Abstract
Background
Type 2 diabetes mellitus (T2DM) is characterized by β cell decline in the pancreas and insulin resistance. This study aimed to investigate the possible pathogenic gene mutation sites of T2DM patients using whole exome sequencing.
Materials and Methods
We recruited a Chinese family with 3-generation history of diabetes. The whole blood genomic DNA of seven members of the family was extracted and sent for whole exome sequencing. Biological information was analyzed with in silico prediction methods, including significance analysis of single nucleotide polymorphism (SNP)/Indel site, and analysis of specific SNP/Indel proteins and their potential mechanisms.
Results
Six out of seven members of the family were diagnosed with diabetes. All DNA samples (23 kb) met quality requirements of library construction. Clean reads of each sample demonstrated high Q20 and Q30 (>80%), indicating good sequencing quality of sequencing data. A total of 130,693 SNPs and 15,928 Indels were found in DNA samples. A total of 22 significant SNPs and Indel mutation sites located on 19 genes were obtained, including ZCCHC3, SYN2, RPL14, SRRD, AMD1, CAMKK2, ZNF787, RNF157, NPIPB15, ALG3, KIAA0040, MAST2, ESRRA, C8orf58, PNLIPRP1, DACH1, MACC1, CAPN9 and DMKN. An rs2305205 mutation of PNLIPRP1 gene and an rs778701848 mutation of CAMKK2 gene may be associated with the pathogenesis of T2DM in this family.
Conclusion
Exons of these diabetic patients demonstrated an rs2305205 mutation in PNLIPRP1 gene and an rs778701848 mutation in CAMKK2 gene. These two mutations might promote T2DM occurrence through reducing sensitivity of peripheral tissue to insulin and reducing insulin secretion.
Keywords: type 2 diabetes, gene mutation, whole exome sequencing, PNLIPRP1, CAMKK2
Introduction
Type 2 diabetes mellitus (T2DM), as a polygenic genetic disease, is mainly characterized by β cell decline in the pancreas and insulin resistance, usually resulting from a combination of genetic and environmental factors. The main clinical manifestations of T2DM are glycolipid metabolism associated disorders. In recent years, with aging of the population and improvement of living standards, the incidence of diabetes is rising year by year worldwide. According to the data of International Diabetes Federation in 2017, there are approximately 451 million diabetes mellitus patients in the whole world, with the potential to achieve 693 million by the year 2045.1 Among these diabetes mellitus patients, T2DM accounts for more than 90%. A previous study2 reported that the occurrence of T2DM is related to genetic susceptibility, with a certain degree of familial aggregation.2 In Arab, Tunisia, France, Sweden, Greece, and China, more than 50% of diabetic patients demonstrated a positive family history of diabetes mellitus.2–7 It is suggested that family history, genetic factors, and similar environmental factors are associated with an increased risk of T2DM.8 In a similar living environment, the prevalence of siblings is 4.2 times higher than that of spouses, indicating that genetic factors play a critical role in the development of T2DM in the home.9 Due to the common incidence of familial diabetes clinically, the screening of disease-associated susceptibility genes in members of a family with T2DM is important for preventing and treating T2DM.
Some previous studies10,11 focusing on genetic associations demonstrated that single nucleotide polymorphisms (SNPs) are obviously associated with T2DM susceptibility. There is a lot of information on genetic architecture of T2DM, such as high levels of polygenicity and small effect-sizes for most genetic risk variants.12 At least 75 independent genetic loci related to T2DM have been discovered until now.9 Some of the variants are associated with the protection against T2DM risk, while some variants are considered as risk markers and correlated with T2DM susceptibility.10 Therefore, it is critical to clarifyi the risk and pathogenesis of T2DM through discovering the genetic variants.
Whole exome sequencing offers a useful strategy for identifying genes involved in human disorders, such as diabetes.13 Whole exome sequencing, in a few studies,14,15 demonstrated that the occurrence of T2DM in family members was associated with low frequency and rare mutations of genes.14,15 In recent years, whole exome sequencing has been widely applied for identifying susceptibility gene variants12 or insulin mutations16 in diabetes patients, and other diseases caused by potential genetic variations in China.17 The incidence of T2DM is high clinically, with an obvious genetic predisposition. Presently, there have been no studies focusing on familial diabetes based on whole exome sequencing method in China until now. Therefore, whole exome sequencing for family members with T2DM and discovery of disease-related susceptibility genes are of great significance for preventing and treating diabetes.
T2DM, as a genetically heterogeneous disease, is commonly characterized by a few specific genes involved in different races or families of the same race. Presently, there is no research focusing on application of whole exome sequencing technology for determining diabetic families in Yunnan province of China. Therefore, the purpose of this study was to discover the possible pathogenetic genes and mutation sites of the pathogenic genes in T2DM patients, using whole exome sequencing method in Han nationality of Kunming in China. This study will provide promising clues for preventing and treating familial diabetes.
Materials and Methods
Study Design and Patients
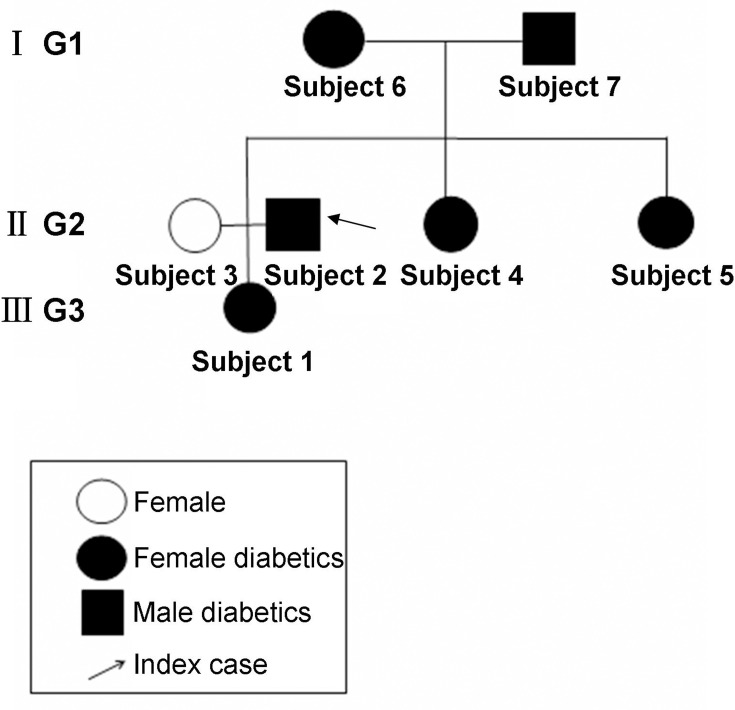
We recruited Chinese patients with a three-generation family history of diabetes (6 members with diabetes and 1 member without diabetes) (Figure 1). The pro-band was a male patient with a ten-year history of diabetes in the second generation. The oral glucose tolerance test (OGTT) confirmed the diagnosis of diabetes. T2DM was considered in all patients except for the wife of pro-band according to the characteristics of medical history and laboratory examinations. There was no information suggesting consanguineous marriage in this family.
Figure 1.
A pedigree with family type 2 diabetes. Subject 1, 2, 3, 4, 5, 6 and 7 represented diabetes patient 1, 2, 3, 4, 5, no-diabetes patient 6 and diabetes patient 7, respectively. I G1, II G2 and III G3 represented the generation 1, generation 2 and generation 3, respectively.
Type 2 Diabetes Mellitus Diagnosis
Diabetes mellitus was diagnosed according to the criteria of diabetes mellitus published by WHO in 1999 including: fasting plasma glucose (FPG) ≥7 mmol/L, plasma glucose 2 hours after meal (2hPG) and OGTT 2hPG>11.1 mmol/L. The diagnostic criteria for T2DM were based on several items: ① the symptoms of polyuria, polydipsia, and without typical weight loss. ② Oral hypoglycemic drugs were effective in controlling blood sugar and there was no spontaneous diabetic ketoacidosis history. ③ Islet cell antibody (ICA), insulin antibody (IAA) and glutamic acid decarboxylase (GAD) antibodies were negative. ④ Fasting serum C-peptide level was within the lower limit of normal reference range. ⑤ The clinical findings of insulin resistance (acanthosis nigricans, hypertension, dyslipidemia). Together with the age of patients (all more than 40 years old), all these supported the diagnosis of T2DM. Moreover, type 1 diabetes mellitus (T1DM) was excluded with presence of autoantibody positivity, severe deficiency and absence of any suggestive symptoms of the other reasons for diabetes.
This study has been approved by The first affiliated hospital of Kunming medical University (Approval No. 2020-L-18). This study was conducted in accordance with the Declaration of Helsinki. All the family members provided written formed consent and approved this study.
Whole Blood Genomic DNA Extraction
Total of 2 mL peripheral blood was collected from each of the seven family members. The DNA samples were extracted from the peripheral blood of these seven family members with whole blood genomic DNA rapid extraction kit (Biomed Corporation, China), as instructed by the manufacturer. Then, the DNA samples from the seven members of this family were sent to BGI-Shenzhen (Shenzhen, China) for whole exome sequencing.
Whole Exome Sequencing
Agilent SureSelect kit V6 was used for the exome capture. Sequencing data were generated using the pair-end 100bp sequencing strategy on the BGISEQ-500 platform.
In silico Prediction Methods for Analyzing SNPs
Biological Information Data Analysis with Significance Analysis of SNP/Indel Site
The probability of exon DNA mutation at any point in the family members can be calculated according to the following binomial distribution probability formula: Pv = pm(1-p)n-m. Where, “p” was the minor allele mutation frequency (MAF) in the normal population (0<p<1),”n” was the total sample, “m” was the number of samples with the mutation observed, “Pv” was the probability of “m” mutations observed in “n” samples. In this study, according to the calculation, n=7, m=6. Mutation frequency of each site in the normal population was analyzed using the corresponding MAF in 1000 human genome database.18 Some single nucleotide polymorphisms (SNP) and Indel had no mutation frequency information in the 1000 human genome database. At this time, rs of SNP and Indel were used to query dbSNP database to obtain the maximum MAF value of this site.19 There was no MAF value corresponding to the new SNP and Indel sites in 1000 human genome database and dbSNP database. A relatively small MAF value was set for Manhattan map and subsequent analysis. In this study, the minimum MAF value was set as 1e-4.
Biological Information Data Analysis with Manhattan Map
The Manhattan map is a general method for displaying and analyzing the significance of each mutation in a genome. In order to obtain significant SNP and Indel site of clustered diabetic members in this family, Manhattan map was used to display information of SNP and Indel site. The specific process was described as follows: ① the binomial distribution probability formula was used to calculate the mutation probability Pv of each SNP and Indel site. ② Manhattan function in qqman,22 a software package used in R language for analyzing SNP, was applied to display information of diabetic exon SNP and Indel in this family. ③ The cutoff value of rs shown in Manhattan map was set as Pv=1e-9. ④ There were many significant SNP and Indel sites. In order to obtain significant and relatively clear biological significance of SNP and Indel sites, the Manhattan map was divided into two categories, including showing all SNP/Indel sites and only showing SNP/Indel sites with the altered protein functions.
Biological Information Data Analysis with Analysis of Specific SNP/Indel Proteins and Their Potential Mechanisms
To determine whether a particular SNP/Indel is directly related to occurrence of diabetes, the SNP/Indel is first mapped into structure of corresponding protein. The effect of SNP/Indel on protein function was predicted using PDB sum.23 The protein with SNP/Indel mutation was then localized in the KEGG pathway to analyze whether this pathway was related to the onset of diabetes.20
Statistical Analysis
All of the statistical analyses for the bioinformatic data were carried out using R software (version: 2.15). Biological information data were analyzed according to the SNP/Indel site and Manhattan map. The average number of original bases, clean bases and average GC content were counted and analyzed. The SNPs and Indel were also counted. The value of Q20 or Q30 more than 80% was assigned as good sequencing quality.
Results
DNA Sample Quality Assessment
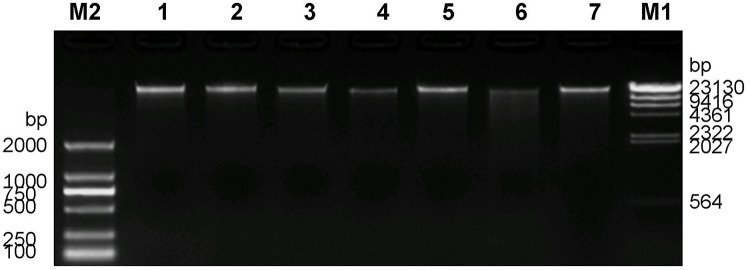
Quality evaluation was conducted on 7 DNA samples. The concentration of DNA samples was detected with Qubit, and then the total amount of DNA was calculated. If the total amount of DNA was more than 1μg, the standard was reached. DNA integrity was then measured by 1% agarose gel electrophoresis. It can be seen from the electro-phoretogram that the main bands of DNA samples were all over 23kb (Figure 2). Although some samples were slightly degraded, they met the quality requirements of the library construction.
Figure 2.
Agarose gel electrophoresis of 7 DNA samples from family members. M1 is λ-Hind III digest DNA Ladder. M2 is D2000 DNA Marker, and 1–7 are DNA samples.
Exon Sequencing Quality
All exon sequencing was carried out on 7 DNA samples, and the average number of original bases was 16065.63Mb, of clean bases was 16024.34Mb. Clean reads of each sample demonstrated high Q20 and Q30 (>80%), indicating that the sequencing data had good sequencing quality. The average GC content was 50.68%. The average sequencing depth of the target region was about 137.37X (Table 1).
Table 1.
Statistical Graph of Exon Sequencing Data from 7 Family Members
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Subject 6 | Subject 7 | |
|---|---|---|---|---|---|---|---|
| Raw reads | 158,603,486 | 156,383,178 | 155,608,204 | 165,373,174 | 158,013,548 | 162,535,482 | 168,076,764 |
| Raw bases | 15860.35 | 15638.32 | 15560.82 | 16537.32 | 15801.35 | 16253.55 | 16807.68 |
| Clean reads | 158,214,218 | 156,000,878 | 155,232,348 | 164,969,052 | 157,635,416 | 162,135,706 | 167,666,246 |
| Clean bases | 15819.51 | 15597.60 | 15520.60 | 16494.42 | 15761.58 | 16211.28 | 16765.36 |
| Clean read1 Q20 (%) | 99.74 | 99.74 | 99.74 | 99.74 | 99.75 | 99.74 | 99.75 |
| Clean read2 Q20 (%) | 96.63 | 96.69 | 96.66 | 96.67 | 96.71 | 96.65 | 96.66 |
| Clean read1 Q30 (%) | 96.51 | 96.63 | 96.89 | 96.65 | 96.72 | 96.55 | 96.52 |
| Clean read2 Q20 (%) | 88.25 | 88.43 | 88.39 | 88.44 | 88.58 | 88.40 | 88.38 |
| Sequencing Depth | 128.99 | 126.18 | 118.85 | 147.66 | 135.58 | 150.89 | 153.44 |
Notes: Raw reads: total number of original sequences. Raw bases: number of original bases. Clean reads: number of sequences obtained after removal of low-quality reads. Clean bases: number of bases obtained after removal of low-mass bases. Clean read1 and Clean read2: Reads obtained by double-end sequencing. Software will give a quality value for each base, which represents the correct recognition rate of sequencing bases. 20 represents the normal recognition rate of bases as 99%, and 30 represents the correct recognition rate of bases as 99.9%. Q20 (%): represents the percentage of bases with a mass value ≥20 (good sequencing quality). Q30 (%): the percentage of bases with a mass value ≥ 20 (good sequencing quality).
Statistical Results of Mutation Detection
SNP Detection Results
A total of 130,693 SNPs were discovered in all samples. Within the coding region, there were 10,367 synonymous mutations, 10,368 missense mutations, 115 stop gain, 38 stop loss, 30 start loss, and 147 splicing.
Indel Detection Results
A total of 15,928 Indel were discovered in all samples. Within the coding region, there were 278 frame shift, 91 non-frame shift insertion, 144 non-frame shift deletion, 0 stop loss, 3 start loss, and 58 splicing.
Bioinformatic Data Analysis
Whole exome sequencing results showed that there were 8383 exon DNA mutations in 6 diabetic family members, while no mutation was found in 1 non-diabetic control member. Among them, there were 7603 single nucleotide polymorphism mutation sites and 780 insertion-deletion mutation sites.
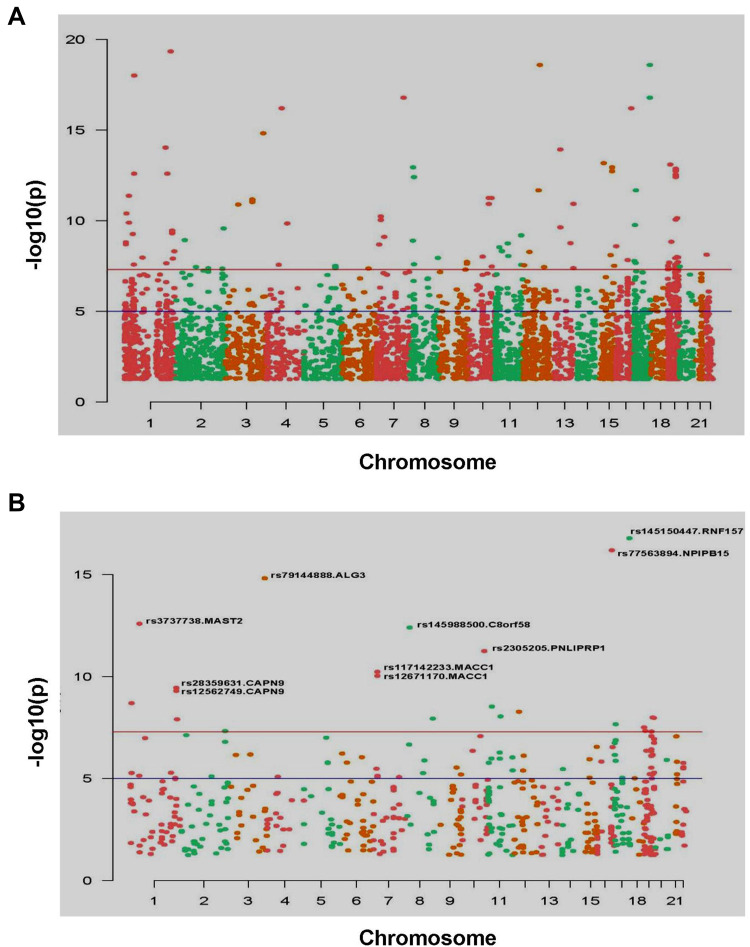
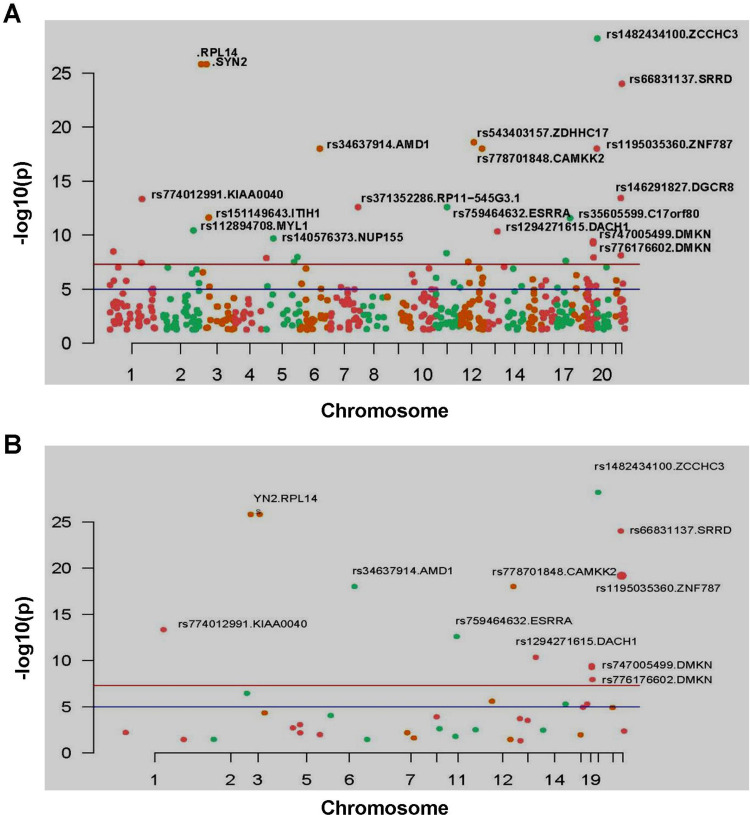
After calculating the significance of each SNP/Indel locus, Manhattan diagram was used to display mutation sites. By setting a specific cutoff value, the significant SNP/Indel sites can be discovered. In Figure 3A, there were 59 SNP sites with high significance (P=1e-9), among which there were 24 SNP sites with moderate and high biological functions. After removing the SNP site without coding function in the intergenic region,10 SNP sites with significant and high biological functions were finally discovered (Figure 3B). Using the same method, 23 Indel mutation sites with high significance (P=1e-9) were discovered (Figure 4A), among which 12 Indel mutation sites with significant and moderately high biological functions were discovered (Figure 4B).
Figure 3.
Manhattan map of all SNP sites (A) and Manhattan map of significant SNP mutation sites, marks the SNP sites with high significance (P<1e-9) (B).
Note: X-axis was the position of the chromosome where the SNP was located. Y-axis was the -log10(P) value corresponding to each SNP site. -log10(P) reflects the degree of association between mutation site and disease occurrence. The greater the value of -log10(P), the stronger the association. The blue horizontal line cutoff value was P=1e-5, and the red horizontal line cutoff value was P=1e-8.
Figure 4.
Manhattan map of all Indel mutation sites (A) and Manhattan map of Indel mutation sites that can change the function of protein, and marks the Indel site with high significance (P<1e-9) (B).
Note: X-axis is the position of the chromosome where the Indel is located. Y-axis is the -log10(P) value corresponding to each Indel mutation site; -log10(P) reflects the degree of association between mutation site and disease occurrence. The greater the value of -log10(P), the stronger the association. The blue horizontal line cutoff value was P=1e-5, and the red horizontal line cutoff value was P=1e-8.
In summary, 22 significant SNP mutation sites and Indel mutation sites in this family were obtained (Table 2). These gene mutation sites were located on 19 genes, including zinc finger CCHC domain-containing protein 3 (ZCCHC3), synapsin 2 (SYN2), ribosomal protein L14 (RPL14), selenite [Se(IV)] reductase D (SRRD), adenosyl methionine decarboxylase 1 (AMD1), calcium/calmodulin dependent protein kinase kinase 2 (CAMKK2), zinc finger protein 787 (ZNF787), ring finger protein 157 (RNF157), nuclear pore complex interacting protein family-B15 (NPIPB15), asparagine-linked glycosylation 3 (ALG3), KIAA0040, microtubule associated serine/threonine kinase 2 (MAST2), estrogen related receptor alpha (ESRRA), chromosome 8 open reading frame 58 (C8orf58), pancreatic lipase-related protein 1 (PNLIPRP1), dachshund homolog 1 (DACH1), metastasis associated colon cancer 1 (MACC1), calpain 9 (CAPN9), and dermokine (DMKN).
Table 2.
List of Significant Mutation Sites in Pedigree
| SNP/Indel | CHR | BP | P value | Func. | Gene | MAF1000G |
|---|---|---|---|---|---|---|
| rs1482434100 | 20 | 298,046 | 6.4E-29 | Disruptive_inframe_insertion | ZCCHC3 | NA |
| Novel SNP | 3 | 12,004,751 | 1.56E-26 | Conservative_inframe_insertion | SYN2 | NA |
| Novel SNP | 3 | 40,462,029 | 1.56E-26 | Conservative_inframe_insertion | RPL14 | NA |
| rs66831137 | 22 | 26,483,980 | 1E-24 | Disruptive_inframe_deletion | SRRD | NA |
| rs34637914 | 6 | 1.11E+08 | 9.99E-19 | Splice_acceptor_variant+intron_variant | AMD1 | NA |
| rs778701848 | 12 | 1.21E+08 | 9.99E-19 | Conservative_inframe_insertion | CAMKK2 | NA |
| rs1195035360 | 19 | 56,088,071 | 9.99E-19 | Disruptive_inframe_insertion | ZNF787 | NA |
| rs145150447 | 17 | 76,173,781 | 1.66E-17 | Missense_variant | RNF157 | 0.001597 |
| rs77563894 | 16 | 74,391,834 | 6.39E-17 | Missense_variant | NPIPB15 | NA |
| rs79144888 | 3 | 1.84E+08 | 1.52E-15 | Missense_variant | ALG3 | 0.003395 |
| rs774012991 | 1 | 1.75E+08 | 4.64E-14 | Conservative_inframe_insertion | KIAA0040 | NA |
| rs3737738 | 1 | 46,035,056 | 2.58E-13 | Missense_variant | MAST2 | 0.007987 |
| rs759464632 | 11 | 64,315,823 | 2.6E-13 | Structural_interaction_variant | ESRRA | NA |
| rs145988500 | 8 | 22,600,944 | 3.97E-13 | Missense_variant | C8orf58 | 0.008586 |
| rs2305205 | 10 | 1.17E+08 | 5.66E-12 | Missense_variant+splice_region_variant | PNLIPRP1 | 0.013379 |
| rs1294271615 | 13 | 71,866,526 | 4.62E-11 | Conservative_inframe_insertion | DACH1 | NA |
| rs117142233 | 7 | 20,161,835 | 5.85E-11 | Missense_variant | MACC1 | 0.019768 |
| rs12671170 | 7 | 20,154,356 | 9.31E-11 | Missense_variant | MACC1 | 0.021366 |
| rs28359631 | 1 | 2.31E+08 | 3.57E-10 | Splice_acceptor_variant+intron_variant | CAPN9 | 0.026757 |
| rs747005499 | 19 | 35,511,478 | 3.77E-10 | Frameshift_variant | DMKN | NA |
| rs12562749 | 1 | 2.31E+08 | 5.05E-10 | Missense_variant | CAPN9 | 0.028355 |
| rs776176602 | 19 | 35,511,479 | 5.78E-10 | Frameshift_variant | DMKN | NA |
Notes: SNP/Indel illustrated the rs number in the dbSNP database, CHR illustrated the chromosome of the SNP/Indel locus, BP illustrated the chromosome position of the SNP/Indel, P value illustrated the probability of SNP/Indel appearing in DNA of 6 diabetic patients simultaneously. This table retained the data with P<1e-9. Func. indicated the mutation type. Gene indicated the gene name corresponding to SNP/indel. MAF1000G represented the frequency of SNP/indel in thousands of human genomes.
Genes Related to Diabetes
In order to analyze how the previously mentioned significant SNP and Indel mutation sites affect the occurrence of diabetes, we mapped the genes corresponding to the 22 mutation sites in KEGG pathway. Finally, the PNLIPRP1 gene and CAMKK2 gene were mapped to the KEGG diabetes-related metabolic pathway.
PNLIPRP1 Gene rs2305205 Site Mutation
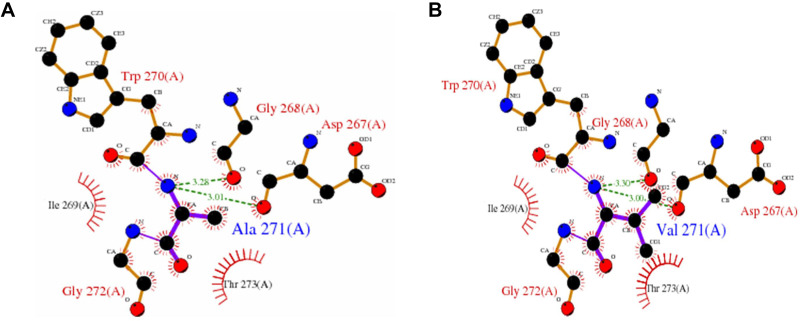
The PNLIPRP1 gene is located on chromosome 10 and contains 13 exons. The rs2305205 mutation site in the 8th exon resulted in the replacement of 271 alanine (Ala) of PNLIPRP1 protein with valine (Val) (Figure 5). This SNP site was located in the N-terminal domain of PNLIPRP1 and was involved in triglyceride metabolism. After the mutation, the surface accessibility of the whole protein increased by 27.4Å2, and the relative surface accessibility increased from 55% to 58.2%. It is suggested that mutations in the rs2305205 site might alter the function of the protein.
Figure 5.
Diagram of the effect of rs2305205 mutation on protein function. (A) Indicates the mode of interaction of Ala271 with other amino acid residues in the unmutated local structure. The hydrogen bond is indicated by a green dotted line, the bond length is identified by a number, and the hydrophobic effect is represented by a semicircle. (B) Mode of interaction of Val271 with other amino acid residues in the local structure after mutation. The effect on protein structure before and after the mutation was obtained from PDB sum, where the PDB ID number of PNLIPRP1 was 2 ppl.
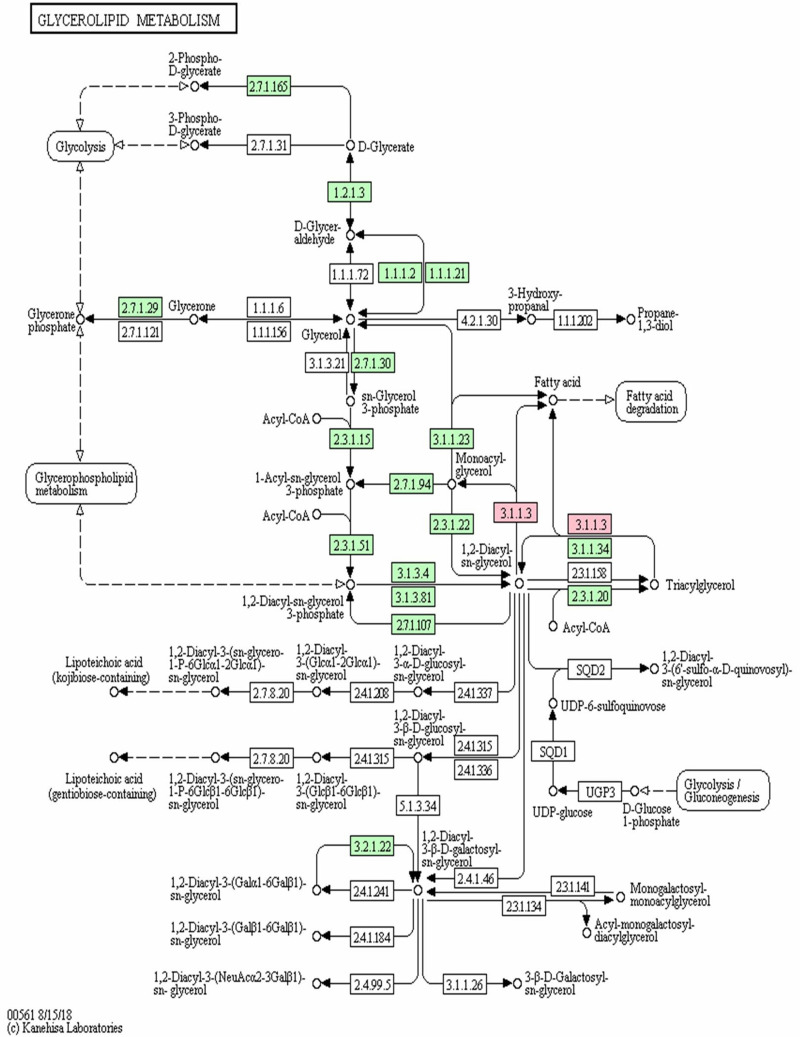
According to the KEGG pathway map for PNLIPRP1 involved triglyceride metabolism (Figure 6), 3.1.1.3 in pink is pancreatic lipase, including pancreatic triglyceride lipase (PLT), PLRP1 and PLRP2. Meanwhile, the 3.1.1.3 in pink mainly involved in metabolism of the fatty acid, monoacyl-glycerol and 1.2-diacyl-sn-glycerol directly (Figure 6).
Figure 6.
The KEGG pathway map of PNLIPRP1 involved in the process of triglyceride metabolism.
Note: The rectangle in the figure indicates the enzyme that catalyzes the reaction. Each enzyme is labeled with the EC (Enzyme Commission) number. The circles in the figure indicate the reaction substrate and product. 3.1.1.3 in pink was pancreatic lipase.
CAMKK2 Gene rs778701848 Site Mutation
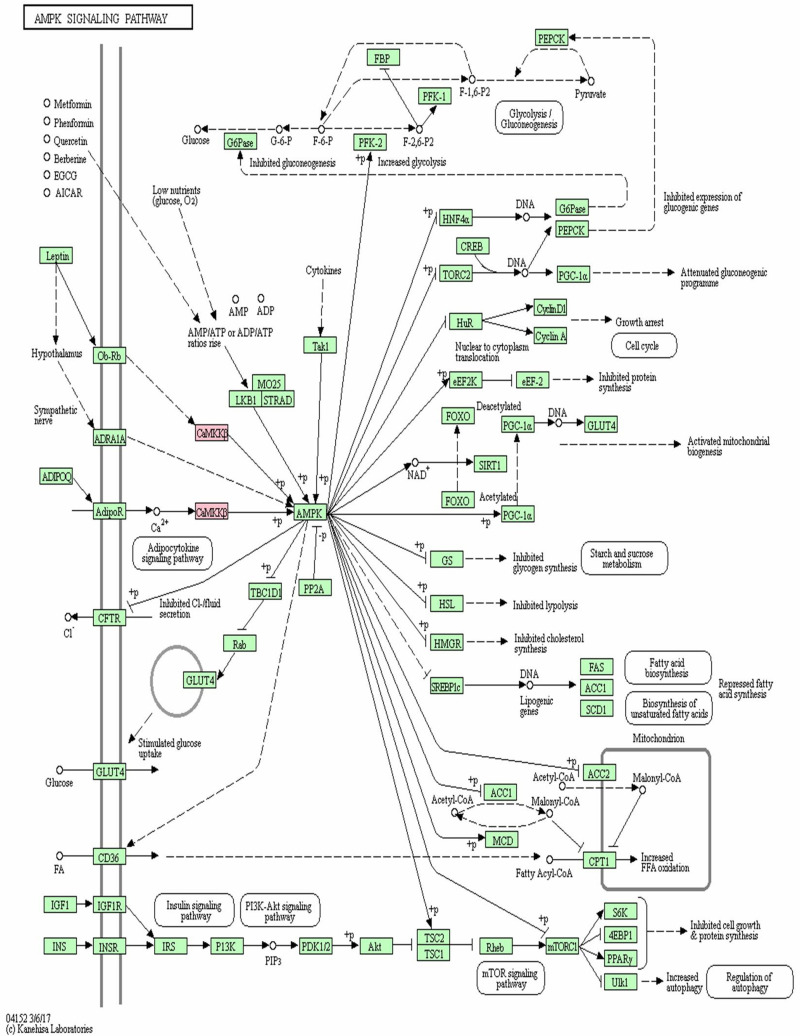
Positioning in KEGG signaling pathway revealed that CAMKK2 was a key node linking endogenous and metabolically related adiponectin (ADIPOQ), leptin and adenylate-activated protein kinase (AMPK) (Figure 7). Adiponectin and leptin activated AMPK through CAMKK2 phosphorylation to regulate glucose metabolism. As Figure 7 shows, AMPK phosphorylation is involved in the gene expression of plenty of genes, including receptor gamma coactivator 1α (PGC-1α), alcohol regulatory element binding protein 1c (SREBP1c), CREB transcriptional coactivator 2 (TORC2), glucose transporter 4 (GLUT4), acetyl-CoA carboxylase (ACC), rapamycin target protein (mTOR), S6 kinase 1 (S6K1) and insulin receptor substrate (IRS). Therefore, mutations in the CAMKK2 gene may be involved in the development and progression of diabetes by inhibiting the downstream AMPK pathway.
Figure 7.
The KEGG pathway map of CAMKK2 participating in AMPK signaling pathway.
Note: The reactions involved in CAMKK2 are marked in light red.
Discussion
The PNLIPRP1 gene was cloned from cDNA in the year 1992,21 consisting of 467 amino acids and localized in the chromosomal region 10q24-q26 in humans.24 PTL could hydrolyze triglyceride into diglyceride, and subsequently convert into mono-glycerides and free fatty acid, which are then absorbed by intestinal epithelial cells. Both PLRP1 and PTL are secreted by pancreatic acinar cells and demonstrated the same affinity with colipase.25,26 In order to study the biological function of PLRP1, a previous study27 showed that food intake could promote the secretion of PLRP1 in the pancreas, therefore, PLRP1 may play a certain role in food digestion. The in vitro studies26,28 showed that PLRP1 could regulate PTL activity by competing for colipase and modulating body fat, obesity, insulin resistance and blood glucose levels in vivo.
In this study, 22 significant SNP mutation sites and Indel mutation sites were discovered, which were then mapped in the KEGG pathway. We found that the PNLIPRP1 gene and CAMKK2 gene were mapped to the KEGG diabetes-related metabolic pathway, therefore, these two variants were discussed. According to the in silico prediction findings, six diabetic patients in this family demonstrated PNLIPRP1 gene rs2305205 site mutation, while normal control did not. The mutation of rs2305205 caused the amino acid encoded by PNLIPRP1 gene from alanine to valine at 271. The mutation of rs2305205 reduced the function of PLRP1, weakened function of colipase competing with PTL, increased activity of PTL, promoted digestion and absorption of fatty acids. Meanwhile, the mutation of rs2305205 could also increase the content of body fat, and then cause obesity, insulin resistance, and increased blood glucose. Therefore, we concluded that PNLIPRP1 gene mutation may promote the development of T2DM through the previously mentioned pathways.
As the KEGG pathway map of PNLIPRP1 involved in triglyceride metabolism, 3.1.1.3 in pink was considered to be a pancreatic lipase, including pancreatic triglyceride lipase (PLT), PLRP1, and PLRP2. Because lipid metabolism disorder can cause triglyceride to deposit in the target tissue of insulin action, reduce insulin sensitivity, and increase the risk of type 2 diabetes,29 it is further speculated that the rs2305205 mutation of PNLIPRR1 was associated with the occurrence of diabetes in this family. Presently, there has been no research on the correlation between PNLIPRP1 gene mutation and occurrence of diabetes at home and abroad, and no research reports on the mutation of this gene in pedigree diabetes.
The calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) gene is located on chromosome 12q24.2 and contains 16 exons.30 The rs778701848 site mutation on the 16th exon results in insertion of amino acid 538. The protein encoded by CAMKK2 gene is involved in energy metabolism and glucose homeostasis through modulating adiponectin, leptin, and insulin.31 The present KEGG pathway map indicated that adiponectin and leptin could activate AMPK through CAMKK2 phosphorylation to regulate glucose metabolism. Meanwhile, AMPK phosphorylation is involved in gene expression of plenty of genes in the process of metabolism. Therefore, loss of CAMKK2 function not only reduced function of adiponectin and leptin, but also reduced levels of adiponectin and leptin in the blood. Moreover, the finding of the present KEGG pathway map for CAMKK2 gene also illustrated plenty of targeting genes and associated signaling pathways, including PGC-1α, SREBP1c, TORC2, GLUT4, ACC, mTOR, S6K1 and IRS, which are consistent with some previous studies.32–46 Therefore, this study further proved that CAMKK2 gene was closely correlated with glucose homeostasis, gluconeogenesis, and hepatic glucose production.
In the present study, we found that six diabetic patients in a family demonstrated CAMKK2 gene rs778701848 site mutation and PNLIPRP1 gene rs2305205 site mutation, while normal family member (control) did not. Both of the mutations rs778701848 and rs2305205 could affect the biofunctions of CAMKK2 gene and PNLIPRP1 gene, and increased risk of diabetes in these family members through the previously mentioned signaling pathways, which have not been discussed in the previous documents. The rs778701848 mutation and rs2305205 mutation were identified as the main causes for the familial diabetes in this study, suggesting that CAMKK2 gene and PNLIPRP1 gene might be promising and novel targets for treating diabetes. Therefore, the mutations at rs2305205 of PNLIPRP1 gene and rs7787018848 of CAMKK2 gene may be related to the occurrence of diabetes in six type 2 diabetic family members.
There are still some shortcomings in this study. Firstly, peripheral blood of the non-diabetic relatives was not collected, which would be more appropriate for clarifying the discovered genome exon. In a following study, we would compare the data in this study with the diabetic patients without a family history or healthy individuals. Secondly, only one family was included in this study, therefore, relevant studies should be carried out to explore diabetes in families. Thirdly, the correlation between PNLIPRP1 gene and CAMKK2 gene in the diabetes patients was not conducted in this study, which is critical for providing the theoretical basis for early prevention and treatment of diabetes. Fourthly, sample size of this study was relatively small (involving only 3 generations and 7 family members). Fifthly, the clinical issues of the diabetes patients, including duration of diabetes, sex, pharmacological treatment for diabetes, overweight/obesity, were not analyzed, which are important for designing a therapeutic strategy. Finally, for the genes identified in this study, we did not determine which gene belongs to a family cluster and which gene belongs to the diabetes disease its own. The analyses for the previously mentioned genes might be critical for the treatment of diabetes.
In conclusion, by conducting whole exome sequencing and bioinformatic data analysis, we found that all exons of diabetic patients in this family demonstrated an rs2305205 mutation in PNLIPRP1 gene and an rs778701848 mutation in CAMKK2 gene. The rs2305205 mutation and rs778701848 mutation might promote occurrence of type 2 diabetes through reducing sensitivity of peripheral tissues to insulin and reducing insulin secretion.
Funding Statement
The study was supported by grants from the National Natural Science Fund of China (Grant No. 81160104, 30760087and 82160165), the Yunnan provincial science and technology department-Kunming medical University applied basic research joint project [Grant No. 2018FE001 (−043)], the Training plan for medical discipline leaders of Yunnan provincial health and family planning commission (Grant No. D-2017039) and The young and middle-aged academic and technical leaders reserve talented person project in yunnan province (Grant NO. 202105AC160093).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cho NH, Shaw JE, Karuranga S, et al. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138(1):271–281. doi: 10.1016/j.diabres.2018.02.023 [DOI] [PubMed] [Google Scholar]
- 2.Albai O, Frandes M, Timar B, Paun DL, Roman D, Rimar R. Long-term risk of malignant neoplastic disorders in type 2 diabetes mellitus patients with metabolic syndrome. Diabetes Metab Syndr Obes. 2020;13(1):1317–1326. doi: 10.2147/DMSO.S243263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peleg O, Hadar E, Cohen A. Individuals with type 2 diabetes: an exploratory study of their experience of family relationships and coping with the illness. Diabetes Educ. 2020;46(1):83–93. [DOI] [PubMed] [Google Scholar]
- 4.Arfa I, Abid A, Malouche D, et al. Familial aggregation and excess maternal transmission of type 2 diabetes in Tunisia. Postgrad Med J. 2007;83(979):348–351. doi: 10.1136/pgmj.2006.053744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wemrell M, Bennet L, Merlo J. Understanding the complexity of socioeconomic disparities in type 2 diabetes risk: a study of 4.3 million people in Sweden. BMJ Open Diabetes Res Care. 2019;7(1):e000749. doi: 10.1136/bmjdrc-2019-000749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souliotis K, Koutsovasilis A, Vatheia G, et al. Profile and factors associated with glycaemic control of patients with type 2 diabetes in Greece: results from the diabetes registry. BMC Endocr Disord. 2020;20(1):16. doi: 10.1186/s12902-020-0496-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui X, Qian DW, Jiang S, Shang EX, Zhu ZH, Duan JA. Scutellariae radix and coptidis rhizoma improve glucose and lipid metabolism in T2DM rats via regulation of the metabolic profiling and MAPK/PI3K/Akt signaling pathway. Int J Mol Sci. 2018;19(11):3634. doi: 10.3390/ijms19113634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kral BG, Becker DM, Yanek LR, et al. The relationship of family history and risk of type 2 diabetes differs by ancestry. Diabetes Metab. 2019;45(3):261–267. [DOI] [PubMed] [Google Scholar]
- 9.Kwak SH, Park KS. Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med. 2016;48(3):e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarhangi N, Sharifi F, Hashemian L, et al. PPARG (Pro12Ala) genetic variant and risk of T2DM: a systematic review and meta-analysis. Sci Rep. 2020;10(1):12764. doi: 10.1038/s41598-020-69363-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachtiar M, Ooi BNS, Wang J, et al. Towards precision medicine: interrogating the human genome to identify drug pathways associated with potentially functional, population-differentiated polymorphisms. Pharmacogenomics J. 2019;19(6):516–527. doi: 10.1038/s41397-019-0096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingelsson E, McCarthy MI. Human genetics of obesity and type 2 diabetes mellitus: past, present, and future. Circ Genom Precis Med. 2018;11(6):e002090. doi: 10.1161/CIRCGEN.118.002090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prudente S, Jungtrakoon P, Marucci A, et al. Loss-of-function mutations in APPL1 in familial diabetes mellitus. Am Hum Genet. 2015;97(1):177–185. doi: 10.1016/j.ajhg.2015.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Yuan K, Ding SX, et al. Hyperglycemia caused by mutation of GCK gene in 10 patients analysis of clinical and mutation characteristics. Zhonghua Er Ke Za Zhi. 2019;57(6):440–444. [DOI] [PubMed] [Google Scholar]
- 15.Meyre D, Andress EJ, Sharma T, et al. Contribution of rare coding mutations in CD36 to type 2 diabetes and cardio-metabolic complications. Sci Rep. 2019;9(1):17123. doi: 10.1038/s41598-019-53388-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J, Jiang F, Zhang R. Whole-exome sequencing identifies a novel INS mutation causative of maturity-onset diabetes of the young 10. J Mol Cell Biol. 2017;9(1):376–383. doi: 10.1093/jmcb/mjx039 [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Liu J, Li X. Identification of a novel mutation site in maturity-onset diabetes of the young in a Chinese family by whole-exome sequencing. Mol Med Rep. 2019;20(1):2373–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abecasis GR, Altshuler D, Auton A, Brooks LD; The 1000 Genomes Project Consortium. A map of human genome variation from population scale sequencing. Nature. 2010;467(7319):1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: theNCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandolfi B, Grahn RA, Gustafson NA, et al. Variant in CMAH is associated with blood typeAB in ragdoll cats. PLoS One. 2016;11(5):e0154973. doi: 10.1371/journal.pone.0154973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laskowski RA, Jablonska J, Pravda L, Varekova RS, Thornton JM. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27(1):129–134. doi: 10.1002/pro.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou YY, Chen LP, Zhang Y, et al. Integrated transcriptomic analysis reveals hub genes involved in diagnosis and prognosis of pancreatic cancer. Mol Med. 2019;25(1):47. doi: 10.1186/s10020-019-0113-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis RC, Diep A, Hunziker W, et al. Assignment of human pancreatic lipase gene (PNLIP) to chromosome 10q24-q26. Genomics. 1991;11(4):1164–1166. doi: 10.1016/0888-7543(91)90048-J [DOI] [PubMed] [Google Scholar]
- 25.CrenonI I, Jayne S, Kerfelec B, Hermoso J, Pignol D, Chapus C. Pancreatic lipase-related protein type 1:a double mutation restores a significant lipase activity. Biochem Biophy Res Commun. 1998;246(2):513–517. doi: 10.1006/bbrc.1998.8651 [DOI] [PubMed] [Google Scholar]
- 26.Lowe ME. The triglyceride lipases of the pancreas. J Lipid Res. 2002;43(12):2007–2016. doi: 10.1194/jlr.R200012-JLR200 [DOI] [PubMed] [Google Scholar]
- 27.Birk RZ, Brannon PM. Regulation of pancreatic lipase by dietary medium chain triglycerides in the weanling rat. Pediatr Res. 2004;55(6):921–926. doi: 10.1203/01.PDR.0000127430.04127.4F [DOI] [PubMed] [Google Scholar]
- 28.Ren J, Chen Z, Zhang W, et al. Increased fat mass and insulin resistance in mice lacking pancreatic lipase-related protein 1. J Nutr Biochem. 2011;22(7):691–698. doi: 10.1016/j.jnutbio.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 29.Athyros VG, Doumas M, Imprialos KP, et al. Diabetes and lipid metabolism. Hormones. 2018;17(1):61–67. doi: 10.1007/s42000-018-0014-8 [DOI] [PubMed] [Google Scholar]
- 30.Hsu LS, Chen GD, Lee LS, Chi CW, Cheng JF, Chen JY. Human Ca2+/Calmodulin-dependent protein kinase kinase beta gene encodes multiple isoforms that display distinct kinase activity. J Biol Chem. 2001;276(33):31113–31123. doi: 10.1074/jbc.M011720200 [DOI] [PubMed] [Google Scholar]
- 31.Marcelo KL, Ribar T, Means CR, et al. Research resource: roles for calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) in systems metabolism. Mol Endocrinol. 2016;30(5):557–572. doi: 10.1210/me.2016-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murao K, Li J, Imachi H, Muraoka T, Masugata H. Exendin-4 regulates glucokinase expression by CaMKK/CaMKIV pathway in pancreatic â-cell line. Diabetes Obes Metab. 2009;11(10):939–946. doi: 10.1111/j.1463-1326.2009.01067.x [DOI] [PubMed] [Google Scholar]
- 33.Li JH, Murao K, Imachi H, et al. Exendin-4 regulates pancreatic ABCA1 transcription via CAMKK/CaMKIVpathway. J Cell Mol Med. 2010;14(5):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyu J, Imachi H, Fukunaga K, et al. Role of ATP-binding cassette transporter A1 in suppressing lipid accumulation by glucagon-like peptide-1 agonist in hepatocytes. Mol Metab. 2020;34(1):16–26. doi: 10.1016/j.molmet.2019.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaac R, Goldstein I, Furth N, et al. TM7SF3, a novel p53-regulated homeostatic factor, attenuates cellular stress and the subsequent induction of the unfolded protein response. Cell Death Differ. 2017;24(1):132–143. doi: 10.1038/cdd.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esser N, Legrand-poels S, Piette J, Scheen AJ, Paquot N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res Clin Pract. 2014;105(2):141–150. doi: 10.1016/j.diabres.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 37.Ward MG, Li G. Apoptotic â-cells induce macrophage reprogramming under diabetic conditions. J Biol Chem. 2018;293(42):16160–16173. doi: 10.1074/jbc.RA118.004565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kam A, Li KM, Razmovski-Naumovski V, et al. Curcumin reduces tumour necrosis factor-enhanced annexin V-Positive microparticle release in human vascular endothelial cells. J Pharm Pharm Sci. 2015;18(4):424–433. doi: 10.18433/J3ZC8G [DOI] [PubMed] [Google Scholar]
- 39.Yang HB, Zhao PJ, Tian SL. Clopidogrel protects endothelium by hindering TNFá-InducedVCAM-1 expression through CaMKKâ/AMPK/Nrf2 pathway. J Diabetes Res. 2016;2016:9128050. doi: 10.1155/2016/9128050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge Q, Zhao L, Ren XM, Ye P, Hu ZY. LCZ696, an angiotensin receptor-neprilysin inhibitor, ameliorates diabetic cardiomyopathy by inhibiting inflammation, oxidative stress and apoptosis. Exp Biol Med (Maywood). 2019;244(12):1028–1039. doi: 10.1177/1535370219861283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwabu M, Yamauchi T, Okada-Iwabu M, et al. Adiponectin and AdipoR1 regulate PGC-1á and mitochondria by Ca2+ andAMPK/SIRT1. Nature. 2010;464(7293):1313–1319. doi: 10.1038/nature08991 [DOI] [PubMed] [Google Scholar]
- 42.Maiese K. Novel nervous and multi-system regenerative therapeutic strategies for diabetes mellitus with mTOR. Neural Regen Res. 2016;11(3):372–385. doi: 10.4103/1673-5374.179032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura FV, Costa CG, Ijlst L, et al. Broad specificity of carnitine palmitoyl transferase II towards long-chain acyl-CoA beta-oxidation intermediates and its practical approach to the synthesis of various long-chain acylacarnitines. J Inherit Metab Dis. 1997;20(3):423–426. doi: 10.1023/A:1005315003913 [DOI] [PubMed] [Google Scholar]
- 44.Maeda A, Shirao T, Shirasaya D, et al. Piperine promotes glucose uptake through ROS-dependent activation of the CAMKK/AMPK signaling pathway in skeletal muscle. Mol Nutr Food Res. 2018;62(11):e1800086. doi: 10.1002/mnfr.201800086 [DOI] [PubMed] [Google Scholar]
- 45.Kim SJ, Quan HY, Jeong KJ, et al. Beneficial effect of betulinic acid on hyperglycemia via suppression of hepatic glucose production. Agr Food Chem. 2014;62(2):434–442. doi: 10.1021/jf4030739 [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Chen S, Shen T, et al. Adropin regulates hepatic glucose production via PP2A/AMPK pathway in insulin-resistant hepatocytes. FASEB J. 2020;34(8):10056–10072. doi: 10.1096/fj.202000115RR [DOI] [PubMed] [Google Scholar]