Abstract
The global increase in transcription of cytoprotective genes induced in response to oxidative challenge has been termed the antioxidant response. Ferritin serves as the major iron-binding protein in nonhematopoietic tissues, limiting the catalytic availability of iron for participation in oxygen radical generation. Here we demonstrate that ferritin is a participant in the antioxidant response through a genetically defined electrophile response element (EpRE). The EpRE of ferritin H identified in this report exhibits sequence similarity to EpRE motifs found in antioxidant response genes such as those encoding NAD(P)H:quinone reductase, glutathione S-transferase, and heme oxygenase. However, the EpRE of ferritin H is unusual in structure, comprising two bidirectional motifs arranged in opposing directions on complementary DNA strands. In addition to EpRE-mediated transcriptional activation, we demonstrate that ferritin is subject to time-dependent translational control through regulation of iron-regulatory proteins (IRP). Although IRP-1 is initially activated to its RNA binding (ferritin-repressing) state by oxidants, it rapidly returns to its basal state. This permits the translation of newly synthesized ferritin transcripts and ultimately leads to increased levels of ferritin protein synthesis following oxidant exposure. Taken together, these results clarify the complex transcriptional and translational regulatory mechanisms that contribute to ferritin regulation in response to prooxidant stress and establish a role for ferritin in the antioxidant response.
Elemental iron is required for normal cell growth and proliferation. However, excess iron is potentially harmful, since it can catalyze the formation of reactive oxygen species via Fenton chemistry. Excess reactive oxygen species have been implicated in damage to DNA, proteins, and lipids and may play a role in cancer and inflammation (13, 20, 38). For these reasons, cells have evolved highly regulated mechanisms for controlling intracellular iron levels. Important among these is the iron storage protein, ferritin. Ferritin is a holoenzyme shell (∼450 kDa) consisting of 24 subunits of two types, H and L, and capable of storing up to 4,500 atoms of ferric iron. The H-to-L ratio within ferritin varies in a tissue-specific manner and is also influenced by pathophysiological conditions, including inflammation and malignancy (3, 9, 41, 46). Ferritin H and L protein sequences are highly conserved among species, suggesting the importance of these proteins in regulating iron homeostasis (10, 40).
The induction of cytoprotective enzymes in cells challenged with chemical carcinogens, toxic electrophiles, and oxidants has been termed the antioxidant response (14). The ability of cells to up-regulate the synthesis of these phase II enzymes is a pivotal cellular defense mechanism that is mechanistically distinct from induction of phase I enzymes mediated by the Ah receptor (6, 34). Expression of enzymes that constitute the antioxidant response is induced at the transcriptional level by a variety of compounds, including tert-butylhydroquinone (t-BHQ), β-naphthoflavone (β-NF), and hydrogen peroxide (14). A cis-acting element responsible for transcriptional activation in response to these compounds has been identified in the 5′-flanking region of the genes encoding mouse (31) and rat (36) glutathione S-transferase (GST)-Ya, rat GST-pi (24), rat (6) and human (19) NAD(P)H quinone reductase, human γ-glutamylcysteine synthetase (23), mouse heme oxygenase (1), mouse ferritin L chain (47), and mouse metallothionein I genes (5). The element, referred to as the electrophile response element (EpRE) or antioxidant response element, has a core AP1-like motif and requires adjacent core-like sequences for its function (14). In animal studies, the levels of both ferritin H and ferritin L mRNA were shown to increase in rat liver in response to the glutathione-depleting agent phorone (2) and the chemopreventive dithiolethione (32). These studies suggest that ferritin induction in response to xenobiotics and oxidants may occur via an EpRE-mediated mechanism. Indeed, Wasserman and Fahl have recently demonstrated that the 5′ upstream region of the ferritin L gene contains a functional EpRE that can be activated by t-BHQ (47).
In addition to transcriptional control, ferritin mRNA is subject to translational control by iron-regulatory proteins (IRPs), proteins that bind to the iron-responsive element (IRE) in the 5′ untranslated region (UTR) of ferritin mRNA and inhibit its translation (10). The activity of IRP-1 is modulated by intracellular iron concentrations, becoming activated as a translational repressor by low iron levels and inactivated by high iron levels (8, 11, 15, 18, 33). Several reports have suggested that IRP-1 is activated in cells treated with hydrogen peroxide (21, 26–28). Although this has not been consistently observed, an anticipated consequence of IRP activation is ferritin repression. Such an effect would be expected to negate transcriptionally mediated increases in ferritin mRNA levels, since the resulting transcripts would not be effectively translated. These unique complexities of ferritin posttranslational regulation, in particular the potential for oxidant-mediated repression of ferritin translation through IRP activation, must be taken into account in models invoking the functional participation of ferritin in the antioxidant response.
To elucidate the role of ferritin in response to oxidant stress, we have asked whether induction of ferritin in response to oxidants occurs at a transcriptional level and whether it proceeds via a definable genetic element. Concomitantly, we have analyzed the effects of cellular exposure to oxidants on IRP activity and ferritin protein synthesis. Our results identify the EpRE of the ferritin H gene as a 75-bp cis-acting element located 4.1 kb upstream of the transcription initiation site. We show an inverse and time-dependent biphasic response of ferritin protein and IRP activation to oxidative stress. These results demonstrate that the relationship between ferritin mRNA, ferritin protein synthesis, and IRP activation can be explained by temporal events that occur in response to prooxidant stress. This sequence of events clarifies discordant observations in the literature, describing both activation and inactivation of the IRP in response to oxygen radicals (2, 29), and identifies ferritin as a component of the antioxidant response.
MATERIALS AND METHODS
Cell culture.
The BNL CL.2 mouse normal liver and Hepa1-6 mouse hepatoma cell lines were obtained from the American Type Culture Collection. They were cultured at 37°C in a humidified 5% CO2 atmosphere in high-glucose Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (Gemini Bioproducts).
Reagents and chemicals.
Hydrogen peroxide, t-BHQ, and β-NF were purchased from Sigma.
Construction of 5′ ferritin H-hGH reporter plasmids. (i) 5′ deletion mutants.
Expression of human growth hormone (hGH) in pGEM7zf(+)−4.8kbFH-hGH was driven by the 5′ flanking region of the mouse ferritin H gene from nucleotides −4819 to +86 (17, 42). A set of 5′ FH deletion mutants was constructed as follows. pGEM7zf(+)−4.8kbFH-hGH was digested with ApaI and AflII, and the 6.3-kb ApaI-AflII pGEM7zf(+)-1.5kbFH-hGH vector fragment was isolated. pBluescript KS(−)−4.0kbFHCAT, pBluescript KS(−)−4.0kbAP1+αFHCAT, pBluescript KS(−)−4.0kbAP1+δFHCAT, and pBluescript KS(−)−4.0kbpAP1+ɛFHCAT (42) were digested with ApaI and AflII, and 2.5 to 2.6 kb of the ferritin H DNA was isolated and ligated to the 6.3-kb ApaI-AflII pGEM7zf(+)-1.5kbFH-hGH vector fragment.
(ii) Insertion of the EpRE into an hGH reporter plasmid.
Synthetic double-stranded 5′-ApaI/blunt-end-3′ DNA; (40-bp FER-1, 47-bp AP1, 59-bp FER-1+AP1, and 75-bp FER-1+AP1 [see Fig. 3]) or the 107-bp FER1+AP1 DNA fragment isolated from pBluescript KS(−)−4.0kbAP1+ɛFHhGH (see Fig. 2C) was cloned into pGEM7zf(+)−0.32kbFH-hGH as follows. Each 5′-ApaI/blunt-end-3′ DNA was ligated to the ApaI-EcoRV 5.1-kb fragment of pGEM7zf(+)-4.0kbFHhGH [this 5.1-kb DNA is equivalent to pGEM7zf(+)−0.32kbFH-hGH]. pGEM7zf(+)−0.32kbFH-hGH (no insertion) was constructed by digestion of pGEM7zf(+)−4.8kbFH-hGH with ApaI and EcoRV followed by generation of blunt ends with T4 DNA polymerase and self-ligation of the 5.1-kb fragment.
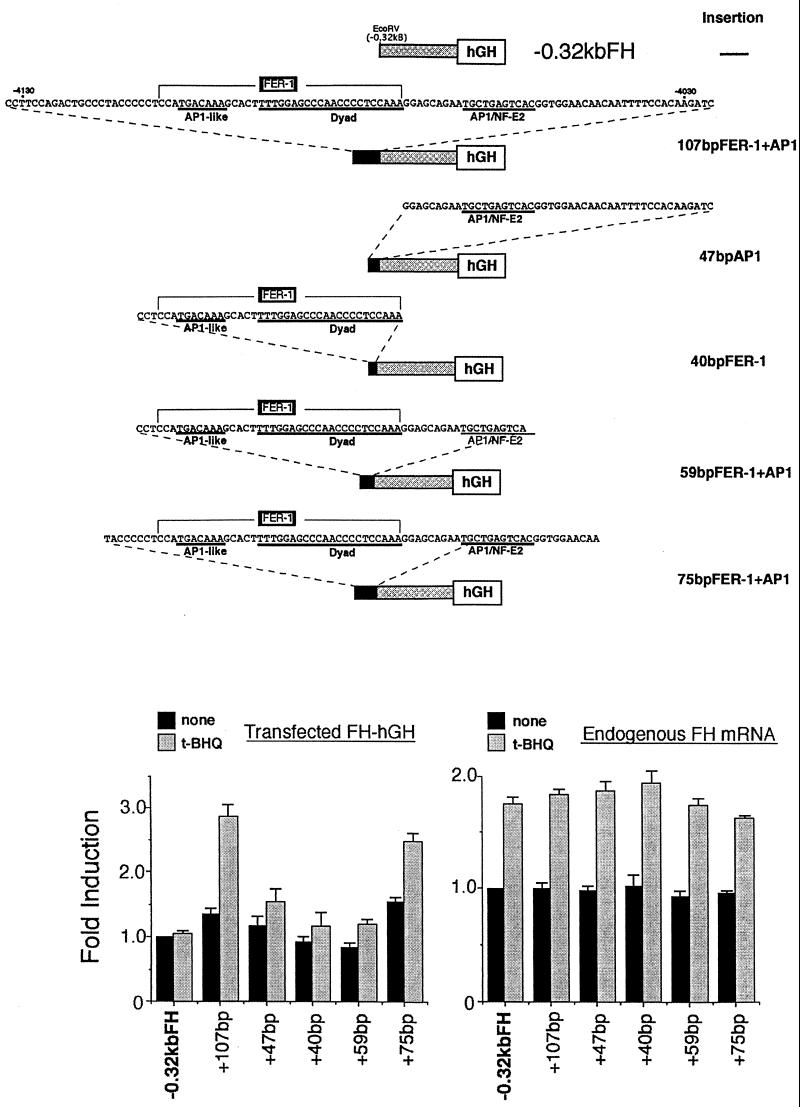
FIG. 3.
The 75-bp cis-acting element responsible for transcriptional activation of the mouse ferritin H gene by H2O2 or t-BHQ contains the FER-1 element and an AP1/NF-E2 site. (Top) Schematic representation of ferritin H-hGH constructs used in this experiment. The cis-acting region in response to H2O2 or t-BHQ was dissected into various pieces, which were inserted into the EcoRV site of −0.32kbFHhGH. −0.32kbFHhGH contains 0.32 kb of 5′-flanking region of the ferritin H gene fused to the hGH reporter gene. 107bpFER-1+AP1 contains the entire cis-acting region (nucleotides −4132 to −4026) defined by the experiments in Fig. 2. 47bpAP1 contains a AP1/NF-E2 site (nucleotides −4072 to −4026). 40bpFER-1 contains the complete FER-1 element (nucleotides −4112 to −4073). 59bpFER-1+AP1 contains the complete FER-1 element plus the AP1/NF-E2 site (nucleotides −4112 to −4054). 75bpFER-1+AP1 contains the complete FER-1 element plus the AP1/NF-E2 site (nucleotides −4117 to −4043). (Bottom) A 15-μg portion of each hGH construct was transiently transfected into Hepa1-6 cells. After 36 to 40 h, the cells were treated with 100 μM t-BHQ for 8 to 10 h. A 10-μg portion of total RNA isolated from each sample was subjected to RNase protection assays to analyze the expression of transfected ferritin H-hGH and endogenous ferritin H mRNA simultaneously. Each transfected and endogenous ferritin H band in the cells transfected with −0.32kbFHhGH without treatment was defined as 1.0. The results from four independent experiments and standard errors are shown.
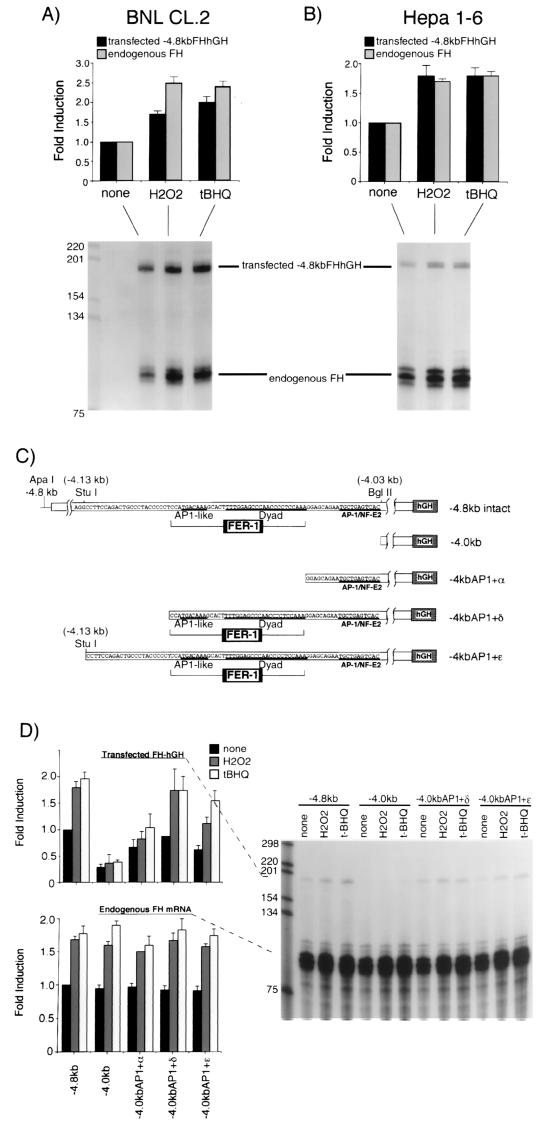
FIG. 2.
The cis-acting element responsible for transcriptional activation of the mouse ferritin H gene by H2O2 or t-BHQ is located 4.1 kb upstream to the transcriptional initiation site. BNL CL.2 (A) or Hepa1-6 cells (B) were transfected with 15 μg of −4.8kbFH-hGH. After 36 to 40 h, the cells were treated with H2O2 (750 μM for BNL CL.2 cells and 250 μM for Hepa1-6 cells) or t-BHQ (250 μM for BNL CL.2 cells and 100 μM for Hepa1-6 cells) for 8 to 10 h. A 10-μg portion of total RNA isolated from each treatment was subjected to RNase protection assays as described in Materials and Methods to analyze the expression of transfected −4.8kbFHhGH (the protected RNA band is 190 bases) and endogenous ferritin H mRNA (the protected RNA band is 86 bases) simultaneously. Each transfected and endogenous ferritin H band without treatment was defined as 1.0, and the results from seven (BNL CL.2) and eight (Hepa1-6) independent experiments and standard errors are shown. (C) Schematic representation of ferritin H-hGH constructs used in this experiment. A basal enhancer element of the mouse ferritin H gene, FER-1 (42), and a proximal AP1/NF-E2 site are indicated in the diagram. (D) A 15-μg portion of each hGH construct was transiently transfected into Hepa1-6 cells. After 36 to 40 h, the cells were treated with 250 μM H2O2 or 100 μM t-BHQ for 8 to 10 h. A 10-μg portion of total RNA isolated from each sample was subjected to RNase protection assays to analyze the expression of transfected ferritin H-hGH and endogenous ferritin H mRNA simultaneously. Each transfected and endogenous ferritin H band in the cells transfected with −4.8kbFHhGH without treatment was defined as 1.0, and the results from six independent experiments and standard errors are shown.
DNA transfection and RNase protection assay.
Transient DNA transfection into BNL CL.2 or Hepa1-6 cells was carried out by the calcium phosphate precipitation method. Calcium phosphate-DNA precipitates were made by mixing 15 μg of each hGH reporter plasmid with 0.2 ml of 0.25 M CaCl2 and adding 0.2 ml of 2× BES-buffered saline [50 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, 280 mM NaCl, and 1.5 mM Na2HPO4 (pH 6.9)]. The resultant 0.4 ml of calcium phosphate-DNA solution was added to the overnight culture of the cells plated in duplicate at a density of 4 × 105 cells per 60-mm dish containing 4 ml of the culture medium. After incubation for 6 h, the cells were washed twice with phosphate-buffered saline, fed with 4 ml of the fresh culture medium, and incubated for 36 to 40 h. The transfected cells were then treated with freshly diluted H2O2 or t-BHQ for 8 to 10 h and harvested for isolation of total RNA. Preparation of total RNA and Northern blotting were carried out as described previously (43). The RNase protection assay was performed essentially as described previously (16) using a chimeric RNA probe spanning the region of the 5′ ferritin H gene between −225 (SmaI site) and +86 and approximately 100 bp of the hGH coding region. RNA hybrids were separated on 6% acrylamide sequencing gels and visualized by autoradiography. Radioactive signal intensities were quantitated using a PhosphorImager analyzer (model 445Si; Molecular Dynamics).
Preparation of RNA probe and measurement of IRP binding to IRE.
IRP RNA probe was synthesized in an in vitro transcription system driven by the T7 promoter using a synthetic DNA template as described previously (22). A 47-base oligonucleotide containing the IRE sequence (5′-GTTCCGTTCAAACACTGTTGAAGCAAGAACTATAGTGAGTCGTATTA-3′) was annealed with an 18-base oligonucleotide containing the T7 promoter sequence (5′-TAATACGACTCACTATAG-3′) at an equal molar ratio. A 2-μg portion of the partially double-stranded oligonucleotide DNA was incubated at 37°C for 1 h with 2 U of T7 RNA polymerase (Promega) in a 20-μl reaction volume containing 10 mM dithiothreitol, 0.05 mM CTP, 0.5 mM ATP, 0.5 mM GTP, and 0.5 mM UTP, and 50 μCi of [α-32P]CTP (400 to 800 Ci/mmol; Amersham). The template DNA was then digested with 250 μg of DNase I per ml for 15 min at 37°C, extracted with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol. A full-length RNA transcript was recovered from a 10% polyacrylamide sequencing gel after the transcripts were stained with ethidium bromide. Preparation of cytosolic cell extracts and the IRP binding assay were performed essentially as described previously (15) with the following modifications. For preparation of cytosolic cell extracts, cells were incubated in 150 μl of extraction buffer (10 mM HEPES [pH 7.6], 3 mM MgCl2, 40 mM KCl, 5% glycerol, 1 mM dithiothreitol, 0.2% NP-40) for 5 min at room temperature. Cell debris was removed by centrifugation at 12,000 × g for 10 min at 4°C, and the protein concentration in each cell extract was measured using the Bio-Rad protein assay kit. A 1-μg portion of cytosolic extract was incubated with 0.5 × 104 to 1 × 104 cpm of the 32P-labeled IRE RNA probe at 25°C for 15 min. To measure total IRP binding, cytosolic extracts were incubated with 2% β-mercaptoethanol prior to addition of the RNA probe. RNA-protein complexes were separated on 5% nondenaturing polyacrylamide gels. The gels were treated with 10% acetic acid–10% isopropanol solution for 5 to 10 min, dried, and subjected to autoradiography.
Immunoprecipitation of ferritin.
For the estimation of ferritin protein synthesis, cell lysates metabolically labeled with Tran35S-label (100 μCi/ml; ICN) for 1 h after H2O2 treatment were immunoprecipitated with rabbit anti-human liver ferritin antibody (Dako Corp.) and protein A-agarose (Calbiochem). The immunoprecipitates were separated on sodium dodecyl sulfate–15%-polyacrylamide gels.
RESULTS
Oxidative stress induces ferritin mRNA.
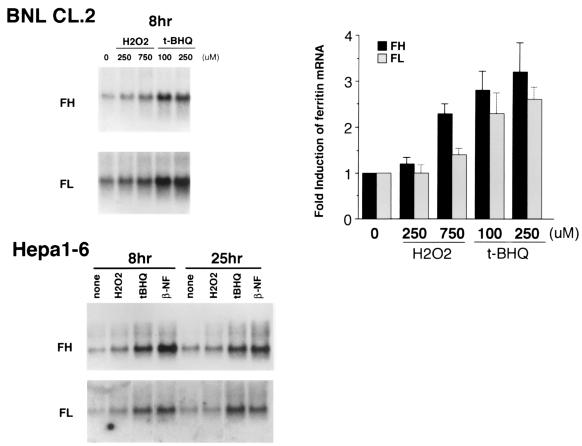
To study ferritin regulation in response to oxidative stress, we first examined ferritin H and L mRNA levels in the nontransformed mouse liver cell line BNL CL.2 following an 8-h exposure to two different oxidants, H2O2 and t-BHQ. Northern blots demonstrated that both reagents induced ferritin H and L mRNA in a dose-dependent manner (Fig. 1). Time course experiments demonstrated that induction of ferritin H mRNA was observed at 4 h after treatment with H2O2 or t-BHQ and sustained for at least 8 h in response to H2O2 and for 24 h following exposure to t-BHQ (data not shown). These concentrations were not cytotoxic, as demonstrated by trypan blue exclusion (the number of viable cells per dish was 5.3 × 106 ± 0.3 × 106 at time zero, 6.3 × 106 ± 0.4 × 106 and 5.6 × 106 ± 0.4 × 106 after 4 and 24 h, respectively, of treatment with 250 μM t-BHQ and 5.1 × 106 ± 0.3 × 106 and 4.6 × 106 ± 0.3 × 106 after 4 and 24 h, respectively, of treatment with 750 μM H2O2). Induction of ferritin mRNA was observed in another mouse liver cell line, Hepa1-6 (Fig. 1), suggesting that the response of ferritin to oxidants is not cell line specific. Increases in the levels of ferritin mRNA were also seen in cells exposed to β-NF (Fig. 1), indicating that induction of ferritin may be a general response to prooxidant stress.
FIG. 1.
H2O2 and t-BHQ induce mRNAs for ferritin H and L in BNL CL.2 and Hepa1-6 mouse liver cells. (Top left) Confluent BNL CL.2 cells were treated for 8 h with H2O2 at 250 or 750 μM or t-BHQ at 100 or 250 μM. A 10-μg portion of total RNA from each treatment was subjected to sequential hybridization with a probe for ferritin H (FH) and ferritin L (FL). Equivalent amounts of RNA loading and transfer to membrane were confirmed by ethidium bromide staining (results not shown). (Top right) The results for ferritin H (FH) and ferritin L (FL) mRNA induction in BNL CL.2 cells (ferritin H and L expression without treatment was defined as 1.0) from five independent experiments are shown, with the standard error indicated. (Bottom) Confluent Hepa1-6 cells were treated with H2O2 at 250 μM, t-BHQ at 100 μM, and β-NF at 50 μM for 8 and 25 h. Ferritin H (FH) and ferritin L (FL) mRNA expression was similarly analyzed by Northern blotting.
Induction of ferritin H by oxidants is transcriptionally mediated.
The induction of ferritin H mRNA by H2O2 or t-BHQ was inhibited by actinomycin D (data not shown), suggesting that it was transcriptionally mediated. To confirm this observation, we transfected −4.8kbFH-hGH, a reporter plasmid containing the ferritin promoter and 5′-flanking sequences fused to the human growth hormone gene, into BNL CL.2 or Hepa1-6 cells. Transfected cells were then treated with H2O2 or t-BHQ to test if these reagents activate hGH transcription driven by the ferritin H gene 5′ region. Total RNA was isolated and subjected to RNase protection assays to detect both endogenous ferritin H and transfected hGH mRNA levels simultaneously (the multiple bands corresponding to endogenous ferritin H mRNA routinely observed in this assay probably result from RNA secondary structure [16]). As shown in Fig. 2A and B, both H2O2 and t-BHQ induced the expression of transfected −4.8kbFH-hGH as well as of endogenous ferritin H in two different liver cell lines. These results indicate that induction of ferritin H mRNA by H2O2 and t-BHQ is mediated by a transcriptional mechanism that targets a response element(s) contained within the 4.8-kb 5′-flanking region of the ferritin H gene.
Identification of sequences in the ferritin H gene that mediate transcriptional regulation by prooxidants.
We have previously identified a 37-bp basal enhancer element (termed FER-1) located 4.1 kb 5′ to the transcriptional initiation site of the mouse ferritin H gene (42). FER-1 comprises two elements, one of which contains an AP1-like sequence (42, 45). Since the EpREs characterized in several genes, including those encoding GST and quinone reductase, contain consensus AP1 and/or AP1-like sequences and serve as basal enhancer elements (14), we asked if a region containing the FER-1 element served as an EpRE of the ferritin H gene. To test this possibility, several hGH reporter constructs with deletions in the 5′ ferritin H gene were constructed (Fig. 2C) and transfected into Hepa1-6 cells. The transfected cells were then treated with H2O2 or t-BHQ for 8 to 10 h and subjected to RNase protection assays. −4.8kb, which has an intact 4.8-kb 5′-flanking region of the ferritin H gene, reproducibly exhibited approximately twofold induction of hGH reporter gene expression in Hepa1-6 cells (Fig. 2D). In contrast, −4.0 kb, deleted in a 0.8-kb region containing the FER-1 element (Fig. 2C), failed to induce hGH expression in response to H2O2 or t-BHQ (Fig. 2D). These results suggest that an EpRE of the ferritin H gene is located between 4.8 and 4.0 kb upstream from the transcription initiation site.
To further elucidate essential elements in the electrophile response region, three additional reporter plasmids with stepwise deletions in the 0.1-kb region containing FER-1 were constructed (Fig. 2C) and examined for their response to H2O2 or t-BHQ. −4.0kbAP1+α, which has a complete consensus AP1/NF-E2 binding sequence and an additional 12 bp upstream sequence but does not contain FER-1, acquired a partial restoration of response to H2O2 and t-BHQ (Fig. 2D). In contrast, when the complete FER-1 element was included (−4.0kbAP1+δ), the basal enhancer activity was restored and was further activated by H2O2 or t-BHQ (Fig. 2D). −4.0kbAP1+ɛ, which contains an additional 23 bp proximal to the FER-1 element, exhibited a similar restoration of response to H2O2 or t-BHQ (Fig. 2D). These results suggest that the functional EpRE of the ferritin H gene is located in the region between −4.03 and −4.13 kb, containing both the FER-1 element and a proximal AP1/NF-E2 site (Fig. 2C).
To verify these results, we dissected elements of the electrophile response region (between −4.03 and −4.13 kb) of the ferritin H gene and inserted them into a minimum ferritin H-hGH construct lacking the ferritin H 5′-flanking region between −0.32 and −4.8 kb (−0.32kbFH-hGH [Fig. 3]). −0.32kbFH-hGH did not show induction of hGH transcription in response to t-BHQ (Fig. 3). This is consistent with the results shown in Fig. 2D because −0.32kbFH-hGH does not contain FER-1 or AP1/NF-E2 (Fig. 3). Five different elements were inserted into −0.32kbFH-hGH (Fig. 3), and each insertion construct was transfected into Hepa1-6 cells and tested for restoration of hGH mRNA induction in response to t-BHQ. As shown in Fig. 3, insertion of 47 bp containing the AP1/NF-E2 site and flanking sequences or insertion of the 40 bp DNA that contains FER-1 alone failed to confer transcriptional activation by t-BHQ. Even insertion of 59 bp of DNA containing FER-1 and the consensus AP1/NF-E2 site was not able to restore t-BHQ responsiveness (Fig. 3). In contrast, insertion of the 107-bp entire electrophile response region or the 75 bp of DNA containing FER-1 and AP1/NF-E2 sites plus 8 and 11 additional 5′ and 3′ nucleotides, respectively, completely conferred induction of hGH mRNA in response to t-BHQ (Fig. 3).
The 75-bp EpRE in the ferritin H gene contains two copies of a canonical electrophile response sequence arranged in opposing orientations.
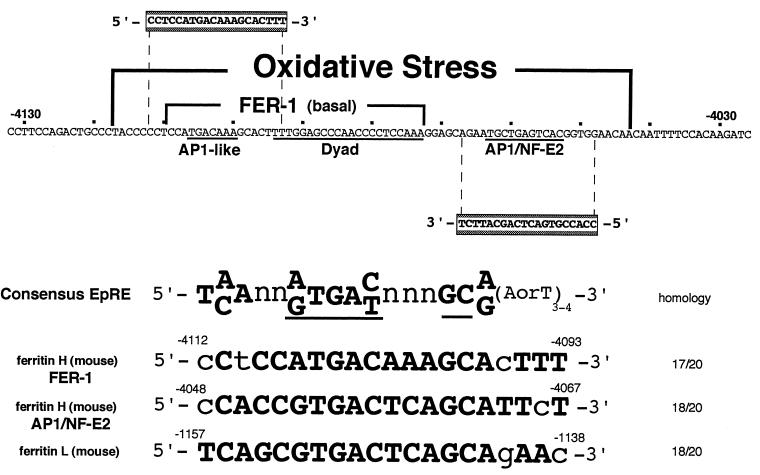
The DNA sequence of the 75-bp EpRE of the mouse ferritin H gene identified in this study is shown in Fig. 4. It contains the basal enhancer element of the ferritin H gene, FER-1, and a proximal consensus AP1/NF-E2 element. Sequence comparison between the 75-bp region of the ferritin H gene and the consensus electrophile response sequence recently reported by Wasserman and Fahl (47) reveal that both the AP1-like element of FER-1 and the AP1/NF-E2 site of the ferritin H gene contain consensus EpRE motifs, arranged in opposite orientations (Fig. 4). Both the ferritin H EpREs and the ferritin L EpRE identified recently (47) conform completely to the core sequence of the consensus EpRE (47). The overall homology of the ferritin H EpREs to the full EpRE consensus was 17 or 18 nucleotides out of 20 (Fig. 4).
FIG. 4.
The FER-1 element and the proximal AP1/NF-E2 site contain a canonical electrophile response sequence. The 75-bp cis-acting element responsible for transcriptional activation of the mouse ferritin H gene by t-BHQ contains the FER-1 element and an AP1/NF-E2 site, whose DNA sequence is shown. The consensus EpRE sequence (47) conserved in this region is shaded. The two EpREs of the ferritin H gene and the EpRE of the ferritin L gene (47) were aligned with the consensus EpRE sequence, in which the core sequence (36) is underlined.
Hydrogen peroxide rapidly but transiently activates IRP-1.
Ferritin mRNA is subject to translational control by IRPs, proteins that bind to the IRE in the 5′ UTR of ferritin mRNA and inhibit its translation. To assess the effect of oxidants on the IRP proteins, we examined the binding of IRP proteins to the IRE under the same experimental conditions as those used above. As shown in Fig. 5, IRP-1 was activated at 0.5 h after H2O2 treatment and remained activated until 2 h in BNL CL.2 mouse liver cells. However, the increased IRP-1 binding was transient, and after 4 h a decline in IRP-IRE interaction to below basal levels was observed (Fig. 5). In contrast to IRP-1, IRP-2 binding to IRE was relatively low and its activation was more modest (Fig. 5), consistent with published results (29). The IRP-1–IRE interaction in the presence of β-mercaptoethanol was unchanged by H2O2 (Fig. 5), suggesting that the dynamic alteration in the IRP-IRE interaction following H2O2 treatment is regulated at a posttranslational level.
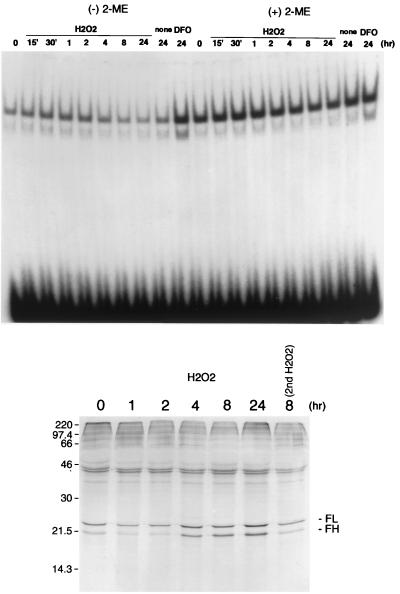
FIG. 5.
Activation of IRP-1 and inhibition of ferritin protein synthesis precede translation of the newly synthesized ferritin mRNA. (Top) BNL CL.2 cells were treated with 750 μM H2O2 or 100 μM desferrioxamine (DFO) for the times indicated, and cytosolic cell extracts were prepared. A 32P-labeled RNA probe for IRE (0.5 × 104 to 1.0 × 104 cpm) was incubated with 1 μg of cell extract at 25°C for 30 min in the presence or absence of β-mercaptoethanol (2-ME) and then subjected to separation on 5% polyacrylamide gels as described in Materials and Methods. (Bottom) BNL CL.2 cells were treated with 750 μM H2O2 for the times indicated and labeled with Tran35S-label for the next 1 h without stimulants. Then 5 × 106 cpm of trichloroacetic acid-insoluble counts from each cell lysate was immunoprecipitated with anti-ferritin antibody and analyzed on a 15% polyacrylamide gel.
Translation of newly synthesized ferritin transcripts is delayed but not prevented following exposure to hydrogen peroxide.
At the level of ferritin protein synthesis, effects of oxidants on the IRP and ferritin mRNA converge. To determine the ultimate effect of modulation of ferritin mRNA and IRP by prooxidants, we measured ferritin protein synthesis in cells treated with hydrogen peroxide. As shown in Fig. 5, consistent with transient IRP activation (0.5 to 2 h) prior to the rise of the ferritin mRNA level (4 to 24 h) seen in cells treated with hydrogen peroxide, ferritin protein synthesis exhibited an initial decline followed by a rise. These results suggest that translation of newly synthesized ferritin mRNA is delayed by the initial activation of IRP-1 activity in response to hydrogen peroxide. When the cells were treated again with H2O2 at 8 h after the first H2O2 stimulation, ferritin protein synthesis was inhibited (Fig. 5, compare lanes 8 hr and 8 hr 2nd H2O2). This suggests that the effects of oxidative stress on ferritin synthesis are reversible. The cumulative effects of oxidative stress on ferritin transcription and translation are depicted in Fig. 6.
FIG. 6.
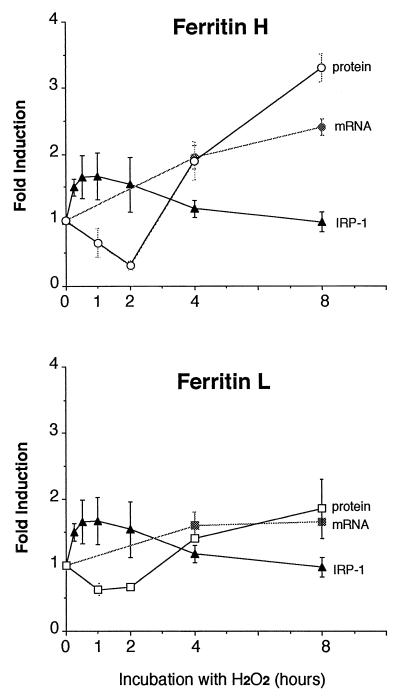
Transcriptional and translational regulation of ferritin. The cumulative effects of hydrogen peroxide on ferritin H and L mRNA synthesis, IRP-1 activity, and ferritin H and L protein synthesis in BNL CL.2 cells are summarized. Ferritin protein synthesis was measured by immunoprecipitation as shown in Fig. 5; mRNA levels were measured by Northern blotting as shown in Fig. 1; and IRP activation was measured by gel mobility shift assays as shown in Fig. 5. The results were quantitated by PhosphoImager analysis, with the signal intensities at time zero being defined as 1.0. Results of two independent experiments (protein and mRNA) or three independent experiments (IRP-1) and standard errors are shown.
DISCUSSION
Transcriptional regulation of the mouse ferritin H gene by oxidative stress.
The results presented in this study demonstrate that ferritin is subject to both transcriptional and translational regulation by oxidative stress and elucidate the mechanism of transcriptional control of ferritin by oxidants. The prooxidants H2O2 and t-BHQ transcriptionally activate the mouse ferritin H gene via a 75-bp region located 4.1 kb 5′ to the transcription initiation site. The 75-bp EpRE contains the FER-1 element we previously identified as a basal enhancer of the ferritin H gene. This result is consistent with the fact that an EpRE generally serves as a basal enhancer element (14). Our results further indicate that a proximal AP1/NF-E2 site is required for full EpRE activity (Fig. 3). Thus, the EpRE of the ferritin H gene is a composite sequence composed of three elements: an AP1-like sequence, an Sp1-like dyad element (45), and an AP1/NF-E2 sequence (Fig. 4). The requirement for a second AP1-like sequence for full functional EpRE activity has been previously reported for human (48) and rat (6) NADP(H):quinone reductase, rat GST-pi (24), and rat and mouse GST-Ya (7, 31, 37) genes. Consistent with these functional similarities, we found that both the AP1-like element of FER-1 and the AP1/NF-E2 site of the ferritin H gene have striking homology to the EpRE consensus sequence reported by Wasserman and Fahl (47) (Fig. 4).
Prooxidant conditions, including treatment with hydrogen peroxide, activate NFκB (39). We and others have suggested that ferritin may function as a cytoprotective protein, whose role in sequestering “free” iron minimizes damage from a variety of oxidative stresses including tumor necrosis factor (TNF) and hydrogen peroxide (2, 4, 41). Our previous experiments have shown that induction of ferritin H in response to TNF is mediated by a tandem NF-κB consensus and NF-κB-like site (16). However, NF-κB does not appear to mediate the response of ferritin H to hydrogen peroxide, since deletion of the upstream region of the ferritin H promoter containing the NF-κB sites had no effect on induction of ferritin by oxidants (Fig. 2). Thus, the response of ferritin to TNF differs from its response to prooxidants: the response to TNF is restricted to the H subunit of ferritin, whereas prooxidant challenge induces both ferritin H and L and the responses are mediated by differing cis-acting elements. These results suggest that multiple independent pathways exist which converge in the augmentation of ferritin synthesis in response to various forms of oxidative insult.
The presence of an EpRE (also termed antioxidant response element) in the mouse ferritin L gene has recently been reported by Wasserman and Fahl as a result of a screen of the GenBank database using a consensus sequence (47). The EpRE in ferritin L was further demonstrated to be functional in reporter assays (47). Since the core sequence of the EpREs identified in ferritin H and L genes is well conserved (Fig. 4), we speculate that a common mechanism may be involved in the coordinate activation of the ferritin H and L EpRE. Functionally, these results reveal that oxidative stress leads to a coordinate increase in the levels of both ferritin H and L subunits, although the increase in ferritin H appeared somewhat greater than that in ferritin L (Fig. 6). The more pronounced activation of ferritin H transcription may be due to a cooperative function of the two bidirectional EpRE motifs in the ferritin H gene (Fig. 4) compared to the single EpRE motif in the ferritin L gene.
The FER-1 component of the EpRE is a composite element of an AP1-like sequence followed by a dyad symmetry, both of which are essential for maximum enhancer activity (42, 45). Previously, we identified the FER-1 element as a target sequence for transcriptional repression of the ferritin H gene by the adenovirus E1A oncogene (42, 44). These results suggest that the EpRE may also be a target for transcriptional repression of the ferritin H gene by E1A. Indeed, our recent studies indicate that E1A inhibits the transcriptional activation of ferritin H gene by H2O2 or t-BHQ and sensitizes cells to cytotoxicity of these oxidative stress inducers (25).
Translational regulation of ferritin by oxidative stress.
Ferritin is subject to translational control by the IRE binding proteins IRP-1 and IRP-2. When activated, these proteins can bind to the IRE in the 5′ UTR of ferritin H or L mRNA and inhibit translation of the mRNA. Importantly, it has been suggested that the activity of IRP-1 is modulated not only by intracellular iron status but also by reactive oxygen species. Thus, to determine the functional consequences of EpRE-mediated ferritin transcription in response to oxidants, we performed time course experiments in which IRP binding activity was measured following exposure of mouse liver cells to oxidative stress. As shown in Fig. 5, IRP binding to the IRE was stimulated rapidly following exposure to hydrogen peroxide. Binding activity peaked at 1 h and remained elevated until 2 h after H2O2 treatment, consistent with results reported by others (26). However, we found in this study that the enhanced IRP-1 binding to IRE gradually declined to below basal levels at later time points, i.e., 8 and 24 h after H2O2 treatment (Fig. 5). Hence, the modulation of IRP activity by H2O2 is reversible. Since transferrin receptor mRNA is stabilized by activated IRP (12), the decline of IRP-1 binding at later time points may, at least in part, be attributable to an increase in transferrin receptor display mediated by the initial wave of enhanced IRP activity. Enhanced iron uptake resulting from this increase in transferrin receptor may in turn function to downregulate IRP activity in a negative feedback loop. Alternatively, regulation of IRP activity by phosphorylation and dephosphorylation may be involved (27). These possibilities are currently under investigation.
Divergent observations on the regulation of IRP by oxidative stress have been reported, suggesting that it can be both activated (21, 27) and inactivated (2) in response to oxidative stress. Results presented here offer a model that partially reconciles these apparently disparate results by suggesting that both occur but do so in a temporal sequence (Fig. 5 and 6). Thus, in cells treated with hydrogen peroxide, we observed an initial activation of the IRP that was sustained for at least 2 h, after which the IRP activity gradually declined. This indicates that the elevated levels of ferritin mRNA induced by oxidative stress gradually become available for translation.
Effects of oxidative stress on ferritin protein synthesis reflect the combined contributions of transcriptional and translational regulatory mechanisms. In accord with our observation that IRP activity is transiently activated following exposure of cells to hydrogen peroxide, we observed an inhibition of ferritin synthesis that was sustained for at least 2 h after exposure to H2O2 (Fig. 5). However, reflecting the rise in ferritin mRNA levels and the gradual inactivation of the IRP, at 4 h after hydrogen peroxide treatment the synthesis of ferritin began to rise above basal levels and was further increased until 24 h (Fig. 5). Interestingly, when a second H2O2 treatment was carried out 8 h after the first H2O2 treatment, the enhanced levels of ferritin translation seen at 8 h were completely abolished, perhaps as a consequence of IRP reactivation (Fig. 5).
Physiologically, it is not clear why treatments with prooxidants should activate the IRP, since this leads to a transient decline in ferritin levels and an attendant decrease in the ability to buffer iron available for participation in oxygen free-radical formation. However, agents that trigger the antioxidant response are often weak oxidants, which may function by generating a level of oxidative stress sufficient to induce the transcription of cytoprotective enzymes but insufficient to elicit major cell damage. By transiently exacerbating oxidative stress, activation of the IRP may contribute to the transient prooxidant state required to trigger a full antioxidant response.
Based on its induction by agents that induce the antioxidant response (H2O2, t-BHQ, and β-NF), as well as its EpRE-dependent mechanism, our results identify ferritin as a constituent of the antioxidant response. Ferritin, with its ability to oxidize and sequester intracellular iron in an internal mineral core, limits the levels of catalytically available iron (30, 35). Since iron is an important contributor to oxygen free-radical toxicity, the inclusion of ferritin within the family of genes that function to reduce intracellular levels of toxic electrophilic species, such as GST, NAD(P)H quinone reductase, and γ-glutamylcysteine synthetase, is concordant with a view of ferritin as a critical cytoprotective protein that constitutes an integral part of the antioxidant response.
ACKNOWLEDGMENTS
We are grateful to Elizabeth Theil and Yaohuang Ke for detailed advice on IRP binding assays.
This work was supported by grants DK-42412 and DK42412-09S1 (to S.P.W.) from the National Institutes of Health. Phosphoimaging analysis was performed in a facility supported by grant CA12197 from the National Institutes of Health and grant 9510-IDG-1006 from the North Carolina Biotechnology Center.
REFERENCES
- 1.Alam J, Camhi S, Choi A M. Identification of a second region upstream of the mouse heme oxygenase-1 gene that functions as a basal level and inducer-dependent transcription enhancer. J Biol Chem. 1995;270:11977–11984. doi: 10.1074/jbc.270.20.11977. [DOI] [PubMed] [Google Scholar]
- 2.Cairo G, Tacchini L, Pogliaghi G, Anzon E, Tomasi A, Bernelli-Zazzera A. Induction of ferritin synthesis by oxidative stress. Transcriptional and post-transcriptional regulation by expansion of the “free” iron pool. J Biol Chem. 1995;270:700–703. doi: 10.1074/jbc.270.2.700. [DOI] [PubMed] [Google Scholar]
- 3.Cairo G, Vezzoni P, Bardella L, Schiaffonati L, Rappocciolo E, Levi S, Arosio P, Bernelli-Zazzera A. Regulation of ferritin synthesis in malignant and non-malignant lymphoid cells. Biochem Biophys Res Commun. 1986;139:652–657. doi: 10.1016/s0006-291x(86)80040-7. [DOI] [PubMed] [Google Scholar]
- 4.Cermak J, Balla J, Jacob H S, Balla G, Enright H, Nath K, Vercellotti G M. Tumor cell heme uptake induces ferritin synthesis resulting in altered oxidant sensitivity: possible role in chemotherapy efficacy. Cancer Res. 1993;53:5308–5313. [PubMed] [Google Scholar]
- 5.Dalton T, Palmiter R D, Andrews G K. Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic Acids Res. 1994;22:5016–5023. doi: 10.1093/nar/22.23.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favreau L V, Pickett C B. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J Biol Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- 7.Friling R S, Bergelson S, Daniel V. Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc Natl Acad Sci USA. 1992;89:668–672. doi: 10.1073/pnas.89.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray N K, Quick S, Goossen B, Constable A, Hirling H, Kuhn L C, Hentze M W. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur J Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 9.Guner G, Kirkali G, Yenisey C, Tore I R. Cytosol and serum ferritin in breast carcinoma. Cancer Lett. 1992;67:103–112. doi: 10.1016/0304-3835(92)90132-f. [DOI] [PubMed] [Google Scholar]
- 10.Harrison P M, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 11.Hentze M W, Caughman S W, Casey J L, Koeller D M, Rouault T A, Harford J B, Klausner R D. A model for the structure and functions of iron-responsive elements. Gene. 1988;72:201–208. doi: 10.1016/0378-1119(88)90145-x. [DOI] [PubMed] [Google Scholar]
- 12.Hentze M W, Kuhn L C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogg N. Free radicals in disease. Semin Reprod Endocrinol. 1998;16:241–248. doi: 10.1055/s-2007-1016284. [DOI] [PubMed] [Google Scholar]
- 14.Jaiswal A K. Antioxidant response element. Biochem Pharmacol. 1994;48:439–444. doi: 10.1016/0006-2952(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 15.Ke Y, Wu J, Leibold E A, Walden W E, Theil E C. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J Biol Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- 16.Kwak E L, Larochelle D A, Beaumont C, Torti S V, Torti F M. Role for NF-kappa B in the regulation of ferritin H by tumor necrosis factor-alpha. J Biol Chem. 1995;270:15285–15293. doi: 10.1074/jbc.270.25.15285. [DOI] [PubMed] [Google Scholar]
- 17.Kwak E L, Torti S V, Torti F M. Murine ferritin heavy chain: isolation and characterization of a functional gene. Gene. 1990;94:255–261. doi: 10.1016/0378-1119(90)90396-9. [DOI] [PubMed] [Google Scholar]
- 18.Leibold E A, Munro H N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Jaiswal A K. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of AP1 binding site contained within human antioxidant response element. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 20.Loft S, Poulsen H E. Cancer risk and oxidative DNA damage in man. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 21.Martins E A, Robalinho R L, Meneghini R. Oxidative stress induces activation of a cytosolic protein responsible for control of iron uptake. Arch Biochem Biophys. 1995;316:128–134. doi: 10.1006/abbi.1995.1019. [DOI] [PubMed] [Google Scholar]
- 22.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulcahy R T, Wartman M A, Bailey H H, Gipp J J. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 24.Okuda A, Imagawa M, Maeda Y, Sakai M, Muramatsu M. Structural and functional analysis of an enhancer GPEI having a phorbol 12-O-tetradecanoate 13-acetate responsive element-like sequence found in the rat glutathione transferase P gene. J Biol Chem. 1989;264:16919–16926. [PubMed] [Google Scholar]
- 25.Orino K, Tsuji Y, Torti F M, Torti S V. Adenovirus E1A blocks oxidant-dependent ferritin induction and sensitizes cells to pro-oxidant cytotoxicity. FEBS Lett. 1999;461:334–338. doi: 10.1016/s0014-5793(99)01443-x. [DOI] [PubMed] [Google Scholar]
- 26.Pantopoulos K, Hentze M W. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 1995;14:2917–2924. doi: 10.1002/j.1460-2075.1995.tb07291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantopoulos K, Hentze M W. Activation of iron regulatory protein-1 by oxidative stress in vitro. Proc Natl Acad Sci USA. 1998;95:10559–10563. doi: 10.1073/pnas.95.18.10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantopoulos K, Mueller S, Atzberger A, Ansorge W, Stremmel W, Hentze M W. Differences in the regulation of iron regulatory protein-1 (IRP-1) by extra- and intracellular oxidative stress. J Biol Chem. 1997;272:9802–9808. doi: 10.1074/jbc.272.15.9802. [DOI] [PubMed] [Google Scholar]
- 29.Pantopoulos K, Weiss G, Hentze M W. Nitric oxide and oxidative stress (H2O2) control mammalian iron metabolism by different pathways. Mol Cell Biol. 1996;16:3781–3788. doi: 10.1128/mcb.16.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picard V, Epsztejn S, Santambrogio P, Cabantchik Z I, Beaumont C. Role of ferritin in the control of the labile iron pool in murine erythroleukemia cells. J Biol Chem. 1998;273:15382–15386. doi: 10.1074/jbc.273.25.15382. [DOI] [PubMed] [Google Scholar]
- 31.Prestera T, Holtzclaw W D, Zhang Y, Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primiano T, Kensler T W, Kuppusamy P, Zweier J L, Sutter T R. Induction of hepatic heme oxygenase-1 and ferritin in rats by cancer chemopreventive dithiolethiones. Carcinogenesis. 1996;17:2291–2296. doi: 10.1093/carcin/17.11.2291. [DOI] [PubMed] [Google Scholar]
- 33.Rouault T A, Hentze M W, Caughman S W, Harford J B, Klausner R D. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988;241:1207–1210. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- 34.Rowlands J C, Gustafsson J A. Aryl hydrocarbon receptor-mediated signal transduction. Crit Rev Toxicol. 1997;27:109–134. doi: 10.3109/10408449709021615. [DOI] [PubMed] [Google Scholar]
- 35.Rucker P, Torti F M, Torti S V. Role of H and L subunits in mouse ferritin. J Biol Chem. 1996;271:33352–33357. doi: 10.1074/jbc.271.52.33352. [DOI] [PubMed] [Google Scholar]
- 36.Rushmore T H, Morton M R, Pickett C B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 37.Rushmore T H, Pickett C B. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 38.Ryan T P, Aust S D. The role of iron in oxygen-mediated toxicities. Crit Rev Toxicol. 1992;22:119–141. doi: 10.3109/10408449209146308. [DOI] [PubMed] [Google Scholar]
- 39.Schreck R, Rieber P, Baeuerle P A. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theil E C. The ferritin family of iron storage proteins. Adv Enzymol Relat Areas Mol Biol. 1990;63:421–449. doi: 10.1002/9780470123096.ch7. [DOI] [PubMed] [Google Scholar]
- 41.Torti S V, Kwak E L, Miller S C, Miller L L, Ringold G M, Myambo K B, Young A P, Torti F M. The molecular cloning and characterization of murine ferritin heavy chain, a tumor necrosis factor-inducible gene. J Biol Chem. 1988;263:12638–12644. [PubMed] [Google Scholar]
- 42.Tsuji Y, Akebi N, Lam T K, Nakabeppu Y, Torti S V, Torti F M. FER-1, an enhancer of the ferritin H gene and a target of E1A-mediated transcriptional repression. Mol Cell Biol. 1995;15:5152–5164. doi: 10.1128/mcb.15.9.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuji Y, Miller L L, Miller S C, Torti S V, Torti F M. Tumor necrosis factor-alpha and interleukin 1-alpha regulate transferrin receptor in human diploid fibroblasts. Relationship to the induction of ferritin heavy chain. J Biol Chem. 1991;266:7257–7261. [PubMed] [Google Scholar]
- 44.Tsuji Y, Moran E, Torti S V, Torti F M. Transcriptional regulation of the mouse ferritin H gene. Involvement of p300/CBP adaptor proteins in FER-1 enhancer activity. J Biol Chem. 1999;274:7501–7507. doi: 10.1074/jbc.274.11.7501. [DOI] [PubMed] [Google Scholar]
- 45.Tsuji Y, Torti S V, Torti F M. Activation of the ferritin H enhancer, FER-1, by the cooperative action of members of the AP1 and Sp1 transcription factor families. J Biol Chem. 1998;273:2984–2992. doi: 10.1074/jbc.273.5.2984. [DOI] [PubMed] [Google Scholar]
- 46.Vaughn C B, Weinstein R, Bond B, Rice R, Vaughn R W, McKendrick A, Ayad G, Rockwell M A, Rocchio R. Ferritin content in human cancerous and noncancerous colonic tissue. Cancer Investig. 1987;5:7–10. doi: 10.3109/07357908709020300. [DOI] [PubMed] [Google Scholar]
- 47.Wasserman W W, Fahl W E. Functional antioxidant responsive elements. Proc Natl Acad Sci USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie T, Belinsky M, Xu Y, Jaiswal A K. ARE- and TRE-mediated regulation of gene expression. Response to xenobiotics and antioxidants. J Biol Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]