down mass spectrometry experiments such as Glypican1 and PlexinA1 (previously characterized Slit binding proteins; Delloye-Bourgeois et al., 2015; Ronca et al., 2001). This could further increase the specificity of a multimeric trans-synaptic molecular complex with a probability constrained of each component being present at axon-dendrite contacts during synapse formation. We also reveal that Neurexin-Robo interaction requires heparin-conjugated moieties by showing the synaptogenic activity of Robo is abolished by treatment with heparinase. Since Neurexin1 was recently shown to require a rare glycan modification, heparan sulfate (Zhang et al., 2018), this posttranslational modification might also participate in increasing the specificity of trans-synaptic interactions between Robo, Slit, and Neurexins. Future experiments will need to provide insights into the structure of such trans-synaptic protein complexes, in order to gain further evidence for this coincidence detection model as a mechanism to increase synaptic specificity.
Input-specific reduction of excitatory drive alters place cell properties
Our results show that cell-autonomous deletion of Robo2 from CA1 PNs reduces the number of inputs they receive from CA2/CA3 onto their basal and apical oblique dendrites by ~40%, which has significant consequences for how and to what extent spatial information is encoded. Our molecular manipulation serves as a tool to precisely reduce the impact of intrahippocampal information on CA1 PNs, while leaving long-range cortical inputs intact. Our cell-type specific molecular manipulation changes the number of CA2/CA3 inputs (but not EC inputs) received by CA1 PNs and thereby changes the CA1 PN output function. The importance of CA2/3 versus EC inputs for CA1 PN spatial tuning is still an area of ongoing research in studies of hippocampal circuits. Interestingly, so far, only a few studies have directly addressed this question, either relying on pharmacological lesion of CA3 (Brun et al., 2002; Mizumori et al., 1989), Tetanus toxin–based suppression of CA3 vesicle release (Middleton and McHugh, 2016; Nakashiba et al., 2008), and, most recently, acute optogenetic silencing of CA3 axon terminals (Davoudi and Foster, 2019). These studies have generated disparate results, with earlier longer-timescale manipulations suggesting only a minor impact on CA1 PN spatial tuning, while the more recent acute manipulation in (Davoudi and Foster, 2019) suggests that CA3 is critical for normal place field responses. Our findings are strongly in line with this latter result— we find that if CA2/3 inputs are reduced in the absence of Robo2, these Robo2-deficient CA1 PNs have decreased firing rates, have smaller place fields, are less stable lap-by-lap, but fire more specifically in the place field. Our results therefore support a critical role for this specific input pathway in driving CA1 PN spatial firing, and suggest that CA3 excitatory drive may be more critical for place field responses than previously thought. More generally, the striking ability of CA1 PNs to spatially regulate their proteome along their dendritic arbor not only mediates anatomically compartmentalized inputs (de Wit and Ghosh, 2016) but also contributes to their unique physiological properties (Piskorowski et al., 2011).
Altogether, our study demonstrates how the precise localization and interactome of trans-synaptic protein complexes underlie the emergence of functional properties of neurons within mature circuits through their ability to regulate synaptic specificity and circuit connectivity.
Limitations of study
Our results reveal a postsynaptic function of Robo2 acting as a synaptogenic cue promoting excitatory (but not inhibitory) synapse formation in a domain-specific way along dendrites of CA1 PNs. Our results also demonstrate that single CA1 PNs deficient for Robo2 display altered place cell properties. By using a combination of slice electrophysiology and in vivo 2-photon Ca2+ imaging in awake behaving mice, our study provides a causal link between the establishment of synaptic specificity and the emergence of functional response properties of a given neuronal subtype within a mammalian neural circuit. The most parsimonious interpretation we propose is that the cell-autonomous reduction of ~40% of the CA3 inputs received by Robo2-deficient CA1 PNs is causally linked to the changes of functional properties we observed in CA1 PNs in vivo given the known role CA3 to CA1 connectivity in place cell emergence ((Davoudi and Foster, 2019); (Brun et al., 2002); (Nakazawa et al., 2002)). However, we cannot exclude the possibility that alterations of other electrophysiological properties of Robo2-deficient CA1 PNs not probed in the current study might contribute to the alterations of their functional properties. These include potential defects in their intrinsic properties such as active dendritic conductances, input resistance or other dendritic and synaptic properties not probed in the current study. Nevertheless, our approach will serve as a template for the exploration of the relationship between the molecular mechanisms patterning neuronal connectivity and the emergence of functional properties of the corresponding circuits.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Franck Polleux (fp2304@columbia.edu).
Materials availability
All unique reagents generated in this study are available from the Lead Contact but we may require a Materials Transfer Agreement.
Data and code availability
The 2P Ca2+ imaging data reported in this paper will be shared by the lead contact upon request.
The SIMA code base is deposited at https://github.com/losonczylab/sima and is publicly available. Suite2p and OASIS are both open source. Our Github repository is archived here: https://doi.org/10.5281/zenodo.5484268.
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were used according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Columbia University and in accordance with National Institutes of Health guidelines. The health and welfare of the animals was supervised by a designated veterinarian. The Columbia University animal facilities comply with all appropriate standards (cages, space per animal, temperature, light, humidity, food, water). Both males and females were used for all experiments, no difference were observed between sexes of individual mice for any of the parameters analyzed in this study.
Timed-pregnant CD1 females were purchased from Charles River. 129/SvJ, C57BL/6J non-transgenic control inbred mice and all other transgenic mice were maintained in a 12-hour light/dark cycle. Timed-pregnant females were obtained by overnight breeding with males of the same strain. Noon the day after the breeding was considered as E0.5. Embryos were electroporated at E15.5 (see below), dendritic morphology analysis was done between P12–21 depending on the study. Electrophysiology studies were done between P21–27, in vivo imaging studies were done between P65–85.
Lentivirus production
Second generation VSV.G pseudotyped lentiviruses were produced as previously described (Dittgen et al., 2004; Kutner et al., 2009). HEK293T cells were cultured in DMEM+10% FBS and maintained in an incubator at 37C and 5% CO2 under passage 20. Cells were transfected with control or shRNA-containing pFUGW (GFP-expressing) vector plasmids and helper plasmids PAX2 and VSVG using Fugene (Promega). Supernatant was collected 65 hr after transfection, spun at 2000 rpm to remove debris and filtered through a 0.45 μm filter (Corning). In order to maximize the purity of viral particle pellets, a small amount of sucrose solution (20% sucrose, 100 mM NaCl, 20 mM HEPES, 1 mM EDTA, at pH 7.4, filtered with a 0.22 μm filter) was placed at the bottom of the centrifuge tubes before adding filtered media. Viral particles were then pelleted by centrifugation at 200.000 rpm for 2 hr at 4C. Final pellet was re-suspended in 200 μL of PBS and stored at −80°C in 10 μL aliquots.
METHOD DETAILS
Immunohistochemistry
Immunofluorescent staining for synaptic proteins in hippocampal sections was performed in 16 μm-thick cryosections from P35 rat brains perfused with 2% paraformaldehyde (PFA) solution. Sections were post-fixed in a 1:1 MeOH:acetone solution at −20°C, and then permeabilized with 0.5% triton in PBS-0.2% gelatin. Blocking solution consisted of 10% normal horse serum and 0.5% triton in PBS-0.2% gelatin. Primary antibodies were the following: goat anti-FLRT2 (R&D Systems) and rabbit anti-Robo2 (Aviva Systems). Primary and secondary antibodies were diluted in 5% normal horse serum and 0.5% triton in PBS-0.2% gelatin. Hoechst was used as a nuclear stain (5nM in PBS). Prolong Gold Antifade (ThermoScientific) was used to mount slides. Confocal images were taken on a Leica TCS SP5 microscope or on a Zeiss LSM880 confocal with an Airyscan detector.
Ex utero electroporation and primary neuron culture
Cortices from E15.5 mouse embryos were dissected followed by dissociation in complete Hank’s balanced salt solution (cHBSS) containing papain (Worthington) and DNase I (100ug/mL, Sigma) for 15 minutes at 37°C, washed three times, and manually triturated in DNase I (100ug/mL) containing neurobasal medium (Life Technology) supplemented with B27 (1x, Thermo Fischer Scientific,), FBS (2.5%, Gibson) N2 (1x, Thermo Fischer Scientific), glutaMAX (2mM, GIBCO). Cells were plated at 10^5 cells per 12 mm glass coverslip pre-coated with Poly-D-Lysine and Laminin (Corning). One-third of the medium was changed every 7 days thereafter with non-FBS containing medium and maintained for 11–21 days in 5% CO2 incubator at 37°C. Ex utero electroporation was performed as previously described (Courchet et al., 2013). In brief, E15.5 embryos were harvested from pregnant dams and decapitated. Plasmids used for ex utero electroporation were all in pCAG vector backbone (Guerrier et al., 2009) expressing the following cDNAs: pCAG-Robo2-pHluorin (Pignata et al., 2019), Homer1c-tdtomato. Plasmids were injected into the lateral ventricles and electroporated using 5 pulses at 40V for 20ms with 200ms intervals. Cortices were dissected and processed as indicated above.
Synaptic fractionation and Western Blotting
Synaptic fractionation was based on a previously described method (Carlin et al., 1980). Briefly, 20 P21 mice brains were homogenized in 10 mL per 2.5 brains with Solution A (0.32 M Sucrose-1 mM NaHC03, 1mM MgCl2, 0.5 mM Cacl2 and protease inhibitors) (homogenate), centrifuged at 1,500 × g for 15 min, and the supernatant was collected (post nuclear supernatant). The supernatant was then centrifuged at 14,000 μ g for 20 min, and the resulting supernatant (cytosol) and pellet (crude membrane) collected. The pellet was re-suspended in 24ml Solution B (0.32 M sucrose- 1mM NaHC03 and protease inhibitors) and loaded onto 0.5 M/1.0 M/1.2 M discontinuous sucrose gradients and centrifuged at 32500 × g for 120 min. The material at the 1.0 M/ 1.2 M interface was collected (synaptosome). The fraction was diluted with Solution B to 60ml. Triton X-100 was added to 0.5% and extracted at 4°C by stirring for 15 min. The extract was centrifuged at 32,500 × g for 25 min, the supernatant collected (soluble synaptosome/Triton-soluble fraction) and the pellet was re-suspended in Solution B, loaded onto a 1.0 M/1.5 M/2.0 M sucrose gradient, and centrifuged at 200,000 × g for 2 hr. Material was collected at the 1.5M/2.0M interface (PSD). 0.5% Triton X-100 was added and detergent-soluble material extracted at 4°C by end-over-end agitation for 10 min. Lastly, the extract was centrifuged at 200,000 × g for 20 min and the pellet re-suspended in Buffer B +5% SDS (purified PSD/Triton-insoluble fraction).
Samples were then prepared for western blotting in Laemmli buffer heated at 60°C for 10mins and 20μg were run on a 4%–20% Acrylamide gel (Biorad.cat no 456–1093). Wet western blotting was carried out using 0.2μm Nitrocellulose membranes (GE Healthcare Amersham, Protean cat no 10600015). Membranes were blocked in TBS-T, 5% Milk and incubated with the following primary antibodies overnight at 4C: mouse anti-PSD-95 (ThermoScientific), rabbit anti-c-Myc (Santa Cruz Biotechnology), rabbit anti-HA (Sigma), rabbit anti-βIII-tubulin (Abcam), mouse anti-Synaptophysin (Sigma), mouse anti-β-actin (Sigma). The following day, blots were washed in TBS-T and incubated with HRP coupled secondary antibodies according to the manufacturer’s instructions. Signals were revealed using ECL Super signal West Femto (ThermoScientific cat no 34095).
In utero hippocampal electroporation and spine analysis
In utero electroporation targeting the hippocampus was performed using a triple-electrode setup as previously described (dal Maschio et al., 2012) to target hippocampal CA1 PN progenitors at E15.5. Plasmids were injected into the lateral ventricle (pCAG-Cre: 0.3μg/μl, pEF1::FLEX-tdTomato or mCherry 0.5μg/μl; Iascone et al., 2020) followed by 5 pulses at 45V (50ms duration, 500ms interpulse interval). Mice were perfused transcardially with PFA 4% at P21 and sectioned at 200μm on a vibratome (Leica). Sections were mounted and imaged on a Nikon W1 spinning disk microscope using a 100x, 1.45 NA silicon-immersion objective. Spine analysis was carried out using vaa3d (detection parameters as in Iascone et al., 2020) by three independent experimenters blinded to experimental condition.
In vitro electrophysiology
Slice preparation
Mice of either sex (total of n = 8, n = 3 female, n = 5 male, between P22-P27) were anesthetized with isoflurane, decapitated and the brain was placed in ice cold dissection solution containing (in mM): 195 sucrose, 10 NaCl, 15 glucose, 26 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 1 CaCl2*2H2O, 2 MgCl2*6H20. 300 or 400 um-thick horizontal slices were cut using a Leica VT1200 vibratome. After the dissection, slices were incubated in an ACSF-containing submerged chamber at 32°C for 30 minutes, then stored at room temperature until used for recording. ASCF contained the following (in mM): 125 NaCl, 10 glucose, 26 NaHCO3, 2 Na-pyruvate, 2.5 KCl, 1.25 NaH2PO4, 2 CaCl2*2H2O, 1 MgCl2*6H20. Brain slices were used until up to 6h post-dissection. Both dissection and ACSF solutions were carbonated to saturation, resulting in a pH of 7.3–7.4 and osmolality of 310 ± 5 mOsm.
Whole-cell recordings
To isolate EPSPs, whole-cell current clamp recordings were performed using a K-gluconate-based internal solution (in mM: 130 K–gluconate, 8 KCl, 10 HEPES, 4 NaCl, 4 Mg-ATP, 0.3 Tris-GTP, 14 (tris-)phosphocreatine) and 6 biocytin with pH = 7.28 and osmolality 295 mOsm) at 32°C in ACSF with the addition of (in uM): 1 SR95531, 2 CGP55845 and 1 TTX to block GABAA receptors, GABAB receptors and voltage-gated sodium channels, respectively. For IPSP recordings, the internal solution contained (in mM) 140 K-gluconate, 2 KCl, 10 HEPES, 5 EGTA, 4 Mg-ATP, 0.5 Na-GTP, 5 creatine phosphate) and 6 biocytin with pH = 7.2 and osmolality 300 mOsm) at 32°C in ACSF with the addition of (in uM): 50 APV, 10 CNQX and 1 TTX to block NMDA receptors, AMPA receptors and voltage-gated sodium channels, respectively. Thick-walled borosilicate glass pipettes (outer diam.: 1.5 mm, inner diam.: 0.86 mm; Sutter Instruments) were pulled with a P-97 pipette puller (Sutter Instruments). Open tip resistance when filled with internal solution was 1.7–5.7 MΩ. Data were acquired with using a MultiClamp 700B amplifier (Molecular Devices) and a CED Micro1401–3 digitizer (Cambridge Electronic Design Limited). Data were digitized at 50 kHz and low-pass filtered at 2 kHz. Series resistance and pipette capacitance were monitored continuously and compensated throughout the 6–10-minute recording sessions. Data were discarded when series resistance exceeded 20 MΩ and the holding current decreased below −100 pA for −60 mV for EPSP recordings. To record IPSPs, positive holding current was injected to keep the cells around −55 mV. After sequentially patching one KO (identified by mCherry fluorescence, see Figure 2C) and a neighboring WT CA1 pyramidal cell, the brain slices were fixed in 4% PFA solution for post hoc histological processing. EPSPs were detected by a template matching algorithm and analyzed in Stimfit (Guzman et al., 2014).
In vitro hemisynapse assay and immunocytochemistry
Mixed culture assays were performed as previously described (de Wit et al., 2013). Briefly, HEK293T cells were transfected with cDNAs for NLG1, CD8, Robo1, Robo2, Robo3, Robo2ΔIg1,2 using Fugene6 (Promega), mechanically dissociated and cocultured with 11 DIV cortical neurons for 8–24 hr, depending on the experiment. Following coculture, the cells were immunostained for Vglut1, Vgat1, GFP, myc and HA.
For the analysis of heparinase III treatment, neurons (11 DIV) were treated with 1 U/ml heparinase III (Sigma-Aldrich) or vehicle (20 mM Tris-HCl [pH 7.5], 0.1 mg/ml BSA, 4mM CaCl2) for 2 hr at 37°C. Cells were then washed twice with neuronal feeding medium and subsequently cocultured with transfected HEK293T cells for an additional 8 hr. Cells were then processed for immunostaining. Coverslips were washed once with 500uL of DPBS and fixed with 4%PFA, 1X PBS, 4% sucrose solution at RT for 15 minutes. Cells were washed with PBS 3X for 5 minutes and permeabilized in 0.2% Triton X-100 in 1xPBS for 15mins. Cells were blocked in 1%BSA, 4%NGS for 30mins at RT. Primary incubation was done overnight at 4C: HA (abcam, sheep), GFP (Aves, chicken), Vglut1 (SySy, guinea-pig), Vgat1 (SySy, mouse), Myc (Thermo Fisher, mouse). The following day, cells were washed in 1xPBS 3x for 5mins followed by incubation with secondary antibodies (Invitrogen-Alexa-dye-coupled) for 1h at RT. Cells were then washed in 1xPBS 3x for 5mins and mounted in Fluoromount-G (Southern Biotech cat no 0100–01). Images were then acquired using a NikonA1R confocal microscope using a 60X, 1.4 NA oil-immersion objective. To test the effect of neuronal Robo1/2 knockout, neurons from Robo1−/−;Robo2F/F mice were infected with lentivirus expressing Cre-recombinase under a synapsin promoter at DIV0. To test the effect of neuronal Neurexin knockdown, neurons were infected with lentivirus expressing a previously validated shRNA against Neurexins (Gokce and Südhof, 2013) at DIV3. For quantification of mixed-culture assays, HEK cell outlines were segmented as ROIs using Magic Cell Wand Tool (Fiji Plugin). Then the total fluorescence intensity of Vglut1 puncta was measured over the total GFP-positive area per cell. Measurements were performed in a minimum of three independent preparations and in each experiment for any given condition a minimum of twenty cells were acquired. Imaging and analysis were conducted blind to the condition.
Protein expression and purification for SPR
We used cDNAs encoding rat Robo1 (NCBI: NP 071524.1), rat Robo2 (NCBI: NP 115289.1), and rat α-neurexin (NCBI: NP 068535.2) as templates for PCR amplification of desired coding regions for expression constructs. Each protein was produced into the pLEXm mammalian cell expression vector proceeded by a BiP signal peptide and in frame with a C-terminal octahistidine tag. The Robo1 construct encoded amino acids for Ig 1–5 (Gly58-Phe545). For Robo2, constructs and corresponding reside ranges were Ig1–2 (Gly21-Glu226), Ig1–3 (Gly21-Ala315), Ig1–4 (Gly21-Asp413) and Ig1–5 (Gly21-Ser509). The α-neurexin construct lacked the splice insertion sequences at splice sites 1, 2 and 4 (Δ1Δ2Δ4) and encoded amino acids (Leu31-Thr1336). β-neurexin1 Δ4 encoded Thr1125-Val1327, and β-neurexin1+4 encoded Thr1125-Val1327 (amino acid numbering based on NCBI: NP 068535.2). All proteins were expressed in Human Embryonic Kidney (HEK) 293 Freestyle cells (Invitrogen) in suspension culture using serum-free media. Plasmid constructs were transfected into HEK293 cells using polyethyleneimine (Polysciences). Cell growths were harvested five days after transfection, and the secreted proteins were purified from supernatant using nickel affinity chromatography followed by size exclusion chromatography in either 10 mM Tris pH 8.0 and150 mM sodium chloride for Robo1/2 or 10 mM Tris pH 8.0, 150 mM sodium chloride and 3 mM calcium chloride for neurexins.
SPR binding experiments
SPR binding experiments were performed using a Biacore T100 biosensor equipped with a Series S CM4 sensor chip. α-Neurexin1(−4), β-Neurexin1(−4), β-Neurexin1(+4) (previously cloned in Koehnke et al., 2008; 2010) were immobilized over individual flow cells using amine-coupling chemistry in HBS-P pH 7.4 (10mM HEPES-OH, 150mM NaCl, 0.005% Tween-20) buffer supplemented with 3mM CaCl2, at 32°C using a flow rate of 20 μL/min. Prior to immobilization, the three Neurexin protein samples were buffer exchanged in HBS pH 7.4, 3mM CaCl2 using Zeba spin desalting columns (Thermo Scientific) prior to immobilization. Dextran surfaces were activated for 7 minutes using equal volumes of 0.1 M NHS(N-Hydroxysuccinimide) and 0.4 M EDC(1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide). Each protein of interest was immobilized at ~40 μg/mL in 10 mM sodium acetate, pH 5.5, 3mM CaCl2 until the desired immobilization level was achieved. The immobilized surface was blocked using a 3-minute injection of 1.0 M ethanolamine, pH 8.5. Each molecule was immobilized at 1600–1700 RU. An unmodified surface was used as a reference flow cell to subtract bulk shift refractive index changes.
Binding experiments were performed at 25°C in a running buffer containing 10 mM Tris-Cl pH 8.0, 150 mM NaCl, 3mM CaCl2, 10 μg/mL heparin, 1 mg/mL BSA and 0.005% (v/v) Tween-20. Binding analysis in the absence of heparin was tested in the same buffer lacking heparin and experiments without CaCl2 were performed in a buffer containing 3mM EGTA instead of CaCl2. Robo2 and 1 (Ig1–5) and Robo2 Ig fragment analytes were prepared in running buffer using a three-fold dilution series at 27, 9, 3, 1 and 0.333 μM, except for the EGTA experiment, where binding was tested at the highest concentration of 27 μM. In each binding cycle, analytes were injected over all immobilized surfaces at 50 μL/min for 45 s, followed by 180 s of dissociation phase, a running buffer wash step and a buffer injection at 100 μL/min for 60 s. Each series was tested in order of increasing concentration and then repeated in the same experiment to confirm the reproducibility of the binding assay. Running buffer was used instead of an analyte every two cycles, to double reference the binding responses by removing systematic noise and instrument drift. The binding signal between 40 and 44 s for each analyte, was fit against the Robo concentration using a 1:1 interaction model to calculate the KD. The data was processed and analyzed using Scrubber 2.0 (BioLogic Software).
Fc-Protein Purification for Mass-spectrometry
Fc protein purification was performed as described previously (Savas et al., 2014). Slit2-Fc (N-(aa1–840) and C-terminal (aa841–1290) fragments) proteins were produced by transient transfection of HEK293T cells using PEI (Polysciences). Six hours after transfection, media was changed to OptiMEM (Invitrogen) and harvested 5 days later. Conditioned media was centrifuged, sterile-filtered and run over a fast-flow Protein-G agarose (Thermo-Fisher) column. After extensive washing with wash buffer (50 mM HEPES pH 7.4, 300 mM NaCl and protease inhibitors), the column was eluted with Pierce elution buffer. Eluted fractions containing proteins were pooled and dialyzed with PBS using a Slide-A-Lyzer (Pierce) and concentrated using Amicon Ultra centrifugal units (Millipore). The integrity and purity of the purified ecto-Fc proteins was confirmed with SDS-PAGE and Coomassie staining, and concentration was determined using a Bradford protein assay.
Affinity Chromatography for Mass-spectrometry
Affinity chromatography experiments were performed as previously described (Savas et al., 2014). Crude synaptosome extracts were prepared from ten P21–22 rat brains, homogenized in homogenization buffer (4 mM HEPES pH 7.4, 0.32 M sucrose and protease inhibitors) using a Dounce homogenizer. Homogenate was spun at 1,000 × g for 10 minutes at 4°C. Supernatant was spun at 14,000 × g for 20 minutes at 4°C. P2 crude synaptosomes were re-suspended in Extraction Buffer (50 mm HEPES pH 7.4, 0.1 M NaCl, 2 mM CaCl2, 2.5 mM MgCl2 and protease inhibitors), extracted with 1% Triton X-100 for 2 hours and centrifuged at 100,000 × g for 1 hour at 4°C to pellet insoluble material. Fast-flow Protein-A Sepharose beads (GE Healthcare) (250 μl slurry) pre-bound in Extraction Buffer to 100 μg human Fc or Slit2-Fc were added to the supernatant and rotated overnight at 4°C.
Beads were packed into Poly-prep chromatography columns (BioRad) and washed with 50 mL of high-salt wash buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 0.1 mM CaCl2, 5% glycerol and protease inhibitors), followed by a wash with 10 mL low-salt wash buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 0.1 mM CaCl2, 5% glycerol and protease inhibitors). Bound proteins were eluted from the beads by incubation with Pierce elution buffer and TCA-precipitated overnight. The precipitate was re-suspended in 8 M Urea with ProteaseMax (Promega) per the manufacturer’s instruction. The samples were subsequently reduced by 20-minute incubation with 5mM TCEP0 (tris(2carboxyethyl)phosphine) at RT and alkylated in the dark by treatment with 10 mM Iodoacetamide for 20 additional minutes. The proteins were digested overnight at 37°C with Sequencing Grade Modified Trypsin (Promega) and the reaction was stopped by acidification. Mass spectrometry analysis was performed by the VIB Proteomics Core (Leuven, Belgium).
Stereotactic virus injection and craniotomy
For imaging experiments, recombinant adeno-associated viruses carrying the GCaMP6f gene were obtained from the Penn Vector Core (AAV1.Syn.GCaMP6f.WPRE.SV40) with titer of 2–4 × 1013. Dorsal CA1 was stereotactically injected at −2.0 mm AP; −1.5 mm ML; and −0.9, −1.2, −1.4 mm DV relative to the cortical surface. Mice were then surgically implanted with an imaging window over the left dorsal CA1 and implanted with a stainless-steel headpost for head fixation during imaging experiments.
In vivo two-photon calcium imaging
We used the same imaging system as described previously (Danielson et al., 2016; Turi et al., 2019). All images were acquired with a Nikon 40 × NIR water-immersion objective (0.8 NA, 3.5 mm WD) in distilled water. For excitation, we used a Chameleon, Ultra II (Coherent) laser tuned to 920 nm, and a Fidelity-2 (Coherent) laser at 1070 nm. We continuously acquired red (mCherry) and green (GCaMP6f) channels separated by an emission cube set (green, HQ525/70 m-2p; red, HQ607/45 m-2p; 575dcxr, Chroma Technology) at 512 × 512 pixels covering 330 μm × 330 μm at 30 Hz with photomultiplier tubes (green GCaMP6f fluorescence, GaAsP PMT, Hamamatsu Model 7422P-40; red mCherry fluorescence, GaAsP PMT Hamamatsu). For four mice, red and green channels were recorded simultaneously with both lasers simultaneously exciting. For three mice, only the green channel was excited/recorded during behavior, while a red-only image for cell identification was acquired immediately prior to recording. A custom dual stage preamp (1.4 × 105 dB, Bruker) was used to amplify signals prior to digitization.
For training, mice were water restricted (> 90% pre-deprivation weight) and trained to run on a cue-poor burlap treadmill belt for a non-operantly delivered water reward over the course of 1–2 weeks. We applied a progressively restrictive water reward schedule, with mice initially receiving 12 randomly placed reward zones per lap and ultimately receiving 1 randomly placed reward zone per lap. Mice were habituated to the optical instrumentation, then trained for 20 min daily until they regularly ran at least one lap per minute. During imaging sessions, mice received one randomly placed water reward per lap, with water delivered for every subsequent lick inside the reward zone for a maximum of 2.5 s. The reward zone position was changed randomly each lap.
RT-qPCR
For mRNA expression analysis, hippocampi were dissected from fresh brains in 1X ice cold PBS and RNA was extracted using the RNeasy Lipid Tissue Mini kit (QIAGEN). RNA (2 μg) was reverse-transcribed using the QuantiTect Reverse Transcription kit (QIAGEN). RT–qPCR was performed using SYBR Green (Applied Biosystems) on RealPlex2 MasterCycler (Eppendorf) and samples ran in triplicates. The primer sets were designed using the NCBI Primer Design Tool. Primer efficiencies were tested using dilution series of cDNA. Data were analyzed using the dCt method normalizing to hypoxanthine-guanine phosphoribosyltransferase (Hprt).
The primers used for RT–qPCR were:
Robo2-Ex4/5
Fwd: AGAGCTCACTGTCTTTGAACG; Rev: GGGATCTCCTTGGACCTGAC
Robo2-Ex5/6
Fwd: GCCAACGGTGAGGTGGAAAA; Rev: AACTGTGGAGGAGCAACAGG
Robo2–27/28
Fwd: AGCAGCCAACTTAGAAGACACA; Rev: CACCAGGGACTCCTCTGATCC
Hprt
Fwd: AGCAGGTGTTCTAGTCCTGTGG, Rev: ACGCAGCAACTGACATTTCTAA
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis, group sizes and p values can be found in the respective figure legends.
Imaging Analysis
Processing of GCaMP6f/Ca2+ Fluorescence Data
Imaging data were processed using the SIMA (Kaifosh et al., 2014), Suite2p (https://www.biorxiv.org/content/10.1101/061507v2), and OASIS (Friedrich et al., 2017) software packages. Motion correction was performed by concatenating all imaging sessions for a field of view and using the Suite2p rigid motion correction strategy. Only the GCaMP6f channel was used for estimating motion artifacts. Regions of interest (ROIs) were then drawn over putative CA1 pyramidal cell somata visible in the cross-session GCaMP6f channel time-average image. To prevent the introduction of any bias, the red mCherry channel time-average image was not viewed when drawing ROIs but only referenced after all ROIs had been drawn in order to tag ROIs over mcherry-expressing cells as Robo2 KO cells. In cases where no red channel was simultaneously recorded, an affine transformation was calculated to align the red-only image acquired just prior to imaging to the GCaMP6f channel time-average image.
Dynamic GCaMP6f fluorescence signals for each imaging session were extracted from the binary ROIs using SIMA. One session with dropped data during the last > 1 minute of recording was removed from analysis. Deconvolved spikes were computed for each ROI for each imaging session according to the following procedure: a baseline was calculated by smoothing the fluorescence trace with a Gaussian filter (std = 5 frames, window = 30 s), calculating the rolling minimum of this smoothed trace, (window = 30 s), and then calculating the rolling maximum of this last trace (window = 30). A preliminary spike train was estimated by deconvolving the baseline-subtracted trace with OASIS according to an AR1 model with penalty and pre-computed decay parameter using 400 ms for the GCaMP6f decay time. Using this spike train, a noise threshold was identified by computing the median and median absolute deviation (MAD) of the baseline-subtracted trace where no spikes were detected (i.e., putative noise-only time points). The noise threshold was defined as the median plus 1.5 MADs. A new spike train was then computed as before, now with the minimum spike size set explicitly to the noise threshold, and sparsity parameter, set to 3.
Spatial Tuning Analysis
When evaluating the spatial tuning of PCs, we restricted our analysis to running-related epochs, defined as consecutive frames of forward locomotion of at least 1 s in duration and with a minimum peak speed of 5 cm/sec. Consecutive epochs separated by < 0.5 s were merged. Position was discretized into 100 2cm bins. Deconvolved spikes were smoothed with a Gaussian filter (std = 1 frame) and binarized. The spatial tuning vector was calculated as, where is the number of running frames with spikes occurring at position i and is the number of running frames acquired at position i. In order to assess the significance of the spatial selectivity, for each cell we generated a null tuning distribution by cyclically permuting the position vector (restricted to running frames) by a random offset and repeatedly recomputing the tuning vector. This process was repeated 1,000 times. The true and null tuning vectors were then smoothed with a Gaussian (std = 3 position bins). Place fields were identified as at least 5 consecutive position bins above the 95th percentile of the null distribution, in which the cell fired on at least 15% of laps.
Statistics
All tests are described in the appropriate figure legends with details in methods section were appropriate. For imaging data, metrics were aggregated across all cells and sessions for a FOV, and FOVs were the unit of analysis for all statistical tests conducted. A paired Student’s t test was used for comparison of means in paired data with n–1 degrees of freedom. For the decoding analysis, a repeated-measures two-way ANOVA with interaction was used to test the difference between subpopulation means across surrogate population sizes.
Supplementary Material
KEY RESOURCES TABLE.
| Reagent or resource | Source | Identifier |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-FLRT2, goat | R&D Systems | Cat #AF2877; RRID: AB_2106600 |
| Anti-Robo2 | Aviva Systems Biology | Cat#: ARP45396_P050, RRID:AB_2047840 |
| Anti-Bassoon, guinea pig | Synaptic Systems | Cat #141 004; RRID: AB_2290619 |
| Anti-Vglutl, guinea pig | Synaptic Systems | Synaptic Systems Cat# 135 304, RRID:AB_887878 |
| Anti-PSD-95, mouse | ThermoFisher Scientific | Cat #MA1 –046; RRID: AB_2092361 |
| Anti-Synaptophysin, mouse | Sigma | Cat #S5768; RRID: AB_477523 |
| Anti-GFP, chicken | Aves | Cat #GFP-1010; RRID: AB_2307313 |
| Anti-Slit2, rabbit | Proteintech | Cat no: 20217–1-AP |
| Anti-HA, rabbit | Sigma-Aldrich | Cat #H6908; RRID: AB_260070 |
| Anti-β-actin, mouse | Sigma | Cat #A2228; RRID: AB_476697 |
| Anti-βIII-tubulin, rabbit | Abcam | Cat #ab18207; RRID: AB_444319 |
| Anti-HA, sheep | Abcam | discontinued |
| Anti-c-myc, mouse | Thermo Fisher | Cat# MA5–11265, RRID:AB_10981641 |
| Alexa 488, goat-anti-mouse | Invitrogen | Cat# A-11001, RRID: AB_2534069 |
| Alexa 555, goat-anti-guinea-pig | Invitrogen | Cat# A-21435, RRID: AB_2535856 |
| Alexa 647, donkey-anti-rabbit | Invitrogen | Cat# A-31573, RRID: AB_2536183 |
| Alexa 488, goat-anti-chicken | Invitrogen | Cat# A32723, RRID: AB_2633275 |
| Alexa 488, donkey-anti-sheep | Invitrogen | Cat# A-11015, RRID: AB_2534082 |
| pAAV.Syn.GCaMP6f.WPRE.SV40 | Addgene/UPenn | Cat#100837-AAV1 |
| FuGENE 6 Transfection Reagent | Promega | Cat# E2691 |
| 5-Fluoro-2′-deoxyuridine | Millipore Sigma | Cat#F0503–100MG |
| Penicillin-Streptomycin | Millipore-GIBCO | Cat#15070063 |
| Polyethylenimine | Polysciences | Cat# 23966 |
|
| ||
| Experimental Models: Cell lines | ||
|
| ||
| HEK293T/17 | ATCC | Cat# CRL-11268, RRID: CVCL_1926 |
| FreeStyle 293-F Cells | Invitrogen | Cat# R79007 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Robo2F/F | Gift from Alain Chédotal | Lu et al., 2007 |
| Robo1−/−;Robo2F/F | Gift from Alain Chédotal | Domyan et al., 2013 |
| Neurod6tm1(cre)Kan | Gift from Klaus-Armin Nave | Goebbels et al., 2006 |
| Tg(Lypd1-cre)NR149Gsat | MMRC/GENSAT | RRID:MMRRC_034612-UCD |
| B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | Stock# 007909 |
| Crl:CD1(ICR) | Charles River | IGS-Mouse, outbred/022 |
| Cloning, sequencing and qPCR primers | EtonBio | N/A |
| shRNA targeting Nrxn1/2/3 | Previously described | Gokce and Sudhof, 2013 |
| pCAG:Cre | Addgene | Cat#13775 |
| pEF1 α:flex-tdtomato | Addgene | Cat#28306 |
| pEF1 α:flex-mcherry | This lab | N/A |
| pCAG:NLG1-HA | This lab | Kwon et al., 2016 |
| pCAG:CD8 | This lab | Kwon et al., 2016 |
| pcDNA3.1:Robo1 | Gift from Alain Chédotal, Institut de la Vision, 17 Rue Moreau, 75012 Paris, France | Zelina et al., 2014 |
| pcDNA3.1:Robo2 | Gift from Alain Chédotal, Institut de la Vision, 17 Rue Moreau, 75012 Paris, France | Zelina et al., 2014 |
| pcDNA3.1:Robo3 | Gift from Dr Alain Chédotal, Institut de la Vision, 17 Rue Moreau, 75012 Paris, France | Zelina et al., 2014 |
| pcDNA3.1:Robo2ΔIg1,2 | Gift from Alain Chédotal, Institut de la Vision, 17 Rue Moreau, 75012 Paris, France | Brose et al., 1999 |
| pCAG-Robo2-pHluorin | Gift from Valérie Castellani, INSTITUT NEUROMYOGENE | Pignata et al., 2019 |
| CNRS UMR 5310 - INSERM U1217 Université de Lyon | ||
| Université Claude Bernard Lyon 1 | ||
| pCAG-Homer1c-tdtomato | This lab | N/A |
| pLEXm-Robo1-Ig1–5 | This lab | N/A |
| pLEXm-Robo2-Ig1–5 | This lab | N/A |
| pLEXm-Robo2-Ig1–2 | This lab | N/A |
| pLEXm-Robo2-Ig1–3 | This lab | N/A |
| pLEXm-Robo2-Ig1–4 | This lab | N/A |
| pLEXm-α-neurexin-Δ1 Δ2Δ4 | This lab | N/A |
| pLEXm-β-neurexin1 Δ4 | This lab | N/A |
| pLEXm-β-neurexin1+4 | This lab | N/A |
| SIMA | Kaifosh et al., 2014 | https://github.com/losonczylab/sima |
| Suite2p | Pachitariu et al., preprint, 2017 | https://github.com/MouseLand/suite2p |
| OASIS | Friedrich et al., 2017 | https://github.com/j-friedrich/OASIS |
| https://zenodo.org/badge/latestdoi/404062875 | ||
| NIS-Elements-Nikon | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements?gclid=Cj0KCQiA0–6ABhDMARIsAFVdQvj1piGkc5l9VPNQlW7D_bbOt8Trve-xeJg0x1-X5nz321nqPi9DPQaAmy3EALw_wcB |
| GraphPadPrism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| ImageStudioLite | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio_lite/ |
| ProLong Gold Antifade Mountant | Thermo Fisher Scientific | Cat#P36930; https://www.thermofisher.com/order/catalog/product/P36930 |
| In-Fusion Cloning Kit | Takara Bio | Cat#638920 https://www.takarabio.com/products/cloning/in-fusion-cloning/in-fusion-hd-cloning-plus-value-bundles |
| Fluoromount-G | Southern Biotech | Cat#0100–01 |
| SYBR-Green | Applied Biosystems | Cat#25742 |
| RNeasy Lipid Tissue Mini Kit | QIAGEN | Cat#74804 |
| QuantiTect Reverse Transcription Kit | QIAGEN | Cat#205311 |
Highlights.
The axon guidance receptors Robo1/2 can function as synaptogenic cues
Postsynaptic Robo forms a trans-synaptic complex with Slit and presynaptic Neurexin
Robo2 is required for formation of a subset of excitatory CA3 to CA1 synapses
Sparse deletion of Robo2 in CA1 pyramidal neurons impairs emergence of place cells
ACKNOWLEDGMENTS
We thank Miyako Hirabayashi and Qiaolian Liu for technical assistance. We also thank Dr Alain Chédotal for providing Robo2 conditional and Robo1 constitutive knockout mice and Dr Klaus-Armin Nave for providing the Nex-Cre mouse line. We thank members of the Polleux and Losonczy labs for stimulating discussions. We would like to thank the Zuckerman Institute Cellular Imaging platform for instrument use and technical advice. This work was supported by NIH-NINDS (RO1NS067557) (F.P.), NIH-NINDS (R21NS109753–01A1) (F.P., A.L., H.B) and NIH-NINDS (K99NS115984–01) (H.B.), an award from the Fondation Roger De Spoelberch (F.P.) and a grant from the Nomis Foundation (F.P.). A.S. was supported by FWO PhD fellowship (11Z3715N/17N); J.d.W. is supported by FWO Odysseus grant, FWO project grant G0C4518N, and FWO EOS grant G0H2818N. Mass spectrometry analysis was performed at the VIB Proteomics Core (Leuven, Belgium). A.L. is further supported by the National Institute of Mental Health (NIMH) 1R01MH124047, 1R01MH124867 National Institute of Neurological Disorders and Stroke (NINDS) 1U19NS104590 and 1U01NS115530, the Zegar Family Foundation Award, and the Kavli Foundation. B.H. is supported by a National Science Foundation grant (MCB-1412472), L.S. by NIH grant R01MH114817.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109828.
REFERENCES
- Barak R, Yom-Tov G, Guez-Haddad J, Gasri-Plotnitsky L, Maimon R, Cohen-Berkman M, McCarthy AA, Perlson E, Henis-Korenblit S, Isupov MN, and Opatowsky Y (2019). Structural Principles in Robo Activation and Auto-inhibition. Cell 177, 272–285.e16. [DOI] [PubMed] [Google Scholar]
- Blockus H, and Chédotal A (2016). Slit-Robo signaling. Development 143, 3037–3044. [DOI] [PubMed] [Google Scholar]
- Blockus H, and Polleux F (2021). Developmental mechanisms underlying circuit wiring: Novel insights and challenges ahead. Curr. Opin. Neurobiol. 66, 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, and Kidd T (1999). Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806. [DOI] [PubMed] [Google Scholar]
- Brun VH, Otnass MK, Molden S, Steffenach HA, Witter MP, Moser MB, and Moser EI (2002). Place cells and place recognition maintained by direct entorhinal-hippocampal circuitry. Science 296, 2243–2246. [DOI] [PubMed] [Google Scholar]
- Bülow HE, Tjoe N, Townley RA, Didiano D, van Kuppevelt TH, and Hobert O (2008). Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Curr. Biol. 18, 1978–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, and Chien CB (2007). Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron 55, 231–245. [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, and Siekevitz P (1980). Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 86, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Sima J, Jin M, Wang KY, Xue XJ, Zheng W, Ding YQ, and Yuan XB (2008). Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11, 36–44. [DOI] [PubMed] [Google Scholar]
- Chen JL, Villa KL, Cha JW, So PT, Kubota Y, and Nedivi E (2012). Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron 74, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchet J, Lewis TL Jr., Lee S, Courchet V, Liou DY, Aizawa S, and Polleux F (2013). Terminal axon branching is regulated by the LKB1-NUAK1 kinase pathway via presynaptic mitochondrial capture. Cell 153, 1510–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dal Maschio M, Ghezzi D, Bony G, Alabastri A, Deidda G, Brondi M, Sato SS, Zaccaria RP, Di Fabrizio E, Ratto GM, and Cancedda L (2012). High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat. Commun. 3, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NB, Zaremba JD, Kaifosh P, Bowler J, Ladow M, and Losonczy A (2016). Sublayer-Specific Coding Dynamics during Spatial Navigation and Learning in Hippocampal Area CA1. Neuron 91, 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoudi H, and Foster DJ (2019). Acute silencing of hippocampal CA3 reveals a dominant role in place field responses. Nat. Neurosci. 22, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2014). Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 37, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2016). Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 22–35. [DOI] [PubMed] [Google Scholar]
- de Wit J, O’Sullivan ML, Savas JN, Condomitti G, Caccese MC, Vennekens KM, Yates JR 3rd, and Ghosh A (2013). Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron 79, 696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delloye-Bourgeois C, Jacquier A, Charoy C, Reynaud F, Nawabi H, Thoinet K, Kindbeiter K, Yoshida Y, Zagar Y, Kong Y, et al. (2015). PlexinA1 is a new Slit receptor and mediates axon guidance function of Slit C-terminal fragments. Nat. Neurosci. 18, 36–45. [DOI] [PubMed] [Google Scholar]
- Dittgen T, Nimmerjahn A, Komai S, Licznerski P, Waters J, Margrie TW, Helmchen F, Denk W, Brecht M, and Osten P (2004). Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl. Acad. Sci. USA 101, 18206–18211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Branchfield K, Gibson DA, Naiche LA, Lewandoski M, Tessier-Lavigne M, Ma L, and Sun X (2013). Roundabout receptors are critical for foregut separation from the body wall. Dev. Cell 24, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorskind JM, and Kolodkin AL (2021). Revisiting and refining roles of neural guidance cues in circuit assembly. Curr. Opin. Neurobiol. 66, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TA, Santiago C, Arbeille E, and Bashaw GJ (2015). Robo2 acts in trans to inhibit Slit-Robo1 repulsion in pre-crossing commissural axons. eLife 4, e08407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Zhou P, and Paninski L (2017). Fast online deconvolution of calcium imaging data. PLoS Comput. Biol. 13, e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo MV, Földy C, Gökce Ö, Rothwell PE, Sun GL, Malenka RC, and Südhof TC (2015). Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron 87, 326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DA, Tymanskyj S, Yuan RC, Leung HC, Lefebvre JL, Sanes JR, Chédotal A, and Ma L (2014). Dendrite self-avoidance requires cell-autonomous slit/robo signaling in cerebellar purkinje cells. Neuron 81, 1040–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SD, Ruthazer ES, and Kennedy TE (2021). Guiding synaptic plasticity: Novel roles for netrin-1 in synaptic plasticity and memory formation in the adult brain. J. Physiol. 599, 493–505. [DOI] [PubMed] [Google Scholar]
- Glasgow SD, Wong EW, Thompson-Steckel G, Marcal N, Séguéla P, Ruthazer ES, and Kennedy TE (2020). Pre- and post-synaptic roles for DCC in memory consolidation in the adult mouse hippocampus. Mol. Brain 13, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, and Nave KA (2006). Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621. [DOI] [PubMed] [Google Scholar]
- Gokce O, and Südhof TC (2013). Membrane-tethered monomeric neurexin LNS-domain triggers synapse formation. J. Neurosci. 33, 14617–14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U, Le Ma, Plump AS, Wang F, Tessier-Lavigne M, and Martin GR (2004). SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev. Cell 6, 709–717. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, and Polleux F (2009). The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman SJ, Schlögl A, and Schmidt-Hieber C (2014). Stimfit: quantifying electrophysiological data with Python. Front. Neuroinform. 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson NT, and Dalva MB (2018). EphBs and ephrin-Bs: Trans-synaptic organizers of synapse development and function. Mol. Cell. Neurosci. 91, 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking JC, Hehr CL, Bertolesi GE, Wu JY, and McFarlane S (2010). Distinct roles for Robo2 in the regulation of axon and dendrite growth by retinal ganglion cells. Mech. Dev. 127, 36–48. [DOI] [PubMed] [Google Scholar]
- Hunyara JL, and Kolodkin AL (2020). Repurposing developmental mechanisms in the adult nervous system. Curr. Opin. Genet. Dev. 65, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iascone DM, Li Y, Sümbül U, Doron M, Chen H, Andreu V, Goudy F, Blockus H, Abbott LF, Segev I, et al. (2020). Whole-neuron synaptic mapping reveals spatially precise excitatory/inhibitory balance limiting dendritic and somatic spiking. Neuron 106, 566–578.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Lovett-Barron M, Turi GF, Reardon TR, and Losonczy A (2013). Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat. Neurosci. 16, 1182–1184. [DOI] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, and Losonczy A (2014). SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform. 8, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Jin X, Trbovic N, Katsamba PS, Brasch J, Ahlsen G, Scheiffele P, Honig B, Palmer AG 3rd, and Shapiro L (2008). Crystal structures of beta-neurexin 1 and beta-neurexin 2 ectodomains and dynamics of splice insertion sequence 4. Structure 16, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke J, Katsamba PS, Ahlsen G, Bahna F, Vendome J, Honig B, Shapiro L, and Jin X (2010). Splice form dependence of beta-neurexin/neuroligin binding interactions. Neuron 67, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropouli E, and Kolodkin AL (2014). Semaphorins and the dynamic regulation of synapse assembly, refinement, and function. Curr. Opin. Neurobiol. 27, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner RH, Zhang XY, and Reiser J (2009). Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 4, 495–505. [DOI] [PubMed] [Google Scholar]
- Kwon SK, Sando R 3rd, Lewis TL, Hirabayashi Y, Maximov A, and Polleux F (2016). LKB1 Regulates Mitochondria-Dependent Presynaptic Calcium Clearance and Neurotransmitter Release Properties at Excitatory Synapses along Cortical Axons. PLoS Biol 14, e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Paratcha G, Sandoval-Guzmán T, and Ibáñez CF (2007). GDNF and GFRalpha1 promote formation of neuronal synapses by ligand-induced cell adhesion. Nat. Neurosci. 10, 293–300. [DOI] [PubMed] [Google Scholar]
- Liu Z, Patel K, Schmidt H, Andrews W, Pini A, and Sundaresan V (2004). Extracellular Ig domains 1 and 2 of Robo are important for ligand (Slit) binding. Mol. Cell. Neurosci. 26, 232–240. [DOI] [PubMed] [Google Scholar]
- Lovett-Barron M, Kaifosh P, Kheirbek MA, Danielson N, Zaremba JD, Reardon TR, Turi GF, Hen R, Zemelman BV, and Losonczy A (2014). Dendritic inhibition in the hippocampus supports fear learning. Science 343, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, van Eerde AM, Fan X, Quintero-Rivera F, Kulkarni S, Ferguson H, Kim HG, Fan Y, Xi Q, Li QG, et al. (2007). Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am. J. Hum. Genet. 80, 616–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton SJ, and McHugh TJ (2016). Silencing CA3 disrupts temporal coding in the CA1 ensemble. Nat. Neurosci. 19, 945–951. [DOI] [PubMed] [Google Scholar]
- Miesenböck G, De Angelis DA, and Rothman JE (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195. [DOI] [PubMed] [Google Scholar]
- Mizumori SJ, McNaughton BL, Barnes CA, and Fox KB (1989). Preserved spatial coding in hippocampal CA1 pyramidal cells during reversible suppression of CA3c output: Evidence for pattern completion in hippocampus. J. Neurosci. 9, 3915–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Young JZ, McHugh TJ, Buhl DL, and Tonegawa S (2008). Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science 319, 1260–1264. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, and Tonegawa S (2002). Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science 297, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet KT, Brose K, Marillat V, Kidd T, Goodman CS, Tessier-Lavigne M, Sotelo C, and Chédotal A (1999). Slit2-Mediated chemorepulsion and collapse of developing forebrain axons. Neuron 22, 463–473. [DOI] [PubMed] [Google Scholar]
- O’Keefe J, and Dostrovsky J (1971). The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 34, 171–175. [DOI] [PubMed] [Google Scholar]
- Pignata A, Ducuing H, Boubakar L, Gardette T, Kindbeiter K, Bozon M, Tauszig-Delamasure S, Falk J, Thoumine O, and Castellani V (2019). A spatiotemporal sequence of sensitization to Slits and semaphorins orchestrates commissural axon navigation. Cell Rep. 29, 347–362.e5. [DOI] [PubMed] [Google Scholar]
- Piskorowski R, Santoro B, and Siegelbaum SA (2011). TRIP8b splice forms act in concert to regulate the localization and expression of HCN1 channels in CA1 pyramidal neurons. Neuron 70, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Giger RJ, Ginty DD, Kolodkin AL, and Ghosh A (1998). Patterning of cortical efferent projections by semaphorin-neuropilin interactions. Science 282, 1904–1906. [DOI] [PubMed] [Google Scholar]
- Polleux F, Morrow T, and Ghosh A (2000). Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature 404, 567–573. [DOI] [PubMed] [Google Scholar]
- Ronca F, Andersen JS, Paech V, and Margolis RU (2001). Characterization of Slit protein interactions with glypican-1. J. Biol. Chem. 276, 29141–29147. [DOI] [PubMed] [Google Scholar]
- Sando R, Jiang X, and Südhof TC (2019). Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, De Wit J, Comoletti D, Zemla R, Ghosh A, and Yates JR 3rd. (2014). Ecto-Fc MS identifies ligand-receptor interactions through extracellular domain Fc fusion protein baits and shotgun proteomic analysis. Nat. Protoc. 9, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, and Serafini T (2000). Neuroligin expressed in non-neuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Savas JN, Herzog E, Brose N, and de Wit J (2017). Synapse biology in the ‘circuit-age’—paths toward molecular connectomics. Curr. Opin. Neurobiol. 42, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, and de Wit J (2018). Leucine-rich repeat-containing synaptic adhesion molecules as organizers of synaptic specificity and diversity. Exp. Mol. Med. 50, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, and Cowan CW (2010). Guidance molecules in synapse formation and plasticity. Cold Spring Harb. Perspect. Biol. 2, a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC (2018). Towards an understanding of synapse formation. Neuron 100, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nguyen AQ, Nguyen JP, Le L, Saur D, Choi J, Callaway EM, and Xu X (2014). Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell Rep. 7, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, and Kolodkin AL (2009). Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature 462, 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turi GF, Li WK, Chavlis S, Pandi I, O’Hare J, Priestley JB, Grosmark AD, Liao Z, Ladow M, Zhang JF, et al. (2019). Vasoactive intestinal polypeptide-expressing interneurons in the hippocampus support goal-oriented spatial learning. Neuron 101, 1150–1165.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chiu SL, Koropouli E, Hong I, Mitchell S, Easwaran TP, Hamilton NR, Gustina AS, Zhu Q, Ginty DD, et al. (2017). Neuropilin-2/PlexinA3 receptors associate with GluA1 and mediate Sema3F-dependent homeostatic scaling in cortical neurons. Neuron 96, 1084–1098.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chédotal A, and Ghosh A (2002). Regulation of cortical dendrite development by Slit-Robo interactions. Neuron 33, 47–61. [DOI] [PubMed] [Google Scholar]
- Wu H, Barik A, Lu Y, Shen C, Bowman A, Li L, Sathyamurthy A, Lin TW, Xiong WC, and Mei L (2015). Slit2 as a β-catenin/Ctnnb1-dependent retrograde signal for presynaptic differentiation. eLife 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelina P, Blockus H, Zagar Y, Péres A, Friocourt F, Wu Z, Rama N, Fouquet C, Hohenester E, Tessier-Lavigne M, et al. (2014). Signaling switch of the axon guidance receptor Robo3 during vertebrate evolution. Neuron 84, 1258–1272. [DOI] [PubMed] [Google Scholar]
- Zhang P, Lu H, Peixoto RT, Pines MK, Ge Y, Oku S, Siddiqui TJ, Xie Y, Wu W, Archer-Hartmann S, et al. (2018). Heparan sulfate organizes neuronal synapses through neurexin partnerships. Cell 174, 1450–1464.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, and Sanes JR (2010). Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell 143, 343–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 2P Ca2+ imaging data reported in this paper will be shared by the lead contact upon request.
The SIMA code base is deposited at https://github.com/losonczylab/sima and is publicly available. Suite2p and OASIS are both open source. Our Github repository is archived here: https://doi.org/10.5281/zenodo.5484268.
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.


