Abstract
A novel clinical assay for the detection and quantitation of antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was adapted from an in-house, research-based enzyme-linked immunosorbent assay (ELISA). Development and validation were performed under regulatory guidelines, and the test obtained emergency use authorization (EUA) from the New York State Department of Health (NYSDOH) and the Food and Drug Administration (FDA). The Mount Sinai coronavirus disease 2019 (COVID-19) antibody assay is an orthogonal, quantitative direct ELISA test which detects antibodies reactive to the receptor binding domain (RBD) and the spike protein of the novel SARS-CoV-2. The assay is performed on 96-well plates coated with either SARS-CoV-2 recombinant RBD or spike proteins. The test is divided into two stages, a qualitative screening assay against RBD and a quantitative assay against the full-length spike protein. The test uses pooled high titer serum as a reference standard. Negative pre-COVID-19 and positive post-COVID-19, PCR-confirmed specimens were incorporated in each ELISA test run, and the assays were performed independently at two different locations.
The Mount Sinai COVID-19 serology performed with high sensitivity and specificity, 92.5% (95% CI: 0.785–0.980) and 100% (CI: 0.939–1.000) respectively. Between-run precision was assessed with a single run repeated over 22 days; and within-run precision was assessed with 10 replicates per day over 22 days. Both were within reported acceptance criteria (CV ≤ 20%).
This population-based study reveals the applicability and reliability of this novel orthogonal COVID-19 serology test for the detection and quantitation of antibodies against SARS-CoV-2, allowing a broad set of clinical applications, including the broad evaluation of SARS-CoV-2 seroprevalence and antibody profiling in different population subsets.
Keywords: Coronavirus disease 2019, Severe acute respiratory syndrome coronavirus-2 (SARSCoV-2), Antibody tests, Diagnosis of COVID-19, ELISA, IgG antibody assay
Coronavirus disease 2019; Severe acute respiratory syndrome coronavirus-2 (SARSCoV-2); Antibody tests; Diagnosis of COVID-19; ELISA; IgG antibody assay.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 in China, and has since then caused a pandemic of coronavirus disease 2019 (COVID-19). SARS-CoV-2 infection induces an antibody response against several viral proteins. Both the nucleoprotein and the spike protein have been used as targets for antibody responses and monitoring of the disease, more recently for monitoring vaccination responses. The nucleoprotein is found within the virion and is therefore unlikely a target of neutralizing antibodies. In fact, for SARS-CoV-1 it has been shown that vaccination with nucleoprotein induces non-neutralizing antibodies and could enhance disease in an animal model (Deming et al., 2006). The trimeric spike protein is found, in part, on the surface of the virus and is responsible for both binding to the cellular receptor angiotensin converting enzyme 2 (ACE2) via its receptor binding domain (RBD), as well as for fusion of viral and cellular membranes, which in turn triggers release of the viral genome into the host cell (Letko et al., 2020; Wrapp et al., 2020). Antibodies that target the RBD can neutralize the virus by preventing its interactions with the host cell (Alsoussi et al., 2020). In addition, epitopes outside the RBD have also been reported to effectively be neutralizing targets.
For certain viruses, correlates of protection have been established (Plotkin, 2010). These correlates of protection are often based on neutralizing antibodies, like for influenza virus or measles virus, or on antibody binding titers that themselves correlate with virus neutralization but are easier to measure (e.g., hepatitis A or B virus). These correlates of protection are of assistance in determining the risk to a person of become infected, informing on vaccine responses, and guiding recommendations on booster doses of a vaccine to remain optimally protected.
Antibodies to the spike protein of SARS-CoV-2 have been shown to neutralize the virus (Amanat and Stadlbauer 2020; Okba et al., 2020). Furthermore, in non-human primate models it has been shown that infection protects from reinfection with SARS-CoV-2, especially in the lower respiratory tract (Chandrashekar et al., 2020; Deng et al., 2020). In addition, a neutralizing monoclonal antibody was able to protect non-human primates from SARS-CoV-2 challenge (Shi et al., 2020), and transfer of serum from convalescent hamsters protected naïve hamsters from SARS-CoV-2 induced disease (Imai et al., 2020). Finally, vaccine-induced neutralizing antibodies have been established as correlate of protection from SARS-CoV-2 infection in a non-human primate model (Yu et al., 2020). We and others have reported that antibody titers measured in enzyme-linked immunosorbent assays (ELISA) against the full-length spike protein correlate with virus neutralization. ELISAs can be performed at high throughput and in a low biosafety level containment (Amanat and Stadlbauer 2020; Okba et al., 2020). A quantitative ELISA against the spike protein would therefore be the ideal tool to establish correlates of protection against SARS-CoV-2. Here we describe the establishment of such a quantitative assay.
2. Methods
Informed consent was obtained from all individuals included in this study. The study was approved by the institutional review board of the Icahn School of Medicine at Mount Sinai (20-1032-00001-01).
2.1. Source for SARS-CoV-2 antigens
Two different versions of the spike (S) protein are used as antigens. The first is a full-length trimeric and stabilized version of the spike protein, the second is the smaller RBD, which is part of the spike. Mammalian cell codon optimized nucleotide sequence coding for the spike protein of SARS-CoV-2 isolate (GenBank: MN908947.3) was commercially synthesized (GeneWiz®). The receptor binding domain (RBD, amino acid 319 to 541, RVQP....CVNF) and full-length spike were cloned into the mammalian expression vector pCAGGS. Recombinant proteins were produced in Expi293F cells (Thermo Fisher®) by transfection with purified DNA using the ExpiFectamine Transfection Kit. Supernatant was harvested three days after transfection, the proteins purified and suspended in phosphate buffered saline (PBS). This methodology is published in greater detail (Amanat and Stadlbauer 2020; Stadlbauer et al., 2020).
2.2. Standard serum and quality control human serum
Negative Controls (NEG) are prepared using remnant pooled serum that was tested negative for SARS-CoV-2 antibodies with optical density at 490 nm (OD490) less than 0.15. Positive Controls are prepared using remnant pooled serum that was tested positive for SARS-CoV-2 antibodies with OD490 more than 0.15 in both RBD screen and spike titration. Low, Medium and High Quality Control (LQC, MQC, HQC) sera were prepared using pooled serum that was tested positive at 1:160, 1:320 and ≥1:2880/2 (1:2 dilution in negative serum pool) in spike titration and resulted in OD490 values equal to 0.38 ± 0.12, 1.0 ± 0.19, and 2.0 ± 0.32 respectively. Standard pools and quality control pools were prepared by pooling serum samples of at least 25 patients who tested positive for a given titer.
2.3. COVID-19 serology ELISA procedure
The Mount Sinai COVID-19 ELISA test is an orthogonal assay usually conducted as a two-day procedure. On the first day, 96-well plates are coated with 50 μL of a 2 μg/mL solution of viral antigen (SARS-CoV-2 RBD protein) in 1X phosphate buffered saline (PBS). Plates are incubated at 4 °C overnight and stored for up to 1 week. Serum samples are heat inactivated in a warm water bath (56 °C) for 1 h. Samples may be stored at 4 °C overnight or until use. On the second day, coated plates are washed 3x with PBS-T (0.1% Tween-20 in PBS). 200 μL of blocking solution, 3% non-fat milk (NFM), in PBS-T is added to each well and the plates are incubated at room temperature (RT) for 1 h. One dilution plate (separate flat-bottomed cell culture plate) per antigen coated plate is prepared. Assay controls and patient samples are diluted 1:50 (5 μL of sample into 245 μL of 1% NFM in PBS-T) and 100 μL of prepared dilution is transferred to the ELISA plates after removing blocking solution. After a two-hour incubation at room temperature, the plates are washed 3x with PBS-T. Anti-human IgG (Fab specific, Sigma A0293) horseradish peroxidase (HRP)-labeled secondary antibody, diluted 1:3000 in 1% NFM in PBS-T is prepared. 50 μl of secondary antibody solution is added to each well and the plate is incubated at room temperature for one hour. After washing, 100 μL of HRP substrate (SigmaFast™ o-phenylenediamine dihydrochloride (OPD)) solution is added for 10 min and enzymatic reaction stopped with 50 μL of 3M HCl. The plates are read in a plate reader (BioTek) at an absorbance of 490 nm. Samples that exceed certain OD490 cutoff value (OD490 ≥ 0.15) are assigned presumptive positive and are tested in confirmatory ELISA using full-length spike protein.
2.4. Quantitation of anti-spike IgG by ELISA
Principles of the ELISA procedure including sample preparation, coating, blocking, incubation, secondary antibody and substrate incubation follow the same scheme as for the above referred screening, with a few modifications. Plates are coated with 50 μL of a 2 μg/mL solution of viral antigen (SARS-CoV-2 full length spike protein) in 1x PBS per well and incubated at 4 °C overnight. Coated plates are washed, blocked and the standard pool as well as sample dilutions are prepared on a separate plate. The standard pool is diluted in an eight-step serial dilution (from 1:8 to 1:4883, in 2.5-fold steps) in duplicates. Sample and quality control dilutions are performed in duplicate by adding 3.75 μL of inactivated patient serum to 296.25 μL of 1% NFM in PBS-T (final dilution 1:80) and incubated for 2h at RT. Further steps of washing, secondary antibody, substrate incubation and plate reading follow exactly the RBD protocol.
2.5. Characterization of the standard pool using a reference curve
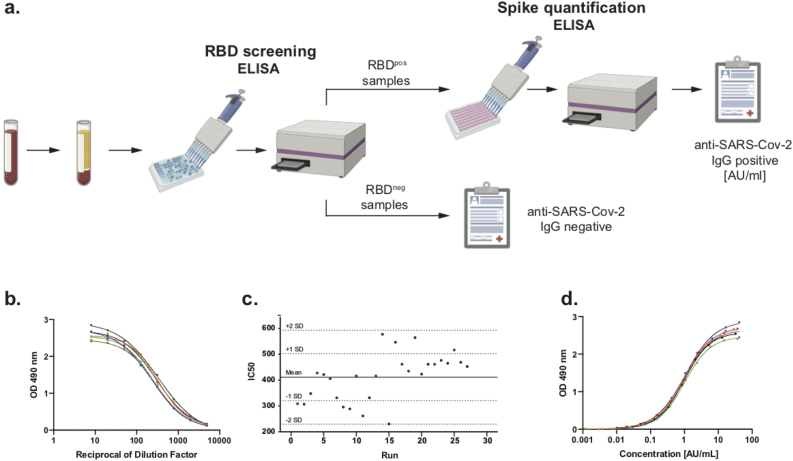
The standard pool was created by combining sera from 25 patients which tested positively for the presence of anti-spike IgG at a titer of ≥1:2880 (2880 pool). To determine baseline arbitrary concentration (BAC) of anti-spike IgG antibodies in the pool, we performed eight-step serial dilutions (from 1:8 to 1:4883) and measured optical density at 490nm (OD490) of each dilution following the ELISA protocol. The results were fitted into a 4-parameter non-linear regression curve (4PL) and the inflection points (IC50) of the resulting curves were determined using GraphPad Prism 8 software (Figure 1b). Longitudinal measurements of the IC50 value of the standard pool provided consistent results (Figures 1c-d) and were assigned as a BAC of the anti-spike IgG antibodies (arbitrary units per milliliter (AU/ml)) of the given standard pool.
Figure 1.
Principle of quantitative COVID-19 IgG ELISA. a. An orthogonal serology ELISA assay was developed to detect anti-SARS-CoV-2 IgG in a two-step test including an RBD screening, where plasma samples are pre-tested for the presence of IgG antibodies against RBD. Next, RBDpos samples undergo quantitative ELISA for presence of anti-spike IgG. Results are reported as amount of arbitrary (Antibody) units per ml (AU/ml). b. Representative reference curves of serial dilutions of ≥1:2880 standard pool used to determine baseline arbitrary concentration (BAC) of this standard pool calculated as inflection point (IC50) of reference curve. c. Measurement of BAC (IC50) of ≥1:2880 reference pool showed persistent consistency over time. d. Representative standard curves plotted based on BAC of a given run. A 4PL equation of the standard curve was used to calculate antibody concentration (AU/ml) in tested samples.
2.6. Standard curve
Once the BAC is determined, the standard curve is created by plotting the concentration in AU/mL of each dilution on the x-axis vs its OD490 readouts on the y-axis on a 4PL curve (Figure 1c). The equation for the standard curve was calculated using GraphPad Prism 8 software and was used to determine antibody concentration in AU/mL of unknown samples using the following equation:
| (1) |
where X = arbitrary antibody concentration (AU/mL), Y = OD490nm, A = lower asymptote, B = hill slope, C = IC50, D = upper asymptote of 4PL curve.
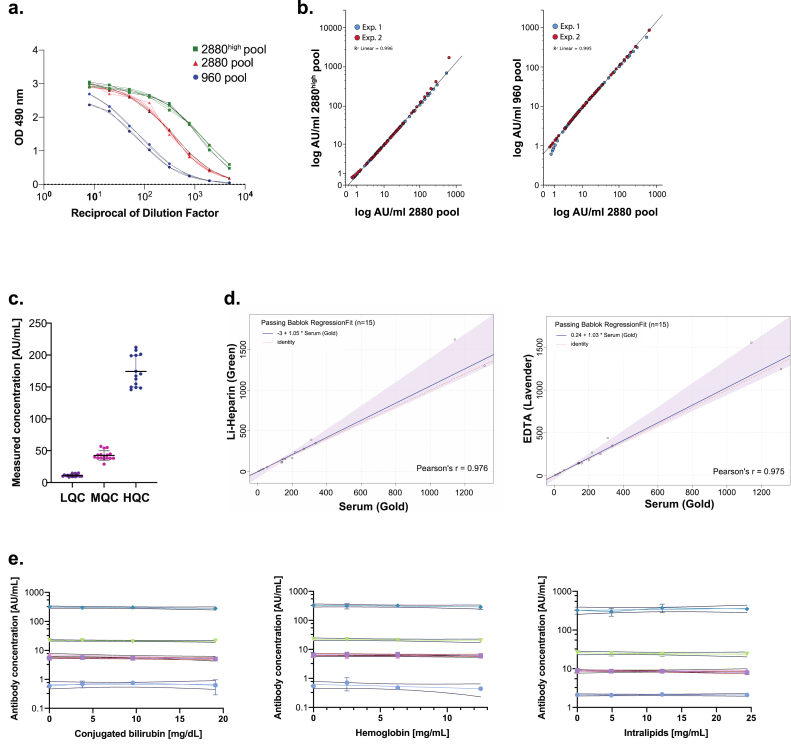
Based on prior literature we hypothesized that using the method of assigning the inflection point of the curve plotted on serial dilutions of a given pool vs OD490nm readouts as a BAC of this pool would allow for accurate characterization of any standard pool created from positive sera of patients (Miura et al., 2008; Vu et al., 2016). To validate this hypothesis, we created two additional standard pools: 960 and 2880high from sera of at least 25 patients positive for the presence of anti-spike IgG at a titer of 1:960 and ≥1:2880, respectively (Figure 2a). Next, we performed an ELISA on samples from 60 patients of various antibody titers and calculated AU/mL using the method described above. Comparison of AU/mL values of each sample calculated using different standard pools showed high consistency (Figure 2b).
Figure 2.
Validation of Mount Sinai COVID-19 Serology Assay. a. Representative reference curves of two additional reference pools: 1:960 pool and ≥1:2880high pool. As expected, a BAC (IC50) of ≥1:2800high pool is higher and of 1:960 pool lower than the original ≥1:2880 pool which corresponds to higher and lower amounts of IgG in these pools in comparison to ≥1:2880 pool. b. 120 patient plasma samples from a range of previously tested titer were run and calculated using two additional reference pools: 1:960 and ≥1:2880high. Antibody concentrations of these samples calculated vs each pool showed high consistency with concentrations calculated using ≥1:2880 pool in two independent experiments. c. Between day precision of the assay of Low, Medium and High Quality Controls measured over 15 days. d. Matrix comparison using Passing–Bablok regression analysis showed no difference in antibody concentration readouts depending on blood sample preparation within serum and lithium-heparin or EDTA plasma. e. Low positive and positive plasma samples were tested in various concentrations of interferent factors (conjugated bilirubin, hemoglobin and intralipids). No interferent affected antibody concentration readouts in tested samples.
2.7. Microneutralization assay
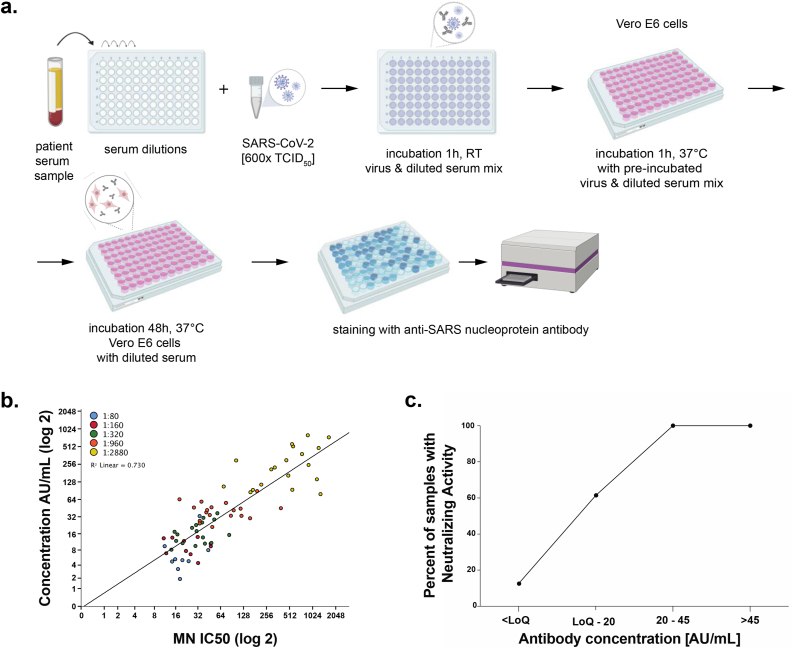
The microneutralization assay was performed as previously reported and described in detail (Amanat and Stadlbauer 2020; Amanat and White 2020). Briefly, serum samples were heat-inactivated at 56 °C for an hour prior to use. Vero E6 cells (ATCC #CRL-1586) were maintained in complete Dulbecco's Modified Eagle's Medium (DMEM; Gibco) and were seeded in 96-well cell culture plates (Corning) at a concentration of 20,000 cells/well (Figure 3a). Serum samples were serially diluted (starting with a 1:10 dilution) in a 96-well cell culture plate in 1X Minimal Essential Medium (MEM; Gibco) supplemented with 2% fetal bovine serum (FBS, Corning). No-virus and virus-only control wells were included on each plate as controls. Next, 80 ul SARS-CoV-2 (USA-WA1/2020 isolate) diluted to 600 tissue culture infectious dose 50 (TCID50) was added to an equal amount of respective serum dilution. The virus-serum mixture was incubated for one hour and the media removed from the Vero cell plates. 120 ul of the virus-serum mixture was added to the cells and incubated for 1 h at 37 °C. The virus-serum mixture was aspirated and replaced with 100 μl MEM and 100 μl of the respective serum dilution. The cells were incubated at 37 °C for two days and the media removed. Next, 150 μl of 10% formaldehyde (Polysciences) was added for 24 h to fix the cells and to inactivate virus. The following day, the formaldehyde was removed, and the cells washed with PBS (Gibco). The cells were permeabilized and stained using monoclonal antibody 1C7 (anti-SARS nucleoprotein antibody generated in-house) as described previously (Amanat and Stadlbauer 2020). The secondary antibody used was an anti-mouse IgG conjugated with horseradish peroxidase (Rockland Immunochemicals) and SIGMAFAST OPD (Sigma–Aldrich) was used to develop the plates. After stopping the reaction with 3M hydrochloric acid, plates were read at an optical density of 490 nm using Synergy 4 plate reader (BioTek). The inhibition of virus replication, compared to the control wells, was calculated and a non-linear regression performed in GraphPad Prism 8 to calculate the 50% inhibitory dilution (ID50). The 50% neutralizing titer was defined as the reciprocal of the ID50.
Figure 3.
Correlation between antibody concentration and SARS-CoV-2 neutralization activity. a. Serial dilutions of patient sera are incubated in the presence of SARS-CoV-2. Next, Vero E6 cells are incubated in the presence of the patient serum-SARS-CoV-2 mix. Neutralizing antibodies have ability to inhibit cellular entry of the virus into the Vero E6 cells. After incubation, an anti-SARS nucleoprotein antibody is used to detect the infected cells, hence, to determine virus neutralization potential of antibodies present in patient's sera b. Correlation of microneutralization with measured quantitative antibody concentration. Points are grouped by color to indicate the corresponding titer previously measured on qualitative assay. c. Percent of samples with neutralizing activity for each antibody concentration range.
2.8. Statistics
Statistical analysis was performed with GraphPad Prism 8.4.2 software and IBM SPSS. Patient demographics and needed information were obtained from the electronic medical record as part of an IRB-approved protocol.
3. Results
3.1. Analytical sensitivity and specificity
There is no standard reference SARS-CoV-2 antigen material available; accordingly, absolute analytical sensitivity cannot be calculated, however, all examined specimens with negative screen results using RBD protein also tested negative against spike protein at 1:80 dilution.
Cross-reactivity of non-SARS-CoV-2 specific Ab against spike proteins was examined using sera with known antibodies against confirmed past infections or elevated gamma globulins. No interference was observed with plasma from patients positive for varicella virus, influenza virus, herpes simplex virus (HSV), rubella virus, cytomegalovirus (CMV), hepatitis B and C virus, human immunodeficiency virus (HIV), multiple myeloma (IgG, IgM, and IgA type), Crohn's disease and systemic lupus erythematosus (SLE) (Supplementary Table 1).
3.2. Assay precision
Assay between-day precision was evaluated using quality control samples from all ranges of antibody concentrations. Serum pools of at least 25 patients tested positive on at 1:160 titer served as Low Quality Control (LQC), 1:320 titer as a Medium Quality Control (MQC) and ≥1:2880 pool diluted 1:2 in negative serum pool as a High Quality Control (HQC) for spike protein ELISA (Figure 2c). Precision was reported as the coefficient of variation (CV). Between-day precision was calculated by analyzing quality controls for 15 different days. The acceptance criterion for between-day precision (%CV) was ≤20% for each control tested (Supplementary Table 2).
3.3. Matrix comparison
To evaluate if performance of the assay remains unchanged among different matrices (serum, lithium heparin plasma, ethylenediaminetetraacetic acid (EDTA) plasma), we tested blood samples collected into three different tubes from the same patients in duplicates. Comparison between serum (gold-top tube) vs EDTA plasma (lavender-top tube) as well as serum vs. lithium heparin plasma (green-top tube) showed a correlation in 15 tested specimens of various antibody concentration levels using Passing–Bablok regression analysis (Supplementary Table 3).
3.4. Interference
To detect the impact of potentially interfering substances on test performance, we performed an analysis of various concentrations of hemoglobin, lipid/triglyceride as intralipid, conjugated bilirubin, on low positive (titer 1:80) and positive (titer 1:320) serum samples. Samples spiked with hemoglobin of a concentration ranging from 2.5 – 12.5 mg/ml, intralipid of a concentration ranging from 4.9 – 24.4 mg/ml and conjugated bilirubin of a concentration ranging from 3.8 – 19.10 mg/dl of different antibody levels showed no difference in antibody concentration readouts, demonstrating no interference of these substances on a test performance (Figure 2e and Supplementary Table 4).
3.5. Correlation with qualitative ELISA and microneutralization studies
120 serum samples were tested for presence of anti-SARS-CoV-2 IgG antibodies using both titer and quantitative method. Twenty samples of each titer (NEG, 1:80, 1:160, 1:320, 1:960 and ≥1:2880) were then tested for their virus neutralization properties (Figure 3a). A correlation of these data with the discrete ELISA titers was already published (Wajnberg et al., 2020). Here, we analyzed the correlation with the arbitrary units. We observed a positive correlation between neutralization titer and arbitrary concentration of anti-SARS-CoV-2 IgG (Figures 3b,c).
3.6. Clinical interpretation of Mount Sinai COVID-19 ELISA test results
A strong positive (>40 AU/mL) result confirms the presence of circulating IgG antibodies specific for SARS-CoV-2 at high levels corresponding to serum titer ranges from 1:960 to above 1:2880 as measured by the Mount Sinai qualitative assay. Moderate positive (16–39 AU/ml) confirms the presence of circulating IgG antibodies specific for SARS-CoV-2 at moderate to high levels, corresponding to serum titers range from 1:320 to 1:960. Weak positive (5–15 AU/ml) results are considered borderline. Most borderline cases correspond to serum titers between 1:80 and 1:160. Follow up testing may be clinically indicated for borderline cases in 10–14 days if recent infection is suspected.
4. Discussion
Quantitation of plasma antibody levels is an important tool in infectious disease research. It allows for the measurement of virus neutralization potency thus a level of protection against a given virus. This is crucial for monitoring vaccine strategies as well as for validation of potential plasma donors. In the case of novel SARS-CoV-2 there is no standard available to which quantification could be performed. To overcome this obstacle, we developed a method to quantitate levels of antibodies in plasma in reference to a created pool of positively tested patient plasma and used it as a standard. The assignment of the IC50 (inflection point) of a 4PL curve as a function of the reciprocal of dilution and the OD490 to the baseline arbitrary concentration of a given standard pool allows for results to be compared while using different standard pools. Therefore, the method described here allows for reliable quantitation of antibody concentration using various reference pools created from samples available to a laboratory.
To date, a discrete titer method has been used to estimate antibody levels and protection against SARS-CoV-2 in patients' plasma. One of the limitations of this method is that it requires serial dilution of each patient's plasma to determine at which dilution the antibodies are still detectable. The qualitative titer-based ELISA to which we compared this assay's performance requires the use of 4 serial dilutions per sample which allows for testing only 14 patient samples on a single ELISA plate. However, by employing a quantitative method, including standard curve and quality control samples we are able to test 33 patient samples in duplicates on a single ELISA plate. Therefore, the described method offers not only a more precise estimation of antibody levels in a plasma sample but is also more time and cost efficient. Additionally, the orthogonal design of the assay allows for rapid identification of negative patient samples which improves turnaround time. Samples which test positive in the RBD screen are then tested in a quantitative assay against full length Spike protein to determine antibody levels. We believe this approach further improves the sensitivity and specificity of the assay.
The Mount Sinai COVID-19 ELISA allows for the use of different standard pools, which is crucial given the fact that a gold standard does not yet exist for measurement of anti-SARS-CoV-2 antibodies, and the pools created from patient's plasma have limited volumes and need to be recreated once used. Therefore, to be able to compare results calculated against different batches of standard pools, other variables of the test (antibody coating, secondary antibody) must remain constant. If the change of secondary antibody is required due to a limited availability, a bridging study must be performed to determine the conversion factor if necessary.
To our best knowledge, this represented, at the time of development, one of the first described quantitative assay to detect IgG antibodies against SARS-CoV-2 spike protein authorized for use on patient specimens in the United States. In this study we performed a neutralization study of the samples previously tested using titer-based method and confirmed that the amount of AU/mL positively correlates with titer as well as with virus neutralization activity of the plasma. Based on these findings, this quantitative assay is highly suitable to establish correlates of protection for SARS-CoV-2.
5. Informed consent
Informed consent was obtained from all individuals included in this study.
6. Ethical approval
Informed consent was obtained from all individuals included in this study. The study was approved by the institutional review board of the Icahn School of Medicine at Mount Sinai (20-1032-00001-01).
Declarations
Author contribution statement
Magdalena M. Żak: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Aryeh Stock: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Daniel Stadlbauer & Damodara Rao Mendu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Wei Zhang, Kirstie Cummings, William Marsiglia, Arsen Zargarov & Monica Tamayo: Performed the experiments.
Fatima Amanat: Performed the experiments; Wrote the paper.
Carlos Cordon-Cardo: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Florian Krammer: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by JPB Foundation and The Open Philanthropy Project (2020-215611).
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare the following conflict of interests: Mount Sinai has licensed serological assays to commercial entities and has filed for patent protection for serological assays. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to the COVID-19 serological assay (“Serology Assay”) and NDV-based SARS-CoV-2 vaccines which list Florian Krammer (“Serology Assay”, vaccines), Daniel Stadlbauer (“Serology Assay”), Damodara Rao Mendu (“Serology Assay”), and Carlos Cordon-Cardo (“Serology Assay”) as co-inventors. The foundational “Serology Assay” intellectual property (IP) was licensed by the Icahn School of Medicine at Mount Sinai to commercial entities including Kantaro Biosciences, a company in which Mount Sinai has a financial interest. Florian Krammer consulted for Merck, Curevac, and Pfizer in the past (before 2020) and is currently consulting for Pfizer, Seqirus, and Avimex. The Krammer laboratory is collaborating with Pfizer on animal models of SARS-CoV-2.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to acknowledge the laboratory staff of the Mount Sinai Clinical Labs without whom none of this would be possible. Special thanks to the post-docs who voluntarily gave their time to assist in the validation. Thank you to the New York Community willing to come in and be tested and donate plasma. Drawings in figures of this manuscript were created with BioRender.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Alsoussi W.B., et al. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 2020;205(4):915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., White K.M., et al. An in vitro microneutralization assay for SARS-CoV-2 serology and Drug screening. Curr. Prot. Microbiol. 2020;58(1) doi: 10.1002/cpmc.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D., et al. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3(12):e525. doi: 10.1371/journal.pmed.0030525. Edited by J. Peiris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. Unit. States Am. 2020:202009799. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., et al. Development and characterization of a standardized ELISA including a reference serum on each plate to detect antibodies induced by experimental malaria vaccines. Vaccine. 2008;26(2):193–200. doi: 10.1016/j.vaccine.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., et al. Severe acute respiratory syndrome coronavirus 2−Specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Stadlbauer D., et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Prot. Microbiol. 2020;57(1) doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu H., et al. Quantitative serology assays for determination of antibody responses to Ebola virus glycoprotein and matrix protein in nonhuman primates and humans. Antivir. Res. 2016;126:55–61. doi: 10.1016/j.antiviral.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.