Abstract
The imprinted mouse gene Gnas produces the G protein α-subunit GSα and several other gene products by using alternative promoters and first exons. GSα is maternally expressed in some tissues and biallelically expressed in most other tissues, while the gene products NESP55 and XLαs are maternally and paternally expressed, respectively. We investigated the mechanisms of Gnas imprinting. The GSα promoter and first exon are not methylated on either allele. A further upstream region (approximately from positions −3400 to −939 relative to the GSα translational start site) is methylated only on the maternal allele in all adult somatic tissues and in early postimplantation development. Within this region lies a fourth promoter and first exon (exon 1A) that generates paternal-specific mRNAs of unknown function. Exon 1A and GSα mRNAs have similar expression patterns, making competition between their promoters unlikely. Differential methylation in this region is established during gametogenesis, being present in oocytes and absent in spermatozoa, and is maintained in preimplantation E3.5d blastocysts. Therefore, this region is a methylation imprint mark. In contrast, differential methylation of the NESP55 and XLαs promoter regions (Nesp and Gnasxl) is not established during gametogenesis. The methylation imprint mark that we identified may be important for the tissue-specific imprinting of GSα.
Genomic imprinting is an epigenetic phenomenon affecting a small number of autosomal genes that results in differences in gene expression between the maternal and paternal allele and explains why both paternal and maternal genomes are required for normal development (3, 12, 38). Mutation, deletion, and dysregulation of imprinted genes have been implicated in several human diseases, such as the Beckwith-Wiedemann, Prader-Willi, and Angelman syndromes and Albright hereditary osteodystrophy, and in carcinogenesis. Virtually all imprinted genes have regions in which CpG dinucleotides are differentially methylated between the maternal and paternal alleles. Loss of imprinting in mice lacking a DNA methyltransferase gene (28, 29) strongly suggests that allele-specific methylation differences are critical for the maintenance of imprinting. The presence of differentially methylated regions (DMRs) in the male or female germline (methylation imprint marks) that are required to establish differential methylation at other loci within the same imprinted region in later development (12) strongly suggests that methylation is important for establishing the maternal and paternal epigenotypes (methylation and expression patterns).
Heterozygous inactivating mutations within GNAS1, the human gene at 20q13 that codes for the heterotrimeric G protein α-subunit GSα, lead to multihormone resistance only when inherited through the maternal germline (13, 42). GSα is a ubiquitously expressed protein that is required for hormone-stimulated cyclic AMP generation. Maternal-specific expression of GSα in hormone target tissues likely explains the hormone resistance that results from maternal inheritance of GNAS1 mutations. Imprinting of GSα has not been confirmed in humans (9, 17, 18). However, we showed, using Gnas knockout mice (Gnas in distal chromosome 2 is the mouse orthologue of GNAS1) that GSα is imprinted in mice in a tissue-specific manner, being maternally expressed in some tissues (e.g., renal proximal tissues and adipose tissue) and biallelically expressed in most other tissues (43, 47, 48).
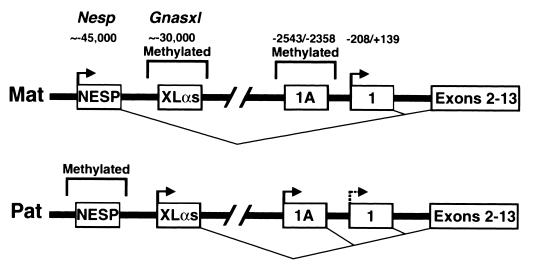
Gnas has at least four alternative promoters and first exons that generate multiple imprinted mRNAs (Fig. 1). The gene was originally defined by 13 coding exons for GSα (exons 1 to 13) (27). Three additional Gnas mRNAs result from splicing of alternative first exons to exon 2. Alternative promoters located 30 and 45 kb upstream of GSα exon 1 are oppositely imprinted and produce mRNAs encoding XLαs, a Golgi-specific isoform of GSα, and the chromogranin-like protein NESP55, respectively (17, 18, 26, 34). XLαs is expressed from the paternal allele, and its promoter region is methylated on the maternal allele, whereas NESP55 is expressed from the maternal allele and its promoter is methylated on the paternal allele (17, 18, 34). Both proteins are expressed primarily in neuroendocrine tissues, and little is known about their function (21, 25). These two promoter regions have been named Gnasxl and Nesp (18). A fourth alternative promoter and first exon (which we call exon 1A) located 2.5 kb upstream of GSα exon 1 generates mRNAs that are probably untranslated and are of unknown function (22, 36). The imprinting status of the exon 1A promoter has not been determined.
FIG. 1.
Schematic diagram showing the maternal (Mat) and paternal (Pat) alleles of Gnas. Alternative first exons which splice into exon 2 to generate alternative mRNAs encoding NESP55, XLαs, an unknown gene product, and GSα are shown as boxes labeled NESP, XLαs, 1A, and 1, respectively. Exons 2 to 13 and the two closely linked exons encoding NESP55 are each shown as a single box. The nucleotide positions of the 5′ and 3′ ends of exons 1 and 1A and the approximate locations of the NESP55 and XLαs exons are indicated (all numbering in Fig. 1 to 7 is relative to the exon 1 translational start site). The NESP55 and XLαs promoter regions have been named Nesp and Gnasxl, respectively (34). Transcriptionally active promoters are designated by horizontal arrows, and regions of differential methylation are outlined above each allele. The splicing of each first exon to exon 2 is indicated below each allele. The dashed horizontal arrow for exon 1 in the paternal allele indicates that this promoter is active in some tissues and inactive in other tissues. Differential methylation and allele-specific expression of Nesp and Gnasxl has been previously reported (17, 18, 34). Maternal-specific methylation and paternal-specific expression of exon 1A is presented in this paper.
Given the close proximity of the oppositely imprinted Gnasxl and Nesp promoters and the similar tissue distribution of their mRNAs, it is possible that they coordinately regulate each other, as has been suggested for other closely linked imprinted genes (2, 3). However two observations make it unlikely that either promoter or their mRNAs are involved in the tissue-specific imprinting of GSα. First, the mRNAs of both promoters are expressed in several human tissues in which the GSα transcript is biallelically expressed (17, 18). Second, we were unable to amplify either transcript in mouse tissues in which the GSα transcript is imprinted (e.g., brown adipose tissue [BAT]) (S. Yu and L. S. Weinstein, unpublished data).
To define the mechanisms that lead to imprinting of the Gnas locus in general and tissue-specific imprinting of GSα in particular, we determined the methylation status of the GSα promoter and of the region immediately upstream of the promoter. In this paper, we show first that imprinting of GSα is not associated with methylation of its promoter. Second, we identify a region upstream of the GSα promoter that is methylated on the maternal allele in all tissues examined. Within this region is exon 1A (22, 36), which generates mRNAs only from the paternal allele. This upstream region is a methylation imprint mark, as methylation in this region is established during gametogenesis and maintained throughout pre- and postimplantation development.
MATERIALS AND METHODS
Genomic DNA clone isolation and sequencing.
A mouse genomic DNA clone (strain 129/SvJ) containing a 12-kb insert including Gnas exons 1 to 3 was isolated as previously described (48). An ∼7.4-kb NotI fragment including exons 1 and 2 and ∼3.0 kb of upstream sequence was subcloned into pBluescript KS(+) (Stratagene) to generate plasmid pSHY46 and sequenced in both directions by automated sequencing.
Mice.
Normal CD1 mice were obtained from Charles River and 129/SvJ mice were provided by Eric Lee (National Institute for Child Health and Human Development, National Institutes of Health [NIH]). 129 × CD1 and CD1 × 129 mice (using the convention female × male) were generated by mating 129/SvJ females with CD1 males and CD1 females with 129/SvJ males, respectively. Mice with insertion of a neomycin resistance cassette into exon 2 of Gnas were created by targeted mutagenesis as previously described (48). The mutant allele was created in the J1 embryonic cell line (strain 129/SvJ) and heterozygotes were continuously crossed with normal CD1 mice. Therefore in heterozygotes (Gnas+/− and Gnas−/+, using the convention Gnasmaternal/paternal), the normal and mutant alleles are derived from CD1 and 129/SvJ, respectively. Animals were cared for according to NIH institutional guidelines.
RNA isolation and primer extension analysis.
Total RNA was isolated by the TRIZol method (Gibco/BRL) and poly(A) selected (Qiagen). Primer extension analysis was performed using a primer extension system (Promega). The oligonucleotide primer 5′-CGGGGAGGGTGGCGGCTCGGACTAAGGCAA-3′ (positions −82 to −111 relative to the exon 1 translational start site) was end labeled with [γ-32P]ATP and hybridized to poly(A)-selected BAT RNA (1 μg) at 58°C for 30 min. Reverse transcription was performed at 42°C for 1 h. The extension products were analyzed by electrophoresis on a 6% denaturing polyacrylamide gel. A sequencing reaction using the same primer was run beside the primer extension reaction.
Ribonuclease protection assays.
Templates for in vitro transcription were generated by PCR using genomic DNA as template. The T7 promoter sequence was included in the 5′ end of the downstream oligonucleotide primer to produce genomic DNA fragments with the T7 promoter attached to the 3′ end. Templates for in vitro transcription were generated using the following primer pairs: 5′-CGCCCTCCCAGCCGCGGCCCT-3′ and 5′-TAATACGACTCACTATAGGGAGGGTGGCGGCTCGGACTAAGGCAA-3′ for exon 1 and 5′-GCCGATTTTTTGCGCGTCCCCTTC-3′ and 5′-TAATACGACTCACTATAGGGAGGCTGGGACAAGGGTTCGCTCCAG-3′ for exon 1A (the T7 promoter sequence is underlined). PCR products were gel purified and used as templates for in vitro transcription to generate 32P-labeled antisense riboprobes (MaxiScript kit; Ambion). Gel-purified riboprobes (4 × 104 to 8 × 104 cpm) were hybridized with RNA samples at 56°C for 24 h and then digested with 100 U of RNase T1 and 0.25 U of RNase A for 30 min at 37°C (RPA II kit; Ambion). The digestion products were precipitated with ethanol and analyzed on a 6% denaturing polyacrylamide gel.
DNA isolation and Southern analysis.
Genomic DNA was isolated from mouse tissues using the QIAamp tissue kit (Qiagen). DNA samples (20 μg) were digested with the indicated restriction enzymes (New England Biolabs), separated by electrophoresis on a 1.5% agarose MS (Boehringer Mannheim) gel, and transferred to nylon filters (Nytran; Schleicher & Schuell). To generate genomic DNA probes, specific restriction fragments were isolated from plasmid pSHY46 and radiolabeled with 32P by random priming (Multiprime DNA labeling kit; Amersham Pharmacia Biotech). Filters were incubated with probes in QuikHyb hybridization solution (Stratagene) at 68°C for 1 h and then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate (SDS) for 30 min at room temperature and once with 0.1× SSC–0.1% (wt/vol) SDS for 1 h at 68°C. Filters were exposed to Kodak Bio-Max MR films. To separate maternal and paternal alleles, genomic DNA (200 μg) from Gnas+/− mice was digested with BglII and separated on a 0.8% agarose gel. Gel slices containing ∼10- and ∼13.5-kb DNA fragments (containing the mutant paternal and normal maternal alleles, respectively) were excised, and DNA was recovered from the gel slices using the QIAEX gel extraction kit (Qiagen).
Collection of sperm and oocytes and DNA isolation.
Mouse spermatozoa were aspirated from the ductus deferens. Spermatozoa were lysed in 20 mM Tris-HCl (pH 8.0)–20 mM EDTA–220 mM NaCl–80 mM dithiothreitol–4% SDS with proteinase K (250 μg/ml) for 1 h at 55°C, and genomic DNA was isolated by phenol-chloroform extractions followed by ethanol precipitation. Unfertilized oocytes were collected 18 h after injection of human chorionic gonadotropin (5 IU; Sigma), as previously described (7), and washed several times in phosphate-buffered saline to remove adhering maternal cells. E3.5d blastocysts were isolated and provided by A. Grinberg, NICHHD, NIH. Pooled oocytes (100 to 150 oocytes/batch) and blastocysts (∼50 blastocysts/batch) were incubated in 20 μl of a solution containing 2 μg of yeast tRNA, 1 mM SDS, and 280 μg of proteinase K per ml for 1 h at 37°C and then incubated for 15 min at 98°C under mineral oil.
Bisulfite treatment.
Bisulfite treatment was carried out essentially as previously described (49). Sperm DNA (0.1 μg) was linearized with the restriction enzyme EcoRI. DNA samples were denatured for 15 min at 37°C by adding 3 M NaOH to a final concentration of 0.3 M. To maximize denaturation, the samples were incubated at 95°C for 3 min and then immediately cooled on ice. Sodium bisulfite (8.1 g; Sigma) was dissolved in 15 ml of water and then mixed with 1 ml of 40 mM hydroquinone (Sigma). The mixture was adjusted to pH 5 by adding 600 μl of 10 N NaOH. Denatured DNA samples (110 μl) were mixed with 1 ml of the bisulfite mixture and incubated at 55°C for 20 h under mineral oil. Samples were desalted using the Wizard DNA Clean-Up system (Promega), and the eluted DNA (in 50 μl of H2O) was desulfonated by the addition of 5.5 μl of 3 M NaOH and incubation at 37°C for 15 min. The samples were then neutralized by the addition of 55 μl of 6 M ammonium acetate (pH 7.0), and the DNA was ethanol precipitated, washed in 70% ethanol, dried, and redissolved in 20 μl of water.
PCR of bisulfite-treated DNA and sequencing.
In each experiment the upper strand was specifically amplified from bisulfite-treated DNA by two rounds of nested PCR. PCR reaction mixtures (total volume, 50 μl) contained a 0.5 mM concentration of each primer, 1.5 mM MgCl2, deoxynucleoside triphosphates (each at a concentration of 200 μM), and 2.5 U of Taq DNA polymerase (Gibco/BRL), and the PCR cycling profile consisted of an initial 5-min denaturation at 94°C, followed by 35 cycles of denaturation (94°C, 45 s), annealing (65°C, 45 s), and extension (72°C, 2 min), with a 10-min extension on the last cycle. In the first PCR the template was 3 to 5 μl of bisulfite-treated genomic DNA, and in the second PCR the template was 0.02 μl of the first PCR reaction mixture. The initial upstream and downstream primers were 5′-GTTTATGGGT(T/C)GGTTTTTTGAAGAGGTT-3′ (positions −2620 to −2593) and 5′-TCTACCCTATCCC(G/A)ACTCTTACCTACT-3′ (positions −2284 to −2310) for the exon 1A DMR, 5′-GTAATTTTATAGGGTTTTATTG-3′ and 5′-ATCCATTCTCTTAAATACTCACC-3′ for Nesp (34), and 5′-GATTTAGATAGTTTGTTGTTGGTGT-3′ and 5′-AAACCCCACTCCCCCCAATCAT-3′ for Gnasxl (34). The nested upstream and downstream primers were 5′-GGGTTGTTTTAGGTGGTTGGTATTAG-3′ (positions −2520 to −2495) and 5′-ACTCTTACCTACTC(G/A)AACACCTC-3′ (positions −2298 to −2320) for the exon 1A DMR, 5′-GAGAGGATTAGTGGAGGTATTTTT-3′ and 5′-ACTCACCCTCTAACTCTACAAAAAAT-3′ for Nesp (34), and 5′-GTGTTGGTGTTTATTTTTTGTGTT-3′ and 5′-ACCCAACAAATTACCCAAAATACCA-3′ for Gnasxl (34). The amplified fragments were gel purified, and then either the fragments were directly sequenced with the nested upstream primer using the Thermo Sequenase kit (Amersham Pharmacia Biotech) or the PCR products were subcloned into pCRII-TOPO by TA cloning (Invitrogen) and individual clones were sequenced using the same primer.
RT-PCR.
Reverse transcription (RT)-PCR was performed on total RNA (1 μg per sample) using a previously described protocol (41). The PCR cycling profile consisted of an initial 4-min denaturation at 95°C, followed by 31 cycles of annealing (56°C, 30 s), extension (72°C, 60 s), and denaturation (95°C, 30 s), with a 10-min extension for the final cycle. The upstream and downstream primers used were 5′-GGACACTCAGTCGCGTCGGCA-3′ and 5′-CTCCGTTAAACCCATTAACAT-3′ for exon 1A mRNAs and 5′-CGTCGACAACGGCTCCGGCATGTGCAAAGC-3′ and 5′-AATAGTGATGACCTGGCCGTCAGGCAGCTC-3′ for β-actin (39). RT-PCR reactions were run on 6% acrylamide gels (Novex). Specific RT-PCR products were isolated from agarose gels and directly sequenced.
Northern analysis.
Northern analysis was performed as described previously (48), except that the final washes were performed at 55°C. The exon 1 (positions +1 to +125)- and 1A (positions −2543 to −2388)-specific probes were generated by PCR using the following primer pairs: 5′-ATGGGCTGCCTCGGCAACAGTAAGACCGAGGACCAGCGC-3′ and 5′-CGGTGCGTGGCCCGGTAGACCTGCTTGTCC-3′ for exon 1 and 5′-CAGTCGCGTCGGCACCGCGGAG-3′ and 5′-GACGCACTCACACGCAAAGCAG-3′ for exon 1A. The mouse multiple-tissue Northern blot, containing poly(A) RNA from various adult mouse tissues (2 μg/lane), was obtained from Clontech. Renal proximal tubules and inner medulla were isolated as previously described (48).
Nucleotide sequence accession number.
The nucleotide sequence discussed in this paper has been deposited in GenBank under accession no. AF 152375.
RESULTS
General organization of Gnas exons 1 and 1A.
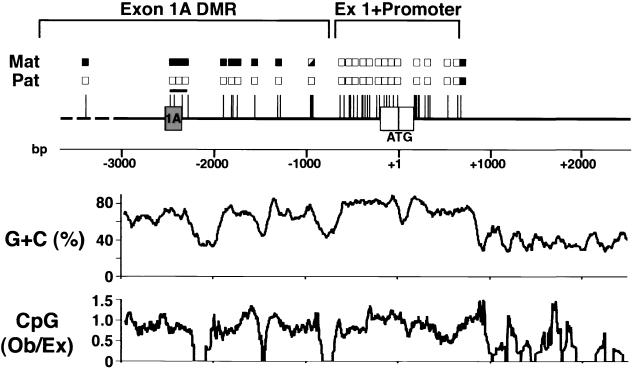
Exon 1 contains the 5′ untranslated region and initial coding sequence for GSα (27). Sequencing of an ∼7.4-kb mouse genomic DNA fragment showed exons 1 and 2 to be separated by a 3,782-bp intron. As in the human gene (27), the sequences just upstream of the ATG initiator codon and downstream of exon 1 are highly GC rich (70 to 80% [see Fig. 5]), with a high frequency of CpG dinucleotides (10 to 20%), and therefore the GSα promoter and first exon are located within a CpG island (4, 15).
FIG. 5.
Summary of the methylation status of Gnas exons 1 and 1A. Methylation of the maternal (Mat) and paternal (Pat) alleles at all sites which we examined are summarized (filled boxes, methylated; open boxes, unmethylated; half-filled boxes, partially methylated). The pSHY46 insert is shown as a solid line, with the region upstream of pSHY46 shown as a broken line. Exons 1A and 1 are shown as grey and white rectangles, respectively. A scale demonstrating positions (in base pairs) relative to the GSα translational start site (ATG) in exon 1 is shown below. The unmethylated GSα promoter and exon 1 and differentially methylated exon 1A region are delineated above. The region analyzed by bisulfite-modified genomic sequencing (as shown in Fig. 6) is delineated with a horizontal line. The GC content (expressed as a percentage) and CpG dinucleotide frequency (expressed as a ratio of observed (Ob)/expected (Ex) is shown graphically below. These were generated with the Genetics Computer Group software package using a 100-base window.
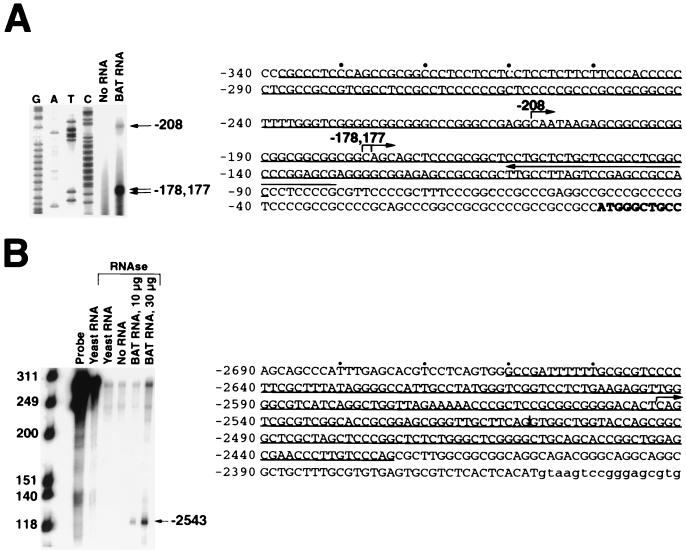
We determined the exon 1 transcriptional start sites by both primer extension analysis and ribonuclease protection assays. Hybridization of a complementary 32P-end-labeled oligonucleotide to BAT poly(A)-selected RNA followed by primer extension generated three major extension products consistent with the presence of three major transcriptional start sites at positions −208, −178, and −177 relative to the translational start site, respectively (Fig. 2A). Ribonuclease protection assays produced three major protected fragments that were consistent with the start sites determined by primer extension (data not shown). There were also several minor bands detected in the ribonuclease protection assay that might represent either minor transcriptional start sites not detected by primer extension or, perhaps, nonspecific digestion products. Based upon sequence similarities, the most upstream start site, at position −208, corresponds to the major transcriptional start site present in the human gene (27).
FIG. 2.
Determination of the exon 1 and 1A transcriptional start sites. (A) Primer extension of a 32P-end-labeled primer complementary to exon 1 (left-pointing arrow above sequence) was performed in the absence of RNA or in the presence of poly(A)-selected BAT RNA (1 μg). A sequencing reaction using the same primer was run beside the primer extension reactions (G, A, T, and C). The major extension products and the corresponding transcriptional start sites are indicated on the right. The exon 1 sequence is shown, with the first several bases of the coding sequence in bold. The major transcriptional start sites are indicated by right-pointing arrows. The extent of the antisense riboprobe used for ribonuclease protection assay (data not shown, see text) is underlined. (B) An exon 1A-specific antisense riboprobe (positions −2661 to −2425 [underlined in sequence]) was hybridized to yeast tRNA (30 μg), no RNA, or total BAT RNA (10 μg or 30 μg) and digested with ribonuclease. Base pairs are shown on the left, and undigested riboprobe in the absence and presence of yeast tRNA is shown in the next two lanes, respectively. The major digestion product and the corresponding transcriptional start site are indicated on the right. The exon 1A sequence is shown, with the first several bases of the downstream intron in lowercase. The position of an alternative donor splice site is indicated by a vertical line, and the major transcriptional start site is indicated with an arrow.
Comparison of our genomic sequence with the nonrepetitive entries in GenBank revealed that an alternative first exon (referred to as A and B in human [36] and 1′ in dog [22], and which we call 1A) is located ∼2.5 kb upstream of exon 1 (Fig. 1), similar to its position in the human and canine genes (22). Splicing of exon 1A to exon 2 generates two major alternative mRNA transcripts by the use of alternative donor splice sites (Fig. 2B). Exon 1A has no ATG codons, and its mRNAs are probably untranslated.
We determined the exon 1A transcriptional start site by ribonuclease protection assay (Fig. 2B). Hybridization of a radiolabeled antisense riboprobe (positions −2661 to −2425) to BAT total RNA followed by ribonuclease digestion produced a single specific ∼119-base-long fragment, consistent with the presence of a single transcriptional start site at about position −2543. Multiple attempts at primer extension analysis were unsuccessful. However, the start site determined by our ribonuclease protection assay corresponds to the exon 1A transcriptional start site in the canine gene which was determined by both S1 nuclease and primer extension analysis (22).
The GSα proximal promoter and exon 1 are within an unmethylated CpG island.
Because GSα is expressed primarily from the maternal allele in several tissues (e.g., BAT and renal cortex), we wanted to determine if there were associated allele-specific methylation differences within the GSα promoter and exon 1. BAT genomic DNA was digested with either BstBI alone, BstBI and BssHII, BstBI and SmaI, or BstBI and SacII, and Southern analysis was performed using a 1,887-bp BstBI genomic fragment (positions −653 to +1234 [Fig. 3]) as the probe. BssHII, SmaI, and SacII only cut when the CpG dinucleotides within their recognition sites are unmethylated. Digestion of the DNA sample with BstBI alone produced the 1,887-bp BstBI fragment. The addition of BssHII, SmaI, or SacII produced only fragments that result from complete digestion by each methylation-sensitive restriction enzyme, with no evidence of partial digestion products. Therefore, none of the sites tested in this region are methylated on either the maternal or paternal allele. Likewise, digestion of genomic DNA with either PstI alone or PstI and the methylation-sensitive enzyme HpaII, followed by hybridization with a 659-bp PstI genomic fragment (positions −567 to +92), demonstrated no evidence for methylation of the HpaII sites (data not shown). While methylation of closely spaced sites could not be ruled out in this experiment, the results of both experiments suggest that neither allele is methylated to a significant degree. Southern analysis using a more downstream EcoRI probe (positions +315 to +1641) revealed that all HpaII sites within this region are unmethylated, except for the most downstream site at position +658, which appears to be methylated on both alleles (data not shown but summarized [see Fig. 5]). There are no further downstream HpaII sites within the intron. HpaII sites as far upstream as position −637 are unmethylated, while one or more sites between positions −960 and −939 are partially methylated on the maternal allele (see below; Fig. 4 and 5). Therefore the GSα promoter and exon 1 are located within an unmethylated CpG island (Fig. 5), and suppression of GSα expression in the paternal allele is not associated with methylation of its promoter.
FIG. 3.
The GSα promoter and exon 1 are unmethylated. A restriction map of the 1,887-bp BstBI fragment (positions −653 to +1234) showing BssHII (Bs), SmaI (Sm), and SacII (S) sites is shown, and the lengths (in base pairs) of the restriction fragments produced by complete digestion with BstBI and each methylation-sensitive enzyme are indicated. For Southern analysis (blot), BAT genomic DNA was digested with BstBI alone (Bst), BstBI and BssHII (Bst/Bs), BstBI and SmaI (Bst/Sm), or BstBI and SacII (Bst/S) and hybridized with the 1,887-bp BstBI fragment. Based on these results all BssHII, SmaI, and SacII sites (both maternal [Mat] and paternal [Pat]) are unmethylated (indicated by unfilled boxes above each site).
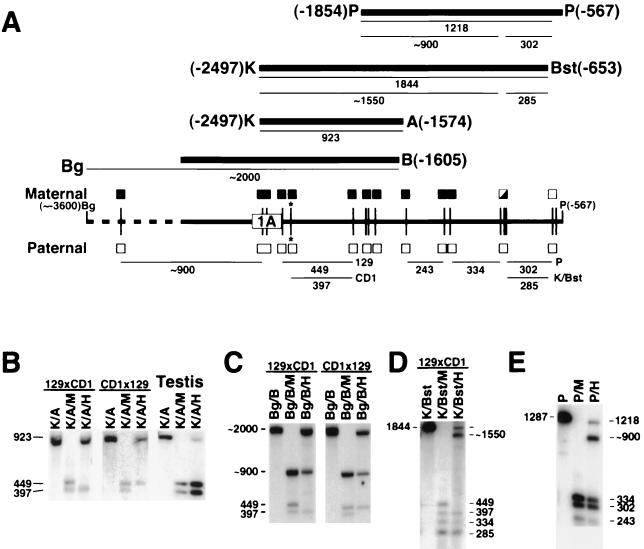
FIG. 4.
Identification of an upstream region with maternal-specific methylation. (A) Restriction map of the region spanning from the BglII (Bg) site at position −3600 to the PstI (P) site at position −567, with HpaII sites shown as vertical lines and exon 1A shown as a rectangle. The polymorphic HpaII site is indicated with an asterisk. The four probes used for Southern analysis are indicated by thick lines (K, KpnI; A, AvaII; B, BamHI; Bst, BstB), with the restriction fragments derived from the maternal allele shown under each probe. The restriction fragments derived from the paternal allele are shown below the map. The methylation status of the maternal and paternal alleles at each HpaII site is shown above and below the map, respectively (filled boxes, methylated; unfilled boxes, unmethylated; half-filled boxes; partially methylated). (B) Genomic DNA from 129 × CD1 (left) and CD1 × 129 (middle) mice was digested with KpnI and AvaII (K/A), KpnI-AvaII and MspI (K/A/M), or KpnI-AvaII and HpaII (K/A/H) and hybridized with the K-A (positions −2497 to −1574) fragment. Identical results were obtained in all tissues except for testis (right) (data derived from 129 × CD1). (C) 129 × CD1 (left) and CD1 × 129 (right) genomic DNA samples were digested with BglII and BamHI (Bg/B), BglII-BamHI and MspI (Bg/B/M), or BglII-BamHI and HpaII (Bg/B/H) and hybridized with the 1.5-kb upstream portion of the pSHY46 insert (positions −3050 to −1605). (D) 129 × CD1 genomic DNA sample was digested with KpnI and BstBI (K/Bst), KpnI-BstBI and MspI (K/Bst/M), or KpnI-BstBI and HpaII (K/Bst/H) and hybridized with the K-Bst (positions −2497 to −653) fragment. The expected 243-bp band after MspI or HpaII digestion is barely detectable in this experiment but was more clearly visible in other experiments. (E) Genomic DNA was digested with PstI alone (P), PstI and MspI (P/M), or PstI and HpaII (P/H) and hybridized with the P (positions −1854 to −567) fragment. All sites in the paternal allele are unmethylated based on the presence of smaller bands of correct size which could be well resolved in some experiments (data not shown).
Identification of a DMR upstream of the GSα promoter.
To look for a DMR upstream of the GSα promoter, we performed Southern analysis using more upstream genomic DNA probes. To facilitate the assignment of parental alleles, we identified a polymorphic HpaII site at position −2298 (Fig. 4) that was present in the CD1 mice used in our studies but absent in 129/SvJ mice, resulting in 397- and 449-bp HpaII restriction fragments in CD1 and 129/SvJ, respectively (Fig. 4). Genomic DNA from 129 × CD1 and CD1 × 129 mice was digested with KpnI and AvaII alone, KpnI-AvaII and MspI, or KpnI-AvaII and HpaII, and the filters were hybridized with a 923-bp KpnI-AvaII fragment (positions −2497 to −1574 [Fig. 4]). This fragment includes the two HpaII sites located within exon 1A. MspI and HpaII recognize the same sequence, but HpaII will only cut if the CpG dinucleotide within the recognition site is unmethylated, while MspI will cut whether or not the site is methylated. Digestion with KpnI and AvaII produced the 923-bp KpnI-AvaII fragment, while addition of MspI produced both the 449- and 397-bp HpaII fragments (Fig. 4B). Digestion of 129 × CD1 DNA with KpnI-AvaII and HpaII produced the 923- and 397-bp fragments, but not the 449-bp fragment, indicating that the maternal allele is methylated at all HpaII sites, while the paternal allele is unmethylated. The CD1 × 129 sample produced the 923- and 449-bp fragments but not the 397-bp fragment, which is also consistent with methylation of the maternal allele.
Identical results were obtained with DNA from multiple adult tissues, including BAT, renal cortex, renal inner medulla, lung, heart, liver, spleen, uterus, ovary, and cerebellum (data not shown). Therefore, the maternal allele is methylated in all tissues, both those in which GSα is imprinted (e.g., BAT and renal cortex) and those in which it is biallelically expressed (e.g., renal inner medulla and lung). The same results were also obtained with DNA from postcoitus (7.5 days) embryos, indicating that maternal-specific methylation is established by the early postimplantation period (data not shown).
The only organ in which the methylation pattern was different was the testis, where the DNA was almost completely digested by HpaII (Fig. 4B). The ability of HpaII digestion to produce both the 449- and 397-bp fragments in roughly equal amounts confirms that both alleles are unmethylated within the testis, which is composed mostly of male germ cells. The faint remaining 923-bp band is most likely due to the presence of testicular somatic cells (e.g., Sertoli and Leydig cells). Presumably, the methylation is erased in primordial germ cells and is not reestablished in the male germ line, consistent with lack of methylation of the paternal allele in later development. Southern analysis (data not shown) and bisulfite-modified genomic sequencing (see below [Fig. 6]) of sperm DNA confirmed that this region is unmethylated.
FIG. 6.
Methylation status of the exon 1A, Nesp, and Gnasxl DMRs in oocytes, sperm, and E3.5d blastocysts. The portions of the exon 1A, Nesp, and Gnasxl DMRs that were analyzed are indicated as filled rectangles in the schematic at the top. The results of methylation analysis by bisulfite-modified genomic sequencing at 16 CpG sites in the exon 1A DMR in oocytes, sperm, and E3.5d blastocysts are shown in the top row. The bar graphs indicate the percent of alleles that were methylated at each site. In blastocysts ∼50% of the alleles were unmethylated (except at site 16), while the rest were methylated at all sites except sites 7, 9, and 14. Similar analysis of 10 CpG sites within the Nesp DMR (34) and 12 CpG sites within the Gnasxl DMR (34) in oocytes, sperm, blastocysts are also shown (n = 9 to 19 clones sequenced).
To examine methylation further upstream, genomic DNA from CD1 × 129 and 129 × CD1 mice was digested with BamHI and BglII, BamHI-BglII and MspI, or BamHI-BglII and HpaII and hybridized with an ∼1.5-kb genomic DNA probe from the upstream portion of pSHY46 (Fig. 4A and C). Digestion with BamHI and BglII produced an ∼2,000-bp fragment, locating a BglII site to about position −3600. Digestion with BamHI-BglII and MspI produced the 449- and 397-bp fragments, as well as an ∼900-bp fragment, locating an HpaII site at about position −3400. Digestion of the CD1 × 129 sample with BamHI-BglII and HpaII produced ∼900- and 449-bp bands from the unmethylated paternal allele and the ∼2,000-bp band from the methylated maternal allele, confirming that the site at position −3400 is methylated in the maternal allele. Analysis of the 129 × CD1 sample confirmed that the maternal allele is the one that is methylated. The faint bands seen with HpaII digestion of the 129 × CD1 sample are probably due to incomplete digestion, although we cannot rule out minimal methylation of the paternal allele. Further studies are required to define the 5′ extent of the DMR.
To determine the 3′ extent of the DMR, 129 × CD1 genomic DNA was digested with KpnI and BstBI, KpnI-BstBI and MspI, or KpnI-BstBI and HpaII and hybridized with a 1,844-bp KpnI-BstBI fragment (positions −2497 to −653 [Fig. 4A and D]). Digestion with KpnI-BstBI and HpaII generated the 397-bp but not the 449-bp fragments, as well as other fragments resulting from complete HpaII digestion, indicating that the paternal (but not the maternal) allele is unmethylated. The methylated maternal allele produced 1,844- and ∼1,550-bp products, consistent with partial methylation of one or more of three closely spaced sites between positions −960 and −939 and complete methylation of all other upstream sites. Genomic DNA was then digested with PstI alone, PstI and MspI, or PstI and HpaII and hybridized with a 1,287-bp PstI fragment (positions −1854 to −567 [Fig. 4A and E]). Digestion with PstI and HpaII produced the complete digestion products from the unmethylated paternal allele and 1,218- and ∼900-bp products from the maternal allele (also consistent with partial methylation of sites between positions −960 and −939), complete methylation of all other upstream sites, and no methylation at the next downstream site at position −636. In summary, we have identified a DMR (which we call the exon 1A DMR) that is densely methylated on the maternal allele over a span of at least 2 kb (Fig. 4 and 5).
The exon 1A DMR is a methylation imprint mark.
Methylation of the exon 1A DMR is established by early postimplantation development and is present in all somatic tissues, making it a possible candidate to be a methylation imprint mark. The hallmark of a methylation imprint mark is that its methylation is established during gametogenesis and maintained throughout pre- and postimplantation development (12). We determined the methylation status of the exon 1A DMR in oocytes, sperm, and E3.5d blastocysts by bisulfite-modified genomic sequencing (10). Bisulfite treatment of genomic DNA mutates unmethylated cytidines to uracils, while methylated cytidines remain unmodified. After subsequent PCR, unmethylated cytidines are converted to thymine (T), while methylated cytidines remain as cytidine (C). PCR products were subcloned and sequenced, and the methylation status of the sense strand at 16 CpG sites located between positions −2478 and −2312 was determined. At each site the percent of methylation is defined as the percent of the individually sequenced PCR products in which the C fails to convert to T. As shown in Fig. 6 (top row), all CpG sites, except for sites 7, 9, and 14, were highly methylated in oocytes. Analysis of the maternal allele in BAT produced similar results (DNA samples containing the maternal and paternal allele were isolated from Gnas+/− mice by taking advantage of the fact that the targeted insertion produces upstream fragments of different length after BglII digestion) (data not shown). In contrast, these sites are unmethylated in sperm DNA, consistent with our results from Southern analysis of testis (Fig. 4B) and bisulfite-modified genomic sequencing of the paternal allele in BAT (except for site 16, which was methylated [data not shown]). Therefore differential methylation of the exon 1A DMR is established in female germ cells. In E3.5d blastocysts, ∼50% of the alleles were methylated and ∼50% were unmethylated (Fig. 6). Therefore the maternal-specific methylation of the exon 1A DMR that is established during oogenesis appears to be maintained during preimplantation development, at a time when the genome is undergoing global demethylation. These results are consistent with the exon 1A DMR being a methylation imprint mark.
Methylation analysis of Nesp and Gnasxl.
We also examined the methylation status of the Nesp and Gnasxl promoter regions in germ cells and blastocysts. In adult tissues, the Nesp promoter region is methylated on the paternal allele (34). Ten CpG sites within the Nesp DMR are unmethylated in sperm and E3.5d blastocysts and minimally methylated in oocytes (Fig. 6, middle row). Lack of methylation in sperm was confirmed by Southern analysis (data not shown). Based on Southern analysis and bisulfite-modified genomic sequencing, methylation in this region is established during postimplantation development by E10.5d (data not shown). These results suggest that the Nesp DMR is not a methylation imprint mark, although we only examined a portion of the DMR and have therefore not ruled out the presence of a methylation imprint mark within other portions of the DMR.
The Gnasxl promoter region is methylated on the maternal allele in adult tissues (34). Analysis of 12 CpG sites within the Gnasxl DMR showed these sites to be methylated in >50% of the alleles in oocytes (Fig. 6, bottom row). However, ∼50% of the alleles were also methylated in sperm and >50% of the alleles were methylated in E3.5d blastocysts. One possible explanation for these findings is that the maternal-specific methylation is not erased in the male or female germline. Further studies are required to clearly define the temporal changes in methylation of this DMR through development. In any case, this region does not show the methylation pattern which is typical of a methylation imprint mark, because the methylation is not specific for female germ cells.
Exon 1A mRNAs are expressed only from the paternal allele.
We predicted that exon 1A mRNAs would only be derived from the paternal allele because its promoter is methylated on the maternal allele. We performed RT-PCR using an exon 1A-specific upstream primer and an exon 2-specific downstream primer on BAT total RNA derived from Gnas−/+, Gnas+/−, and wild-type (Gnas+/+) mice (Fig. 7A). In Gnas knockout mice, the targeted insertion is located between the upstream and downstream primers and therefore prevents amplification of the RT-PCR product. Two major products were amplified from Gnas+/+ and Gnas−/+ mice, but not from Gnas+/− mice, indicating that these mRNAs are only expressed from the paternal allele. Simultaneous amplification with β-actin-specific primers ruled out differences in RNA amount or integrity. Similar results were also obtained with renal proximal tubule RNA (data not shown). Northern analysis of BAT RNA using an exon 1A-specific probe was also consistent with paternal-specific expression (data not shown). Sequencing of the two major RT-PCR products demonstrated the use of two donor splice sites (Fig. 2B). The position of the downstream splice site is identical in mouse, dog, and human, while the upstream splice site is unique to mouse (22, 36).
FIG. 7.
Expression studies of exon 1A mRNAs. (A) Paternal-specific expression of exon 1A mRNAs. RT-PCR was performed on BAT total RNA using an exon 1A-specific upstream primer and an exon 2-specific downstream primer. The position and size (in base pairs) of two major products are indicated on the left. Mouse genotypes and the presence or absence of enzyme in the RT reaction are indicated above. The results of RT-PCR analysis performed using β-actin-specific primers are shown underneath. The two major mRNAs result from the use of alternative donor splice acceptor sites, which are shown in Fig. 2B. (B) Tissue distribution of exon 1A mRNAs. Mouse multiple-tissue Northern blot obtained from Clontech (left) and a second blot prepared by us (right) (2 μg of poly(A) RNA per lane in both panels) were hybridized with exon 1 and 1A probes, which recognize 1.8- and 1.7-kb bands, respectively. The results shown do not reflect the proportion of the two mRNAs, as the exposure time for the exon 1A probe was more than 10 times greater than that for the exon 1 probe. Sk muscle, skeletal muscle; WAT, white adipose tissue; renal IM, renal inner medulla; renal PT, renal proximal tubules.
Exon 1A mRNAs are ubiquitously expressed.
One possible model to explain tissue-specific imprinting of GSα would be a promoter competition model in which the GSα and exon 1A promoters are reciprocally regulated. Suppression of the maternal exon 1A promoter would allow GSα to be maternally expressed in all tissues. The paternal GSα promoter would be suppressed in tissues where the exon 1A promoter is active and would remain active in tissues where the exon 1A promoter is silent. This model would predict that exon 1A mRNAs are expressed only in tissues where GSα is imprinted (e.g., BAT and renal proximal tubules). We determined the tissue distribution of GSα and exon 1A mRNAs in multiple mouse tissues by Northern analysis using exon 1- and 1A-specific cDNA probes, which recognize specific 1.8- and 1.7-kb bands, respectively (Fig. 7B). We estimate that, as was previously found in dog (22), the levels of GSα mRNA are generally at least 10-fold higher than those of exon 1A mRNA (the data in Fig. 7B do not reflect equal exposure times). Like GSα, exon 1A mRNAs are expressed in most tissues. Moreover there is a strong correlation between the expression of GSα and exon 1A mRNAs in most tissues, suggesting that their promoters might be regulated by common mechanisms. Although we have not formally ruled out competition between the GSα and exon 1A promoters, our results suggest that this is not the major mechanism for tissue-specific imprinting of GSα.
DISCUSSION
As in the human homolog GNAS1 (27), the GSα promoter and first exon in Gnas are highly GC rich with a large number of CpG dinucleotides. CpG dinucleotides are underrepresented in the genome except in highly GC-rich regions called CpG islands (4, 15, 23). While CpG dinucleotides throughout most of the genome are methylated, CpG islands which correspond to promoters of ubiquitously expressed proteins (such as GSα) remain unmethylated (4). Promoter methylation is generally associated with transcriptional repression (23), and the promoters of several imprinted genes are methylated on the inactive allele (3, 12). In both the Igf2 (31) and Igf2r (20) genes, tissue-specific differences in imprinting are correlated with tissue-specific differences in allele-specific promoter methylation. However, we found that in mice the GSα promoter region remains unmethylated on both alleles in all tissues. Therefore, allele-specific expression of GSα is not due to promoter methylation. Studies of humans suggest that the GSα promoter in GNAS1 is also unmethylated (18). It is interesting to note that this CpG island has 17 putative Sp1 binding sites, as these sites may be important for protecting CpG islands from de novo methylation (6, 30).
We have identified a new DMR (the exon 1A DMR) located upstream of the GSα promoter that is methylated only on the maternal allele and have recently determined that the same region in human GNAS1 is methylated in a similar manner (J. Liu and L. S. Weinstein, unpublished data). This DMR also appears to be a CpG island based on its GC content and the density of CpGs (Fig. 5). Differentially methylated CpG islands have been identified in other imprinted genes (14, 45). Tandem direct repeats have been found in the vicinity of other DMRs and have been implicated in establishing allele-specific methylation (1, 12, 32). However, we did not identify any tandem direct repeat elements within or near the exon 1A DMR in mouse or human. We also did not locate the de novo methylation signal sequence that appears to be required to establish methylation of the Igf2r imprint mark in the female germline (5).
Allele-specific methylation is generally erased in primordial germ cells. DMRs whose methylation is reestablished during gametogenesis and maintained throughout pre- and postimplantation development are presumed to be critical in establishing the maternal and paternal epigenotypes and have therefore been termed methylation imprint marks or core DMRs (12). Other DMRs, whose methylation is established later in postimplantation development, are often located within inactive promoters and are probably important for maintaining (or possibly are the result of) allele-specific differences in expression. Maternal-specific methylation of the exon 1A DMR is established in female germ cells and maintained through pre- and postimplantation development, and, therefore, this region is a methylation imprint mark. A high density of methylated CpG dinucleotides within CpG-rich DMRs may protect them from the genome-wide demethylation that occurs during preimplantation development (19). Based on the results of bisulfite-modified genomic sequencing, at least a portion of the exon 1A DMR is densely (although not totally) methylated prior to fertilization.
Consistent with maternal-specific methylation of its promoter, exon 1A mRNA transcripts are only expressed from the paternal allele. These RNAs are probably not translated, as exon 1A lacks an ATG translational start site and there is no evidence for the existence of translation products in vivo. Several other imprinted genes encode untranslated RNAs (e.g., Xist, H19, and Snrpn) (3). The potential role of these untranslated RNAs in the imprinting mechanism are poorly understood, although there is evidence that the Xist untranslated RNA might inactivate the X chromosome in cis by remaining attached to and coating the chromosome (11). It remains to be determined what roles, if any, the exon 1A mRNAs play in Gnas imprinting or in preventing the exon 1A DMR from being methylated in male germ cells.
Two additional regions of the Gnas transcriptional unit are also differentially methylated in somatic cells, namely, the Nesp and Gnasxl DMRs that include the NESP55 and XLαs promoters, respectively (Fig. 1) (17, 18, 34). Our studies examining a portion of the Nesp DMR show that its methylation is not established until postimplantation development, and therefore this DMR does not appear to be a methylation imprint mark. However, further studies will be required to rule out the presence of a methylation imprint mark within a different portion of the Nesp DMR. A paternal-specific antisense RNA transcript that overlaps the Nesp DMR that may be important in establishing the paternal epigenotype of Nesp has been recently identified (44). Similarly, the methylation imprint mark of the Igf2r gene includes a promoter for a paternal-specific antisense transcript that is important for establishing the imprinting of the Igf2r promoter in postimplantation development (45). Our observation that Nesp methylation is not established until postimplantation development is consistent with Nesp imprinting being regulated by a similar mechanism.
The Gnasxl DMR is unusual in that it is partially methylated in both sperm and oocytes and is resistant to demethylation during preimplantation development. This suggests that either the maternal-specific methylation is not erased in primordial germ cells or that de novo methylation occurs in both male and female gametes. In any case, this is not the pattern typical for a methylation imprint mark. Studies of humans suggest that the paternal antisense transcript originates from a promoter located at the 5′ end of the Gnasxl DMR; therefore, this region may be important for establishing the imprinting of Nesp (16). Further studies will be required to determine when and how differential methylation is established in the Gnasxl DMR and its role in establishing the imprinting of Gnas.
In various other imprinted regions methylation imprint marks are required to establish the methylation of other distant DMRs within the same imprinted region, suggesting that these regions are imprinting centers from which the maternal and paternal epigenotypes of the whole imprinted region are established (8, 14, 37, 45, 46). As the exon 1A DMR appears to be a methylation imprint mark, it is possible that this region is required to establish the maternal and paternal epigenotypes throughout the Gnas locus. It remains to be determined if and how the various Gnas DMRs regulate each other.
In humans with pseudohypoparathyroidism type Ib, a disorder characterized by renal resistance to parathyroid hormone that is likely due to abnormal imprinting of GSα (24), the exon 1A DMR is unmethylated on both alleles (J. Liu and L. S. Weinstein, unpublished data), strongly suggesting that this DMR is important for the tissue-specific imprinting of GSα. Models to explain how methylation of the exon 1A DMR leads to imprinting of GSα must account for the fact that the exon 1A DMR is methylated in all somatic tissues, while GSα is only imprinted in some tissues. One possibility is that the exon 1A DMR contains binding sites for tissue-specific repressors that can bind to the paternal allele but are unable to bind to the maternal allele because its binding site is methylated. A second possibility is that the exon 1A DMR contains a boundary element that blocks activation of the GSα promoter by an upstream tissue-specific enhancer in the paternal allele but does not block promoter activation in the maternal allele because it is methylated. A similar mechanism most likely explains how the H19 DMR produces the reciprocal imprinting of H19 and Igf2 (35). Finally, it is possible that the exon 1A and GSα promoters on the paternal allele are reciprocally regulated, due to competition for common enhancers or negative regulation of the GSα promoter by exon 1A mRNAs. Our finding that exon 1A mRNA expression does not correlate with allele-specific expression of GSα makes the promoter competition model less likely. Further studies of mice in which the exon 1A DMR is deleted or mutated will define the mechanisms by which its differential methylation is established, the roles of the DMR and its mRNAs in establishing and maintaining the complex imprinting pattern of Gnas, and the mechanism by which GSα is imprinted in a tissue-specific manner.
ACKNOWLEDGMENTS
We thank Karl Pfeifer and Marc Reitman for reviewing the manuscript and helpful discussions, Eric Lee for providing 129/SvJ mice, Alex Grinberg for providing blastocysts, and Ruth Vinitsky for technical assistance.
REFERENCES
- 1.Barlow D P. Methylation and imprinting: from host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- 2.Barlow D P. Competition—a common motif for the imprinting mechanism? EMBO J. 1998;16:6899–6905. doi: 10.1093/emboj/16.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 4.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 5.Birger Y, Shemer R, Perk J, Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- 6.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 7.Brinster H L, Chen N Y, Trumbauer M E, Yagle M K, Palmiter R D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls R D, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 9.Campbell R, Gosden C M, Bonthron D T. Parental origin of transcription from the human GNAS1 gene. J Med Genet. 1994;31:607–614. doi: 10.1136/jmg.31.8.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemson C M, McNeil J A, Willard H F, Lawrence J B. Xist RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constancia M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 13.Davies S J, Hughes H E. Imprinting in Albright's hereditary osteodystrophy. J Med Genet. 1993;30:101–103. doi: 10.1136/jmg.30.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls R D, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 16.Hayward B E, Bonthron D T. An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet. 2000;9:835–841. doi: 10.1093/hmg/9.5.835. [DOI] [PubMed] [Google Scholar]
- 17.Hayward B E, Kamiya M, Strain L, Moran V, Campbell R, Hayashizaki Y, Bonthron D T. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc Natl Acad Sci USA. 1998;95:10038–10043. doi: 10.1073/pnas.95.17.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward B E, Moran V, Strain L, Bonthron D T. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95:15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell C Y, Steptoe A L, Miller M W, Chaillet J R. cis-acting signal for inheritance of imprinted DNA methylation patterns in the preimplantation mouse embryo. Mol Cell Biol. 1998;18:4147–4156. doi: 10.1128/mcb.18.7.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J F, Oruganti H, Yu T H, Hoffman A R. Tissue-specific imprinting of the mouse insulin-like growth factor II receptor gene correlates with differential allele-specific DNA methylation. Mol Endocrinol. 1998;12:220–232. doi: 10.1210/mend.12.2.0062. [DOI] [PubMed] [Google Scholar]
- 21.Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R. Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem. 1997;272:11657–11662. doi: 10.1074/jbc.272.17.11657. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa Y, Bianchi C, Nadal-Ginard B, Homcy C J. Alternative promoter and 5′ exon generate a novel GSα mRNA. J Biol Chem. 1990;265:8458–8462. [PubMed] [Google Scholar]
- 23.Jones P A, Laird P W. Cancer epigenetics come of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 24.Juppner H, Schipani E, Bastepe M, Cole D E, Lawson M L, Mannstadt M, Hendy G N, Plotkin H, Koshiyama H, Koh T, Crawford J D, Olsen B R, Vikkula M. The gene responsible for pseudohypoparathyroidism type Ib is paternally imprinted and maps in four unrelated kindreds to chromosome 20q13.3. Proc Natl Acad Sci USA. 1998;95:11798–11803. doi: 10.1073/pnas.95.20.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kehlenbach R H, Matthey J, Huttner W B. XLαs is a new type of G protein. Nature. 1994;372:804–809. doi: 10.1038/372804a0. [DOI] [PubMed] [Google Scholar]
- 26.Kelsey G, Bodle D, Miller H J, Beechey C V, Coombes C, Peters J, Williamson C M. Identification of imprinted loci by methylation-sensitive representational difference analysis: application to mouse distal chromosome 2. Genomics. 1999;62:129–138. doi: 10.1006/geno.1999.6022. [DOI] [PubMed] [Google Scholar]
- 27.Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gs alpha gene. Proc Natl Acad Sci USA. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li E, Beard C, Jaenisch R. Role of DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 29.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 30.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 31.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann B, Kubicka P, Barlow D P. Characteristics of imprinted genes. Nat Genet. 1995;9:12–13. doi: 10.1038/ng0195-12. [DOI] [PubMed] [Google Scholar]
- 33.Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 34.Peters J, Wroe S F, Wells C A, Miller H J, Bodle D, Beechey C V, Williamson C M, Kelsey G. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt J V, Levorse J M, Tilghman S M. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc Natl Acad Sci USA. 1999;96:9733–9738. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swaroop A, Agarwal N, Gruen J R, Bick D, Weissman S M. Differential expression of novel Gsα signal transduction protein cDNA species. Nucleic Acids Res. 1991;19:4725–4729. doi: 10.1093/nar/19.17.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorvaldsen J L, Duran K L, Bartolomei M S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tilghman S M. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 39.Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. Nucleotide sequence of a full-length cDNA for mouse cytoskeletal β-actin mRNA. Nucleic Acids Res. 1986;14:2829. doi: 10.1093/nar/14.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warner D R, Gejman P V, Collins R M, Weinstein L S. A novel mutation adjacent to the switch III domain of GSα in a patient with pseudohypoparathyroidism. Mol Endocrinol. 1997;11:1718–1727. doi: 10.1210/mend.11.11.0013. [DOI] [PubMed] [Google Scholar]
- 42.Weinstein L S. Albright hereditary osteodystrophy, pseudohypoparathyroidism and GS deficiency. In: Spiegel A M, editor. G proteins, receptors, and disease. Totowa, N.J: Humana Press; 1998. pp. 23–56. [Google Scholar]
- 43.Weinstein L S, Yu S, Ecelbarger C A. Variable imprinting of the heterotrimeric G protein GS α-subunit within different segments of the nephron. Am J Physiol. 2000;278:F507–F514. doi: 10.1152/ajprenal.2000.278.4.F507. [DOI] [PubMed] [Google Scholar]
- 44.Wroe S F, Kelsey G, Skinner J A, Bodle D, Ball S T, Beechey C V, Peters J, Williamson C M. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc Natl Acad Sci USA. 2000;97:3342–3346. doi: 10.1073/pnas.050015397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 46.Yang T, Adamson T E, Resnick J L, Leff S, Wevrick R, Francke U, Jenkins N A, Copeland N G, Brannan C I. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 47.Yu S, Gavrilova O, Chen H, Lee R, Liu J, Pacak K, Parlow A F, Quon M J, Reitman M L, Weinstein L S. Paternal versus maternal transmission of a stimulatory G protein α subunit knockout produces opposite effects on energy metabolism. J Clin Investig. 2000;105:615–623. doi: 10.1172/JCI8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein L S. Variable and tissue-specific hormone resistance in heterotrimeric GS protein α-subunit (GSα) knockout mice is due to tissue-specific imprinting of the GSα gene. Proc Natl Acad Sci USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeschnigk M, Schmitz B, Dittrich K, Horsthemke B, Doerfler W. Imprinted segments in the human genome: different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum Mol Genet. 1997;6:387–395. doi: 10.1093/hmg/6.3.387. [DOI] [PubMed] [Google Scholar]