Abstract
Background
NAC (NAM, ATAF and CUC) transcription factors (TFs) play vital roles in plant development and abiotic stress tolerance. Salt stress is one of the most limiting factors for rice growth and production. However, the mechanism underlying salt tolerance in rice is still poorly understood.
Results
In this study, we functionally characterized a rice NAC TF OsNAC3 for its involvement in ABA response and salt tolerance. ABA and NaCl treatment induced OsNAC3 expression in roots. Immunostaining showed that OsNAC3 was localized in all root cells. OsNAC3 knockout decreased rice plants’ sensitivity to ABA but increased salt stress sensitivity, while OsNAC3 overexpression showed an opposite effect. Loss of OsNAC3 also induced Na+ accumulation in the shoots. Furthermore, qRT-PCR and transcriptomic analysis were performed to identify the key OsNAC3 regulated genes related to ABA response and salt tolerance, such as OsHKT1;4, OsHKT1;5, OsLEA3–1, OsPM-1, OsPP2C68, and OsRAB-21.
Conclusions
This study shows that rice OsNAC3 is an important regulatory factor in ABA signal response and salt tolerance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03333-7.
Keywords: Salinity, Abscisic acid, Salt tolerance, Transcription factor, Rice
Background
Rice (Oryza sativa L.) is the major staple cereal food for over one-third of the world’s population. Unfortunately, rice yield and grain quality are often affected by adverse environmental factors such as salinity, drought, or inappropriate temperature [1–3]. Especially, high salinity severely impacts rice growth and harvest [4]. During evolution, plants developed numerous sophisticated but effective strategies to counter such adverse conditions. For instance, plant transcription factors (TFs) can activate a series of stress-related genes to synthesize diverse functional proteins that enable plant survival. TFs WRKY [5], bZIP [6], MYB [7], and NAC [8] have been well characterized for their important roles in plant stress or defense responses.
Among the stress-related TFs, the NAC (NAM, ATAF, CUC1/2) family of TFs has acquired great attention for their role in abiotic stress resistance [9]. So far, 151 rice and 117 Arabidopsis NAC proteins have been identified [10]. Normally, NAC proteins consist of a conserved DNA-binding domain (NAC domain) in the N-terminal region; on the contrary, the transcriptional regulating C-terminal domain usually varies in both amino acid composition and biological function [11]. NAM from petunia [12] and CUC2 from Arabidopsis [13] are one of the first characterized NAC genes, which participate in the shoot apical meristem development. NAC genes have received significant attention for their regulatory role in plant organ development and stress resistance [11]. For example, AtNAC1 regulates lateral root development in Arabidopsis via auxin signal transduction [14]. OsNAC2 gets significantly upregulated during leaf senescence and dramatically accelerates the process in rice [15]. Downregulation of ONAC122 or ONAC131 enhances rice susceptibility to Magnaporthe grisea [16]. ABA, salt, drought and cold stresses induced SNAC2 conferring drought and salt tolerance in transgenic rice [17]. The RD26 gene is an important regulator of ABA-dependent stress response in Arabidopsis [18].
Apart from stress-related TFs that directly regulate the expression of abiotic stresses related genes, phytohormones such as ABA also play a critical role in plant abiotic resistance [19]. Stress conditions upregulate the ABA-biosynthesis genes to increase the ABA content [20]. Notably, under abiotic stresses, NAC genes can also regulate the ABA signal transduction pathway. For example, in rice, over-expression of OsNAC2 confer to drought and salt resistance and ABA biosynthesis gene OsNCED3 is upregulated [21]. OsNAP expression profile in rice showed that ABA and abiotic stresses such as high salinity, drought, and low temperature significantly induced the expression level of OsNAP [22]. Transgenic plants overexpressing OsNAC52 showed high sensitivity to ABA [23].
Previously, OsNAC3 was suggested to be involved in rice stress responses [24]. However, the exact functions of OsNAC3 were not investigated. Here, we examined the expression profile of OsNAC3 and physiological phenotypes using its knockout and overexpressing transgenic rice lines. High throughput RNA-seq assay was performed to find the downstream target genes of OsNAC3. We show that OsNAC3 positively regulates the ABA pathway and salt tolerance in rice via the regulation of OsHKT1;4, OsHKT1;5, OsLEA3–1, OsPM-1, OsPP2C68, and OsRAB-21. These findings can help the development of better salt-tolerant crops.
Results
Sequence analysis of OsNAC3
The full-length open reading frame of OsNAC3 (Os07g0225300) was cloned based on the Rice Annotation Project Database (RAP-DB, http://rapdb.dna.affrc.go.jp/). We used the OsNAC3 amino acid sequence as a query to search for its homologs in rice and Arabidopsis, and 17 homologs with more than 75% identity were obtained. These proteins, together with OsNAC3 and OsNAC45, were used to construct the phylogenetic tree. Phylogenetic analysis showed that OsNAC3 is closely related to the NAC family of TFs (SNAC1 and OsNAC4) in rice (Additional file 1 Fig. S1 A). It contains a single exon encoding the 277 amino acids protein with a highly conserved N-terminal NAC domain, which can be divided into five subdomains(A-E)(Additional file 1 Fig. S1 B). Additionally, many stress-related cis-acting elements were found in the promoter region (2 kb upstream from the start codon) of the OsNAC3 gene, including nine MYB binding sites, six MYC binding sites, three W-boxes, two ABRE(ABA-responsive element)sites, and three As-1 sites (Additional file 1 Fig. S1 C).
Expression pattern of OsNAC3
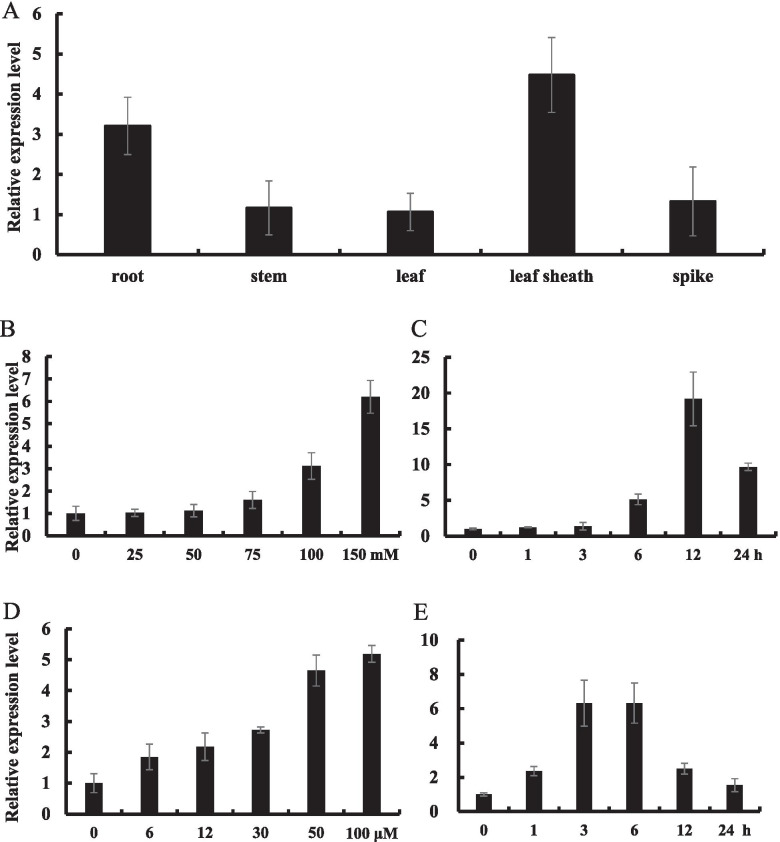
The expression level of OsNAC3 in root, stem, leaf, leaf sheath, and spike were investigated with qRT-PCR. The results showed that the highest expression of OsNAC3 was in leaf sheath, and the lowest expression was in stem and leaf (Fig. 1A). To investigate the possible role of OsNAC3 in abiotic stress resistance, we examined its expression in rice roots after salt (NaCl) treatment in a time and dose gradient manner. Notably, OsNAC3 expression was the highest at 12 h and showed a positive correlation with the concentration of NaCl (Fig. 1B, C). Considering ABA’s role in plant abiotic stress response, we investigated OsNAC3 expression after rice treatment with exogenous ABA. We found that OsNAC3 expression was the highest during 3–6 h treatment of 100 μM ABA, and showed a positive correlation with ABA concentration (Fig. 1D, E).
Fig. 1.
Expression profile of OsNAC3. (A) OsNAC3 expression in the root, stem, leaf, leaf sheath, and spike of heading stage rice. (B) Dose-dependent expression level of OsNAC3 in rice roots under salt treatment. 5-day-old rice seedlings were exposed to different concentrations of NaCl (0, 25, 50, 75, 100, and 150 mM) for 12 h. (C) Time-course expression level of OsNAC3 in rice roots under 100 mM NaCl treatment. 5-day-old rice seedlings were exposed to 100 mM NaCl for different times (0, 1, 3, 6, 12, and 24 h). (D) Dose-dependent expression level of OsNAC3 in rice roots under ABA treatment. 5-day-old rice seedlings were exposed to different concentrations of ABA (0, 6, 12, 30, 50, and 100 μM) for 12 h. (E) Time-course expression level of OsNAC3 in rice roots under 50 μM ABA treatment. 5-day-old rice seedlings were exposed to 50 μM ABA at different times (0, 1, 3, 6, 12, and 24 h). Histone H3 was used as an internal standard. Data represent means ± SD (n = 3)
Subcellular and cellular localization of OsNAC3
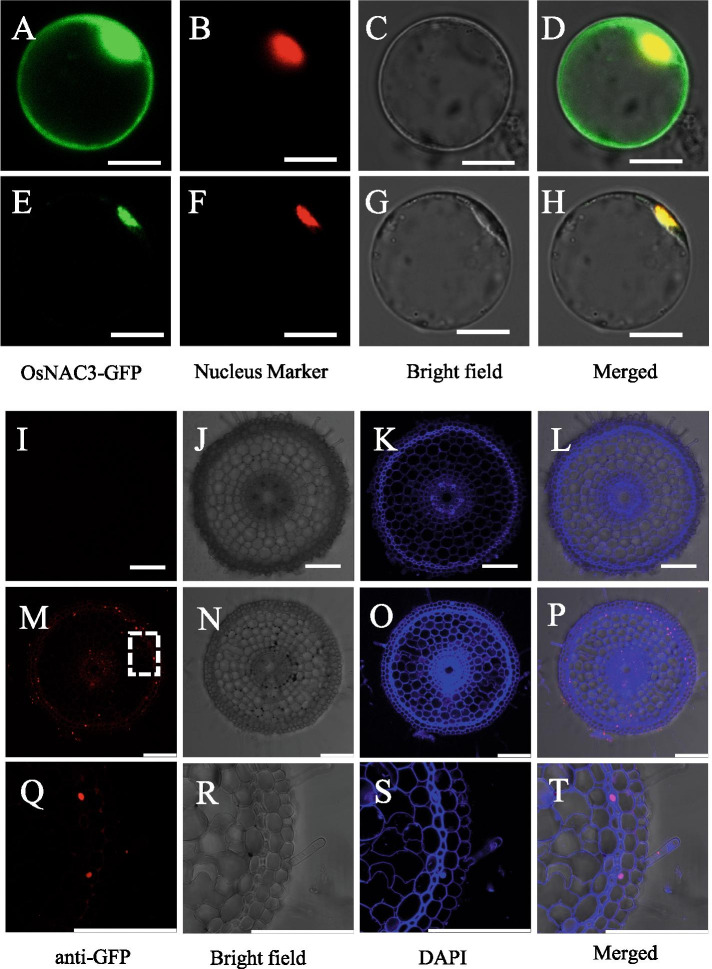
To investigate the subcellular localization of OsNAC3, we fused its coding region with green fluorescence protein (GFP) and introduced it into rice protoplasts along with nuclear marker OsGhd7-RFP. The green fluorescence of the control vector (35S: GFP) was observed in the whole cell, while the green signal of OsNAC3 nicely overlapped with the red fluorescence of OsGhd7-RFP (Fig. 2A-H), indicating nuclear localization of OsNAC3.
Fig. 2.
Subcellular and cellular localization of OsNAC3. (A-H) Rice protoplast co-expressing GFP-OsNAC3 or GFP with OsGhd7 (nuclear marker) under the control of CaMV35S promoter. The protoplasts were isolated from the leaf sheaths of 14-day-old rice seedlings (9311, Oryza sativa L. ssp.indica). Scale bar = 10 μm. (I-T) Immunostaining of the roots of 7-day-old Nipponbare rice (upper panels) and ProOsNAC3-OsNAC3-GFP transgenic plants (middle panels) with anti-GFP antibodies. High-magnification images (Q-T) of the dotted part of the middle panels (M-P). The red and blue colors are the signal of anti-GFP antibody and DAPI staining of cell walls and nuclei, respectively. Scale bar = 20 μm
To further investigate the cellular localization of OsNAC3 in rice roots, ProOsNAC3-OsNAC3-GFP transgenic rice was constructed. Immunostaining assay was performed with GFP and DAPI was used as a nuclear stain. Antibodies against GFP (red signal) were detected in all the cell layers of transgenic roots, while it nicely overlapped with DAPI stain (blue signal) in the nucleus (Fig. 2M-T). This confirmed the nuclear localization of OsNAC3 in root cells. Meanwhile, the red signal was absent in wild-type roots indicating the antibody specificity (Fig. 2I-L).
Transcriptional activation analysis of OsNAC3
To examine the transcriptional activation activity of OsNAC3, we fused its coding region with the GAL4 DNA binding domain in the pGBK-OsNAC3 vector, which was then transformed into yeast strain AH109. If OsNAC3 has transcriptional activation activity, GAL4-OsNAC3 binding to the GAL1 upstream sequence will activate the HIS3 reporter gene and restore the growth of the host yeast strain on a histidine-deficient plate. The results of the assay showed that the yeast cells carrying pGBK-OsNAC3 and positive control cells grew normally on the SD/Trp- and SD/Trp−/His−/Ade- plates, while the negative control (pGBK-T7) only grew on the SD/Trp-, but not SD/Trp−/His−/Ade- plates (Additional file 1 Fig. S2). These results indicated that full-length OsNAC3 has self-activation activities in yeast.
ABA affect the growth of OsNAC3-transgenic rice
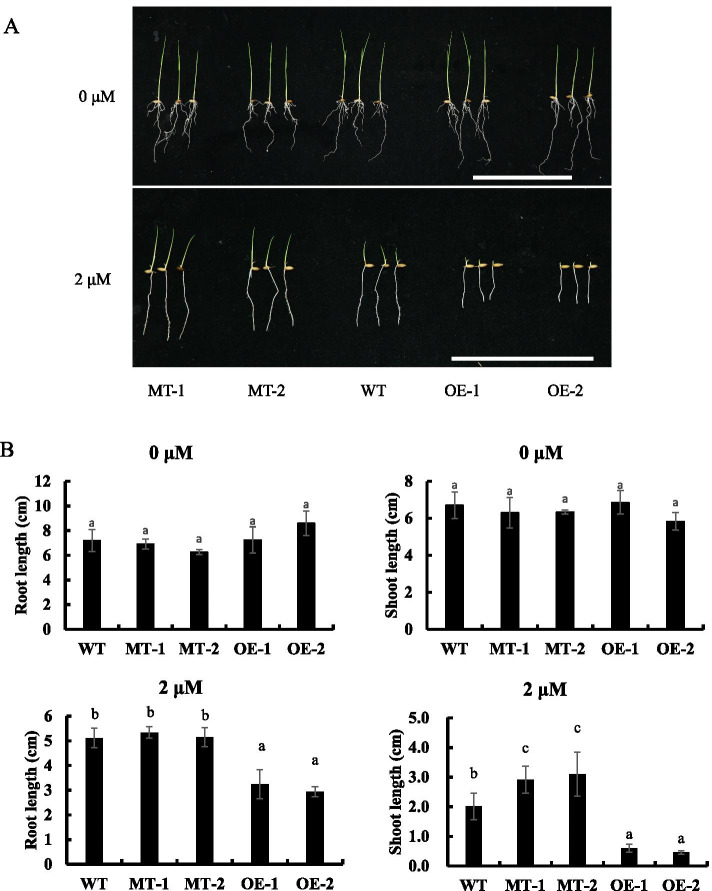
Given that exogenous ABA induces the expression of OsNAC3, we speculate that exogenous ABA treatment may affect the growth of OsNAC3-transgenic rice. To test this hypothesis, we established an OsNAC3-knockout line (MT-1, MT-2) with CRISPR/Cas9 method and a maize ubiquitin promoter-driven OsNAC3-overexpression line (OE-1, OE-2) (more details in Additional file 1 Fig. S3, S4).
The seeds of OsNAC3-knockout, OsNAC3-overexpression, and wild-type rice were germinated in 1/2 MS medium for 2 days. Next, seedlings of similar size were transferred to a new 1/2 MS medium containing 0 or 2 μM ABA; after 5 days of cultivation, seedlings were photographed, and the lengths of roots and shoots were recorded. Results showed that in the absence of ABA, the growth of WT and OsNAC3-transgenic lines showed no obvious difference, indicating no effect of change in OsNAC3 expression level under normal conditions. However, in the presence of exogenous ABA, the shoot and root growth of OE lines were severely inhibited compared with WT lines. The shoot length of MT lines was longer than WT lines, while root length was similar between WT and MT lines (Fig. 3A, B). These results demonstrated that overexpression of OsNAC3 increased the ABA sensitivity of rice plants, while OsNAC3 knock-out decreased the sensitivity of rice shoots but not roots in the presence of ABA.
Fig. 3.
The growth of MT and OE plants under ABA treatment. (A) Growth performance of WT, MT, and OE seedlings under normal conditions for 5 days (upper panels). Growth performance of WT, MT, and OE seedlings under ABA treatment for 5 days (lower panels). Scale bar = 10 cm. (B) Shoot and root length of WT, MT, and OE seedlings under 0 or 2 μM ABA treatment for 5 days. All seeds were germinated on 1/2-strength MS medium without ABA for 2 days before the ABA treatment. Data represent means ± SD (n = 10). Different letters indicate significant differences (Tukey test, p < 0.05)
OsNAC3 positively regulates rice tolerance to salt stress
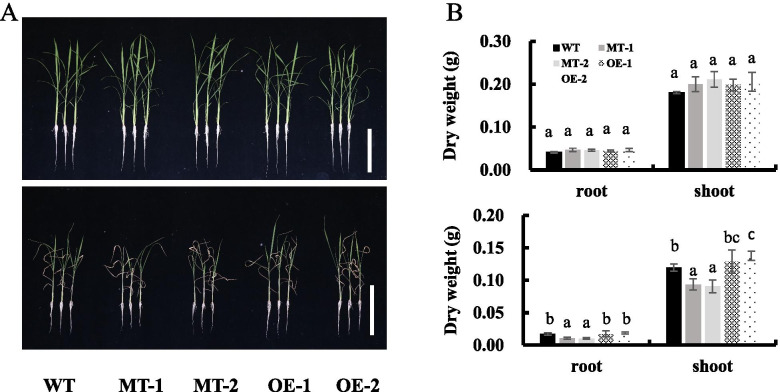
To investigate the role of OsNAC3 in rice salt tolerance, the seedlings of MT, OE, and WT plants were treated with different concentrations of NaCl (0 or 75 mM) for two weeks. We found that in the absence of NaCl, the growth of WT and OsNAC3- transgenic plants did not show a significant difference. However, in the presence of 75 mM NaCl, MT lines showed more withered leaves and less dry weight compared with WT lines; on the contrary, OE lines displayed attenuated chlorosis symptoms and greater dry weight (Fig. 4A, B). This phenotypic data suggest that OsNAC3 positively regulates salt tolerance in rice.
Fig. 4.
Loss of OsNAC3 decreases while overexpression increases salt tolerance in rice. (A) Growth performance of WT, MT, and OE plants after treatment with 0 (upper panels), 75 (lower panels) mM NaCl for 10 days. Scale bar = 10 cm. (B) Dry weight of shoot and root in WT, MT, and OE plants after treatment with 0 (upper panels), 75 (lower panels) mM NaCl. 4-leaf-stage transgenic and Nipponbare rice were treated with 0 or 75 mM NaCl for 10 days. Data represent means ± SD (n = 3). Different letters indicate significant differences (Tukey test, p < 0.05)
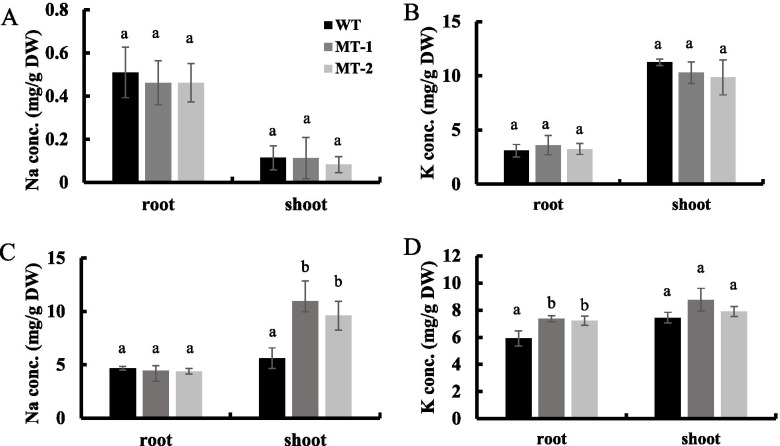
OsNAC3 knockout increases Na accumulation in shoots of transgenic rice
To test the effect of OsNAC3 on tissue-specific K+/Na+ homeostasis, we measured Na+ and K+ content in WT and OsNAC3-knockout plants under control (without NaCl) and salt (75 mM NaCl) conditions. Under control conditions, tissue Na+ and K+ content showed no differences between the WT and osnac3 lines (Fig. 5A, B). However, in the presence of 75 mM NaCl, Na+ concentration in shoots of two MT lines was about twice (on average) that of WT plants, while no significant difference was found in the roots. Besides, K+ concentration in roots of two MT lines was only slightly higher than that of the WT plants, while showed no significant difference in the shoots (Fig. 5C, D). These results suggest that OsNAC3 mainly regulates the Na+ homeostasis of rice shoot under salt stress conditions.
Fig. 5.
Tissue Na+ and K+ concentration in WT and OsNAC3 knockout lines under normal condition or 75 mM NaCl treatment for 10 days. (A) Na+ concentration in the roots and shoots under normal conditions. (B) K+ concentration in the roots and shoots under normal conditions. (C) Na+ concentration in the roots and shoots under 75 mM NaCl treatment. (D) K+ concentration in the roots and shoots under 75 mM NaCl treatment. 14-day-old transgenic and Nipponbare rice were treated with 0 or 75 mM NaCl for 10 days. Data represent means ± SD (n = 3). Different letters indicate significant differences (Tukey test, p < 0.05)
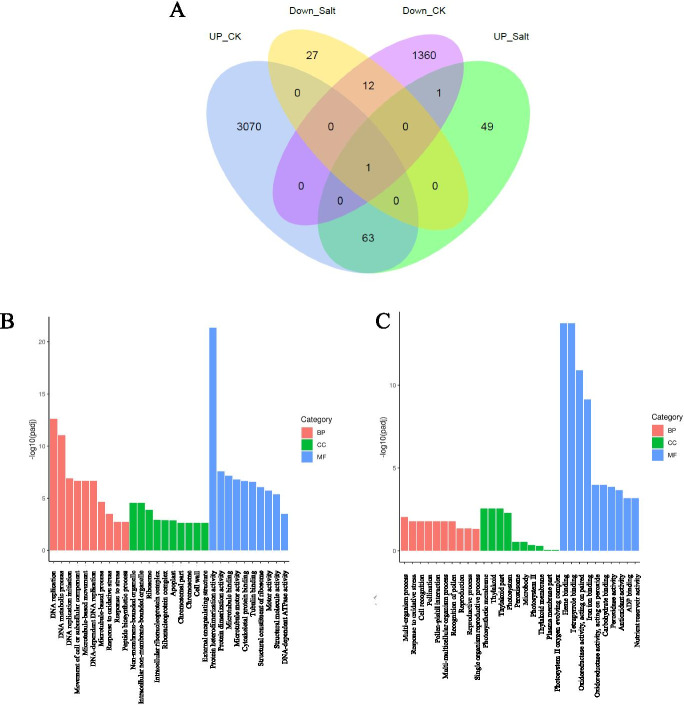
Knockout of OsNAC3 affects the expression profiles of several key genes in rice
TFs at large regulate the expression of many genes. To investigate the regulatory function of OsNAC3 in rice, we used high-throughput RNA-seq to analyze the transcriptomes of Nipponbare (WT) and mutant osnac3 (MT) rice roots under normal and salt conditions. A two-fold change in expression was selected as the threshold to determine the DEGs between the WT and MT lines under normal and salt conditions. We found that there were 3184 up-regulated and 1401 down-regulated DEGs between the WT and MT lines under normal and salt conditions, respectively (Additional file 2). Under the two conditions, 63 and 12 genes were found to be steadily up- and down-regulated, respectively. Therefore, we hypothesized that these genes may be regulated by OsNAC3 (Fig. 6A).
Fig. 6.
Transcriptome analysis of OsNAC3-regulated genes. (A) Venn diagram showing the OsNAC3 regulated genes in MT and WT lines. UP_CK: upregulated genes in MT compared with WT under normal conditions (fold-change > 2). UP_SALT: upregulated genes in MT compared with WT under high salinity (fold-change > 2). DOWN_SALT: the downregulated genes in MT compared with WT under high salinity (fold-change < 0.5). DOWN_CK: the downregulated genes in MT compared with WT under normal conditions (fold-change < 0.5) (B, C) GO analysis of downregulated (left) and upregulated (right) genes in MT compared with WT. BP: biological process. CC: cellular component. MF: molecular function. Three biological replicates (n = 3) were performed for each treatment
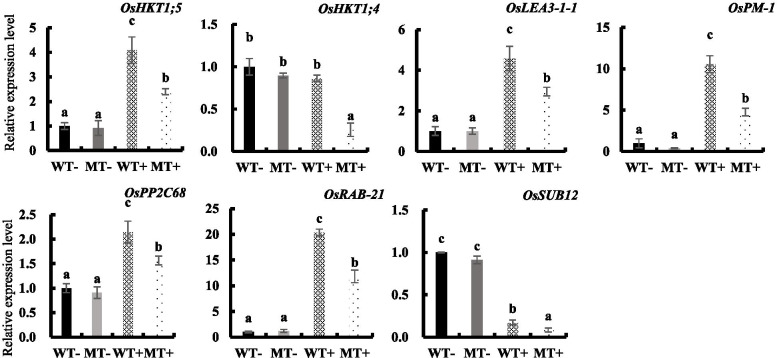
Under normal conditions, the up-regulated genes are mainly involved in DNA replication, DNA metabolic process, and protein heterodimerization activity, while down-regulated genes are mainly related to heme binding, tetrapyrrole binding, and oxidoreductase (Fig. 6 B, C). To verify the RNA-seq data, we performed qRT-PCR to validate the expression level of DEGs and their homologs in rice. As expected, some genes showed different expression levels between WT and MT under two conditions. Under salt conditions, the expression level of seven genes, including OsRAB21(Os11g0454300), OsPP2C68 (Os09g0325700), OsLEA3–1(Os05g0542500), OsPM1 (Os05g0381400), OsSUB12 (Os02g0198700), OsHKT1;4 (Os04g0607600) and OsHKT1;5 (Os01g0307500), was lower in the MT than in WT plants (Fig. 7). These seven genes are considered to be involved in abiotic stress response. Overall, these results indicate that OsNAC3 potentially regulated the stress-related genes in rice to resist salt stress.
Fig. 7.
Relative expression levels of potential DEGs under normal and high salinity conditions. WT- and MT- indicate Nipponbare and OsNAC3-knockout lines under control conditions. WT+ and MT+ represent Nipponbare and OsNAC3-knockout lines treated with 100 mM NaCl for 24 h. 14-day-old OsNAC3-knockout and Nipponbare rice were treated with 0 or 100 mM NaCl for 24 h. Data are means ± SD of three biological replicates. Different letters represent significant differences (Tukey test, p < 0.05)
Discussion
In this study, we isolated and functionally characterized a novel NAC transcriptional factor OsNAC3 in rice. OsNAC3 is a nuclear protein with self-activation activities and expresses in root cells. RT-qPCR showed that ABA and NaCl treatment enhanced the root expression of OsNAC3. Disruption of OsNAC3 by CRISPR/Cas9 reduced rice sensitivity to ABA but increased sensitivity to salt stress, while OsNAC3 overexpression did the opposite. Furthermore, under salt conditions, Na+ homeostasis in the shoots and transcript profiles in the roots were altered in the MT plants compared with the WT plants. This indicates a significant role of rice OsNAC3 in ABA response and salt tolerance.
Many rice NAC TFs have been reported to play important roles in ABA response and salt tolerance. For example, ABA and salt stress rapidly induced the expression of SNAC1 [8]. Overexpression of SNAC1 significantly enhanced the rice tolerance to drought and salinity through the transcription regulation of many stress-related genes such as OsSRO1c and OsPP18 [8, 25–27]. OsNAC2 was shown to enhance rice salt tolerance through promoting the expression of ABA biosynthesis genes such as OsNCED1 and OsNCED3 [21]. OsNAC5 and OsNAC6 were strongly induced by ABA treatment [28–31]. Overexpression of OsNAC5 and OsNAC6 improved the tolerance of rice to high salt stress [17, 29, 30, 32]. Phylogenetic analysis showed a close association between OsNAC3 and SNAC1. The amino acid alignment showed that the two proteins shared the conserved domains. Similar to SNAC1, we found that OsNAC3 expression was also rapidly induced by ABA and salt stress. Notably, OsNAC3 overexpression improved the salt tolerance of rice. These results suggested that OsNAC3 and SNAC1 may have similar regulatory mechanisms and functional redundancy in ABA response and salt tolerance.
To further investigate the role of OsNAC3 in rice ABA response, phenotypes of MT and OE seedlings were analyzed after exogenous ABA treatment. We found that the shoot and root length of OE plants was significantly suppressed compared with WT; on the contrary, the shoot length of MT plants was significantly more than that of WT plants. These results demonstrated that OsNAC3 plays important role in ABA signal response. In a previous study, we showed that another rice NAC transcription factor OsNAC45 was involved in ABA response. Under exogenous ABA treatment, OsNAC45-OE plants displayed a longer length of the shoots and roots, but OsNAC45-MT plants showed a shorter length of the shoots and roots compared to the WT plants [33]. Similar phenotypes of OsNAC3 transgenic plants suggest that it may use the similar ABA response pathway of OsNAC45. Additionally, we found that OsNAC3 and OsNAC45 jointly regulate the expression of OsPM1 (Plasma Membrane Protein 1) encoding an ABA influx carrier, which is reported to mediate ABA influx functioning in the ABA signaling pathway [34]. This somewhat explains the reasons for similar phenotypes between OsNAC3 and OsNAC45 transgenic rice under ABA treatment.
OsNAC3 knockout changed rice sensitivity to salt stress. Under 75 mM NaCl treatment, MT plants exhibited more wilting leaves compared to WT plants. Moreover, in the absence of NaCl, the roots and shoots concentrations of Na+ and K+ were similar between the WT and MT plants. In the presence of 75 mM NaCl, shoot Na+ concentration significantly increased in MT than in WT, while root K+ concentration was only slightly higher in MT. However, under the same treatments, tissue Na+ and K+ concentrations were similar between WT and osnac45 lines although OsNAC45 knockout lines also showed more sensitivity to salt stress than the WT plants. In osnac45 mutants, more accumulation of reactive oxygen species (ROS) in roots was detected [33]. These differences suggested that OsNAC3 and OsNAC45 might be involved in rice salt tolerance through distinct regulatory mechanisms. The RNA-seq and qRT-PCR analysis showed that the expression of some genes encoding Na+/K+ transporters such as OsHKT1;4 and OsHKT1;5 was regulated by OsNAC3. It is well known that HKT (High-affinity K+ Transporter) gene family mediate Na+ and K+ transport in rice [35]. Under salinity stress conditions, OsHKT1;4 and OsHKT1;5 perform Na+ unloading in xylem to prevent over accumulation of Na+ in shoot [36–38]. Therefore, compared to WT, increased Na+ accumulation in MT shoots under salt conditions can be attributed to the lower expression of OsHKT1;4 and OsHKT1;5. These results indicated that OsNAC3 mutation changes the expression of some Na+/K+ transporters in roots causing less Na exclusion in roots and more Na root-to-shoot translocation, which in turn increases rice sensitivity to salt stress.
Furthermore, transcriptome sequencing analysis revealed 1401 down-regulated DEGs in the roots after OsNAC3 knockout under normal and salt conditions. Go enrichment analysis showed that most DEGs were related to heme binding, tetrapyrrole binding, and oxidoreductase. To validate some of the DEGs and their homologs in rice, we performed qRT-PCR and found that OsLEA3–1, OsRAB-21, OsPM-1, OsPP2C68, OsHKT1;4, OsHKT1;5, OsSUB12 might be regulated by OsNAC3. OsLEA3–1 is an abiotic stress-induced gene, which encodes for a late embryogenesis abundant (LEA) protein. Overexpression of OsLEA3–1 significantly increases the rice tolerance to salt and drought stress [39, 40]. Rice RAB (response to abscisic acid) proteins belong to another subgroup of the LEA protein family. In tobacco and rice plants, OsRAB-21 is upregulated to counter the salinity, drought, and ABA stresses [41, 42]. OsPM1 is an ABA influx carrier. A previous report suggested that OsPM1 expression is regulated by OsbZIP46, which then plays important role in response to drought stress [34]. Recently, we showed that OsPM1 expression is regulated by OsNAC45, indicating the regulatory role of OsNAC3 in the ABA signal transduction pathway along with OsbZIP46 and OsNAC45 [33]. OsPP2C68, a member of the PP2C family, is one of the key components of the ABA signal transduction pathway and regulates abiotic stress [43]. OsHKT1;4 and OsHKT1;5 mediating Na+ and K+ transport are proposed to be major players in root-to-shoot Na+ partitioning and K+/Na+ homeostasis [36–38]. These imply that OsNAC3 involved in ABA response and salt tolerance might be due to regulate the expression of above-mentioned genes.
Interestingly, OsSUB12 (Submergence Tolerance 12), encoding an ethylene-responsive transcription factor, is homologous to OsSUB1A that mediates rice submergence stress tolerance [44]. Furthermore, analysis of cis-acting elements showed that there are many abiotic stress response-related elements, such as MYB binding sites, W-boxes, and ABRE (ABA-responsive element), in the promoter region of OsNAC3. These results suggest that OsNAC3 might play important roles in multiple abiotic stress responses in rice.
Conclusion
In summary, OsNAC3, a novel nuclear-localized transcription factor, participates in ABA response and salt tolerance through regulating the expression of stress-responsive genes and shoot Na+ homeostasis in rice.
Methods
Plant materials and growth conditions
We used the wild-type rice (Oryza sativa cv Nipponbare), two OsNAC3-knockout lines, and two OsNAC3 overexpression lines in this work. The wild type rice was obtained from rice resources conservation center of Guangxi University. The OsNAC3 mutants and overexpression lines were constructed in our laboratory (see below). The cultivation of plants conforms to China’s legislation on genetically modified plants. The formal identification of the OsNAC3-knockout and overexpression lines was conducted by Xiang Zhang and Yan long. Seeds of the wild-type, OsNAC3-knockout and overexpression lines were preserved in our lab, but not in a publicly available herbarium.
Rice seeds were germinated in water at 28 °C for 2 days in darkness. The germinated seeds were then placed on a floating net containing 0.5 mM CaCl2 solution at 28 °C and a 12/12 h light/dark cycle. After growing for 4–7 days at 28 °C, plants were cultured in a 4-L plastic spot filled with one-half-strength Kimura B solution (pH 5.6) as described previously [45].
Root, stem, leave, leave sheath, and spike of heading stage rice were harvested for tissue-specific expression analysis of OsNAC3. 5-day-old Nipponbare was used to study the induction of OsNAC3. For salt stress, rice seedlings were exposed to different concentrations of NaCl (0, 25, 50, 75, 100, and 150 mM) for 12 h or 100 mM NaCl for different times (0, 1, 3, 6, 12, and 24 h). For ABA treatment, rice seedlings were exposed to different concentrations of ABA (0, 6, 12, 30, 50, and 100 μM) for 12 h or 50 μM ABA at different times (0, 1, 3, 6, 12, and 24 h).
Generation of transgenic rice plants
To create the OsNAC3-OE construct, total RNA, extracted from Nipponbare, was reverse transcribed to cDNA by RT-PCR, which was then used as a template to amplify the OsNAC3 coding region. The full-length cDNA of OsNAC3 was inserted into the pCAMBIA1300-Ubi vector between the Ubiquitin promoter and nopaline synthase terminator. To create the OsNAC3-MT construct, CRISPR/Cas9 gene-editing system was used. The pCRISPR-OsNAC3 construct with two OsNAC3-specific target sites was constructed as reported previously [46]. These constructs were transferred to Agrobacterium tumefaciens strain EHA101, which was then transformed into rice cv. Nipponbare. The primers used in this work are listed in Supplementary Table 1.
Bioinformatics analysis
Amino acid sequences of several stress-response NAC genes were compared with OsNAC3 using MEGA7 with 1000 bootstrap replicates and the neighbor-joining tree (NJT) method. Alignment of OsNAC3, ANAC102, SNAC1, and OsNAC4 proteins was performed with CLUSTAL OMEGA (http://www.clustal.org/omega/) and graphical representation was created with Espript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/).
For promoter analysis, 2000 bp upstream of the transcription start site were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) and putative cis-elements were searched with Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Transactivation activity analysis
For transactivation activity analysis, the coding region of OsNAC3 was fused in-frame to yeast GAL4 DNA binding domain of vector pGBK-T7, producing the pGBK- OsNAC3. pGBK -OsNAC45 (positive control), pGBK -OsNAC3, or pGBK-T7 empty vector (negative control); these vectors were then transformed into yeast strain AH109 according to the Matchmaker Gold Yeast Two-Hybrid System user manual (Clontech). Yeast cells carrying the pGBK- OsNAC3, pGBK -OsNAC45 or empty vector were spotted on SD/Trp- or SD/Trp−/His−/Ade- medium. The plates were photographed after incubation for 3 days at 30 °C.
Protoplast isolation
The protoplasts were isolated based on the procedures described by Zhang et al. [47] with some modifications. Briefly, seeds of the indica rice 9311 were grew on MS medium in the dark in a growth chamber at 28 °C for two weeks. Leaf sheaths of 50 rice seedlings were harvest and cut into 1 mm pieces using a fresh sharp razor blade. The leaf sheath pieces were quickly transferred into 0.6 M mannitol for a quick plasmolysis treatment, followed by enzymatic digestion in the dark with gentle shaking. After digestion, the protoplasts were collected by filtration through a 50-μm cell strainer. Finally, the protoplasts were resuspended gently in MMG solution (4 mM MES, pH 5.7, 0.6 M mannitol, and 15 mM MgCl2). Approximately 1 × 106 cells were used for the following transformation.
Subcellular localization of OsNAC3
To study the subcellular localization of OsNAC3, the coding region of OsNAC3 was fused in-frame to GFP of vector pYL322-GFP, producing the pYL322-OsNAC3-GFP construct. pYL322-OsNAC3-GFP, or vector control; these were co-transformed with nucleus marker (RFP-OsGhd7) into rice protoplast as described previously [48]. After 12 h of incubation, fluorescence images were captured by a confocal laser scanning microscope (TCS SP8; Leica).
Cellular localization of OsNAC3
A 2000 bp region upstream of the transcription start site was PCR amplified from Nipponbare genomic DNA; the coding region of OsNAC3 without stop codon was PCR amplified from Nipponbare cDNA. Promoter and coding region were cloned into the pCAMBIA1300-GFP vector to generate the ProOsNAC3-OsNAC3-GFP construct, which was transfected into Agrobacterium tumefaciens strain EHA101 for transformation into rice cv. Nipponbare.
Immunofluorescence assays were performed to detect cellular localization of OsNAC3 as described previously [33]. Briefly, roots of transgenic Nipponbare plants carrying ProOsNAC3-OsNAC3-GFP were embedded in 5% agarose. Next, cross-sections were generated with a micro slicer (VT1000 S, Leica), and the sections were incubated with the rabbit anti-GFP polyclonal antibodies, followed by secondary antibodies (Alexa Fluor 555 goat anti-rabbit IgG; Molecular Probes) at room temperature (RT). Fluorescence images were captured by confocal laser scanning microscope (TCS SP8; Leica); DAPI was used to stain nuclei.
Seedling growth assay
To examine the growth rate of transgenic rice under ABA treatment, Nipponbare and transgenic rice seeds were germinated on 1/2 MS medium at 28 °C for 2 days in darkness. Then, the seedlings of similar size were transferred to 1/2 MS medium containing 0 or 2 μM ABA in a growth chamber at 28 °C and a 12/12 h light/dark cycle. Shoot and root length were measured after 5 days of incubation.
Salt stress treatment assay
To investigate the salt tolerance of transgenic rice, 4-leaf-stage transgenic and Nipponbare rice (14-days old) were treated with 0 or 75 mM NaCl for 2 weeks. Then, the roots and shoots were sampled, dehydrated at 70 °C for 3 days, and weighed. The samples were digested with 65% HNO3 at 130 °C [49]. The Na+ and K+ contents in the digested solution were measured by ICP-MS (Plasma Quant MS; Analytik Jena AG).
RNA-sequencing
Four-leave stage rice (MT and WT) was treated with 0 or 100 mM NaCl for 24 h, and then rice roots were harvested to extract the total RNA for the synthesis of cDNA [50]. RNA-seq was performed on an Illumina Nova Seq platform and the Deseq2 method was used to compare the expression profile of downstream genes in WT and MT. DEGs (differentially expressed genes) were sorted using the criteria |log2_ratio| > 2. DEGs were subjected to GO (Gene Ontology; http://geneontology.org/) analysis and p-values were used to assess the significant enrichment of the corresponding category.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- NAC
NAM ATAF and CUC
- TFs
transcription factors
- RT-PCR
reverse transcription-PCR
- qRT-PCR
Quantitative real-time PCR
- RFP
red fluorescent protein
- RNA-Seq
RNA sequencing
- DEGs
Differentially expressed genes
- GO
Gene ontology
- HKT
High-affinity K+ Transporter
Authors’ contributions
DZ and JX conceived the study and designed the experiments. XZ and YL performed most of the experiments. XC, BZ, YX, LL, SC, FL, ZW, and HW participated in the research. XZ, YL, DZ, and JX wrote the manuscript. All authors read and approved the final manuscript.
Funding
The research was funded by Guangxi Natural Science Foundation (2016GXNSFFA380013), Hundred-Talent Program of Guangxi (2014), and Guangxi innovation-driven development special funding project (Grant no. Guike-AA17204070). The funders did not play any role in the design of the study, the collection, analysis, and interpretation of the relevant data, and writing the manuscript.
Availability of data and materials
All the data supporting the conclusions of this article are provided within the article and in its additional files. All data and materials are available upon reasonable request from the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiang Zhang and Yan Long contributed equally to this work.
Contributor Information
Degui Zhou, Email: zhoudegui@gdaas.cn.
Jixing Xia, Email: xiajx@gxu.edu.cn.
References
- 1.M M Sachs a, Ho THD: Alteration of gene expression during environmental stress in Plants. 1986, 37(1):363–376.
- 2.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.VanWallendael A, Soltani A, Emery NC, Peixoto MM, Olsen J, Lowry DB. A molecular view of plant local adaptation: incorporating stress-response networks. Annu Rev Plant Biol. 2019;70:559–583. doi: 10.1146/annurev-arplant-050718-100114. [DOI] [PubMed] [Google Scholar]
- 4.Ismail AM, Horie T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol. 2017;68:405–434. doi: 10.1146/annurev-arplant-042916-040936. [DOI] [PubMed] [Google Scholar]
- 5.Shi WY, Du YT, Ma J, Min DH, Jin LG, Chen J, Chen M, Zhou YB, Ma YZ, Xu ZS et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int J Mol Sci. 2018;19(12):4087. [DOI] [PMC free article] [PubMed]
- 6.Kim S, Kang JY, Cho DI, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40(1):75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 7.Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143(4):1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103(35):12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17(6):369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465(1–2):30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 13.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9(6):841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F. A Rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017;174(3):1747–1763. doi: 10.1104/pp.17.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, Nelson RS, Zhou X, Song F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol. 2013;81(1–2):41–56. doi: 10.1007/s11103-012-9981-3. [DOI] [PubMed] [Google Scholar]
- 17.Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67(1–2):169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- 18.Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004;39(6):863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 19.Ye N, Jia L, Zhang J. ABA signal in rice under stress conditions. Rice (New York, NY) 2012;5(1):1. doi: 10.1186/1939-8433-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 21.Jiang D, Zhou L, Chen W, Ye N, Xia J, Zhuang C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice (New York, NY) 2019;12(1):76. doi: 10.1186/s12284-019-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, Zhang X, Ma H, Ming F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014;55(3):604–619. doi: 10.1093/pcp/pct204. [DOI] [PubMed] [Google Scholar]
- 23.Gao F, Xiong A, Peng R, Jin X, Xu J, Zhu B, Chen J, Yao Q. OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Culture (PCTOC) 2010;100(3):255–262. [Google Scholar]
- 24.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research. 2003;10(6):239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- 25.You J, Zong W, Du H, Hu H, Xiong L. A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol Biol. 2014;84(6):693–705. doi: 10.1007/s11103-013-0163-8. [DOI] [PubMed] [Google Scholar]
- 26.You J, Zong W, Hu H, Li X, Xiao J, Xiong L. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 2014;166(4):2100–2114. doi: 10.1104/pp.114.251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Chang Y, Ma S, Shen J, Hu H, Xiong L. Genome-wide identification of SNAC1-targeted genes involved in drought response in Rice. Front Plant Sci. 2019;10:982. doi: 10.3389/fpls.2019.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, Grusak MA, Fett JP. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta. 2009;230(5):985–1002. doi: 10.1007/s00425-009-1000-9. [DOI] [PubMed] [Google Scholar]
- 29.Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics. 2010;284(3):173–183. doi: 10.1007/s00438-010-0557-0. [DOI] [PubMed] [Google Scholar]
- 30.Song SY, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta. 2011;234(2):331–345. doi: 10.1007/s00425-011-1403-2. [DOI] [PubMed] [Google Scholar]
- 31.Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J. 2013;11(1):101–114. doi: 10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- 32.Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun. 2009;379(4):985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Long Y, Huang J, Xia J. OsNAC45 is involved in ABA response and salt tolerance in Rice. Rice (New York, NY) 2020;13(1):79. doi: 10.1186/s12284-020-00440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao L, Cheng X, Gu Z, Huang W, Li S, Wang L, Wang YF, Xu P, Ma H, Ge X. The AWPM-19 family protein OsPM1 mediates Abscisic acid influx and drought response in Rice. Plant Cell. 2018;30(6):1258–1276. doi: 10.1105/tpc.17.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A. Sodium transport and HKT transporters: the rice model. Plant J. 2003;34(6):788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, et al. OsHKT1;4-mediated Na(+) transport in stems contributes to Na(+) exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016;16:22. doi: 10.1186/s12870-016-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37(10):1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 38.Hauser F, Horie T. A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K(+)/Na(+) ratio in leaves during salinity stress. Plant Cell Environ. 2010;33(4):552–565. doi: 10.1111/j.1365-3040.2009.02056.x. [DOI] [PubMed] [Google Scholar]
- 39.Xu D, Duan X, Wang B, Hong B, Ho T, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic Rice. Plant Physiol. 1996;110(1):249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. TAG Theoretical Appl Genet. 2007;115(1):35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- 41.RoyChoudhury A, Roy C, Sengupta DN. Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 2007;26(10):1839–1859. doi: 10.1007/s00299-007-0371-2. [DOI] [PubMed] [Google Scholar]
- 42.Ganguly M, Datta K, Roychoudhury A, Gayen D, Sengupta DN, Datta SK. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal Behav. 2012;7(4):502–509. doi: 10.4161/psb.19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics. 2010;11:435. doi: 10.1186/1471-2164-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma N, Dang TM, Singh N, Ruzicic S, Mueller-Roeber B, Baumann U, Heuer S. Allelic variants of OsSUB1A cause differential expression of transcription factor genes in response to submergence in rice. Rice (New York, NY) 2018;11(1):2. doi: 10.1186/s12284-017-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaji N, Ma JF. Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 2007;143(3):1306–1313. doi: 10.1104/pp.106.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7(1):30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu S, Huang J, Chen Z, Xia J. C2 domain plays critical roles in localization of novel C2 domain-containing protein OsC2DP. Plant Signal Behav. 2019;14(11):1667208. doi: 10.1080/15592324.2019.1667208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Q, Fu L, Dai F, Jiang L, Zhang G, Wu D. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genomics. 2016;17(1):889. doi: 10.1186/s12864-016-3242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L, Kuang L, Li X, Wu L, Wu D, Zhang G. Metabolomic and transcriptomic analyses reveal the reasons why Hordeum marinum has higher salt tolerance than Hordeum vulgare. Environ Exp Bot. 2018;156:48–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the conclusions of this article are provided within the article and in its additional files. All data and materials are available upon reasonable request from the corresponding author.