Abstract
Bone marrow houses a multifunctional stromal cell population expressing C-X-C motif chemokine ligand 12 (CXCL12), termed CXCL12-abundant reticular (CAR) cells, that regulates osteogenesis and adipogenesis. The quiescent pre-adipocyte-like subset of CXCL12+ stromal cells (“Adipo-CAR” cells) is localized to sinusoidal surfaces and particularly enriched for hematopoiesis-supporting cytokines. However, detailed characteristics of these CXCL12+ pre-adipocyte-like stromal cells and how they contribute to marrow adipogenesis remain largely unknown. Here we highlight CXCL12-dependent physical coupling with hematopoietic cells as a potential mechanism regulating the adipogenic potential of CXCL12+ stromal cells. Single-cell computational analyses of RNA velocity and cell signaling reveal that Adipo-CAR cells exuberantly communicate with hematopoietic cells through CXCL12-CXCR4 ligand-receptor interactions but do not interconvert with Osteo-CAR cells. Consistent with this computational prediction, a substantial fraction of Cxcl12-creER+ pre-adipocyte-like cells intertwines with hematopoietic cells in vivo and in single-cell preparation in a protease-sensitive manner. Deletion of CXCL12 in these cells using Col2a1-cre leads to a reduction of stromal-hematopoietic coupling and extensive marrow adipogenesis in adult bone marrow, which appears to involve direct conversion of CXCL12+ cells to lipid-laden marrow adipocytes without altering mesenchymal progenitor cell fates. Therefore, these findings suggest that CXCL12+ pre-adipocyte-like marrow stromal cells prevent their premature differentiation by maintaining physical coupling with hematopoietic cells in a CXCL12-dependent manner, highlighting a possible cell-non-autonomous mechanism that regulates marrow adipogenesis.
Keywords: BONE–FAT INTERACTIONS, GENETIC ANIMAL MODELS, PRECLINICAL STUDIES, STROMAL/STEM CELLS
Introduction
Bone marrow stromal cells expressing C-X-C motif chemokine ligand 12 (CXCL12) are one of the most important cell types constituting the interface between bone and blood.(1–4) CXCL12+ stromal cells, universally marked by a Cxcl12-GFP allele,(5) are located in proximity to bone marrow vasculatures, particularly in a perisinusoidal space associated with a characteristic reticular morphology; these cells are therefore termed CXCL12-abundant reticular (CAR) cells.(6) Further, these cells overlap with Lepr-cre-marked cells in adult bone marrow;(7) therefore, these cells are also termed CXCL12+LepR+ cells.(8) These CXCL12+ cells exert two major functions to maintain bone and blood. First, these cells provide a microenvironment for blood cells by expressing a repertoire of cytokines supporting bone marrow hematopoiesis,(6,9,10) and second, these cells provide a source for bone cells by encompassing precursor cell populations for osteoblasts, marrow stromal cells, and adipocytes.(8)
Recent studies postulate that that these undifferentiated CXCL12+ stromal cells are composed of two functionally distinct cellular subsets of pre-osteoblast-like cells and pre-adipocyte-like cells. Combined single-cell and spatial transcriptomics analyses demonstrate that CAR cells are composed of Osteo-CAR and Adipo-CAR cells localized to arteriolar and sinusoidal surfaces, respectively.(11) Similarly, large-scale scRNA-seq studies revealed fundamental diversity of CXCL12+ cells.(12–15) Our recent scRNA-seq analysis demonstrates that Cxcl12-GFP+ cells are composed of two major groups of cells expressing pre-osteoblast and pre-adipocyte markers; the latter group of pre-adipocyte-like cells are preferentially marked by Cxcl12-creER and particularly enriched for a variety of genes encoding hematopoiesis-supporting cytokines, including Cxcl12 (encoding CXCL12) and Kitl (encoding stem cell factor [SCF]).(16) Further, in vivo cell-fate analyses demonstrate that Osteo-CAR and Adipo-CAR cells represent functionally distinct entities. Although Ebf3-creER+ cells, encompassing both Osteo-CAR and Adipo-CAR cells, provide a robust source of both osteoblasts and CAR cells themselves in physiological conditions,(8) Cxcl12-creER+ cells, coinciding with Adipo-CAR cells, provide a source of cortical bone osteoblasts only under regenerative conditions.(16) Therefore, a pre-adipocyte-like subset of CXCL12+ cells (Adipo-CAR cells) with highly abundant CXCL12 expression is dormant and might be more committed to marrow adipogenesis and less prone to osteogenesis under normal conditions.
Functionally, CXCL12 plays essential roles in maintaining bone marrow hematopoiesis by regulating retention, quiescence, and repopulation of hematopoietic stem cells (HSCs) and B-lymphoid progenitors;(9,10,17,18) in fact, these cells express CXCL12’s cognate receptor, C-X-C chemokine receptor type 4 (CXCR4), and are histologically in contact with CAR cells.(19,20) Additionally, CXCL12 released by bone marrow stromal cells regulates osteogenesis and adipogenesis; while osteogenesis is controlled by a cell-autonomous mechanism mediated by CXCL12-CXCR4 signaling, adipogenesis is dictated by a cell-non-autonomous mechanism involving an unidentified cell type.(21)
In this study, we set out to uncover unique properties of CXCL12+ pre-adipocyte-like stromal cells and define their contribution to marrow adipogenesis, using integrative single-cell transcriptomic analyses and mouse genetic models. Our findings demonstrate that many CXCL12+ pre-adipocyte-like stromal cells are physically coupled with hematopoietic cells in vivo and in single-cell preparation, and loss of CXCL12 in these cells leads to a reduction of stromal-hematopoietic coupling and extensive marrow adipogenesis in adult bone marrow. These findings demonstrate the interactive nature of the adipogenic subset of CXCL12+ stromal cells and a potential link between CXCL12-dependent stromal-hematopoietic coupling and marrow adipogenesis.
Materials and Methods
Mice
Cxcl12GFP/+(5) and Cxcl12-creER(16) mice have been described previously. Col1a1(2.3 kb)-GFP (JAX013134), Col2a1-cre (JAX003554), Rosa26-CAG-loxP-stop-loxP-ZsGreen (Ai14: R26R-tdTomato, JAX007907), and Cxcl12-floxed (JAX021773) mice were acquired from the Jackson Laboratory (Bar Harbor, ME, USA). All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by the University of Michigan’s Institutional Animal Care and Use Committee (IACUC), protocol 7681 and 9496 (Ono). All mice were housed in a specific pathogen-free condition and analyzed in a mixed background. Mice were housed in static microisolator cages (Allentown Caging, Allentown, NJ, USA). Access to water and food (irradiated LabDiet 5008, Richmond, IN, USA) was ad libitum. Animal rooms were climate controlled to provide temperatures of 22°C to 23°C on a 12-hour light/dark cycle (lights on at 6:00 a.m.). Mice were identified by micro-tattooing or ear tags. Tail biopsies of mice were lysed by a HotShot protocol (incubating the tail sample at 95°C for 30 minutes in an alkaline lysis reagent followed by neutralization) and used for PCR-based genotyping (GoTaq Green Master Mix [Promega, Madison, WI, USA] and Nexus X2 [Eppendorf, Framingham, MA, USA). Mice were euthanized by overdosage of carbon dioxide or decapitation under inhalation anesthesia in a drop jar (Fluriso, Isoflurane USP, VetOne, Boise, ID). Tamoxifen (Sigma T5648, St. Louis, MO, USA) was mixed with 100% ethanol until completely dissolved. Subsequently, a proper volume of sunflower seed oil (Sigma S5007) was added to the tamoxifen-ethanol mixture and rigorously mixed. The tamoxifen-ethanol-oil mixture was incubated at 60°C in a chemical hood until the ethanol evaporated completely. The tamoxifen-oil mixture was stored at room temperature until use. To label Cxcl12-creER+ cells, a single dose of tamoxifen (1 mg) was injected to Cxcl12-creER; R26RZsGreen or R26RtdTomato mice at postnatal day (P) 21.
Cell preparation
Soft tissues and epiphyses were carefully removed from dissected femurs. After removing distal epiphyseal growth plates and cutting off proximal ends, femurs were cut roughly and incubated using three different protocols: (i) without enzymatic digestion; (ii) 2 Wunsch units of Liberase TM (Sigma/Roche 5401127001) digestion; and (iii) 2 Wunsch units of Liberase TM and 1 mg of Pronase (Sigma/Roche 10165921001) digestion in 2 mL Ca2+, Mg2+-free Hank’s balanced salt solution (HBSS; Sigma H6648) at 37°C for 60 minutes on a shaking incubator (ThermomixerR, Eppendorf). After cell dissociation, cells were mechanically triturated using an 18-gauge needle with a 1 mL Luer-Lok syringe (BD, Franklin Lakes, NJ, USA) and a pestle with a mortar (CoorsTek, Golden, CO, USA), and subsequently filtered through a 70 μm cell strainer (BD) into a 50 mL tube on ice to prepare single-cell suspension. These steps of mechanical trituration and filtration were repeated five times, and dissociated cells were collected in the same tube. Cells were pelleted and resuspended in Flow Cytometry Staining Buffer (Invitrogen/eBioscience, Thermo Fisher Scientific, Waltham, MA, USA) for subsequent applications. For cell culture experiments, cells were resuspended in 10 mL culture medium and counted on a hemocytometer.
Flow cytometry and ImageStream analysis
Dissociated cells were incubated with the following antibodies (1:500) in Flow Cytometry Staining Buffer on ice for 30 minutes. Allophycocyanin (APC)-conjugated CD45 (30F-11, eBioscience [San Diego, CA, USA] 17–0451-82), phycoerythrin (PE)-conjugated CD48 (HM48-1, BioLegend [San Diego, CA, USA] 103405), CD41 (MWReg30, BioLegend 133905), CD79b (HM79-12, BioLegend 132803), F4/80 (BM8, BioLegend 123109), Gr-1 (RB6-8C5, BioLegend 108407), brilliant violet 650 (BV650)-conjugated CD150 (TC15-12F12.2, BioLegend 115931), CD19 (6D5, BioLegend 115541), CD11b (M1/70, BioLegend 101239). Flow cytometry analysis was performed using a four-laser BD LSR Fortessa (Ex. 405/488/561/640 nm) and FACSDiva software. Acquired raw data were further analyzed on FlowJo software (TreeStar, Ashland, OR, USA). Representative plots of at least three independent biological samples are shown in the figures. ImageStream analysis was performed using Amnis ImageStreamX Mark II (MilliporeSigma, Burlington, MA, USA). Acquired raw data were further analyzed on Amnis IDEAS software.
Bulk RNA-seq analysis of fluorescence-activated cell sorting (FACS)-isolated cells
Dissociated bone marrow cells harvested from P21 littermate mice were pooled based on the genotype (Cxcl12GFP/fl; R26RtdTomato/+ [Control] or Cxcl12GFP/fl; Col2a1-cre; R26RtdTomato/+ [Col2a1-CXCL12 cKO]). Cell sorting was performed using a four-laser BD FACS Aria III (Ex. 407/488/561/640 nm) or a six-laser Sony Synergy SY3200 (Ex. 350/405/488/561/594/685 nm) high-speed cell sorter with a 100 μm nozzle. Cxcl12-GFP+ cells from Control or Cxcl12-GFP+tdTomato+ cells from Col2a1-CXCL12 cKO were directly sorted into ice-cold Dulbecco’s phosphate-buffered saline (DPBS) with 10% fetal bovine serum (DPBS/10% FBS) and pelleted by centrifugation. Total RNA was isolated using PicoPure RNA Isolation Kit (KIT0204, Thermo Fisher Scientific), followed by DNA-free DNA removal kit (AM1906, Thermo Fisher Scientific) to remove contaminating genomic DNA. RNA integrity number (RIN) was assessed by Agilent (Santa Clara, CA, USA) 2100 Bioanalyzer RNA 6000 Pico Kit. Samples with RIN > 8.0 were used for subsequent analyses. Complementary DNAs were prepared by SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Clontech [Mountain View, CA, USA] 634888). Post-amplification quality control was performed by Agilent TapeStation DNA High Sensitivity D1000 Screen Tape system. DNA libraries were prepared by Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and submitted for NextGen sequencing (Illumina HiSeq 4000). DNA libraries were sequenced using the following conditions: six samples per lane, 50-cycle single-end read. Reads files were downloaded and concatenated into a single .fastq file for each sample. The quality of the raw reads data for each sample was checked using FastQC to identify quality problems. Tuxedo Suite software package was subsequently used for alignment (using TopHat and Bowtie2), differential expression analysis, and post-analysis diagnostics. FastQC was used for a second round of quality control (post-alignment). HTSeq/DESeq2 was used for differential expression analysis using UCSC mm10 as the reference genome sequence. Only non-ambiguously mapped reads were counted. Meta-analysis of RNA-seq data was performed using iPathwayGuide software (Advaita, Ann Arbor, MI, USA). The bulk RNA-seq data sets presented herein have been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO), and are accessible through GEO Series accession numbers GSE160158 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160158).
Integrative single-cell computational transcriptomic analysis
The data set GSE136970 has been published previously and deposited at NCBI’s GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136970). We filtered out cells with less than 1000 genes per cell and with more than 15% mitochondrial read content.
LIGER data integration
LIGER (version 0.5.0)(22) was used for sample integration, normalization, clustering, and visualization. A joint LIGER object was created from the two Seurat objects (Cxcl12CE_P21-28_1 and Cxcl12CE_P21-28_2), including 2000 highly variable genes, with function “seuratToLiger.” Gene expression count data for the combined sample was scaled with function “normalize,” followed by combining 3000 informative genes across samples with function “selectGenes.” We carried out integrative non-negative matrix factorization (iNMF) with function “optimizeALS,” and used “quantile_norm” to align the data sets. Two-dimensional visualization and clustering were carried out with functions “louvainCluster” at resolution 0.25 and “runUMAP” with cosine distance, nearest neighbors set to 30 and minimum distance to 0.35. To identify the differentially expressed genes among clusters, Wilcoxon rank sum test was performed, with function “runWilcoxon.” We filtered and ranked statistically significant (adjusted p value <0.05) and highly differentially expressed (logFC >3) genes for each cluster. Genes that were ranked in the top 200 were selected for comparison and were used for cluster annotation. Using known gene markers associated with particular biological processes, we elucidated the cell type of each cluster, including Adipo-CAR-1 and −2, Stromal-cell-1 and −2, Osteo-CAR, Osteoblast-like-cell, Myeloid-1 and −2, B-cell-1 and −2, Endothelial-1 and −2, Erythroid, and Pan-hematopoietic. The cluster identities are shown in Fig. 1C.
Fig 1.

Computational lineage inference and intercellular communications of CXCL12+ stromal cell subsets. (A, B) Single-cell RNA-seq analysis of fluorescently sorted single cells gated on a GFP+ fraction harvested from Cxcl12GFP/+; Cxcl12-creER; R26RtdTomato femur bone marrow at P28 (pulsed at P21). (A) UMAP-based visualization of two biological replicate data sets (Cxcl12CE_P21-28_1 and Cxcl12CE_P21-28_2) merged by LIGER. (B) Feature plots. Purple = high expression. n = 7326 cells. (C) RNA velocity analysis. Dynamic velocity vectors superimposed on UMAP plot. Arrows = dynamic velocity vectors inferring future states. (D, E) CellPhoneDB intercellular communication analysis. (D) Summary of putative ligand-receptor interactions. (E) Split-dot-based visualization of ligand-receptor interactions among CXCL12+ stromal, hematopoietic, and endothelial cell subsets. Top row: Cxcl12-Cxcr4; middle row: Kitl-Kit; bottom row: Il7-Il7r pairs. Columns = ligand-expressing (bottom) and receptor-expressing (top) clusters.
RNA velocity analysis (velocyto and scVelo)
The aligned bam files were used as input for velocyto(23) to derive the counts of unspliced and spliced abundances in loom format. Furthermore, the two sample-wise loom files were merged with function “adata.concatenate.” To infer gene-specific RNA velocities (rates of transcription, splicing, and degradation), we utilized scVelo (version 0.2.2)(24) for exploring future cell trajectories. The combined loom file was normalized and log transformed with function scvelo.pp.filter_and_normalize(min_shared_counts = 20, n_top_genes = 5500, flavor = “seurat”). The first and second order moments for each cell across its nearest neighbors were computed with function scvelo.pp.moments(method = ‘umap’, n_neighbors = 30, n_pcs = 30, knn = True). Then, the velocities were estimated by running the likelihood-based dynamical model with function scvelo.tl.velocity(mode = “dynamical”) and the velocity graph was constructed with scvelo.tl.velocity_graph (). To test for the presence of differential kinetics across cell types that could not be well explained by a single model of the overall dynamics, we applied the function scvelo.tl.differential_kinetic_test. RNA velocities were then recomputed with functions scvelo.pp.neighbor(method = ‘umap’, n_neighbors = 30, n_pcs = 30, knn = True), scvelo.tl.velocity (diff_kinetics = True), and scvelo.tl.velocity_graph(). To visualize the RNA velocities, the UMAP coordinate information and cluster assignments were extracted from the LIGER analysis output. Velocities were projected into 2D on top of the previously computed UMAP coordinates, using function scvelo.tl.velocity_embedding_stream(basis = ‘umap’). The resulting visualization can be found in Fig. 1C.
CellPhoneDB, intercellular communication analyses
To explore the intercellular communication between the three major classes of cells (CXCL12+ mesenchymal stromal, endothelial, and hematopoietic populations), we performed CellPhoneDB analysis.(25) CellPhoneDB v.2.0 was employed with default parameters. First, the cell types were defined by the previously annotated clusters from LIGER analysis. To better visualize the relative expression of the interaction pairs, the mean expression was normalized to zero mean and unit variance. Significant (p < 0.05) interactions of three pairs, Cxcl12-Cxcr4, Kitl-Kit, and Il7-Il7r, were displayed in Fig. 1E.
Histology and immunohistochemistry
Samples were fixed in 4% paraformaldehyde for a proper period, typically ranging from 3 hours to overnight at 4°C, then decalcified in 15% EDTA for a proper period, typically ranging from 3 hours to 14 days. Decalcified samples were cryoprotected in 30% sucrose/PBS solutions and then in 30% sucrose/PBS:OCT (1:1) solutions, each at least overnight at 4°C. Samples were embedded in an OCT compound (Tissue-Tek, Sakura Finetek USA, Torrance, CA, USA) and cryosectioned at 14 μm using a cryostat (Leica Biosystems [Wetzlar, Germany] CM1850) and adhered to positively charged glass slides (Fisherbrand ColorFrost Plus, Thermo Fisher Scientific). Sections were postfixed in 4% paraformaldehyde for 15 minutes at room temperature. For immunostaining, sections were permeabilized with 0.25% TritonX/TBS for 30 minutes, blocked with 3% BSA/TBST for 30 minutes, and incubated with rat anti-CD45 monoclonal antibody (1:100, Invitrogen 14–0451-81) or rabbit anti-CD79b monoclonal antibody (1:200, Abcam [Cambridge, MA, USA] ab134147) overnight at 4°C, and subsequently with Alexa Fluor 633-conjugated goat anti-rat IgG (A21049) for 3 hours at room temperature. Sections were further incubated with DAPI (4′,6-diamidino-2-phenylindole, 5 μg/mL, Invitrogen D1306) to stain nuclei before imaging. For lipid staining, cryosections were immediately coverslipped with LipidTOX Deep Red (1:200, Invitrogen H34477) and 5 μg/mL DAPI solution and incubated for 2 hours at 43°C. Stained samples were mounted in TBS with No. 1.5 coverslips (Thermo Fisher Scientific).
Imaging
Images were captured by an automated inverted fluorescence microscope with a structured illumination system (Zeiss [Thronwood, NY, USA] Axio Observer Z1 with ApoTome.2 system) and Zen 2 (blue edition) software. The filter settings used were: FL Filter Set 34 (Ex. 390/22, Em. 460/50 nm), Set 38 HE (Ex. 470/40, Em. 525/50 nm), Set 43 HE (Ex. 550/25, Em. 605/70 nm), Set 50 (Ex. 640/30, Em. 690/50 nm), and Set 63 HE (Ex. 572/25, Em. 629/62 nm). The objectives used were: Fluar 2.5×/0.12, EC Plan-Neofluar 5×/0.16, Plan-Apochromat 10×/0.45, EC Plan-Neofluar 20×/0.50, EC Plan-Neofluar 40×/0.75, Plan-Apochromat 63×/1.40. Images were typically tile-scanned with a motorized stage, Z-stacked, and reconstructed by a maximum intensity projection (MIP) function. Differential interference contrast (DIC) was used for objectives higher than 10×. Representative images of at least three independent biological samples are shown in the figures.
Colony-forming assay and trilineage differentiation
Nucleated bone marrow cells were plated into tissue culture 6-well plates (BD Falcon) at a density of <105 cells/cm2, and cultured in low-glucose DMEM with GlutaMAX supplement (Gibco 10567022) and 10% mesenchymal stem cell-qualified FBS (Gibco 12662029) containing penicillin–streptomycin (Sigma P0781) for 10 ~ 14 days. Cell cultures were maintained at 37°C in a 5% CO2 incubator. Representative images of at least three independent biological samples are shown in the figures. Colonies marked by tdTomato were individually subcloned. A tile-scanned virtual reference of tdTomato+ colonies and cloning cylinders (Bel-Art) were used to isolate an individual colony. Single-cell-derived clones of tdTomato+ cells were cultured in a basal medium. Cells at passage 4–7 were used for trilineage differentiation. To induce chondrocyte differentiation, cells were transferred into a non-tissue culture-coated V-bottom 96-well plate (Corning [Corning, NY, USA] 3896). The plate was centrifuged at 150g for 5 minutes at room temperature to pellet the cells, and the supernatant was carefully aspirated. StemPro Chondrogenesis medium (Thermo Fisher A1007101) was gently added, and the plate was centrifuged at 150g for 5 minutes at room temperature to pellet the cells. The pellet was cultured in the differentiation medium with exchanges into fresh media every 3 to 4 days for up to 3 weeks, each time with centrifugation at 150g for 5 minutes at room temperature to repellet the cells. Cell pellets were fixed with 70% ethanol for 5 minutes and stained for Alcian-Blue Staining Solution (Millipore TMS-010-C) for 30 minutes. To induce osteoblast differentiation, cells were plated on a 48-well plate and cultured with αMEM with GlutaMAX supplement (Gibco 32571036) with 10% FBS containing 1 μM dexamethasone (Sigma D4902), 10 mM β-glycerophosphate (Sigma G9422), and 50 μg/mL ascorbic acid (Sigma A5960) with exchanges into fresh media every 3 to 4 days for up to 3 weeks. Cells were fixed with 4% paraformaldehyde for 5 minutes and stained for 2% Alizarin Red S (Sigma A5533) for 30 minutes. To induce adipocyte differentiation, cells were plated on a 48-well plate and cultured with αMEM with GlutaMAX supplement with 10% FBS containing 1 μM dexamethasone (Sigma D4902), 0.5 mM IBMX (3-Isobutyl-1-methylxanthine, Sigma I5879), and 1 μg/mL insulin (Sigma I6634) with exchanges into fresh media every 3 to 4 days for up to 2 weeks. Cells were stained with LipidTOX Green (Invitrogen H34475, 1:200 in basal medium) for 3 hours at 37°C, or fixed with 4% paraformaldehyde for 5 minutes and stained for Oil Red O (Sigma O0625). Five independent clones of each genotype were used in this study to test their trilineage differentiation potential. For outcome assessment, the presence of Oil Red O+, Alizarin Red+, and Alcian Blue+ cells in each well was used as the criterion for adipocyte, osteoblast, and chondrocyte differentiation, respectively.
Statistical analysis
Results are presented as mean values ± SD. Statistical evaluation was conducted using the Mann–Whitney U test. A p value of <.05 was considered significant. No statistical method was used to predetermine sample size. Sample size was determined on the basis of previous literature and our previous experience to give sufficient standard deviations of the mean so as not to miss a biologically important difference between groups. The experiments were not randomized. All of the available mice of the desired genotypes were used for experiments. The investigators were not blinded during experiments and outcome assessment. One femur from each mouse was arbitrarily chosen for histological analysis. Genotypes were not particularly highlighted during quantification.
Results
Computational analysis of RNA velocity and intercellular communications in CXCL12+ stromal cell subsets
We first set out to define detailed characteristics of the pre-adipocyte-like subset of CXCL12+ stromal cells (Adipo-CAR cells) using integrative single-cell RNA-seq analyses (Supplemental Fig. S1A). For this purpose, we performed new computational analyses on previously published single-cell RNA-seq data set(16) (Supplemental Fig. S1B). This data set includes two biological replicates (Cxcl12CE_P21-P28_1 and Cxcl12CE_P21-P28_2) of fluorescently sorted cells gated on a GFPhigh fraction of bone marrow cells isolated from Cxcl12GFP/+; Cxcl12-creER; R26RtdTomato/+ mice at P28 after a tamoxifen pulse at P21. To identify a common set of cell types, we integrated the two replicates using LIGER(22) (Fig. 1A, B). The 7326 cells include both Cxcl12-GFP+ and Cxcl12-creER-marked tdTomato+ (Cxcl12CE-tdTomato+) cells, as well as contaminating hematopoietic cells that we and others have observed in single-cell RNA-seq data sets from FACS-isolated cells bone marrow stromal cells.(11–15) In fact, approximately 20% of the cells in the data set were myeloid, lymphoid, and erythroid cells without detectable eGFP expression. eGFP (marked by Cxcl12-GFP) and tdTomato (marked by Cxcl12-creER) were expressed by non-hematopoietic cells, predominantly by mesenchymal stromal cells expressing Pdgfrb (encoding PDGFβ, CD140b) (~95%) but also by a small number of endothelial cells (~5%) (Fig. 1B, Supplemental Fig. S1C). As we reported previously, tdTomato was preferentially expressed by a cluster abundantly expressing adipocyte-related genes such as Lpl, Adipoq, and Lepr; cells in this cluster (Adipo-CAR cells(11)) also expressed a number of genes encoding secreted and membrane-bound niche factors such as Cxcl12,(9,10) Kitl,(26) Gas6,(27) Fgf7,(28) and Il7.(29) These analyses confirm that Cxcl12-GFP+ CAR cells, as well as the pre-adipocyte-like Adipo-CAR subset and pre-osteoblast-like Osteo-CAR subset are non-hematopoietic, and predominantly belong to the mesenchymal stromal cell lineage.
The developmental relationships among CXCL12+ stromal cell subsets remain undefined. We subsequently analyzed the data set with RNA velocity, a computational approach that infers whether genes are actively being up- or downregulated in individual cells based on the ratio of spliced to unspliced transcripts.(23,24) Knowing which genes are being induced or repressed, in turn, yields “RNA velocity” vectors that predict the routes by which cell populations develop into each other. In our data set, this analysis successfully projected dynamic velocity vectors onto individual cells and predicted their future states, not only in hematopoietic cells but also in stromal cells, allowing us to infer the dynamic developmental relationships among identified subsets (Fig. 1C). Importantly, RNA velocity analysis indicates that Adipo-CAR and Osteo-CAR cells do not interconvert. This suggests that each CXCL12+ stromal cell subset may be independent and constitute its own mini-lineage without much contributing one another.
CXCL12+ stromal cells constitute the bone marrow microenvironment and interact with a range of hematopoietic cells. Because our single-cell data set contains expression information for ligands and receptors from both types of cells, it provides an opportunity to characterize these intercellular signaling interactions. Accordingly, we analyzed the data set with CellPhoneDB,(25) a computational approach that uses cell type–specific expression of annotated ligand-receptor gene pairs to quantify the evidence for signaling interactions between cell types. We particularly focused on three ligand-receptor pairs with known significant roles in stromal-hematopoietic interactions, Cxcl12-Cxcr4, Kit ligand (Kitl)-cKit (Kit), and Il7-Il7r pairs (Supplemental Fig. S1D). First, Cxcl12-Cxcr4 interactions were most notable between Adipo-CAR cells and hematopoietic cells (including myeloid cells, B cells, and erythroid cells), as well as endothelial cells (Fig. 1D, red arrows; Fig. 1E, first row); importantly, Cxcl12-Cxcr4 interactions were not enriched in other stromal subsets. Second, Kitl-Kit interactions showed a pattern overlapping with but different from that of Cxcl12-Cxcr4 pairs. Notably, Kitl-Kit interactions were observed not only between Adipo-CAR cells and hematopoietic or endothelial cells but also between Adipo-CAR cells and other stromal cell subsets (Fig. 1D, purple arrows; Fig. 1E, 2nd row). Kitl-Kit interactions were also enriched between the end-state Stromal-cell-2 cells and hematopoietic cells (Fig. 1D, orange arrows; Fig. 1E, purple). Third, Il7-Il7r interactions were observed between a broad range of stromal cell subsets and hematopoietic or endothelial cells (Fig. 1E, third row). Interestingly, only Il7-Il7r interactions were notable in Osteo-CAR cells (Fig. 1E, green).
In summary, these integrative single-cell RNA-seq analyses predict the pre-adipocyte-like subset of CXCL12+ stromal cells (Adipo-CAR cells) as a self-contained group of cells that exuberantly communicate with hematopoietic cells through Cxcl12-Cxcr4 ligand-receptor interactions, in a more specific manner than through Kitl-Kit or Il7-Il7r interactions.
Interactive nature of CXCL12+ pre-adipocyte-like stromal cells in vivo
The computational prediction that CXCL12+ pre-adipocyte-like stromal cells interact with hematopoietic cells through ligand-receptor relationships prompted us to further examine their interactions in vivo at a single-cell level. To this end, we first interrogated individual collagenase-digested Cxcl12-GFP+ cells under the ImageStream imaging cytometer. Cells with a reticular morphology, marked with cytoplasmic GFP, were clearly distinguishable from round hematopoietic cells marked by membranous signals emanating from APC-conjugated CD45 antibodies (Fig. 2A). A substantial fraction of Cxcl12-GFP+ with an irregular reticular morphology physically intertwined with CD45+ hematopoietic cells in a way indistinguishable from purely single cells (295/629 cells observed; Fig. 2A). Therefore, many Cxcl12-GFP+ cells are physically coupled with hematopoietic cells in single-cell preparation, underscoring their close association with and robust adherence to hematopoietic cells.
Fig 2.

Interactive nature of CXCL12+ pre-adipocyte-like stromal cells in vivo. (A) ImageStream analysis of Control cells isolated from Cxcl12GFP/+ bone marrow at P28, stained for CD45 antibodies. Leftmost panels: bright field; left center panels: Cxcl12-GFP; right center panels: CD45-APC; rightmost panels: merged. Scale bar = 7 μm. (B) Flow cytometry analysis of bone marrow cells without enzymatic digestion (leftmost), with collagenase (Liberase) digestion (left center) and with collagenase (Liberase) and protease (Pronase) digestion (right center). Rightmost panels: percentage of GFP+ or ZsGreen+ cells. Upper panels: Cxcl12GFP/+ femur bone marrow at P28. Center panels: Cxcl12-creER; R26RZsGreen femur bone marrow at P28 (pulsed at P21). Lower panels: Col1a1(2.3 kb)-GFP femur bone marrow at P28. n = 4 (Cxcl12-GFP+: No Dig, Cxcl12-ZsG+: No Dig, Lib and Lib + Pro, Col1a1-GFP+: No Dig, Lib and Lib + Pro), n = 7 (Cxcl12-GFP+: Lib and Lib + Pro) mice. (C) Percentage of CD45+GFP+/ZsG+ cells among total GFP+/ZsG+ cells. Lib = Liberase (collagenase) only. Lib + Pro = combined Liberase and Pronase (protease). (D) Percentage of HSCs (upper left), B-cell precursors (upper right), and granulo/monocytes (bottom) in total GFP+/ZsG+ fraction (collagenase digestion). (E) CD79b staining of Cxcl12GFP/+; Cxcl12-creER; R26RtdTomato bone marrow at P28 (pulsed at P21). Boxed areas = magnified views of representative GFP+tdTomatoneg (left) and GFP+tdTomato+ (middle) cells. Right: Percentage of CD79b+ B-cell-attaching CAR cells, GFP+tdTomatoneg (blue) and GFP+tdTomato+ (red) cells. Two-tailed, one-way ANOVA followed by Tukey’s post hoc test. All data are presented as mean ± SD.
We subsequently asked whether the pre-adipocyte-like subset of CXCL12+ stromal cells adhere to hematopoietic cells in single-cell preparation using flow cytometry. We consistently observe that the quantity and the type of marrow stromal cells released into single-cell suspension vary depending on the digestion protocol. We, therefore, tested three different protocols: (i) without enzymatic digestion; (ii) collagenase (Liberase) digestion; and (iii) collagenase (Liberase) and protease (Pronase) digestion, to harvest bone marrow cells from P28 Cxcl12GFP/+ reporter mice (Cxcl12-GFP+ cells) and Cxcl12-creER; R26RZsGreen/+ mice (pulsed at P21, Cxcl12CE-ZsGreen+ cells). As a control, we simultaneously analyzed bone marrow cells marked by Col1a1(2.3 kb)-GFP that is specifically active in osteoblastic cells (Col1a1-GFP+ cells). Bone marrow cells isolated from above mentioned mice and protocols were stained for a pan-hematopoietic marker CD45.
Collagenase digestion is essential for harvesting these stromal cells for flow cytometry, as the percentage of fluorescently marked cells by Cxcl12-GFP, Cxcl12CE-ZsGreen, and Col1a1-GFP among total forward scatter (FSC)/side scatter (SSC)-gated single cells increased significantly by Liberase digestions (Fig. 2B, left two panels). Surprisingly, at odds with the non-hematopoietic nature of CXCL12+ cells identified by scRNA-seq analyses, a substantial fraction of collagenase-digested Cxcl12-GFP+ and Cxcl12CE-ZsGreen+ cells appeared in a CD45+ fraction in flow cytometry (CD45+ cells: 13.1 ± 4.6% and 29.8 ± 6.8% of Cxcl12-GFP+ and Cxcl12CE-ZsG+ cells, respectively, compared with 4.9 ± 1.4% of Col1a1-GFP+ cells; Fig. 2C, Lib). In particular, Cxcl12CE-ZsGreen+ cells demonstrated the highest fraction of cells in a CD45+ fraction among the three cell types (Fig. 2C, red). Therefore, Cxcl12-GFP+ stromal cells, particularly those in the Cxcl12-creER+ Adipo-CAR subset, have a strong propensity to adhere to CD45+ hematopoietic cells in collagenase-dissociated single-cell preparation.
Protease digestion improved the harvest of Cxcl12-creER+ Adipo-CAR cells, as the percentage of Cxcl12CE-ZsGreen+ cells increased significantly with addition of Pronase in the digestion cocktail (Fig. 2B, second row). Importantly, addition of Pronase significantly decreased the percentage of Cxcl12-GFP+ and Cxcl12CE-ZsGreen+ cells in a CD45+ fraction (Fig. 2C, Lib + Pro), indicating that adherence of CD45+ hematopoietic cells to CXCL12+ stromal cells is protease-sensitive.
To define identities of CD45+ hematopoietic cells conjugating with CXCL12+ cells in collagenase-dissociated single-cell preparation, we further stained bone marrow cells for a panel of cell surface markers for hematopoietic stem and progenitor cells (CD150, CD48, and CD41), B cell precursors (CD19 and CD79b), and granulocytes and monocytes (CD11b, Gr-1,and F4/80). Virtually none of the CD45+ cells in Cxc12-GFP+, Cxcl12CE-ZsGreen+, or Col1a1-GFP+ fractions were in a CD150+CD41negCD48neg fraction (Fig. 2D, left upper panel), indicating that HSCs do not adhere to these mesenchymal cells. In contrast, approximately 20% to 30% of these CD45+ cells were in a CD11b+F4/80+Gr1+ fraction, indicating that monocytes and granulocytes robustly attach to these mesenchymal cells (Fig. 2D, left lower panel). Strikingly, more than 30% of CD45+Cxcl12CE-ZsGreen+ cells were in a CD19+CD79b+ fraction (Fig. 2D, right panel), demonstrating that B-cell precursors might preferentially adhere to Cxcl12-creER+ Adipo-CAR cells, as expected from the previous study.(20)
Mature bone marrow adipocytes extensively intertwine with cells of the myeloid and lymphoid lineages at the electron microscopy level.(30) To confirm if Cxcl12-creER+ pre-adipocyte-like stromal cells adhere to B cells in vivo, we further stained P28 bone marrow sections of Cxcl12GFP/+; Cxcl12-creER; R26RtdTomato/+ mice (pulsed at P21, Cxcl12CE-tdTomato+ cells) with CD79b, a canonical B cell marker. Quantification of CXCL12+ stromal cell subsets—Cxcl12-GFP+Cxc12CE-tdTomatoneg for Osteo-CAR cells and Cxcl12-GFP+Cxc12CE-tdTomato+ for Adipo-CAR cells—adherent to CD79b+ B cells revealed that a significantly higher fraction of Adipo-CAR cells was attached to B cells in vivo than that of Osteo-CAR cells (Adipo-CAR cells: 62.3 ± 6.9%; Osteo-CAR cells: 45.8 ± 7.8% of cells adherent to CD79b+ B cells) (Fig. 2E). Therefore, these findings indicate that a substantial fraction of CXCL12+ pre-adipocyte-like stromal cells are physically coupled with hematopoietic cells, particularly with B-cell precursors and monocytes/granulocytes, in vivo and in single-cell preparation.
CXCL12 is functionally important for maintaining stromal-hematopoietic coupling
We subsequently set out to investigate how CXCL12 functionally affects physical coupling between CXCL12+ stromal cells and hematopoietic cells that broadly express its cognate receptor, CXCR4. To this end, we conditionally deleted CXCL12 from bone marrow stromal cells and their precursor cells, using Col2a1-cre(31) and Cxcl12-floxed alleles.(9) A great majority of Cxcl12-GFP+ cells are derived from mesenchymal precursor cells expressing Col2a1-cre.(32) Conditional deletion of CXCL12 using Prrx1-cre and Osx-cre results in a marked loss of HSCs and B-lymphoid progenitors, respectively, in bone marrow.(9,10) We analyzed littermate mice with corresponding genotypes of Cxcl12GFP/fl; R26RtdTomato/+ (Control) and Cxcl12GFP/fl; Col2a1-cre; R26RtdTomato/+ (Col2a1-CXCL12 cKO) at P21. In the latter cKO mice, Cxcl12-GFP+ cells fail to express functional CXCL12 but continue to express GFP in a Cxcl12 locus-dependent manner (ΔCXCL12 cells, Fig. 3A). Histologically, a reduced cellularity in hematopoietic cells was observed in Col2a1-CXCL12 cKO bone marrow without any conspicuous change in Cxcl12-GFP+ cells (Fig. 3B). Further, flow cytometry analysis revealed that CD45+ cells were significantly reduced in Col2a1-CXCL12 cKO bone marrow (CD45+ cells, Control: 46.5 ± 1.0%; Col2a1-CXCL12 cKO: 21.1 ± 1.8% of total single cells; Fig. 3C, left three panels). Correspondingly, ΔCXCL12 Cxcl12-GFP+ cells had a significantly lower percentage of cells in the CD45+ fraction than Control Cxcl12-GFP+ cells (CD45+ %, Control: 7.3 ± 2.1%; Col2a1-CXCL12 cKO: 3.9 ± 0.6%; Fig. 3C, right two panels). Therefore, functional CXCL12 is important for maintaining physical coupling between CXCL12+ stromal cells and CD45+ hematopoietic cells.
Fig 3.

CXCL12 is functionally important for maintaining stromal-hematopoietic coupling. (A) Conceptual diagram of CXCL12 conditional deletion experiment. Col2a1-cre; Cxcl12GFP/+; R26RtdTomato mice were mated with Cxcl12fl/fl mice to generate Col2a1-cre; Cxcl12GFP/fl; R26RtdTomato (Col2a1-CXCL12 cKO) and Cxcl12GFP/fl; R26RtdTomato (Control) mice. In Col2a1-CXCL12 cKO mice, Cxcl12-GFP+ cells lose functional CXCL12 expression but still express GFP in a Cxcl12 promoter/enhancer-dependent manner. (B) CD45 staining of Control (left panels) and Col2a1-CXCL12 cKO (right panels) bone marrow. Dotted area = magnified view of Cxcl12-GFP+ reticular stromal cells and neighboring CD45+ hematopoietic cells. Green = Cxcl12-GFP; red = tdTomato; magenta = CD45-Alexa647; gray = DAPI. Scale bar = 10 μm. n = 4 mice per group. (C) Flow cytometry analysis of Control and Col2a1-CXCL12 cKO bone marrow cells. The percentage of CD45+ cells in total bone marrow cells (center) and CD45+GFP+ cells in whole GFP+ fraction (right). n = 4 mice per group. Two-tailed, Mann-Whitney U test. All data are presented as mean ± SD.
Paradoxical overrepresentation of hematopoietic signature in CXCL12+ stromal cells
We further defined molecular changes associated with loss of functional CXCL12. To achieve this goal, we performed a comparative bulk RNA-seq analysis of Cxcl12-GFP+ cells isolated by collagenase digestion from Control and Col2a1-CXCL12 cKO bone marrow using fluorescence-activated cell sorting (FACS) (Fig. 4A, Control and ΔCXCL12 cells, respectively). An unsupervised clustering analysis demonstrated that loss of CXCL12 induced a consistent change in the transcriptome of Cxcl12-GFP+ cells (Supplemental Fig. S2A). Importantly in ΔCXCL12 cells, a significant reduction of reads was observed in loxP-flanked exon 2 as well as in its downstream exons (93%, 86%, and 60% reduction for exon 2, 3, and 4, respectively, Fig. 4B), confirming that a significant functional loss of CXCL12 occurred in ΔCXCL12 cells.
Fig 4.

Paradoxical overrepresentation of hematopoietic signature in CXCL12+ stromal cells. (A) FACS-sorting of Control (green box) and ΔCXCL12 (cKO) cells (red box). Left three panels: gating strategy for bone marrow cells isolated from Cxcl12GFP/fl; R26RtdTomato (Control) and Col2a1-cre; Cxcl12GFP/fl; R26RtdTomato mice (Col2a1-CXCL12 cKO). Right panels: x axis: Col2a1-cre-tdTomato; y axis: Cxcl12-GFP. Cells within the colored boxes were individually sorted. (B, C) Comparative bulk RNA-seq analysis of ΔCXCL12 cells. (B) Structure of Cxcl12-floxed allele (left) and expression level of each Cxcl12 exon in Col2a1-cre; Cxcl12GFP/fl; R26RtdTomato mice (Col2a1-CXCL12 cKO) (right). (C) Heat map of differentially expressed genes (DEGs). Left panel: Heat map of 1206 DEGs with hierarchical clustering based on DESeq2 algorithm. Each column represents an individual biological sample; each row represents a DEG. Red in heat map = higher expression; blue in heat map = lower expression. Center and right panels: extract of heat map data. Star = statistically significant difference between Control and ΔCXCL12. n = 3 biological replicates per group. (D) Venn diagram of highly expressed genes among Adipo-CAR cells, Osteo-CAR cells, and ΔCXCL12 cells from the combinational data of scRNA-seq (Cxcl12GFP/+; Cxcl12-creER; R26RtdTomato) and bulk RNA-seq ΔCXCL12).
Notably, 1195 genes were differentially expressed between the two groups (Fig. 4C, Supplemental Fig. S2B, C). Surprisingly, the Pathway analysis revealed the most significant enrichment of hematopoietic cell lineage (KEGG:04640), phagosome (KEGG:04145), and chemokine signaling pathway (KEGG:04062) (Supplemental Table S1A). The Gene Ontology analysis revealed significant enrichment of a number of terms related to immune responses and hematopoiesis pertinent to myeloid cell and B-cell functions (Supplemental Table S1B). Particularly, marker genes associated with hematopoietic cells, such as B-cell precursors and myeloid cells, including Cxcr4, Ptprc (CD45), Cd34, Cd79b, and Itgam (CD11b), were unanimously downregulated in ΔCXCL12 cells (Fig. 4C, right panel). We assume that this unusual transcriptomic change occurring in ΔCXCL12 cells might be the consequence of loss of intertwining hematopoietic cells, which could result in underrepresentation of the hematopoietic markers in this group.
We next compared 235 genes upregulated in ΔCXCL12 cells with cell type–specific marker genes for the pre-adipocyte-like subset (Adipo-CAR, 662 genes) and the pre-osteoblast-like subset (Osteo-CAR, 305 genes) that were identified in the above-described scRNA-seq analysis. ΔCXCL12 genes overlapped substantially with Adipo-CAR genes (79/235, 34%), but not with Osteo-CAR genes (3/235, 1.3%); the overlapping genes included representative pre-adipocyte marker genes, such as Sfrp4, Adipoq, Ebf3, and Lepr (Fig. 4D). Therefore, Adipo-CAR signatures become significantly enriched in Cxcl12-GFP+ cells upon loss of functional CXCL12. This result supports the scenario that Adipo-CAR cells lose their physical coupling with hematopoietic cells upon loss of functional CXCL12, which results in overrepresentation of Adipo-CAR signatures in the ΔCXCL12 transcriptome. Taken together, these findings demonstrate that loss of CXCL12 causes significant impairment in stromal-hematopoietic coupling of CXCL12+ pre-adipocyte-like stromal cells in bone marrow.
CXCL12 loss causes rampant adipogenesis by direct conversion of stromal pre-adipocytes
Lastly, we set out to define the functional consequence of loss of CXCL12 in the postnatal bone marrow stromal compartment. For this purpose, we analyzed above-described littermate mice (Control: Cxcl12GFP/fl; R26RtdTomato/+, and Col2a1-CXCL12 cKO: Cxcl12GFP/fl; Col2a1-cre; R26RtdTomato/+) at the juvenile stage of P21 and the adult stage of 3 M. Histological sections of distal femurs were stained for LipidTOX to visualize marrow adipocytes. Previously reported models of conditional CXCL12 deletion by Prrx1-cre and Osx-cre develop marrow adiposity early in the postnatal stage, particularly in the metaphyseal marrow space.(21) In contrast, Col2a1-CXCL12 cKO bone marrow did not develop premature adiposity at the juvenile stage of P21 (Fig. 5A). However, there was a sudden onset of marrow adiposity in the Col2a1-CXCL12 cKO bone marrow at the adult stage of 3 M. Remarkably, LipidTOX+ marrow adipocytes were extensively formed throughout the marrow space, not only in the metaphysis but also in the diaphysis (Fig. 5B), in a distinct pattern from marrow adiposity observed in the previous models.(21) Quantification of LipidTOX+ cells revealed a significant increase in marrow adipocytes in Col2a1-CXCL12 cKO bone marrow at 3 M but not at P21 (Fig. 5C). At higher magnification, both Cxcl12-GFP+tdTomato+ΔCXCL12 cells and Cxcl12-GFPnegtdTomato+ΔCXCL12 cells developed into marrow adipocytes (Fig. 5B, lower panel), suggesting that CXCL12-negative cells could also develop into marrow adipocytes in the absence of CXCL12.
Fig 5.
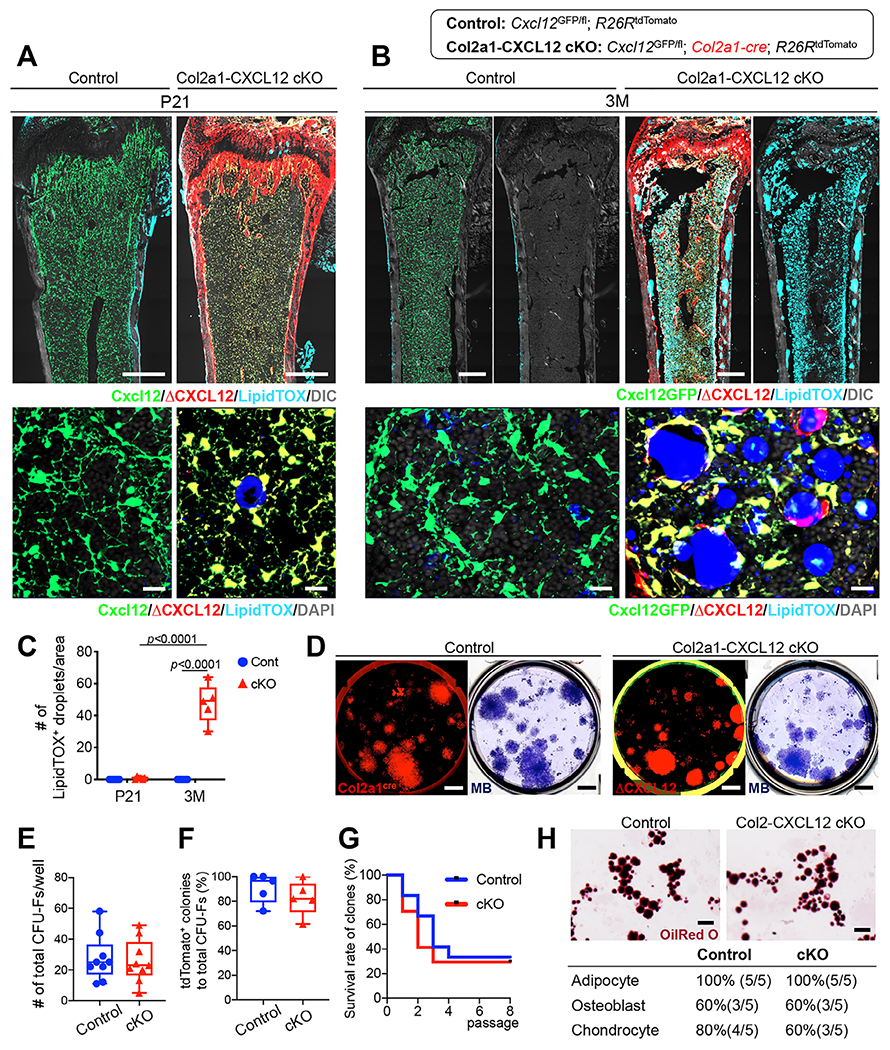
CXCL12 loss causes rampant adipogenesis by direct conversion of stromal pre-adipocytes. (A, B) Conditional deletion of CXCL12 using Control and Col2a1-CXCL12 cKO. Femur bone marrow sections with distal epiphysis on top (upper panels) and magnified view of marrow space (lower panels), at 21 days of age (P21; A) and 3 months of age (3 M; B). Scale bars = 500 μm (upper), 20 μm (lower). (C) Number of LipidTOX+ droplets per field. n = 6 (Control at P21 and 3 M, cKO at P21), n = 5 (cKO at 3 M) per group. Two-tailed, Mann–Whitney U test. (D–H) CFU-F assay of bone marrow cells isolated from Control and Col2a1-CXCL12 cKO mice at 8 W. Red = Col2a1-cre-tdTomato (Control) and Col2a1-cre-tdTomato (ΔCXCL12, cKO). Purple = methylene blue staining. Scale bars = 5 mm. (E) Number of total CFU-Fs. n = 9 mice per group. (F) Percentage of tdTomato+ colonies to total CFU-Fs per well. n = 5 mice per group. (G) Survival curve of Control and Col2a1-CXCL12 cKO clones. n = 12 (Control), n = 17 (ΔCXCL12, cKO) clone. (H) Trilineage differentiation assay of Control and Col2a1-CXCL12 cKO clones. Upper panels: Oil red O staining. Lower panels: Rate of trilineage differentiation. Scale bars = 100 μm. All data are presented as mean ± SD.
The pre-adipocyte-like subset of CXCL12+ stromal cells marked by Cxcl12-creER possesses limited colony-forming activity ex vivo;(16) therefore, colony-forming activities are likely to be highly enriched in the pre-osteoblast-like subset of CXCL12+ stromal cells. To determine whether this observed rampant adipogenesis induced by CXCL12 deficiency involves mesenchymal progenitor cell compartments, we further examined ex vivo properties of colony-forming unit fibroblasts (CFU-Fs) isolated from Control and Col2a1-CXCL12 cKO bone marrow at P21 (Fig. 5D). The frequency of CFU-Fs was not significantly different between Control and Col2a1-CXCL12 cKO bone marrow (Control: 27.6 ± 15.0 colonies/well, Col2a1-CXCL12 cKO: 25.7 ± 14.0 colonies/well; Fig. 5E). Furthermore, both Control and ΔCXCL12 cells contributed to an equally large fraction of CFU-Fs (tdTomato+ CFU-Fs: Control: 91.0 ± 12.0%; Col2a1-CXCL12 cKO: 82.7 ± 14.0%; Fig. 5F), indicating that loss of CXCL12 does not induce inherent defects in colony-forming activities of bone marrow mesenchymal progenitor cells.
Subsequently, to determine whether Control and ΔCXCL12 clones can self-renew in vitro, we isolated individual primary tdTomato+ colonies and subcultured them further up to eighth generations, using the method we previously developed.(33) Both Control and ΔCXCL12 clones could survive for multiple passages. While 33.3% (4/12) of Control clones could be passaged for eight generations, 29.4% (5/17) of ΔCXCL12 clones could be passaged to the same generations, without any statistical significance between the two groups (Fig. 5G). We further performed trilineage differentiation assays of Control and ΔCXCL12 clones. Both Control and ΔCXCL12 clones exhibited a similar differentiation potential to adipocytes, osteoblasts, and chondrocytes in vitro, with all of the tested clones possessing an adipogenic potential (Fig. 5H).
Thus, loss of CXCL12 does not appear to impair in vitro self-renewal and differentiation potential of bone marrow mesenchymal progenitor cells. We postulate that mild defects in bone marrow mesenchymal progenitor activities are unlikely to account for massive adiposity occurring in the Col2a1-CXCL12 cKO adult bone marrow. These findings support the notion that CXCL12 deficiency induces rampant adipogenesis in adult bone marrow through direct conversion of CXCL12+ pre-adipocyte-like stromal cells to mature lipid-laden adipocytes.
Discussion
Taken together, our findings demonstrate that CXCL12+ pre-adipocyte-like bone marrow stromal cells have a propensity to adhere to hematopoietic cells, particularly to B-cell precursors and monocytes and granulocytes, and loss of such physical coupling due to CXCL12 deficiency may lead to extensive marrow adipogenesis by directly converting these pre-adipocytes to lipid-laden mature adipocytes. This process appears to involve a non-cell-autonomous mechanism as proposed by the previous study;(21) in fact, CXCL12-deficient mesenchymal progenitor cells do not exhibit apparent cell-intrinsic defects in their cell fates. We believe that our study reveals an interactive nature of an adipogenic subset of CXCL12+ stromal cells and highlights a possible mechanism that regulates marrow adipogenesis through intercellular physical coupling.
The prevailing view is that marrow adiposity is induced by a cell fate shift of bone mesenchymal progenitor cell populations due to cell-intrinsic changes. For example, loss of Wnt/β-catenin signaling,(34) intracrine VEGF signaling,(35) or PTH/PTHrP receptor signaling(36) and its downstream Gsα signaling(37) induces an imbalance in differentiation of bone mesenchymal progenitor cells and promotes adipocyte differentiation. We believe that our study sheds light on a previously unappreciated mechanism of marrow adipogenesis, in which pre-adipocyte-like Adipo-CAR cells are directly converted into mature adipocytes without involving mesenchymal progenitor cells. Whether this non-cell-autonomous anti-adipogenic effect is unique to CXCL12 remains to be determined. It is possible that other abundantly expressed hematopoiesis-supporting cytokines, such as SCF, similarly possess anti-adipogenic effects by regulating stromal-hematopoietic physical coupling.
CXCL12+ marrow stromal cells regulate the three major functions of bone marrow, which are hematopoiesis, osteogenesis, and adipogenesis. These three functions are closely interrelated;(38) the most described is the relationship between osteogenesis and hematopoiesis. In fact, osteoblasts directly or indirectly support marrow hematopoiesis by regulating hematopoietic stem and progenitor cells.(39–42) In addition, marrow adipocytes either promote the regeneration of hematopoiesis by supporting primitive hematopoietic cells(43) or negatively regulate hematopoietic progenitors in the bone marrow microenvironment.(38) Inverse correlation between marrow osteogenesis and adipogenesis has long been described.(44,45) However, the least described is how hematopoiesis regulates osteogenesis and adipogenesis. Our findings provide preliminary evidence that hematopoietic cells may directly or indirectly regulate marrow adipogenesis in a manner possibly dependent on CXCL12-mediated physical coupling, although details need to be clarified by further experimentation.
Technically, our findings call attention to the important fact that an adipogenic subset of CXCL12+ stromal cells can be often co-purified with hematopoietic cells in single-cell preparation, which may impact the outcome of single-cell studies of bone marrow stromal cells. Our data clearly demonstrate that a combination of collagenase (Liberase) and protease (Pronase) can effectively dissociate stromal-hematopoietic physical coupling and reduce undesirable co-purification in flow cytometry. In fact, our laboratory consistently experiences reduced hematopoietic contamination in the single-cell RNA-seq data sets of FACS-isolated marrow stromal cells upon the combinatory use of collagenase and protease. We therefore recommend to use both collagenase and protease for future single-cell studies aiming to understand these highly interactive bone marrow stromal cells.
Our study highlights the interactive nature of CXCL12+ pre-adipocyte-like reticular stromal cells that constitutes an important component of the bone marrow environment. These CXCL12+ pre-adipocyte-like stromal cells marked by Cxcl12-creER are likely to overlap with the recently described marrow adipocyte lineage precursor (MALP) cells marked by adiponectin (Adipoq)-cre.(15) Interestingly, however, the outcome of conditional cell ablation experiments differs between these two cell types; Cxcl12-creER+ cell ablation causes impaired bone regeneration, whereas Adipoq-cre+ cell ablation induces increased osteogenesis. Therefore, these two types of marrow pre-adipocytes are functionally different. Further studies are needed to decipher the relationship among different classes of pre-adipocytes in bone marrow and their functions in physiological and pathological conditions.
In conclusion, we find that stromal-hematopoietic physical coupling plays important roles in regulating murine marrow adipogenesis by directly converting marrow fat precursor cells to mature fat cells. This may provide important insight into mechanisms underlying age-related increase in marrow adiposity commonly observed in humans.
Supplementary Material
Acknowledgments
This research was supported by NIH R01DE026666 to NO, R03DE027421 to WO, and R01HG010883 to JDW. We thank T Nagasawa (Osaka University) for Cxcl12-GFP mice, DC Link (Washington University) for Cxcl12-floxed mice, M Pihalja and K Saiya-Cork (University of Michigan Flow Cytometry Core), and R Tagett (University of Michigan Bioinformatics Core) for supporting this study.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/jbmr.4282.
References
- 1.Crane GM, Jeffery E, Morrison SJ. Adult haematopoietic stem cell niches. Nat Rev Immunol. 2017;17(9):573–590. [DOI] [PubMed] [Google Scholar]
- 2.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Q, Frenette PS. Niches for hematopoietic stem cells and their progeny. Immunity. 2018;48(4):632–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ara T, Tokoyoda K, Sugiyama T, Egawa T, Kawabata K, Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19(2):257–267. [DOI] [PubMed] [Google Scholar]
- 6.Omatsu Y, Sugiyama T, Kohara H, et al. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33(3):387–399. [DOI] [PubMed] [Google Scholar]
- 7.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seike M, Omatsu Y, Watanabe H, Kondoh G, Nagasawa T. Stem cell niche-specific Ebf3 maintains the bone marrow cavity. Genes Dev. 2018;32(5–6):359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenbaum A, Hsu YM, Day RB, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495(7440):227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495(7440):231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baccin C, Al-Sabah J, Velten L, et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol. 2020;22(1):38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baryawno N, Przybylski D, Kowalczyk MS, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915–32.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tikhonova AN, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolock SL, Krishnan I, Tenen DE, et al. Mapping distinct bone marrow niche populations and their differentiation paths. Cell Rep. 2019;28(2):302–11.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong L, Yao L, Tower RJ, et al. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. Elife. 2020;9:e54695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita Y, Nagata M, Kozloff KM, et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat Commun. 2020;11(1):332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzeng YS, Li H, Kang YL, Chen WC, Cheng WC, Lai DM. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood. 2011;117(2):429–439. [DOI] [PubMed] [Google Scholar]
- 18.Asada N, Kunisaki Y, Pierce H, et al. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat Cell Biol. 2017;19(3):214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25(6):977–988. [DOI] [PubMed] [Google Scholar]
- 20.Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20(6):707–718. [DOI] [PubMed] [Google Scholar]
- 21.Tzeng YS, Chung NC, Chen YR, Huang HY, Chuang WP, Lai DM. Imbalanced osteogenesis and adipogenesis in mice deficient in the chemokine Cxcl12/Sdf1 in the bone mesenchymal stem/progenitor cells. J Bone Miner Res. 2018;33(4):679–690. [DOI] [PubMed] [Google Scholar]
- 22.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. 2019;177(7):1873–87.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature. 2018;560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. [DOI] [PubMed] [Google Scholar]
- 25.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. Cell-PhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481(7382):457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiozawa Y, Pedersen EA, Patel LR, et al. GAS6/AXL axis regulates prostate cancer invasion, proliferation, and survival in the bone marrow niche. Neoplasia. 2010;12(2):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishino R, Minami K, Tanaka S, et al. FGF7 supports hematopoietic stem and progenitor cells and niche-dependent myeloblastoma cells via autocrine action on bone marrow stromal cells in vitro. Biochem Biophys Res Commun. 2013;440(1):125–131. [DOI] [PubMed] [Google Scholar]
- 29.Cordeiro Gomes A, Hara T, Lim VY, et al. Hematopoietic stem cell niches produce lineage-instructive signals to control multipotent progenitor differentiation. Immunity. 2016;45(6):1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robles H, Park S, Joens MS, Fitzpatrick JAJ, Craft CS, Scheller EL. Characterization of the bone marrow adipocyte niche with three-dimensional electron microscopy. Bone. 2019;118:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26(2):145–146. [PubMed] [Google Scholar]
- 32.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16(12):1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuhashi K, Ono W, Matsushita Y, et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563(7730):254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song L, Liu M, Ono N, Bringhurst FR, Kronenberg HM, Guo J. Loss of wnt/β-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res. 2012;27(11):2344–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Berendsen AD, Jia S, et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J Clin Invest. 2012;122(9):3101–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y, Hanai JI, Le PT, et al. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab. 2017;25(3):661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha P, Aarnisalo P, Chubb R, et al. Loss of Gsα early in the osteoblast lineage favors adipogenic differentiation of mesenchymal progenitors and committed osteoblast precursors. J Bone Miner Res. 2014;29(11):2414–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, Daley GQ. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460(7252):259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M, Tao F, Venkatraman A, et al. N-cadherin-expressing bone and marrow stromal progenitor cells maintain reserve hematopoietic stem cells. Cell Rep. 2019;26(3):652–69.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. [DOI] [PubMed] [Google Scholar]
- 41.Christodoulou C, Spencer JA, Yeh SA, et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature. 2020;578(7794):278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Méndez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou BO, Yu H, Yue R, et al. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat Cell Biol. 2017;19(8):891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fazeli PK, Horowitz MC, MacDougald OA, et al. Marrow fat and bone—new perspectives. J Clin Endocrinol Metab. 2013;98(3)935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadie-Van Gijsen H, Crowther NJ, Hough FS, Ferris WF. The interrelationship between bone and fat: from cellular see-saw to endocrine reciprocity. Cell Mol Life Sci. 2013;70(13):2331–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


