Abstract
E2F-mediated transcription is thought to involve binding of an E2F-pocket protein complex to promoters in the G0 phase of the cell cycle and release of the pocket protein in late G1, followed by release of E2F in S phase. We have tested this model by monitoring protein-DNA interactions in living cells using a formaldehyde cross-linking and immunoprecipitation assay. We find that E2F target genes are bound by distinct E2F-pocket protein complexes which change as cells progress through the cell cycle. We also find that certain E2F target gene promoters are bound by pocket proteins when such promoters are transcriptionally active. Our data indicate that the current model applies only to certain E2F target genes and suggest that Rb family members may regulate transcription in both G0 and S phases. Finally, we find that a given promoter can be bound by one of several different E2F-pocket protein complexes at a given time in the cell cycle, suggesting that cell cycle-regulated transcription is a stochastic, not a predetermined, process.
The E2F family of transcription factors plays an important role in the regulation of gene expression at the G1/S-phase transition of the mammalian cell cycle (see reference 16 for an extensive review of the E2F regulatory pathway). E2F binding sites are found in the promoters of genes whose products are required for nucleotide synthesis (e.g., dihydrofolate reductase [DHFR] and thymidine kinase [TK]), for DNA replication (e.g., DNA polymerase α and cdc6), and for cell cycle progression (e.g., cyclin E, cyclin D1, c-myc, b-myb, and cdc2). Transcription from each of these promoters increases during late G1 or early S phase, and this regulation is mediated by protein binding to one or more E2F binding sites (3, 11, 22, 30, 34, 43, 44). To date, eight members of the E2F family have been identified: six E2F proteins (E2F1 to -6) and two DP proteins (DP1 and DP2). The E2F and DP proteins bind to DNA as a heterodimer and can function as activators of transcription. Alternatively, E2F-DP heterodimers can also repress transcription when complexed with members of the Rb family of pocket proteins (pRb, p107, and p130) due to the ability of the pocket proteins to bind to and mask the E2F transactivation domain and to recruit histone deacetylases (6, 17, 25). Individual E2Fs preferentially bind to different pocket proteins. E2F1, E2F2, and E2F3, for example, bind to pRb, while E2F4 predominantly binds to p130 and p107 and E2F5 binds to p130.
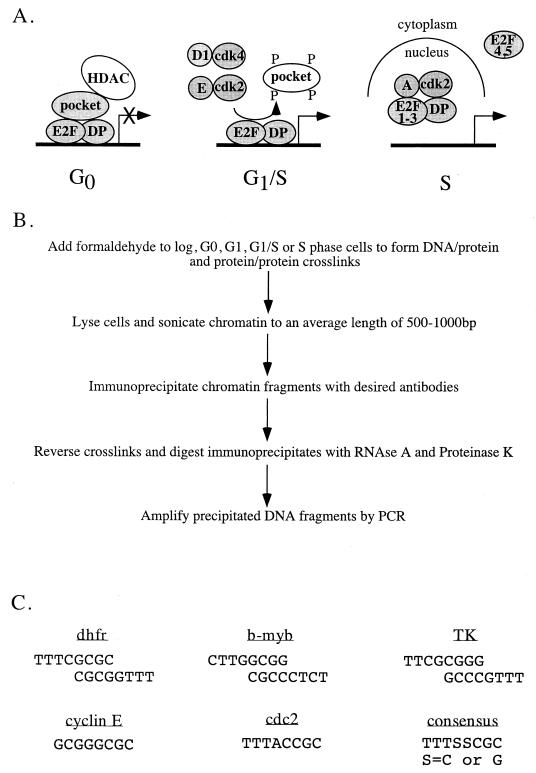
A popular model for how E2F family members regulate G1/S-phase-specific gene expression invokes a complex pattern of protein-protein and protein-DNA interactions that change as cells progress through the cell cycle (Fig. 1A). As depicted, transcription from E2F site-containing promoters is thought to be repressed in G0 phase due to the binding of a trimolecular E2F-DP-pocket protein complex and recruitment of histone deacetylase activity by the pocket protein component (6, 17, 25). As cells progress through the cell cycle, various cyclin–cyclin-dependent kinase (cdk) complexes phosphorylate the pocket proteins, causing release of the hyperphosphorylated pocket protein and associated proteins from the DNA-bound E2F-DP heterodimer (1, 2, 7). Finally, traversal through S phase is thought to be accompanied by cyclin-cdk-mediated phosphorylation of the DP subunit of E2F1-3–DP complexes, resulting in release of the heterodimers from the promoter DNA (15, 21, 42). In some cells, E2F4-containing complexes are thought to be inactivated by relocation to the cytoplasm (28, 38).
FIG. 1.
E2F-mediated transcriptional regulation. (A) The current model of E2F-mediated transcriptional regulation is thought to involve binding of an E2F-pocket protein complex to promoters in G0 phase of the cell cycle and release of the pocket protein in late G1, followed by release of E2F in S phase, either as a result of the action of cyclin-cdk's or due to changes in subcellular localization of E2F. (B) Shown is the protocol for formaldehyde cross-linking and immunoprecipitation used to detect E2F and pocket protein binding to endogenous target genes. (C) Shown are the E2F sites present in the promoters of the different target genes analyzed in this study. The DHFR, b-myb, and TK promoters have a set of inverted, overlapping E2F recognition sites whereas the cyclin E and cdc2 promoters have a single E2F recognition site.
Although this model is attractive, it is largely based upon circumstantial data, and several important questions remain unanswered. For example, the transcriptional activity of complexes containing E2F4 and E2F5 may be shut off during mid- to late S phase by association with unphosphorylated p107 or p130, rather than by relocation to the cytoplasm (10). Additionally, it is not known if E2F target gene specificity exists or if all six E2Fs bind to and regulate every target gene or if Rb family members display target gene specificity. Determination of target gene specificity has been difficult due to the fact that most cells studied to date contain all of the E2Fs and pocket proteins. Thus, most analyses of E2F and pocket protein binding specificity have been performed using in vitro systems or by overexpression of an individual E2F or pocket protein in cells. In vitro systems, however, cannot recapitulate the complex environment of living cells, and altering the relative amounts of individual proteins through overexpression may abolish important protein-protein interactions. Therefore, we wished to determine which E2Fs and pocket proteins bind to and regulate expression of specific target genes in intact cells under physiological conditions. We felt that the use of an unperturbed in vivo system was of particular importance in the analysis of E2F target gene specificity since regulation occurs in the context of cell cycle progression which cannot be mimicked in vitro and which is often altered when individual E2F proteins are overexpressed. Toward this goal, we have used a formaldehyde cross-linking and immunoprecipitation system to monitor protein-DNA and protein-protein interactions on E2F target genes in living cells. Importantly, this procedure has allowed us to determine the in vivo patterns of E2F and pocket protein binding to specific E2F target genes as cells progress through a cell cycle. Our results indicate that, while some promoters fit the proposed model for E2F-mediated transcriptional regulation, others do not, necessitating individualization of the current model. Furthermore, our data suggest that cell cycle-regulated activity is a stochastic event, as cells have the ability to form several different E2F-pocket protein complexes on a given promoter at each stage of the cell cycle.
MATERIALS AND METHODS
Cell culture and synchronization.
Cells were maintained in Dulbecco's modified Eagle medium (BRL-Life Technologies) supplemented with 5% bovine calf serum (BCS; BRL-Life Technologies) and 1% penicillin-streptomycin (BRL-Life Technologies) and grown in a 5% CO2 incubator. NIH 3T3 cells which have been stably transfected with a plasmid containing a 290-bp region of the murine DHFR promoter in which the E2F binding site has been mutated from TTTCGCGCCAAA to CCCTATATCAAA have been described previously (26). For synchronization, cells were trypsinized (BRL-Life Technologies) and plated in starvation medium, Dulbecco's modified Eagle medium supplemented with 0.5% BCS and 1% penicillin-streptomycin. After 60 h of growth in starvation medium, cells were either collected as G0-phase cells or stimulated to reenter the cell cycle by the addition of serum to a final concentration of 10%. G1-phase cells were harvested at 8 h after serum stimulation, G1/S-phase cells were harvested at 12 h, and S-phase cells were harvested at 16 h.
Replicate cultures of cells were trypsinized, fixed in ethanol, and stained with propidium iodide. Stained cells were analyzed on a FACS Caliber flow cytometer (Becton Dickinson) using CellQuest acquisition and analysis software. Pulse width and area allowed the exclusion of doublets. Cell cycle percentages were calculated with ModFit 2.0 software (Verity Software House).
RNA preparation and RT-PCR analysis.
Cytoplasmic RNA was prepared from either asynchronously growing or replicate plates of synchronized NIH 3T3 cells as previously described (34). For reverse transcription-PCR (RT-PCR) analysis, each reaction mixture contained 100 ng of RNA, 1× EZ buffer (Perkin-Elmer), 0.3 mM (each) deoxynucleoside triphosphates, 5 U of rTth DNA polymerase (Perkin-Elmer), 2.5 mM manganese acetate, and 4 nmol of each primer in a final reaction volume of 50 μl. Reaction mixtures were amplified for 1 cycle of 60°C for 30 min and 95°C for 2 min and 35 cycles of 95°C for 1 min, melting temperature of the primers for 2 min, and 60°C for 1 min followed by incubation at 60°C for 10 min. PCR products were resolved by electrophoresis through a 1.5% agarose gel and visualized by ethidium bromide staining. The sequences of the primers used for RT-PCR analysis are as follows: TK1a, 5′-CAGCATCTTGAACCTGGTGC-3′; TK1b, 5′-CTGAGAGGCAAAGAGCTTCC-3′; GAPDH1, 5′-TGGCCAAGGTCATCCATGAC-3′; GAPDH2, 5′-ATGTAGGCCATGAGGTCCAC-3′; cdc2a, 5′-CTTACACCAAATGCTCCAGG-3′; cdc2b, 5′-CGTTTGGCAGGATCATAGAC-3′; dhfr1a, 5′-CCAGCATATGCACAGGGTAC-3′; dhfr1b, 5′-CTCTCGTCTCCATGGAACAC-3′; cyclin E1a, 5′-GCAGAAGGTCTCAGGTTATC-3′; cyclin E1b, 5′-GTGGCCTCCTTAACTTCAAG-3′; b-myb 1560, 5′-CTCTCCAGCTCCAGGGTATC-3′; and b-myb 1211, 5′-GCACTGCAGTCATCCCAGCA-3′.
Formaldehyde cross-linking and immunoprecipitation.
Cells were formaldehyde cross-linked essentially as described previously (5). In brief, formaldehyde (Fisher Scientific) was added directly to tissue culture medium to a final concentration of 1%. Cross-linking was allowed to proceed for 10 min at room temperature and was then stopped by the addition of glycine to a final concentration of 0.125 M. Cross-linked cells were trypsinized, scraped, washed with 1× phosphate-buffered saline, and swelled in RSB buffer (3 mM MgCl2, 10 mM NaCl, 10 mM Tris-chloride [pH 7.4], and 0.1% IGEPAL CA-330 [Sigma]). Nuclei were pelleted by microcentrifugation and lysed by incubation in nuclear lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-chloride [pH 8.1], 0.5 mM phenylmethylsulfonyl fluoride, 100 ng of leupeptin per ml, and 100 ng of aprotinin per ml). The resulting chromatin solution was sonicated for three 30-s pulses at maximum power. After microcentrifugation, the supernatant was precleared with blocked protein A-positive Staph cells (Boehringer Mannheim), diluted 1:5 with dilution buffer (0.01% sodium dodecyl sulfate, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-chloride [pH 8.1], 167 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 100 ng of leupeptin per ml, and 100 ng of aprotinin per ml), and divided into aliquots. One microgram of antibody was added to each aliquot of chromatin and incubated on a rotating platform for 12 to 16 h at 4°C. Antibodies against E2F and pocket proteins, E2F-1 (sc-193), E2F-2 (sc-633), E2F-3 (sc-879), E2F-4 (sc-866), E2F-5 (a cocktail of sc-1083 and sc-999), pRb (sc-1538), p107 (sc-318), and p130 (sc-317), were purchased from Santa Cruz. Antibody-protein-DNA complexes were isolated by immunoprecipitation with blocked protein A-positive Staph A cells. Following extensive washing, bound DNA fragments were eluted and analyzed by subsequent PCR.
PCR analysis and Southern blotting.
Immunoprecipitates were dissolved in 30 μl of water, except for input samples which were diluted in 100 μl and then further diluted 1:100. Each reaction mixture contained 3 μl of immunoprecipitated chromatin, 1× Taq reaction buffer (Promega), 1.5 mM MgCl2, 50 ng of each primer, 1.7 U of Taq polymerase (Promega), 200 μM (each) deoxynucleoside triphosphates (Boehringer Mannheim), and 1 M betaine (Sigma) in a final reaction volume of 20 μl. PCR mixtures were amplified for 1 cycle of 95°C for 5 min, annealing temperature of the primers for 5 min, and 72°C for 3 min and 34 to 36 cycles of 95°C for 1 min, annealing temperature of the primers for 2 min, and 72°C for 1.5 min. PCR products were separated by electrophoresis through a 1.5% agarose gel and visualized by ethidium bromide intercalation. Alternatively, immunoprecipitates were amplified for 16 cycles of PCR, separated by electrophoresis on a 1.5% agarose gel, transferred to Hybond-N membranes (Amersham Life Science), and visualized by hybridization with a radiolabeled probe as previously described (4). Radiolabeled probes were generated by nick translating PCR products created by amplification of genomic DNA using the same primers as those used for analysis of the immunoprecipitates. Each experiment was performed a minimum of three times, and representative results are shown in Fig. 2, 3, 5, and 6. All of the log-phase results shown in Fig. 2A and 3 were generated using chromatin from the same experiment. Similarly, the results of Fig. 5 were generated using chromatin from one experiment, and the results of Fig. 6 were generated by PCR analysis of chromatin from a second experiment. The sequences of the primers used are as follows: myb+446, 5′-CAGAGCCAGGCCTCGCGCCTCATTG-3′; myb+858, 5′-TCAGGACTCAGGCTGCTCGAGCCGC-3′; dhfr+962, 5′-CGGCAATCCTAGGGTGAAGGCTGGT-3′; dhfr+1360, 5′-GGCTCCATTCAGCGACGAAAGGTGC-3′; cycE−134, 5′-AAGAACACGCCCCCCGGGAGGCCAC-3′; cycE+202, 5′-AAGCTGTGTCCGCCGCAGGCAGGCG-3′; cdc2−20, 5′-GGTAAAGCTCCCGGGATCCGCCAAT-3′; cdc2−358, 5′-GTGGACTGTCACTTTGGTGGCTGGC-3′; mAlb A, 5′-GGACACAAGACTTCTGAAAGTCCTC-3′; and mAlb B, 5′-TTCCTACCCCATTACAAAATCATA-3′. All primers were synthesized at the University of Wisconsin Biotechnology Center.
FIG. 2.
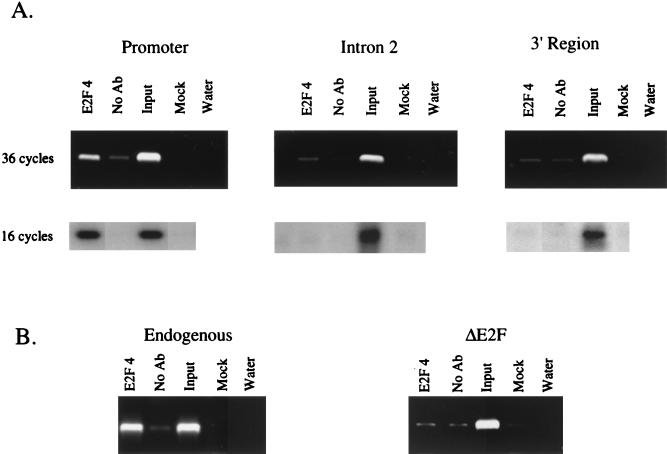
E2F4 binding to the DHFR gene is promoter specific and requires an intact E2F binding site. As a demonstration of the specificity of our formaldehyde cross-linking assay, we monitored binding of E2F4 to three regions of the DHFR gene in NIH 3T3 cells (A) and to both the endogenous and integrated DHFR promoter construct in NIH 3T3 NW luc cells (B). Cross-linked chromatin from asynchronously growing cells was incubated with an antibody against E2F4 or in the absence of antibody (No Ab). Immunoprecipitates from each sample were analyzed by PCR using primers specific for different regions of the DHFR promoter. As a positive control, a sample representing 0.03% of the total input chromatin (input) was included in the PCRs. Additional controls include a precipitation lacking both antibody and chromatin (Water).
FIG. 3.
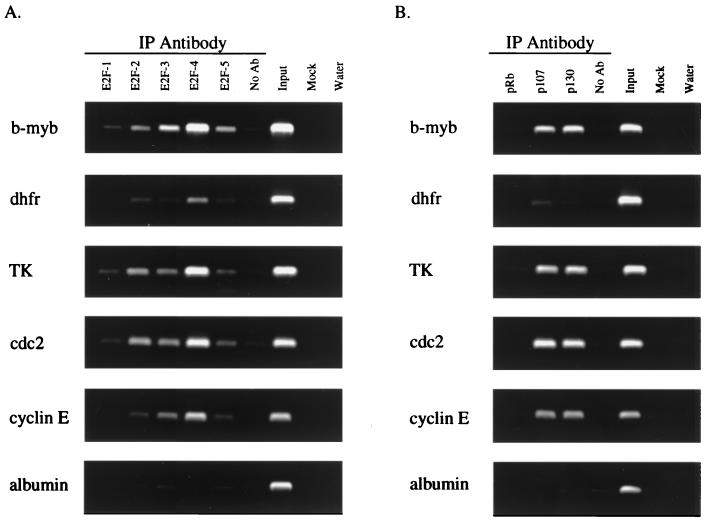
E2F target genes lack E2F and pocket protein binding specificity in asynchronously growing NIH 3T3 cells. The figure shows analysis of E2F (A) and pocket protein (B) binding to E2F target genes in asynchronously growing NIH 3T3 cells. Cross-linked chromatin from asynchronously growing cells was incubated with antibodies against E2F1-5, pRb, p107, or p130 or in the absence of antibody (No Ab). Immunoprecipitates from each sample were analyzed by PCR using primers specific for the different promoters. As a control, a sample representing 0.03% of the total input chromatin (Input) was included in the PCRs. This ensures that a low signal (as in the case of the DHFR samples precipitated with pocket protein antibodies) is not due to failure of the PCRs. Additional controls included a precipitation lacking both antibody and chromatin (Mock) and a PCR control to which water was added instead of template DNA (Water). IP, immunoprecipitation.
FIG. 5.
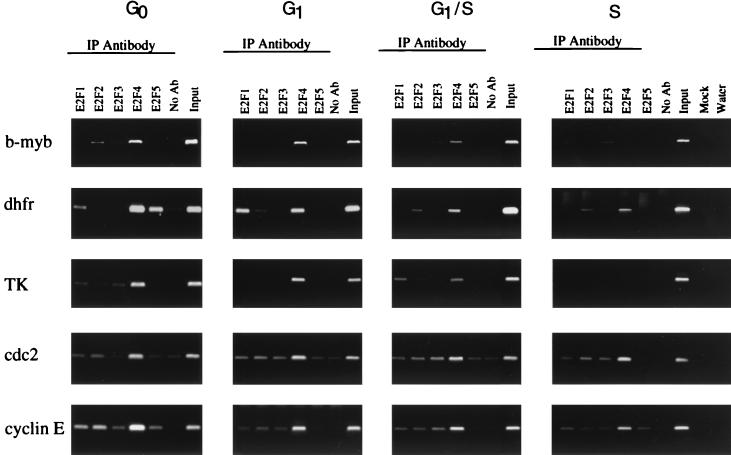
Analysis of E2F binding to target genes in NIH 3T3 cells synchronized by a serum starvation and stimulation procedure. Cross-linked chromatin from synchronized cell populations was incubated with antibodies to E2F1-5 and analyzed as described in the Fig. 2 legend.
FIG. 6.
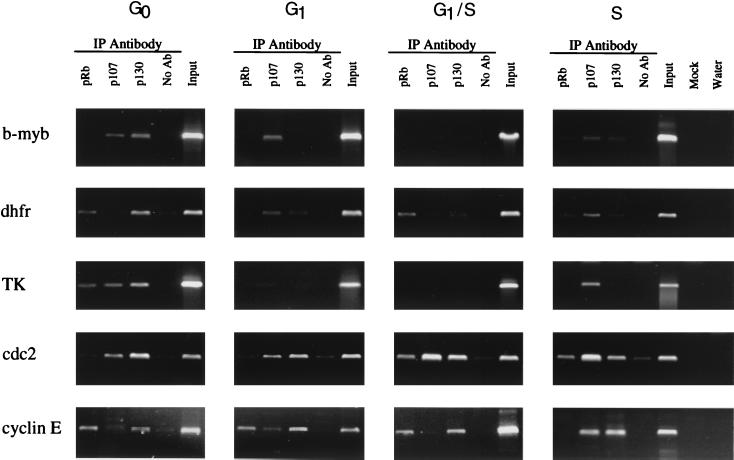
Analysis of pocket protein binding to target genes in synchronized cell populations. Cross-linked chromatin from synchronized cell populations was incubated with antibodies to the pocket proteins and analyzed as described in the Fig. 2 legend. IP, immunoprecipitation.
RESULTS
E2F4 binding is localized to the DHFR promoter and requires an intact E2F binding site.
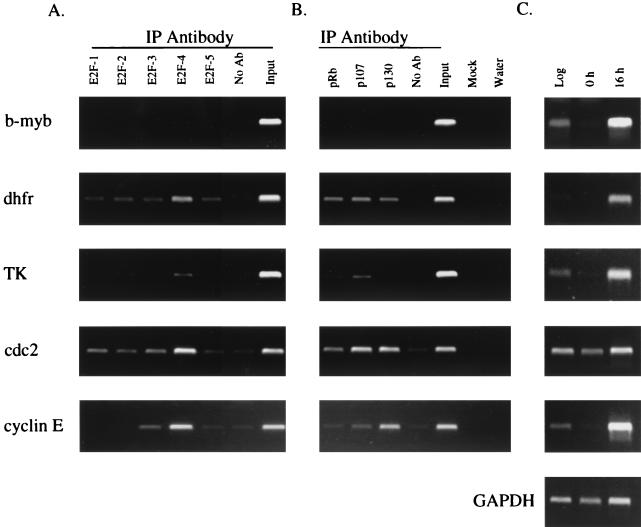
In an effort to determine if specific E2Fs bind to specific target genes in vivo, we used a formaldehyde cross-linking assay (Fig. 1B) to monitor binding to the promoters of several E2F target genes in murine NIH 3T3 cells. Briefly, this protocol involves treating cells with formaldehyde to cross-link the transcription complexes to the promoter DNA and immunoprecipitation of the protein-DNA complexes with an antibody against an individual E2F or pocket protein followed by analysis of the immunoprecipitated DNA by PCR using promoter-specific primers (see Fig. 1C for the sequences of analyzed E2F binding sites). Before analyzing the transcriptional complexes bound to target gene promoters of interest, we first wished to demonstrate the specificity of our cross-linking assay. We began by analyzing binding of E2F4, the most abundant E2F protein in most cell types studied to date, to several regions of the DHFR gene (Fig. 2A). E2F4 binding was visualized by ethidium bromide staining of the products obtained after 36 cycles of PCR amplification and by Southern blotting of the PCR products obtained after 16 cycles of PCR amplification. The results from both analyses are nearly identical and demonstrate that E2F4 bound to the DHFR promoter region but not to either intron 2 or the 3′ untranslated region of the gene. Since binding of E2F4 was localized to the promoter region of the DHFR gene, we next wished to determine if binding of E2F4 was dependent upon the presence of an intact E2F binding site. Toward this goal, we utilized a line of NIH 3T3 cells which have been stably transfected with a plasmid containing a 290-bp region of the DHFR promoter in which the sequence of the overlapping E2F binding sites has been mutated from TTTCGCGCCAAA to CCCTATATCAAA (Fig. 2B). PCR analysis of E2F4-immunoprecipitated chromatin from these cells showed that E2F4 bound to the endogenous DHFR promoter but not to the integrated DHFR promoter construct bearing the mutated E2F binding sites. Thus, formaldehyde cross-linking demonstrates that binding of E2F4 is localized to the DHFR promoter and requires an intact E2F binding site.
E2F target genes can be bound by multiple E2Fs and pocket proteins.
Satisfied with the specificity of our cross-linking assay, we next determined which E2F proteins were bound to the b-myb, DHFR, TK, cdc2, and cyclin E promoters in asynchronously growing NIH 3T3 cells. We also analyzed E2F binding to the albumin promoter, which does not contain a consensus E2F binding site, nor is expression of albumin thought to be regulated by E2F. As expected, we did not detect binding of any E2F protein to the albumin promoter, as indicated by the lack of PCR signals in all lanes except for the input positive control sample (Fig. 3A). However, we did detect E2F protein bound to each of the E2F target gene promoters. Our results suggest that very little binding specificity exists in asynchronous cells, as all five E2F proteins bound to a variety of binding site sequences. For example, the binding patterns on the TK and cdc2 promoters are similar and yet the TK promoter contains two inverted overlapping E2F binding sites while the cdc2 promoter contains one binding site with a sequence which differs from both sites of the TK promoter. Interestingly, the intensity of the signals for the individual E2F proteins relative to the input signal for the DHFR promoter was considerably weaker than those for the other E2F target genes. This result was unexpected, since the DHFR E2F site is the only site which is composed of two perfect matches to the consensus binding site (Fig. 1C) and is a high-affinity site when analyzed using gel mobility shift assays.
Examination of pocket protein binding to the E2F target genes indicated that p107 and p130 were bound to the TK, cdc2, b-myb, and cyclin E promoters (Fig. 3B). In fact, the pocket protein binding patterns on these four promoters are indistinguishable. However, similar to the E2F results, we detected very weak signals for pocket protein binding to the DHFR promoter. The same immunoprecipitated chromatin samples were used to monitor E2F and pocket protein binding to the DHFR promoter as were used to monitor binding to the other target gene promoters. Therefore, the low signals on the DHFR promoter are not due to variation between experiments or technical difficulties with the cross-linking assay. As expected, we did not detect binding of any of the pocket proteins to the albumin promoter.
E2F binding patterns on target genes change as cells progress through the cell cycle.
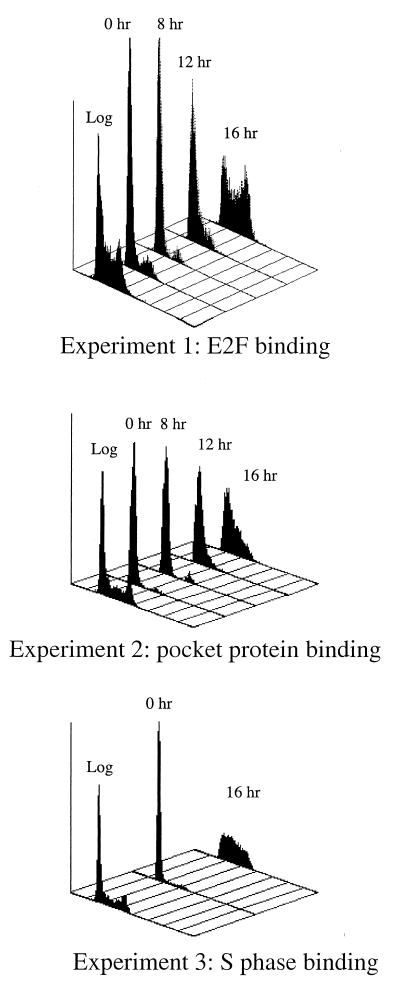
Genes containing E2F sites display cell cycle-stage-specific transcriptional regulation. Therefore, it was possible that we observed multiple E2Fs bound to given target genes in asynchronously growing cells because different E2Fs bound to the same promoter during different stages of the cell cycle. To test this hypothesis, we synchronized mouse 3T3 cells using a serum starvation and stimulation protocol and then treated the cells with formaldehyde at 0, 8, 12, and 16 h after serum stimulation. For each experiment, replicate cultures of cells were ethanol fixed and stained with propidium iodide and the DNA content of each population used in formaldehyde cross-linking assays was determined by flow cytometry analysis. As shown in Fig. 4, NIH 3T3 cells were arrested and became quiescent after 48 to 60 h of growth in low-serum (0.5%)-containing medium. Following stimulation with 10% serum, cells entered a cell growth cycle as indicated by the lateral movement of the fluorescence peak. Although flow cytometry analysis is not able to distinguish quiescent and early G1 cells from mid- to late-G1-phase cells, the 16-h time points clearly indicate that cells reentered the cell cycle following the addition of serum and entered S phase. Immunoprecipitation of the chromatin using antibodies against E2F1-5 was then performed, and the samples were assayed by PCR using primers specific for the different target genes. In contrast to the data from asynchronous cells, we found that each E2F target gene displayed a unique pattern of binding of the E2F family members (Fig. 5). For example, the b-myb promoter was occupied by E2F4 after serum starvation with reduced signal intensity for E2F4 at the G1/S-phase boundary and no detectable binding during S phase. These cross-linking results corroborate the previous in vivo footprinting studies of b-myb (44) and extend the analyses by suggesting that E2F4 is the sole regulator of b-myb as quiescent cells are induced to proliferate. We found that the DHFR promoter was occupied by E2F4 from quiescence through S phase but that additional E2Fs also bound to DHFR at specific times in the cell cycle. In quiescent cells, E2F5 bound to the DHFR promoter while E2F1 binding was prominent only 8 h later. Again, these cross-linking data corroborate previous in vivo footprinting studies which showed that one strand of the E2F site was constitutively occupied but that the pattern of binding to the other strand changed as cells progressed through the cycle (41). Similar to b-myb, we observed that E2F4 was the predominant E2F binding activity detected on the TK, cdc2, and cyclin E promoters although faint signals from the other E2F proteins were visible.
FIG. 4.
Synchronization of NIH 3T3 cells by serum starvation and stimulation. Cells were serum starved (0.5% BCS) for 48 to 60 h and then stimulated to reenter the cell cycle by the addition of 10% serum. At 0, 8, 12, and 16 h after serum stimulation, cells were trypsinized, ethanol fixed, and stained with propidium iodide. The DNA content of each population of cells was measured by flow cytometry analysis. As a reference, analysis of a population of asynchronous cells which were not starved and stimulated (log) is also included. IP, immunoprecipitation.
E2F target genes can be bound by Rb family members in all stages of the cell cycle.
Inspection of the E2F binding pattern on the different promoters indicated that most promoters were predominantly bound by E2F4, suggesting that most E2F target genes could be regulated by any of the three pocket proteins. To test this hypothesis, 3T3 cells were treated with formaldehyde at various times in the cell cycle and then the chromatin was immunoprecipitated using antibodies against Rb, p107, or p130 (see Fig. 6). Since previous gel shift analysis of extracts from NIH 3T3 (9) and other murine (18) cells detected pocket protein-containing complexes mainly during G0 and early G1 phase, we expected that the target promoters would be immunoprecipitated with antibodies against the pocket proteins predominantly from quiescent cells. Accordingly, we found that all of the E2F target genes tested were bound by pocket proteins in quiescent cells. However, the particular pocket protein bound during quiescence as well as the pattern of pocket protein binding to the promoter during the subsequent stages of the cell cycle varied for each target gene. For example, we detected binding of p130 and p107 to the b-myb promoter during quiescence and binding of p107 during early G1 but did not detect binding of any pocket proteins during subsequent stages of the cell cycle. In contrast, we detected binding of p107 and p130 to the cdc2 promoter from quiescence through S phase in addition to binding of pRb from late G1 through early S phase. We detected binding of p130 to the cyclin E promoter from quiescence through S phase. We also observed binding of pRb to the cyclin E promoter during quiescence, and binding persisted until cells entered S phase, at which point p107 replaced pRb. Analysis of pocket protein binding to the DHFR promoter revealed binding of pRb and p130 in quiescent cells. As cells reentered the cell cycle, we detected binding of p107 and p130 to the DHFR promoter during G1, binding of only pRb 4 h later, and binding of all three pocket proteins in S-phase cells. In contrast to the other promoters tested, we observed binding of all three pocket proteins to the TK promoter during quiescence and loss of binding in mid-G1. Once cells entered S phase, we again detected binding of p107 to the TK promoter. As for Fig. 5, all of the results in Fig. 6 were obtained from analysis of chromatin from the same immunoprecipitation experiment. Therefore, the differences we detected were promoter specific and not due to experimental variation.
E2F and pocket proteins are bound to target gene promoters when the genes are being transcribed.
Our results clearly showed binding of E2Fs and pocket proteins to certain promoters during S phase. Although E2F-pocket protein complexes have previously been observed for extracts prepared from S-phase cells (19, 27, 31, 32), it has remained unclear as to whether these complexes functioned to activate or to repress transcription of target genes. To address this question, we serum starved NIH 3T3 cells and stimulated them to enter the growth cycle by the addition of serum. Flow cytometric analysis indicated that during serum starvation >94% of the cells had a 2N DNA content and entered S phase by 16 h following serum stimulation (the S-phase profile is shown in Fig. 4). We prepared cytoplasmic RNA from asynchronously growing cells and from cells collected at 0 and 16 h following serum stimulation. RT-PCR analysis revealed that expression of b-myb, DHFR, TK, cdc2, and cyclin E was low in serum-starved cells and increased during S phase (Fig. 7C). Therefore, as expected based on previous studies, there appeared to be higher transcriptional activity of all five target genes in S phase than in quiescent cells. In this same serum starvation and stimulation experiment, we also treated the cells with formaldehyde at 16 h and repeated the immunoprecipitation assay using antibodies against the five E2Fs and the three pocket proteins. In the experiments shown in Fig. 5 and 6, the E2F and pocket protein binding had been analyzed using chromatin that was immunoprecipitated in separate experiments due to the large number of different cell cycle stages examined. However, the use of all eight antibodies in a single experiment eliminates any possible variation in cell synchrony between experiments and also allows a comparison of the S-phase binding patterns in two separate experiments. In agreement with the results shown in Fig. 5 and 6, we detected S-phase binding of E2F and pocket protein complexes to all of the promoters tested except for b-myb. Comparison of the results shown in Fig. 5, 6, and 7 demonstrates that the cross-linking and immunoprecipitation assay is consistent and reproducible. Therefore, our results support the conclusion that some E2F target promoters are bound by E2F-pocket protein complexes when they are transcriptionally active.
FIG. 7.
Analysis of protein binding to and expression of target genes in S-phase cells. Cross-linked chromatin from synchronized S-phase cells was immunoprecipitated with antibodies against E2F1-5 (A) or pocket proteins (B) and analyzed as described in the Fig. 2 legend. Replicate plates of cells were harvested, and cytoplasmic RNA was prepared from these cells. Expression levels of target genes were measured by RT-PCR analysis of 100 ng of RNA from asynchronous log-phase cells, quiescent serum-starved cells (0 h), or S-phase cells (16 h) (C). IP, immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
Previous studies have investigated E2F target gene specificity by overexpressing exogenous E2F family members using either retroviruses (13) or transfection assays (33, 39). Although such studies have, in some cases, shown differential patterns of gene expression by the different E2Fs, the experimental design precludes a conclusive interpretation of these studies. For example, exogenously introduced E2F can drive quiescent cells into S phase (33). Since E2F target genes show increased expression at the G1/S-phase boundary, there is no way to determine if overexpression of a particular E2F leads to direct activation of a particular target gene or if target gene activation is an indirect response due to cell proliferation. An alternative method for examining E2F target gene specificity has been to prepare nuclear extracts and perform in vitro gel shift assays using antibodies against the E2F and pocket proteins to determine which E2F-pocket protein complex binds to an isolated E2F site (37, 41). Unfortunately, such studies often identify the most abundant E2F and pocket protein but do not necessarily reflect protein-DNA specificity. Due to the shortcomings of overexpression and/or in vitro systems, we have used an in vivo formaldehyde cross-linking assay to determine which endogenous E2F family members bind to specific E2F target genes at different stages of the cell cycle (summarized in Fig. 8). Several conclusions can be drawn from our studies.
FIG. 8.
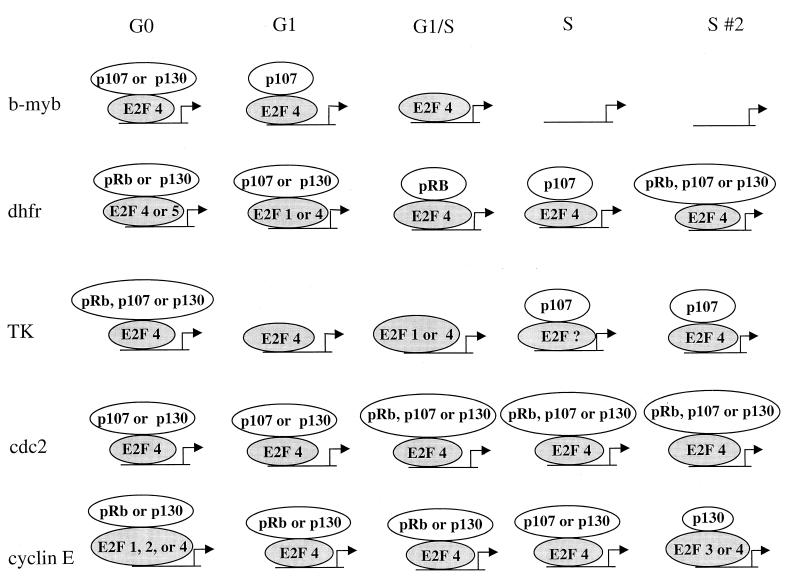
Molecular snapshots of the transcription complexes formed on E2F target genes. Shown is a schematic summarizing the predominant E2F and pocket proteins detected on the b-myb, DHFR, TK, cdc2, and cyclin E promoters at different stages of the cell cycle. As noted in the text, the presence of multiple E2F or pocket proteins bound to the same promoter at a specific stage of the cell cycle does not necessitate that all proteins are bound to the same DNA site simultaneously or that all complexes are transcriptionally active. For simplicity, we have depicted the pocket proteins as being recruited by the E2F proteins. However, it remains possible that pocket proteins can be recruited to promoters via interaction with different DNA binding proteins.
(i) Binding patterns of E2F and pocket proteins are both promoter and cell cycle phase specific.
Very little difference in binding site specificity has been observed between the different E2F proteins using in vitro assays. Accordingly, we found little difference in the E2F binding patterns of the individual promoters tested in log-phase cells, as most promoters were bound by several E2F proteins. In contrast to log-phase cells, our results clearly show different E2F binding patterns on different target gene promoters during the first cell cycle following serum starvation and stimulation. On the b-myb promoter, for example, we detected only binding of E2F4 in synchronized cells, whereas on the DHFR promoter, we detected binding of E2F1, E2F4, and E2F5 and binding patterns changed as cells progressed from quiescence through S phase. Unexpectedly, we detected very little binding of E2F2 or E2F3 to any of the promoters tested in synchronized cells despite our observation that these E2F proteins bound to several promoters in asynchronous cells. Using the same chromatin samples analyzed in Fig. 5, we have recently detected robust E2F2 and E2F3 binding to the retinoblastoma gene promoter (data not shown). Thus, our inability to detect binding of E2F2 and E2F3 in the experiment shown in Fig. 5 does not appear to be the result of a technical problem with the assay. At present, we cannot rule out the possibility that E2F2 or E2F3 is bound to one of the promoters we tested but in a conformation that is not recognized by the antibodies we used. Our results also raise the possibility that E2F binding patterns during the first cell cycle after serum starvation and stimulation may be different from the E2F binding patterns during the cell cycles of proliferating cells. Preliminary results of experiments comparing G1-phase binding patterns in cells synchronized by serum starvation and stimulation with G1-phase cells isolated by fluorescence-activated cell sorting suggest that binding patterns of certain E2F proteins in subsequent cell cycles are different for some promoters (data not shown), as previously proposed by Leone et al. (23).
Analogous to the E2F proteins, we observed that the pocket proteins also displayed promoter- and cell cycle phase-specific binding patterns. For example, we detected binding of p107 and p130 to the b-myb promoter in G0 phase but not during S phase. In contrast, we detected binding of pocket proteins to the DHFR, cdc2, and cyclin E promoters from quiescence through S phase. Thus, each promoter tested showed cell cycle variation in pocket protein binding. We also found that p107 and p130 bound to all E2F target genes tested here (at some stage of the cell cycle) but that pRb did not bind to the b-myb promoter. Previously, Hurford et al. studied the cell cycle-regulated expression of several E2F target genes, including those tested here, in primary mouse embryo fibroblasts lacking pRb, p107, p130, or both p107 and p130 (18). Our results are consistent with several aspects of that previous study. For example, Hurford et al. found that loss of p107 or p130 alone did not alter the expression of any E2F target gene analyzed but that expression of several E2F target genes was altered by loss of both p107 and p130. Accordingly, we have found that both p107 and p130 were bound to all E2F target genes examined. Hurford et al. also found that cyclin E was one of only two E2F target genes whose expression was deregulated in Rb null cells, and we have shown that the cyclin E promoter is bound by pRb in vivo. However, our results are not completely consistent with those of Hurford et al. For example, we found that the TK promoter is bound by p107 and p130, but the previous studies did not show an alteration in regulation of TK gene expression in the p107-p130 null cells. It is possible that pocket proteins may bind to some promoters, such as TK, and yet not play a critical role in regulating expression of such genes. Alternatively, discrepancies between our results and those of Hurford et al. could be due to differences in E2F and pocket protein binding patterns in primary mouse embryo fibroblasts versus cultured NIH 3T3 cells, a possibility which we are currently testing.
(ii) The standard model for E2F-mediated regulation does not reflect the E2F-pocket protein binding pattern for most E2F target genes.
Previous studies have shown that the promoters analyzed in this study all show low transcriptional activity in G0-phase cells and high activity in late-G1- and early-S-phase cells (3, 11, 22, 30, 34, 43, 44). In agreement with previous studies, we found that mRNA levels of all of the genes that we tested here were higher in S-phase cells than in serum-starved cells (Fig. 7C). It has been commonly believed that the promoters tested here were bound by E2F-pocket protein complexes in G0 phase whereas the E2F site was unoccupied in S phase. Many of the data used to support this model of E2F-mediated regulation come from studies of the b-myb promoter. Accordingly, we show that the b-myb promoter does show release of all DNA-bound E2F and pocket proteins during S phase, the time at which the b-myb promoter is most active. However, various aspects of the binding patterns of the other E2F target gene promoters analyzed here contradict elements of the current model. For example, the DHFR, cdc2, and cyclin E promoters are bound by E2F complexes both at the G1/S-phase boundary and during mid-S phase. Previously, it has been proposed that E2F4 complexes would not be bound to promoters during S phase due to translocation of E2F4 to the cytoplasm during S phase in some cell types (38). In contrast, others (24) have found that the amount of E2F4 in nuclear extracts is fairly constant during progression from G0 to S phase. One possibility is that, while non-DNA-bound E2F4 is translocated to the cytoplasm during S phase, E2F4 that is part of a DNA-bound transcriptional complex remains in the nucleus. In support of this hypothesis, Leone et al. found that the abundance of E2F4 DNA binding activity in nuclear extracts from synchronized REF52 cells did not change as cells entered S phase despite their finding that >80% of E2F4 DNA binding activity was cytoplasmic at this time (23).
Given the current view that hyperphosphorylation of pocket proteins during late G1 phase leads to dissociation of pRb-, p107-, and p130-containing complexes from DNA, our finding that pocket proteins were bound to certain promoters through S phase was, at first, surprising. Although it could be argued that the pocket protein binding that we detected during S phase is due to inefficient cell synchronization, we do not favor this explanation for several reasons. First, flow cytometry analysis revealed that >95% of the cells analyzed in the experiments shown in Fig. 5 and 6 had entered S phase by 16 h after serum stimulation (profiles shown in Fig. 4). Second, RT-PCR analysis showed that all of the genes tested were highly expressed in our S-phase population of cells. Accordingly, we have also recently performed transfection studies in NIH 3T3 cells which confirm that, under identical synchronization conditions, the DHFR, b-myb, and cdc2 promoters are all more active in G1/S- and S-phase cells than in G0-phase cells (M. J. Oberley and P. J. Farnham, unpublished data). Finally, we stress that the same chromatin samples used to show loss of E2F and pocket protein binding to the b-myb promoter as cells entered S phase were also used to show retention of E2F and pocket protein binding to the other promoters. Our results are in agreement with those of Moberg et al., who detected complexes containing pRb and E2F4 in human T cells only after cells had reached the G1/S-phase boundary (27) as well as with the results of others who have detected E2F-pRb complexes during S phase (19, 31). Additionally, our observation that pRb is bound to the cdc2, DHFR, and cyclin E promoters at the G1/S-phase boundary and during S phase is consistent with data showing increased Rb promoter activity at these same time points (14). Thus, we detected binding of pRb to the promoters of E2F target genes at a time when pRb protein is maximally abundant. The p107 promoter is also activated in late G1 phase, resulting in peak p107 protein levels during late G1 and early S phase (35). In contrast, p130 protein levels are maximal in G0 phase, and yet we can detect binding of p130 to certain promoters throughout the cell cycle. Although the decrease in p130 levels after serum stimulation of starved cells may prevent new p130-containing transcriptional complexes from forming on promoter DNA, the complexes that we detected at later times in the cell cycle may represent stable transcription complexes which were formed during quiescence or G1. Evidence in support of the idea that, once formed, an E2F-pocket protein complex can be long-lived comes from experiments showing that transcription complexes can be stable even after several rounds of DNA replication (40). Thus, it is possible, perhaps even likely, that a majority of the E2F-p130 complexes that we detected during S phase represent stable complexes that were formed earlier in the cell cycle.
Our demonstration that most of the promoters tested here were bound by E2F-pocket protein complexes during mid-S phase raises several interesting possibilities. For example, gene expression may be activated in only a small portion of S-phase cells (i.e., those which do not have pocket proteins bound to the promoters). The strict requirement for the products of these genes for cell cycle progression, however, makes this scenario unlikely. An alternative hypothesis is that pocket proteins may not always serve as transcriptional repressors when bound to promoters. In support of this hypothesis, others have shown that Rb and p107 can interact with Sp1 (8, 12), and evidence suggests that pRb may be an important activator of certain promoters that contain Sp1 sites (8, 20, 36). Therefore, promoter-bound pocket proteins may act as docking platforms for repressors in quiescent cells and activators in S-phase cells. Alternatively, pocket proteins bound to promoters in S phase may have a neutral effect on transcription, simply remaining bound to recruit repressors, such as histone deacetylases, during G1 phase of the following cell cycle.
(iii) Multiple complexes can be formed on an E2F target gene promoter.
Clearly, E2F target genes are not regulated by a single, static transcription complex (Fig. 8). Rather, the E2Fs and pocket proteins bound to a given promoter during one stage of the cell cycle are not necessarily identical to the E2F and pocket proteins bound to the same promoter in a different stage of the cell cycle. Interestingly, some promoters are bound by one of several different E2F and pocket proteins at a given time during the cell cycle. For example, the DHFR promoter can be bound by either E2F4 or E2F5 in G0-phase cells and by either E2F1 or E2F4 in mid-G1-phase cells. At present, we cannot eliminate the possibility that more than one E2F can bind to the same DNA site simultaneously. However, we favor the alternate interpretation that each cell in a synchronized population of cells has the potential to form several different transcription complexes and the actual complex which does form is a result of stochastic, not predetermined, events. Therefore, during mid-G1 phase, the DHFR promoter in one cell may be occupied by E2F1 whereas the DHFR promoter in an adjacent cell may be occupied by E2F4. The presence of alternative transcription complexes on a given promoter at a given time in the cell cycle raises the very intriguing possibility that different cells in a synchronized population may have different transcriptional profiles. Support for a stochastic model of transcriptional regulation comes from previous studies showing that a given promoter is activated only in a subset of the nuclei of multinucleated myofibers at a given time, despite the fact that all the nuclei share a common cytoplasm (29). The ability to separate transcriptionally active complexes from transcriptionally inactive complexes prior to immunoprecipitation is required before an understanding of the role of the different E2F-pocket complexes can be completely understood.
In summary, analysis of in vivo DNA-protein interactions has allowed us to develop a molecular snapshot of the transcription complexes bound to different E2F target genes at different stages of the cell cycle. Our results indicate that the accepted model for E2F-mediated gene regulation is applicable to only a subset of E2F target genes. We are currently extending our use of the formaldehyde cross-linking assay toward the refinement of additional models which more closely represent the molecular mechanisms by which cell cycle regulation of a variety of E2F target genes is achieved.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant CA45240 (to P.J.F.) and training grants CA09681 (J.W.) and CA09135 (K.E.B. and C.J.F.) from the National Institutes of Health.
We thank Kathleen Schell, Kristin Elmer, and Janet Lewis for excellent technical assistance with flow cytometry analysis; members of the Farnham lab for helpful discussions; and Rick Maser for critical reading of the manuscript. We also express our gratitude to the laboratories of Mark Biggen, David Allis, and Richard Treisman for sharing cross-linking protocols.
ADDENDUM
While the manuscript was under review, a similar study reporting different results was published by Takahashi et al. (35a). It is possible that the discrepancies between the two studies are due to promoter-specific variations. However, the b-myb promoter was analyzed in both studies, and we detected robust binding of only E2F4 to the b-myb promoter in serum-starved and G1-phase cells which disappeared as cells entered S phase. In contrast, Takahashi et al. observed very low levels of E2F binding to the b-myb promoter in G0 and early G1 cells. We would like to emphasize that our cross-linking results are in agreement with previous in vivo footprinting analyses showing occupancy of the E2F site within the b-myb promoter from G0 through G1 phase in serum-synchronized NIH 3T3 cells (44). Strikingly, Takahashi et al. reported no binding of E2F4 to promoters of target genes during S phase, while we observed robust S-phase binding of E2F4 to several target gene promoters. Since different antibodies had been used in the two studies, we performed an additional cross-linking experiment to directly compare the immunoprecipitation efficiencies of the E2F4 antibody which we used for the results presented here (sc-866X; Santa Cruz) and of that used by Takahashi et al. (sc-1082X; Santa Cruz). Our results indicated that both antibodies detected binding of E2F4 to several different promoters in synchronized NIH 3T3 cells but that the sc-1082X antibody generated a noticeably weaker signal on some promoters, and this difference was most pronounced in S-phase cells (data not shown). Finally, the differences in our results and those of Takahashi et al. could be species specific, as we used immortalized murine cells (NIH 3T3) while they used a human glioblastoma cell line (T98G). Recent cross-linking experiments (data not shown) have confirmed binding of E2F proteins, particularly E2F4, to multiple promoters during S phase in aphidicolin-synchronized HeLa and Raji cells, both of which are human tumor cell lines. It remains possible that expression of E2F target genes in T98G cells may be mediated by different E2F proteins than those in other human or murine cell lines.
REFERENCES
- 1.Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-1, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- 2.Bandara L R, Adamczewski J P, Hunt T, La Thangue N B. Cyclin A and the retinoblastoma gene product complex with a common transcription factor. Nature. 1991;352:249–251. doi: 10.1038/352249a0. [DOI] [PubMed] [Google Scholar]
- 3.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Dürr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd K E, Farnham P J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Chellappan S P, Hiebert S W, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 8.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y-H F, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Prywes R. Serum-Induced expression of the cdc25A gene by relief of E2F-mediated repression. Mol Cell Biol. 1999;19:4695–4702. doi: 10.1128/mcb.19.7.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobrinik D, Whyte P, Peeper D S, Jacks T, Weinberg R A. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 11.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta P K, Raychaudhuri P, Bagchi S. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol Cell Biol. 1995;15:5444–5452. doi: 10.1128/mcb.15.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiFiore B, Palena A, Felsani A, Palitti F, Caruso M, Lavia P. Cytosine methylation transforms an E2F site in the retinoblastoma gene promoter into a binding site for the general repressor methylcytosine-binding protein 2 (MeCP2) Nucleic Acids Res. 1999;27:2852–2859. doi: 10.1093/nar/27.14.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dynlacht B D, Flores O, Lees J A, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 16.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurford R K J, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda M-A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S-J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G J, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 22.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 26.Means A L. Ph.D. Thesis. University of Wisconsin, Madison; 1991. [Google Scholar]
- 27.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller H, Moroni M, Vigo E, Peterson B O, Barteck J H, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newlands S, Levitt L K, Robinson C S, Karpf A B C, Hodgson V R M, Wade R P, Hardeman E C. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogris E, Rotheneder H, Mudrak I, Pichler A, Wintersberger E. A binding site for transcription factor E2F is a target for trans activation of murine thymidine kinase by polyomavirus large T antigen and plays an important role in growth regulation of the gene. J Virol. 1993;67:1765–1771. doi: 10.1128/jvi.67.4.1765-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz J K, Devoto S H, Smith E J, Chellappan S P, Jakoi L, Nevins J R. Interactions of the p107 and Rb proteins with E2F during the cell proliferation response. EMBO J. 1993;12:1013–1020. doi: 10.1002/j.1460-2075.1993.tb05742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 33.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. In: Farnham P J, editor. Transcriptional control of cell growth: the E2F gene family. New York, N.Y: Springer-Verlag; 1996. pp. 1–30. [DOI] [PubMed] [Google Scholar]
- 34.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. . (Author's correction, 13:7201.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith E J, Leone G, Nevins J R. Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 1998;9:297–303. [PubMed] [Google Scholar]
- 35a.Takahashi Y, Rayman J B, Dynlacht B D. Analysis of promoter binding by the E2F and pRB families in vivo: proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 36.Udvadia A J, Rogers K T, Higgins P D R, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Sman J, Thomas N S B, Lam E W-F. Modulation of E2F complexes during G0 to S phase transition in human primary B-lymphocytes. J Biol Chem. 1999;274:12009–12016. doi: 10.1074/jbc.274.17.12009. [DOI] [PubMed] [Google Scholar]
- 38.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe G, Albanese C, Lee R J, Reutens A, Vairo G, Henglein B, Pestell R G. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci USA. 1988;85:5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells J, Held P, Illenye S, Heintz N H. Protein-DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster ovary cell cycle. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M, Sheppard K A, Peng C Y, Yee A S, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zacksenhaus E, Phillips R A, Gallie B L. Molecular cloning and characterisation of the RB1 promoter. Oncogene. 1993;8:2343–2351. [PubMed] [Google Scholar]
- 44.Zwicker J, Liu N, Engeland K, Lucibello F C, Müller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]