Abstract
Exogenous chicken anemia virus (CAV) has been detected in commercial poultry vaccines in various countries of the world. The presence of unwanted CAV in vaccines not only influences the epidemiology of chicken infectious anemia disease, but may also lead to vaccine failure and confusing results when vaccine responses are monitored. To detect CAV in contaminated vaccines, nucleic acid testing (unlike conventional testing) has a shorter processing time and does not require cell culture or live animals. The aim of the current study was to develop a TaqMan real-time polymerase chain reaction (PCR) assay to detect and quantify CAV in poultry vaccines and investigate CAV contamination in Razi live Newcastle disease vaccines. The TaqMan real-time PCR assay was set up, optimized, and validated in successive experiments. A standard plasmid pUC-VP2 containing viral protein 2 of CAV was constructed and used in the assay to generate a standard curve to quantify CAV genomes. A clear linear correlation was observed between threshold cycle (Ct) values and plasmid copy numbers in the amplification plots of 10-fold serial dilution of the plasmid. Total DNA of three samples of each of four different Razi live Newcastle disease vaccines, namely LaSota, B1, clone.12IR, and thermo-resistant strains, were extracted and subjected to real-time PCR assay. No CAV contamination was detected in the Razi Live Newcastle vaccines. The developed TaqMan real-time PCR assay provides a quick, specific, and sensitive method for use in detecting CAV in quality control vaccine testing and viral load studies.
Keywords: CAV, real-time PCR, Newcastle disease, vaccine contamination, exogenous virus
1. Introduction
Chicken anemia virus (CAV) is the sole member of genus Gyrovirus which belongs to the family Anelloviridae. It induces a disease in young chickens which can be distinguished by aplastic anemia and generalized lymphoid atrophy with concomitant immunosuppression ( 1 ). CAV is not only a threat to specific pathogen free (SPF) egg production, but also of economic importance in the broiler industry, where a reduction in net income is observed because of decreased weight gain and increased mortality ( 2 ).
CAV can be transmitted both horizontally and vertically, and reports have stated that viral DNA may remain in reproductive and splenic tissues in mature commercial and SPF chickens and be transmitted vertically to their progeny, whether or not antibodies against CAV are detectable in the parents ( 3 - 5 ).
CAV contamination of various live poultry vaccines has been reported in many parts of the world ( 6 - 9 ). It has been suggested that the epidemiology of the chicken infectious anemia disease in poultry flocks have been affected by contaminated vaccines. The presence of unwanted CAV in vaccines may also result in vaccine failure and confusing results when vaccine responses are monitored (8). Because of the importance of vaccine contamination with CAV, the European Union necessitated that eggs used for the production of poultry vaccines be free of CAV if those vaccines are to be administrated in birds less than seven days of age ( 10 ).
Nucleic acid testing for vaccine batch control was introduced in the late nineteen-nineties ( 11 ). Over the past few years, several polymerase chain reaction (PCR) formats, including nested-PCR, real-time quantitative PCR, and droplet digital PCR, have been developed and used to detect CAV contamination in poultry vaccines.
The current study aimed to develop a TaqMan real-time PCR assay to detect and quantify CAV in poultry vaccines and to investigate CAV contamination of Razi live Newcastle disease vaccines which are used extensively on poultry farms in Iran.
2. Material and Methods
2.1. Virus Strain
The infectious clone of the Iranian strain of CAV, IR ( 12 ), originating from a commercial broiler flock, was used for this study.
2.2. Design of Primers
GenBank database was searched to retrieve complete and partial sequences of the CAV genome. The nucleotide sequences were then aligned using MEGA7 software ( 13 ) and subsequently examined by searching for highly conserved regions. Oligo 7 software ( 14 ) was used to design primers and probe for real-time PCR in the highly conserved region (Table 1). The specificity of primers and probes was tested using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) website.
Table1.
Primers and probe sequences
| Primer/probe | Sequence (5' to 3') |
|---|---|
| CAV-F | AGCTCGTCTTGCCATCTTACA |
| CAV-R | AAAGCTTGATTACCACTACTCCCA |
| CAV Probe | 6FAM-ACCTTCTTGCGTTCGGGGTC-0TMR |
2.3. Production of the Standard Curve
The CAV IR sequence (accession number KT276305.1) was used to amplify CAV ORF 2 (encoding VP2) by PCR assay which has already been described ( 15 ). The PCR product, which was a DNA band of 677 bp, was cut out from 1% agarose gel and ligated into the pUC18 vector using T4 DNA ligase and ligase buffer in an InsTAclone™ PCR cloning kit (Fermentas). The result of ligation was used to transform E. coli Top 10.
A Plasmid Midi Kit (Qiagen) was used to purify the cloned plasmid, pUC-VP2, from transformants. To obtain the linearized plasmid, the purified pUC-VP2 was then digested with EcoRI. Serial 10-fold dilutions of the linearized plasmid were used in duplicate to generate a standard curve for real-time PCR. The plasmid concentration was calculated using the following formula:
Number of molecules of DNA/ml = A260 × 4.56 × 1016/N
where A260 is the DNA concentration at 260 nm, and N is the number of base pairs in the molecule of pUC18-VP2.
2.4. Vaccine Samples and DNA Extraction
Three samples of each of four different Razi live Newcastle disease vaccines, namely LaSota, B1, clone.12IR, and thermo-resistant strains, were used in this study. These vaccines are widely used for immunizing chickens against Newcastle disease on poultry farms in Iran. Total DNA was extracted from 200 µl of each reconstituted Newcastle disease vaccine suspension and a CAV vaccine (Circomune, Ceva), for positive control using the Viral Nucleic Acid Kit (Roche) according to manufacturer's instructions. After quantification with Nanodrop, the extracted DNA from vaccines was diluted to 1 dose of vaccine to be used in each reaction. The DNA samples were kept at -20 ℃ until they were used in real-time PCR reaction.
2.5. Quantitative PCR Assay
The concentrations of each forward and reverse primer and probe were optimized. The optimized annealing temperature was determined using gradient PCR and analysis on 1% agarose gel. The CAV genome in DNA extracted from vaccines were quantified in reaction volumes of 20 µl using 1 µl of template containing extracted DNA of vaccine, 10 µl of Real Q Plus 2x Master Mix for Probe Without ROXTM (Ampliqon), 100 nM of each forward and reverse primers and 200 nM of probe. The amplification was performed in a real-time thermal cycler (Corbett Research) under the following conditions: 95 ℃ for 15 min for activation of the TEMPase hot start enzyme, followed by 40 cycles of amplification (20 sec at 95 ℃ and 60 sec at 62 ℃).
2.6. Specificity of Real-Time RT-PCR Assay
The specificity of the primers and probe of the developed real-time PCR assay was evaluated using cDNA related to Newcastle disease virus (LaSota strain), infectious bronchitis virus (variant strain), avian influenza virus H9N2, avian metapneumovirus virus (vaccine strain), and avian reovirus (strain 1133).
3. Results
3.1. Real-Time PCR Development
The calculated concentration of the plasmid pUC-VP2 in 10-fold serial dilution ranged from 21 to 210,000/µl. Amplification plots of this serial dilution produced a standard curve which showed a clear linear correlation between threshold cycle (Ct) values and the number of copies of the plasmid (Figure 1a). The linear relationship was between 100.3 and 105.3 copies/𝜇l , and the lowest detection limit of the assay was 21 copies per reaction (Figure 1b). Further analysis demonstrated that the square of the coefficient of regression (R2) and efficiency of amplification were within the ranges of 0.987 to 0.994 and 93% to 1.02%, respectively.
Figure 1.
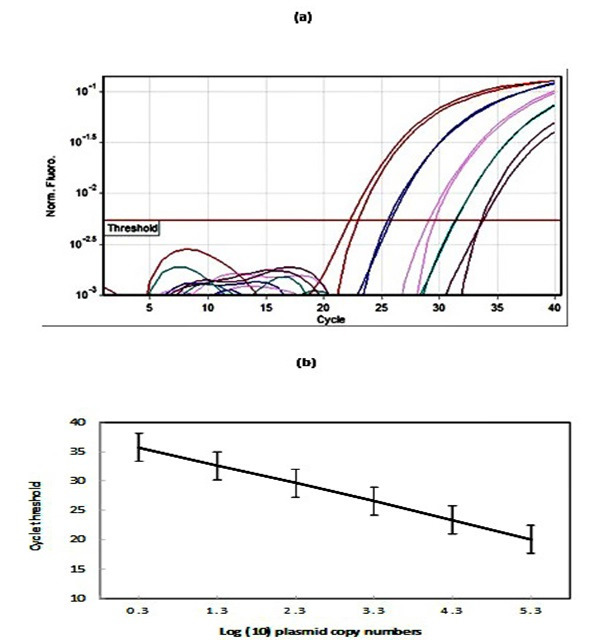
The production of a standard curve using the the plasmid pUC-VP2. (a) An amplification curve obtained using a dilution series of the plasmid pUC-VP2. (b) Reproducibility of the pUC-VP2 standard curve. The standard deviation of the cycle threshold at each dilution of the plasmid for 3 qPCR runs are shown.
The accuracy and reproducibility of the assay was proven to be satisfactory as the coefficient of variation (CV) for the intra- and intergroup replicates ranged from 0.22 to 0.5%. The developed real-time PCR assay was shown to be specific, as it did not amplify cDNA of the viruses used for evaluation of specificity (above).
3.2. Detection of CAV Contamination in Vaccine
No CAV contamination was seen in the Razi Live Newcastle vaccines. The detected CAV genome copies in the tested vaccine were below the detection limit threshold of the real-time PCR assay.
4. Discussion
The potential contamination of vaccines with CAV has become a major concern for vaccine manufacturers ( 1 ). This study is a response to this concern and describes the development of a TaqMan real-time PCR assay which can detect exogenous CAV in vaccines. The methods recommended for detection of CAV contamination which include virus isolation in cell culture, chicken embryos and chickens, are laborious, expensive, and time-consuming. The assays such as immunoassay staining, enzyme immunoassay, and serum neutralization are also recommended for testing sera against CAV ( 16 ). However, the specificity, sensitivity, and applicability of these assays are changeable, and they indirectly detect the presence of CAV. If injected animals are unable to develop an antibody response, it is possible that unwanted viruses may not be detected ( 11 ).
PCR is more sensitive and specific, has a shorter processing time, and does not require cell culture or live animals ( 11 ); however, there is very small quantity of CAV genome loads in live vaccines. For this reason, real-time PCR has been applied for the detection of exogenous CAV in poultry vaccines ( 9 , 17 ).
Compared with PCR, real-time PCR has much higher sensitivity. A previous study showed that the exogenous genome of 100, 10, and 1 EID50 CAV per vial of the reconstituted vaccine can be detected by real-time PCR assay, while conventional PCR can only detect the presence of 10 EID50 CAV or higher in contaminated vaccine as the positive target band. The lowest detection limit of standard recombinant plasmid was 102 copies/𝜇L for conventional PCR, while that of real-time PCR was 20 copies of standard plasmid ( 17 ). Our developed TaqMan real-time PCR had a sensitivity of 21 copies/𝜇L, which is comparable with previous studies.
Real-time PCR technology can be not only automated and used for quantification of template concentration, but also used to co-detect several pathogens in the same test tube. One study has already reported the development of duplex quantitative real-time PCR (dqPCR) which can detect and determine viral loads of both avian gyrovirus 2 and CAV in a single assay in commercially available vaccines ( 9 ).
In the current study, Razi live Newcastle disease vaccines were tested for possible contamination with CAV. These vaccines are used extensively on poultry farms in Iran. The contamination of live Newcastle vaccines with CAV has already been detected using PCR ( 6 - 8 ). Although PCR does not show infectious virus exclusively, the detection of the extraneous viral genomes in vaccines can also be important. This was shown by a study in which vaccine samples which had tested positive for CAV by PCR were inoculated into SPF chickens. The sera of inoculated chickens tested positive for CAV-specific antibody by ELISA, and CAV was also detected in the extracted DNA of these sera by PCR ( 7 ). In a simulation study in which exogenous CAV isolated from the contaminated Newcastle disease vaccine was used, synergetic pathogenicity with the LaSota strain in the vaccine was observed and resulted in severe disease with a relatively low dose through oral infection ( 18 ).
In conclusion, a TaqMan real-time PCR as a rapid, specific, and sensitive assay was successfully developed in the current study. The method used to detect CAV in live Newcastle disease vaccines in this study can also be applied in quality control testing of other vaccines and in viral load studies. The developed real-time PCR has the potential to be used for co-detection of other exogenous viruses in one test tube.
Authors' Contribution
Study concept and design: A. K.
Acquisition of data: A. K. and M. M.
Analysis and interpretation of data: A. K.
Drafting of the manuscript: A. K.
Critical revision of the manuscript for important intellectual content: A. K.
Statistical analysis: A. K.
Administrative, technical, and material support: A. K., M. M. and S. A. K.
Ethics
All the procedures and animal handling were approved by the Animal Ethics Committee at the Razi Vaccine and Serum Research Institute under the project number of 2020-558845-1.
Acknowledgement
The authors would like to thank Dr. M. H. Fallah, Deputy of Research and Technology at the Razi Vaccine and Serum Research Institute, for facilitating the performance of the current study.
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Schat KA, van Santen VL. Chicken Infectious Anemia and Circovirus Infections in Commercial Flocks. In: Swanye DE, Boulianne M, Logue CM, McDougald LR, N. V, D.L. S, editors. Diseases of Poultry. 14 ed. Hoboken, NJ, USA: Wiley Blackwell; 2020. p. 284-32 [Google Scholar]
- 2.McNulty MS. Chicken anaemia agent: a review. Avian Pathol: J WVPA. 1991;20(2):187–203. doi: 10.1080/03079459108418756. [DOI] [PubMed] [Google Scholar]
- 3.Brentano L, Lazzarin S, Bassi SS, Klein TA, Schat KA. Detection of chicken anemia virus in the gonads and in the progeny of broiler breeder hens with high neutralizing antibody titers. Vet Microbiol. 2005;105(1):65–72. doi: 10.1016/j.vetmic.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Cardona C, Lucio B, O'Connell P, Jagne J, Schat KA. Humoral immune responses to chicken infectious anemia virus in three strains of chickens in a closed flock. Avian Dis. 2000;44(3):661–7. [PubMed] [Google Scholar]
- 5.Cardona CJ, Oswald WB, Schat KA. Distribution of chicken anaemia virus in the reproductive tissues of specific mpathogen-free chickens. J Gen Virol. 2000;81(Pt 8):2067–75. doi: 10.1099/0022-1317-81-8-2067. [DOI] [PubMed] [Google Scholar]
- 6.Barrios PR, Marín Gómez S, Rios R, Pereira CG, Resende M, Resende JS, et al. A retrospective PCR investigation of avian Orthoreovirus, chicken infectious anemia and fowl Aviadenovirus genomes contamination in commercial poultry vaccines in Brazil. Arq Bras Med Vet Zootec. 2012;64:231–5. [Google Scholar]
- 7.Li Y, Hu Y, Cui S, Fu J, Wang Y, Cui Z, et al. Molecular characterization of chicken infectious anemia virus from contaminated live-virus vaccines. Poult Sci. 2017;96(5):1045–51. doi: 10.3382/ps/pew406. [DOI] [PubMed] [Google Scholar]
- 8.Marín Gómez S, Barrios P, Rios R, Resende M, Resende J, Santos B, et al. Molecular Characterization of Contaminating Infectious Anemia Virus of Chickens in Live Commercial Vaccines Produced in the 1990s. Avian Dis. 2013;57:15–21. doi: 10.1637/10056-011212-Reg.1. [DOI] [PubMed] [Google Scholar]
- 9.Varela AP, Dos Santos HF, Cibulski SP, Scheffer CM, Schmidt C, Sales Lima FE, et al. Chicken anemia virus and avian gyrovirus 2 as contaminants in poultry vaccines. Biologicals. 2014;42(6):346–50. doi: 10.1016/j.biologicals.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 10.European Pharmacopoeia. Chicken flocks free from specified pathogens for the production and qulaity control of vaccines. 1. Strasbourg Cedex, France: Directorate for the Quality of Medicines & HealthCare, Council of Europe; 2013. p. 579-82 [Google Scholar]
- 11.Ottiger HP. Development, standardization and assessment of PCR systems for purity testing of avian viral vaccines. Biologicals. 2010;38(3):381–8. doi: 10.1016/j.biologicals.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Kaffashi A, Eshratabadi F, Shoushtari A. Full-length infectious clone of an Iranian isolate of chicken anemia virus. Virus Genes. 2017;53(2):312–6. doi: 10.1007/s11262-016-1417-8. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rychlik W. OLIGO 7 primer analysis software. Methods Mol Biol. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- 15.Kaffashi A, Noormohammadi AH, Allott ML, Browning GF. Viral load in 1-day-old and 6-week-old chickens infected with chicken anaemia virus by the intraocular route. Avian Pathol. 2006;35(6):471–4. doi: 10.1080/03079450601028837. [DOI] [PubMed] [Google Scholar]
- 16.European Pharmacopoeia. Avian viral vaccines: tests for extraneous agents in seed lots. 1. 8th ed. Strasbourg Cedex, France: Directorate for the Quality of Medicines & HealthCare, Council of Europe; 2013. p. 209-12 [Google Scholar]
- 17.Li Q, Zhang Y, Meng F, Jiang H, Xu G, Ding J, et al. A New Strategy for the Detection of Chicken Infectious Anemia Virus Contamination in Attenuated Live Vaccine by Droplet Digital PCR. Biomed Res Int. 2019;2019 doi: 10.1155/2019/2750472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Q, Wang T, Meng F, Cui Z, Chang S, Zhao P. Synergetic pathogenicity of Newcastle disease vaccines LaSota strain and contaminated chicken infectious anemia virus. Poult Sci. 2019;98(5):1985–92. doi: 10.3382/ps/pey555. [DOI] [PubMed] [Google Scholar]


