Summary
Teredinibacter turnerae is an intracellular bacterial symbiont in the gills of wood-eating shipworms, where it is proposed to use antibiotics to defend itself and its animal host. Several biosynthetic gene clusters are conserved in T. turnerae and their host shipworms around the world, implying that they encode defensive compounds. Here, we describe turnercyclamycins, lipopeptide antibiotics encoded in the genomes of all sequenced T. turnerae strains. Turnercyclamycins are bactericidal against challenging Gram-negative pathogens, including colistin-resistant Acinetobacter baumannii. Phenotypic screening identified the outer membrane as the likely target. Turnercyclamycins and colistin operate by similar cellular, although not necessarily molecular, mechanisms, but turnercyclamycins kill colistin-resistant A. baumannii, potentially filling an urgent unmet clinical need. Thus, by exploring environments that select for the properties we require, we harvested the fruits of evolution to discover compounds with potential to target unmet health needs. Investigating the symbionts of shipworms is a powerful example of this principle.
Keywords: Antibiotics, Natural Products, Nonribosomal Peptide, Symbiosis, Chemical Biology, Acinetobacter, Lipopeptide
eTOC Blurb
The symbiotic bacteria associated with marine shipworms are prolific producers of bioactive metabolites Miller et al. use an ecology-guided approach to isolate the turnercyclamycins, proposed to be important for this association. Turnercyclamycins are described as a group of lipopeptide antibiotics with selective activity against gram-negative pathogens, including drug resistant Acinetobacter.
Graphical Abstract
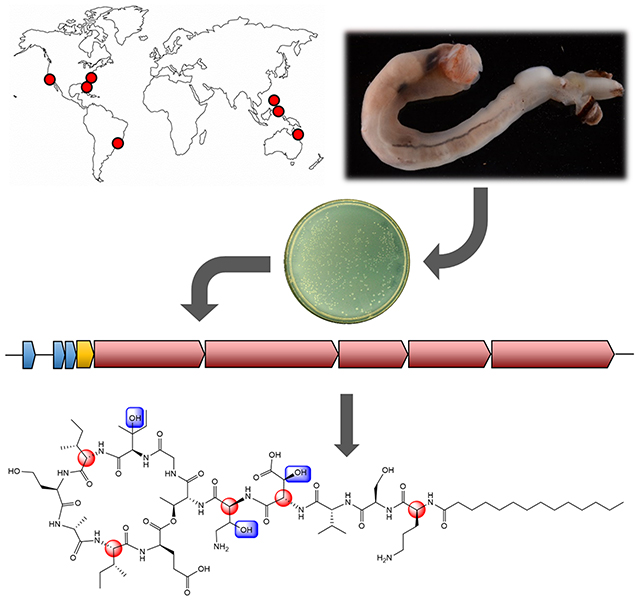
Introduction
Wood eating shipworms (Teredinidae), a family of marine bivalve mollusks, have a close symbiotic relationship with the intracellular bacteria living in their gills. These bacteria, largely represented by the gammaproteobacterial genus Teredinibacter, support their hosts both through the fixation of atmospheric nitrogen and by contributing cellulolytic enzymes necessary for wood degradation (Distel et al., 2002; Ekborg et al., 2007; Lechene et al., 2007). While these microbes are located in bacteriocytes within the gill filaments of the shipworm, their enzymes are transported to and can be detected in the cecum, where wood is digested (O’Connor et al., 2014). Interestingly, this organ has been shown to be nearly devoid of bacterial inhabitants, despite a presumed abundance of nutritionally accessible carbon released by digested lignocellulose (Betcher et al., 2012). Thus, in contrast to other wood-eating symbioses wherein the microbes consume energy rich glucose and secrete lower-energy metabolites, this strategy allows the host shipworm to directly consume glucose by hosting the bacteria in the gills and not in the digestive organ. In order to make this remarkable lifestyle possible, it has been hypothesized that shipworm symbionts secrete potent antibiotics into the host, where they would have to be compatible with animal metabolism (Elshahawi et al., 2013; Yang et al., 2009). If so, the symbiosis might provide a ready source of new antibiotics to target the rise of multidrug resistance, a crucial problem in human health. Typically, intracellular symbiotic bacteria are difficult to cultivate, making it hard to discover the important antibiotics that result. However, a number of Teredinibacter turnerae strains have been successfully cultured, facilitating genome sequencing and large-scale fermentation for natural product isolation. Significantly, some of the potent compounds isolated from Teredinibacter in culture are also found in host shipworm animals, where they presumably help to keep out competing microbes (Han et al., 2013; Elshahawi et al., 2013). The compounds known so far include tartrolon, an exceptionally potent antiparasitic compound (Elshahawi et al., 2013; O’Connor et al., 2020), and a siderophore, turnerbactin (Han et al., 2013). However, the key antibacterial compounds, which would potentially be responsible for shaping the shipworm gill communities and the axenic cecum, have yet to be described.
To more rapidly hone in on ecologically important antibacterial compounds, we analyzed the genomes of cultivated symbiont strains and the metagenomes of shipworm gills, focusing on antibiotic-like biosynthetic gene clusters (BGCs) (Altamia et al., 2020). These symbionts and hosts were collected from around the world, including tropical and temperate waters, implying that highly conserved compounds are important for the symbiotic relationship. Supporting this idea, BGCs encoding the important antiparasitic tartrolon and siderophore turnerbactin were found in the metagenomes of all Teredinibacter-containing shipworm gills, as well as in all cultivated Teredinibacter genomes (Altamia et al., 2020). In addition, a small number of uncharacterized BGCs were observed which we suspected might encode antibacterial compounds. One of these, BGC_1, was extremely widespread in shipworms, encoding a nonribosomal peptide synthetase (NRPS) that had features leading us to suspect that it encoded an antibiotic lipopeptide.
Here, we describe the identification of the chemical products of BGC_1 as potent antibacterial agents. The turnercyclamycins represent a group of lipopeptides that kill several challenging Gram-negative bacterial species, including Acinetobacter baumannii, which has been designated by the World Health Organization as one of the most successful and serious of the ESKAPE pathogens (Boucher et al., 2009), and yet are not toxic to mammalian cells. Colistin (polymyxin E) is the last-line therapy for otherwise lethal A. baumannii infections, but increasing instances of resistance threaten to leave the disease without viable treatment options. Here, we show that turnercyclamycins target the outer membrane like colistin, but they retain efficacy against colistin-resistant Acinetobacter, making them promising leads for antimicrobial development.
The turnercyclamycins were not previously observed in T. turnerae cultures in part because the crude mixture of turnercyclamycins is nearly insoluble in common solvents used in natural products purification methods, although they are well behaved when purified. Bioinformatics analysis of the tur pathway to turnercyclamcyins reveals that the pathway is very highly conserved across shipworm isolates and gills, so that the same or a very similar suite of compounds should be found in T. turnerae-containing shipworms globally. These results reinforce the utility of symbiosis and chemical ecology in providing new solutions to multidrug resistant infections.
Results
Conservation of NRPS gene cluster across Teredinibacter turnerae
A previous systematic analysis of shipworm symbiont isolate genomes and animal gill metagenomes identified a NRPS BGC, where it was defined as “GCF_1”, as being highly conserved in T. turnerae, and portions of the cluster were detected in all T. turnerae-containing shipworms (Altamia et al., 2020). Here, multigene BLAST analysis of 8 sequenced T. turnerae strains found this cluster to be ubiquitous (Figure 1A); however, the region including turD and turE appeared to be disrupted in 6 of these strains. In correctly assembled sequences, this region contained two repeats of 6k bp in length that were 98% identical, while in the misassembled sequences this region was scrambled. Therefore, it is likely that this is a result of bioinformatic assembly error, and the gene cluster is intact and functional in all strains. All of the intact NRPSs were further compared by BLASTp, and it was found that each megasynthetase was greater than 85% similar to all of the other corresponding proteins from the homologous clusters, demonstrating a very high degree of conservation (Figure 1B).
Figure 1.
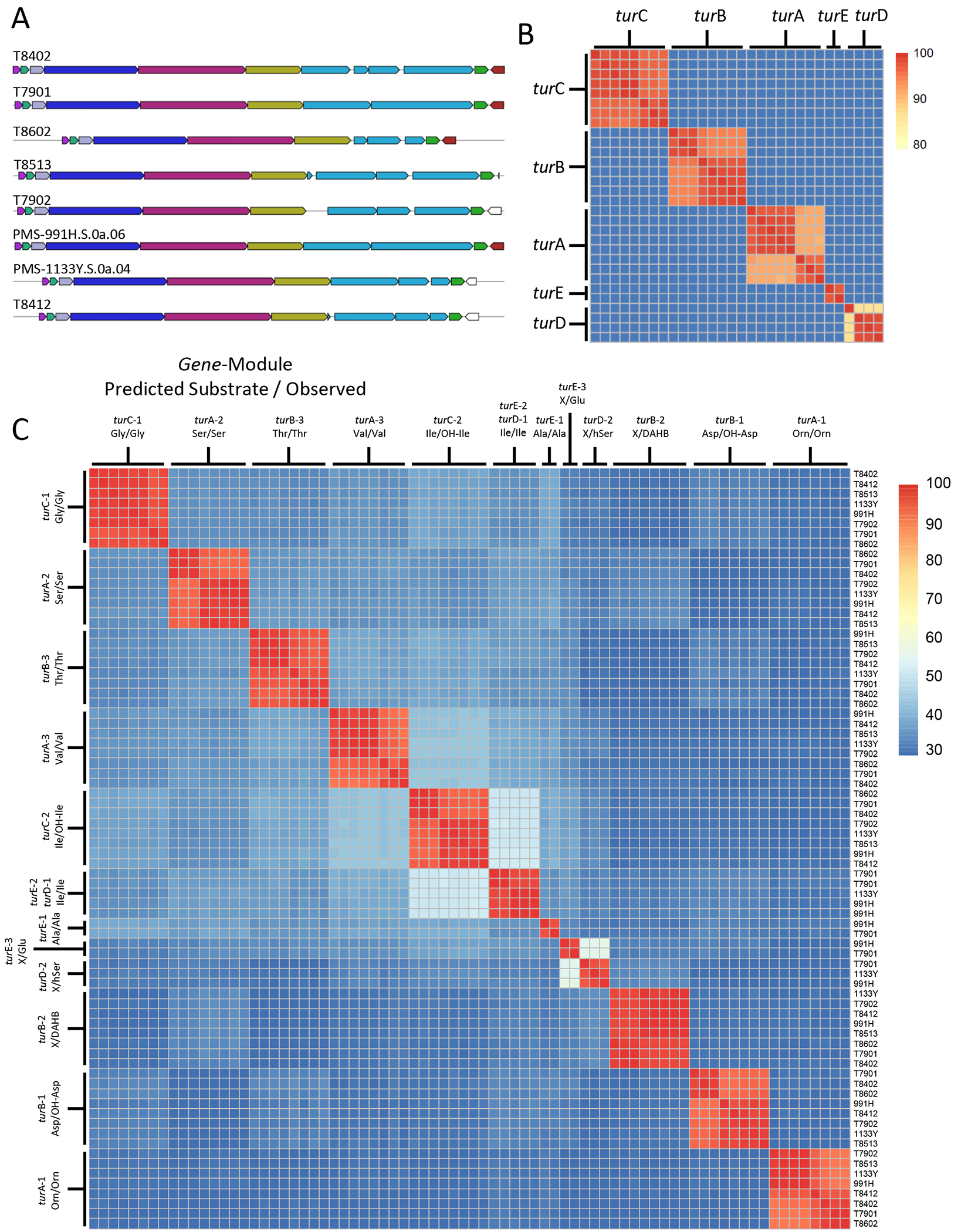
Conservation of the turnercyclamycin biosynthetic gene cluster. A) Multigene BLAST identified a conserved NRPS cluster present in eight sequenced T. turnerae strains. B) The proteins encoded by each NRPS gene are greater than 85% identical when compared to corresponding proteins in all eight clusters. C) BLASTp identity comparison of extracted adenylation domains from all identified tur gene clusters. Corresponding adenylation domains retain greater than 90% identity, indicating that the peptide backbone of the product is likely highly conserved across all T. turnerae strains. Note the much higher sequence conservation of TurE and TurD adenylation domains in comparison to the rest of the TurE/TurD proteins. In addition, within single NRPS proteins, there is very little similarity between NRPS adenylation domains, even when they are activating very similar or identical amino acids (such as 2 x Ile and OH-Ile).
In the T. turnerae T7901 genome, the five genes within this cluster include turA (3 modules, 11.2 kbp), turB (3 modules, 12.8 kbp), turC (2 modules, 6.6 kbp), turD (2 modules, 8.1 kbp), and turE (3 modules, 12.1 kbp). Each module contains a condensation (C), adenylation (A), and thiolation (T) domain, and 5 of the modules also contain epimerization domains. The first domain begins with a so called C-starter domain, which are known to acylate the initial amino acid in a process known as lipoinitiation (Rausch et al., 2007). The final module terminates in a thioesterase domain, which is responsible for product release, often by means of macrocyclization (Keating et al., 2001). This domain architecture indicates that the product of the tur cluster is a cyclic lipopeptide, a known class of antibacterial scaffolds.
Often, NRPS genes are naturally combinatorialized to create families of compounds that consist of structural analogs distributed throughout related producing organisms (Zan et al., 2019). In order to look for the potential of variants with divergent peptide backbones, the adenylation domains for the cluster in each of the 8 sequenced strains were compared by BLASTp. In some of the misassembled sequences, the A domains in turD and turE were not intact and were thus not included in the analysis. The results show that tur is an exceptionally well-conserved cluster, with each A domain retaining greater than 90% identity to all of the analogous domains across all 8 strains (Figure 1C). The substrate prediction, as determined by AntiSmash 5.0, was also consistent for each domain (Blin et al., 2019). Importantly, the strains included in this analysis were isolated from a variety of shipworm genera and species collected in disparate locations around the world, indicating that this gene cluster, and thus its biosynthetic product, are well preserved, and thus likely important to the association between host and symbiont (Table S4).
Based upon the ubiquitous nature of this potential antibiotic, which is proposed to function in the symbiosis to defend the host and symbiont from other bacteria, we prioritized discovering the products of the tur pathway, seeking a lipopeptide containing 13 amino acids in the cultures of T. turnerae T7901.
Isolation and Planar Structure Elucidation
The liquid culture of T. turnerae T7901 (6.6L) was extracted and subjected to a series of partitions. Initially, the expected peptides were not observed in crude extracts, and indeed the crude extract was not toxic to Acinetobacter. However, after several trials we observed a particularly robust boundary layer between the water and ethyl acetate fractions, which was enriched in a series of lipopeptides. Semi-preparative HPLC yielded two major compounds 1 and 2, along with minor analogs 3 and 4.
Turnercyclamycin A (1) had a molecular formula of C71H125N15O24 based upon high resolution mass spectrometry ([M+2H]2+ m/z786.9618), inherent in which are 17 degrees of unsaturation. The 1H and 13C spectra were consistent with a lipopeptide based on numerous amide NH resonances between δH 7.6-8.5, alpha proton signals from δH 4.03-5.05, and the presence of a large methylene envelope centered at δH 1.23 (Figure S1). The gHSQC experiment revealed the presence of 11 methyl groups, 17 distinct methylene proton pairs outside of the lipid envelope, and 19 methine protons (Table S1).
The amide NH and alpha methine protons for eight amino acids could clearly be correlated through well separated gCOSY cross peaks, and verified by gHMBC experiments. A zTOCSY experiment further confirmed these eight correlations and identified an additional five residues. A combination of gCOSY, zTOCSY, and gHMBC experiments were used to elucidate the side chain from each alpha methine proton (Figure 2A), revealing the presence of ornithine, serine, valine, threonine, glycine, two isoleucines, glutamine, alanine, homoserine, β-hydroxyaspartic acid, β-hydroxyisoleucine, and an intriguing 2,4-diamino-3-hydroxybutanoic acid (DAHB) moiety. The presence of all amino acids was confirmed through subsequent Marfey’s analysis, which also revealed masses consistent with the FDLA-derivatized β-hydroxyisoleucine and DAHB moieties, despite the lack of standards for retention time comparison.
Figure 2:
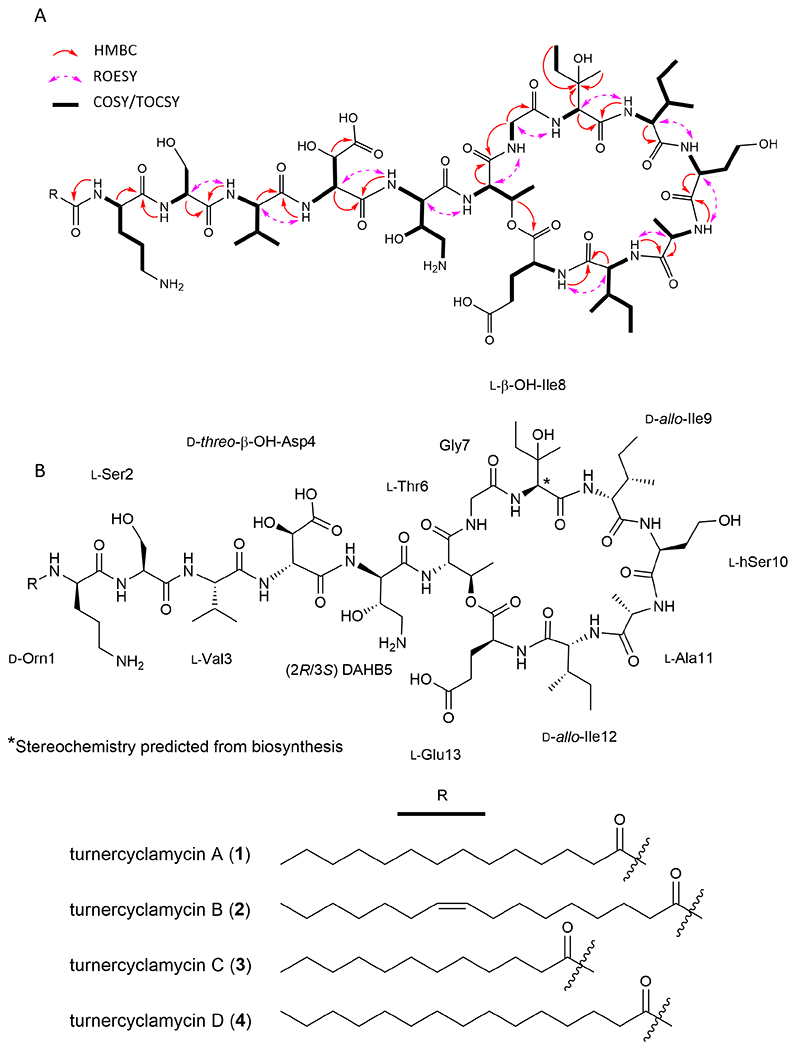
Structure elucidation of turnercylamycins A-D. A) COSY/TOCSY (bold bonds), key ROESY (purple, dashed arrows), and key HMBC (red, solid arrows) correlations used to solve the planar structure. B) Structures of turnercyclamycins with assigned absolute configurations.
gHMBC correlations from amide NH protons to carbonyls, alpha methine protons to carbonyls, and ROESY correlations from amide NH protons to alpha methine protons were used to determine the sequence of residues in the peptide portion of the molecule. These data converged on the linear sequence of Orn—Ser—Val—β-OH-Asp—DAHB—Thr—Gly—β-OH-Ile—Ile—hSer—Ala—Ile—Glu, which was consistent with the order of substrate predictions for the adenylation domains of each NRPS module in the proposed biosynthetic pathway.
The formula of the peptide backbone left a remainder of C14H21O, indicating a tetradecanoic acid, and two degrees of unsaturation. TOCSY and gHMBC correlations verified the fatty acid moiety, and gHMBC and ROESY correlations confirmed it was linked through the terminal ornithine residue. The final remaining degree of unsaturation indicated macrocylization, a common feature of lipodepsipeptides. A key HMBC correlation from the β-H of the threonine residue (δH 5.06) to the carbonyl of the glutamic acid moiety (δC 170.7) indicated that macrocyclization is via ester linkage through the threonine oxygen. This was further supported by the downfield shifted β-oxymethine carbon at δC 70.2.
Turnercyclamycin B (2) was obtained as the second major compound. Based on the observed [M+2H]2+ ion at m/z=799.9693, its molecular formula was C73H127N15O24, differing from that of 1 by C2H2 and an additional degree of unsaturation. Comparison of the proton and HSQC spectra showed that all chemical shifts and correlations associated with the peptide portion of the molecule were identical to those of 1 (Figure S2, Table S1). However, an HSQC correlation between δH 5.32 (m, 2H) and δC 129.6, as well as a distinct TOCSY spin system including a methylene envelop and the olefinic protons, indicated the presence of an unsaturation within the fatty acid side chain. The 1H and 13C chemical shifts of both olefinic protons were coincident. Both allylic carbons were observed at δC 26.7, a value that is consistent with a cis configuration. Typically, trans olefins in fatty acids result in a chemical shift approximately 5 ppm downfield of this value (Gunstone et al., 1977).
To determine the location of the double bond in the lipid side chain of 2, ozonolysis was performed on intact 2 followed by a reductive workup and LCMS. A major detected [M+2H]2+ ion of m/z = 758.91, corresponding to a monoisotopic mass of 1515.80, matches the mass of the intact peptide with a 9-formyl-nonoic acid moiety (Figure S5H-I). Therefore, 2 was determined to contain a cis-9-hexadecanoic fatty acid tail.
The minor analogs turnercyclamycin C (3) and D (4) were obtained as pure compounds, but in insufficient yield for complete NMR-based structural characterization. However, the observed [M+2H]2+ ions at m/z = 772.9473 for 3 and m/z = 793.9621 for 4 indicated formulas of C69H121N15O24 and C72H127N15O24, respectively. The 1H NMR and COSY spectra for both compounds are superimposable with those of 2 (Figures S3). MSMS fragmentation localized the differences in mass to the lipid tail (Figure S4A-B), indicating that 3 and 4 are identical to 1 and 2, except that they contain unbranched C12:0 and C15:0 fatty acid tails, respectively.
A number of other minor analogs were identifiable by LCMS analysis but were inseparable by HPLC. MSMS fragmentation revealed the inclusion of a number of other lipid tails on the same peptide core, as well as two structural modifications to the peptide portion of the molecule. One appears to be an ornithine to lysine substitution, which is well supported by multiple fragment ions (Figure S4D). The other is a loss of 14 amu that is localized to the cyclic portion of the peptide, but the fragmentation pattern did not clearly identify which amino acid was altered. The explanation most consistent with the data is that an isoleucine residue is substituted by valine.
Lastly, we detected by LCMS an assortment of analogs that represent ring-open linear peptides, methyl esters, and linear methyl esters. We propose that these represent hydrolysis artifacts and transesterification byproducts of the methanolic extraction and isolation work up (Figure S4K).
Proposed biogenesis
The adenylation domain substrate predictions of each NRPS module map very well to the NMR-elucidated structure. The only adenylation domains for which no substrate was predicted correspond to the incorporation of DAHB and homoserine, both unusual amino acids. The prediction for the modules incorporating β-OH-Asp and β-OH-Ile were aspartic acid and isoleucine, respectively. Additionally, the presence or absence of epimerase domains perfectly matched the results of the Marfey’s analysis, with E domains present in all D-configuration amino acid extensions for which standards were available (Figure 3).
Figure 3.
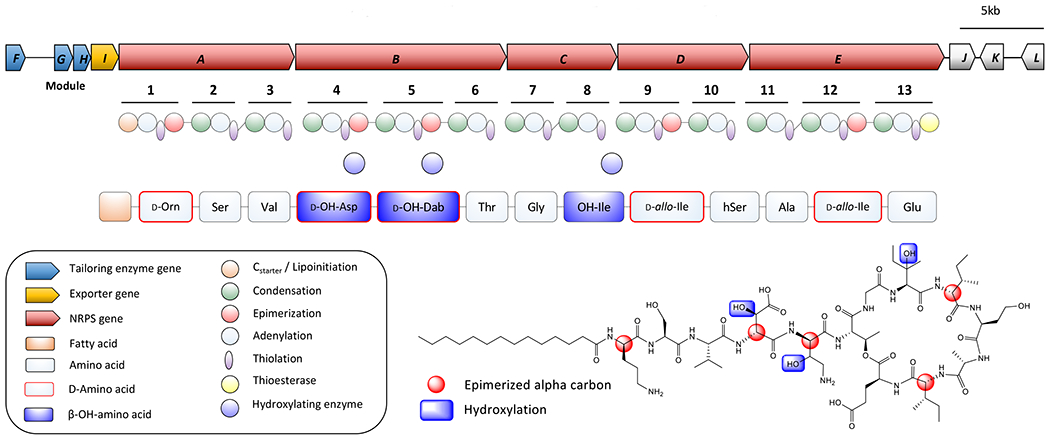
Proposed biogenesis. Red amino acid labels indicate d-configuration, green indicates l-configuration, and blue hydroxyl groups are proposed to be installed by tailoring enzymes.
Upstream of the five NRPS megasynthase genes are four ORFs that are predicted to be involved in the tailoring and export of the compound (Table S3). Two, turF and turH, encode alpha-ketoglutarate dependent dioxygenases (Fe/αKG), which are well characterized for performing hydroxylation reactions (Wu et al., 2016). Another, turG, encodes a cupin domain-containing protein with similarity to the protein JmjC. This class of enzyme also uses non-heme Fe and αKG to demethylate histones via hydroxylation, and related enzymes are responsible for the β-hydroxylation of lysine (Markolovic et al., 2018; Tsukada et al., 2006). It is likely that these three enzymes are performing the three β-hydroxylation reactions for the final product (Figure 3).
Stereochemical assignment by advanced Marfey’s analysis
Compound 1 was hydrolyzed, and the constituent amino acids were derivatized with l-FDLA then analyzed by RP-UPLC-HRESIMS. The result was compared against derivatized standards, when available, and d-FDLA derivatized l-configuration standards were used to confirm retention times for d-configuration amino acids. This analysis confirmed the serine, valine, threonine, homoserine, alanine, and glutamic acid residues are all l-configuration. The ornithine residue was determined to be in the d-configuration, and both isoleucine residues were confirmed to be d-allo-configured by comparison to D- and l-FDLA derivatized standards of l-Ile and l-allo-Ile, which all separated by UPLC. These results were in accordance with the presence or absence of an epimerase domain within their associated NRPS modules in the proposed biosynthetic gene cluster.
The β-hydroxyaspartic acid was resolved based on comparison of the elution order to previously reported values (Fujii et al., 1997) and the elution pattern from the potashchalins (Li et al., 2020). The hydrolysate of 1 was derivatized with both l-FDLA and d-FDLA, and after considerable LC method development, a small but distinct change in retention time was observed. The l-FDLA derivatized peak elutes earlier, which corresponds to the d-configured alpha carbon. This is further supported by the presence of an epimerase domain within the module incorporating this residue into the growing peptide chain. The l- and d-FDLA derivatized hydrolysate peaks were then compared to the l-FDLA derivatized d/l-threo-β-hydroxyaspartic acid standard. The elution of the standard matches well with that of the hydrolysate, and thus the absolute configuration is deemed to be d-threo-β-hydroxyaspartic acid. EICs and retention times for the derivitized hydrolaste and amino acid standards are supplied in Figure S5A-G and Table S2.
As no standards are available for 3-hydroxy-2,4-diaminobutanoic acid (DAHB), the amino acid was synthesized following the methods employed in the total synthesis of odilorhabdin, in which the base hydrolysis and subsequent deacetylation of hydroxyectoine was reported to result in a 70:30 mixture of the 2S,3S and 2R,3S diastereomers, respectively (Sarciaux et al., 2018). The crude reaction mixture was derivatized with l- and d-FDLA, and the 4 diastereomers were clearly distinguishable based on retention time and peak height. The minor peak in the l-FDLA derivitized reaction mixture, corresponding to 2R,3S DAHB, matched the l-FDLA derivitized peak in the hydrolysate of 1.
Stereochemical prediction by biosynthetic logic
The remaining residue, β-hydroxyisoleucine, presented a challenge due to the lack of any commercially available standards. β-Hydroxyisoleucine is rarely found in natural products; it was reported without stereochemical assignments in stalobactin (Matsui et al., 2020), and the C2 and C3 configurations of this residue were only resolved in phomopsin A through X-ray crystallography (Culvenor et al., 1989). Attempts to grow diffraction-quality crystals were unsuccessful, so investigation of the NRPS domain architecture for the proposed biosynthesis of the turnercyclamycins was used to predict the C2 stereochemistry. The l- or d-configuration of all 11 residues that were assigned via Marfey’s analysis perfectly correlated to the absence or presence of an epimerase domain. Thus, it is predicted that the β-hydroxyisoleucine is in the l- configuration due to the lack of an epimerase domain. The C3 position, however, remains ambiguous due to a lack of knowledge of the stereoselectivity of the hydroxylating enzyme for this position.
Antibiotic activity of 1 and 2
Compounds 1 and 2 were initially tested against a panel of Gram-positive and Gram-negative pathogens in liquid broth assays. Both 1 and 2 were significantly active against Escherichia coli (MIC 1 μg/mL), Acinetobacter baumannii (MIC 8 μg/mL), and Klebsiella pneumoniae (MIC 2 μg/mL), and neither compound showed activity against Enterococcus faecalis up to 64 μg/mL (Table 1). Neither 1 nor 2 were bactericidal to Staphylococcus aureus at concentrations up to 64/μg/mL, but at 4 μg/mL and above they slowed growth, decreasing it by 50% during the period of the assay (Figure S7A). Since many antibiotics are relatively inactive against A. baumannii, we further investigated this activity, substituting the nonpathogenic relative A. baylyi. 1 and 2 showed the same MIC against A. baylyi, with similar phenotypic effects on the organism, validating the use of the strain. Plating of inhibited wells from A. baylyi broth cultures determined that the growth inhibition was a result of bactericidal, not bacteriostatic, activity (Figure S6).
Table 1.
Minimum inhibitory concentrations (MICs) of 1 and 2 bacterial and human (HEK-293) cells.
| Pathogen or Cell Line | MIC (μg/mL) |
|
|---|---|---|
| 1 | 2 | |
|
| ||
| Acinetobacter baumannii | 8 | 8 |
| Escherichia coli | 1 | 1 |
| Enterococcus faecalis | >64 | >64 |
| Klebsiella pneumoniae | 2 | 2 |
| Staphylococcus aureus | >64 | >64 |
| HEK-293 | >64 | >64 |
Lipopeptide antibiotics often meet hurdles in development due to toxicity against mammalian cells and the lysing of erythrocytes owing to their amphipathic nature (Agner et al., 2000). Thus, 1 and 2 were tested for cytotoxicity against the human kidney-derived HEK-293 cell line and for hemolytic activity in freshly harvested murine erythrocytes. Both compounds showed no activity in either assay up to 64 μg/mL, in comparison to control compounds that behaved as expected.
The combination of bactericidal activity against Acinetobacter and a lack of observed toxicity led us to further investigate the mechanism of action. A phenotypic assay was employed in which A. baylyi was cultivated in the presence or absence of drug, and the morphology and growth characteristics were compared to knockout mutants that are deficient in various essential proteins (Bailey et al., 2019; Gallagher et al., 2020). A total of 43 genes corresponding to 14 essential processes have been deleted, and the terminal morphologies of the mutant cells are distinct and depend on the process inactivated (Figure 4). In the presence of either 1 or 2 at the MIC, cells would divide a few times, then arrest growth as clumps of rounded cells. This death phenotype was strikingly similar to what is observed in mutants deficient in outer membrane protein and LPS synthesis (bamA, lptAB, tamAB, and lolCD) (Figure 4). Treatment with colistin, a clinically used lipopeptide targeting the outer membrane, also produced clumped, rounded cells. At concentrations 4 times the MIC, both 1 and 2 killed cells immediately and without the same morphological changes, potentially indicating a second mechanism at higher concentrations. Overall, the findings imply that the turnercyclamycins compromise outer membrane integrity.
Figure 4.
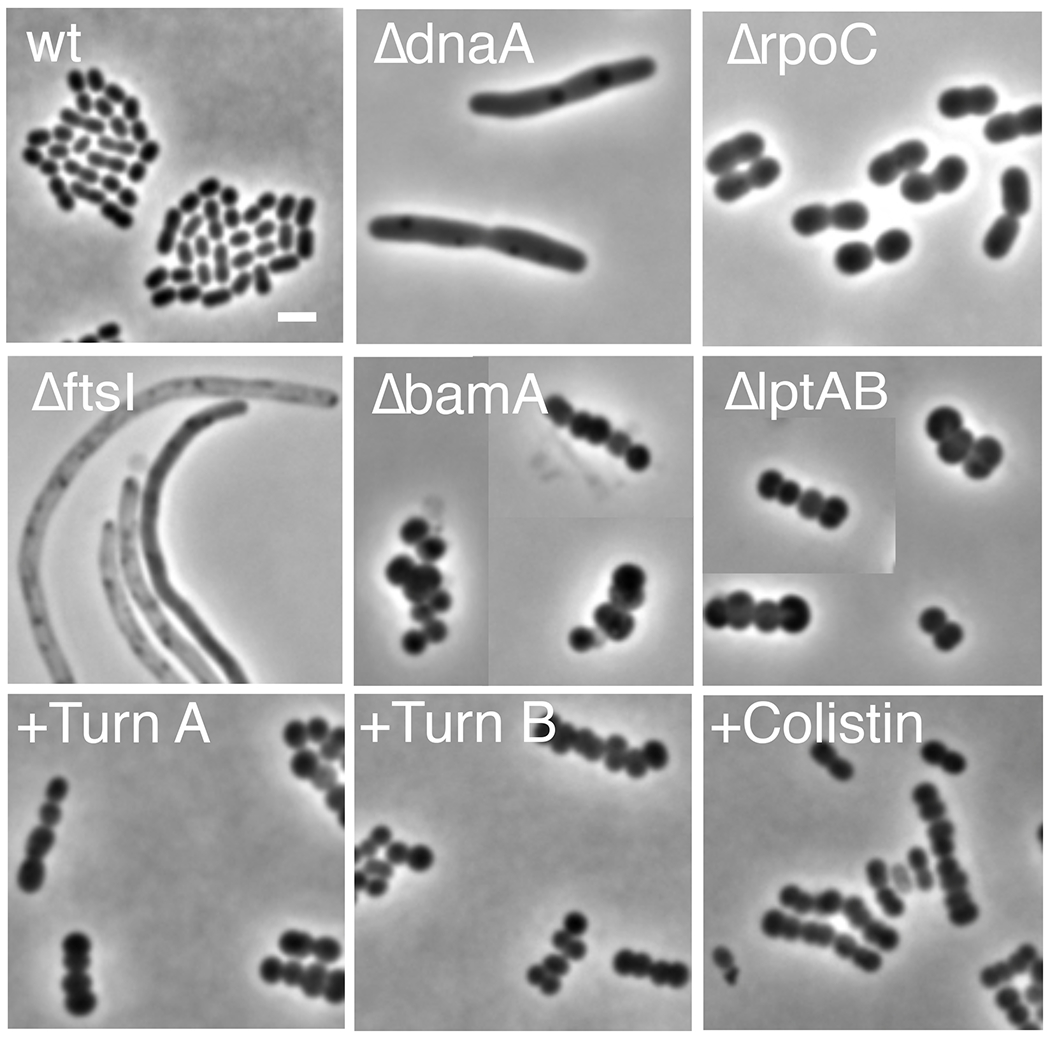
Terminal morphologies of mutant and antibiotic-treated A. baylyi. Each panel is a compilation of representative cells from the same condition. The images show bacteria whose growth has been inhibited by essential gene deletion or treatment with antibiotics at approximately the MICs (turnercyclamycin A and B, 8 μg/ml; colistin, 0.5 μg/ml). The deletion mutations inactivate the following processes: dnaA, DNA replication; rpoC, transcription; ftsI, cell division; bamA, outer membrane protein localization; lptAB, LPS localization. Scale bar, 2 μm. wt, wild-type; TurnA, turnercyclamycin A (1); Turn B, turnercyclamycin B (2).
If colistin and turnercyclamycins have the same molecular mechanism, it might be expected that colistin-resistant strains would also evade turnercyclamycins. To test this, we obtained clinical A. baumannii complex (ABC) isolates from the Associated Regional and University Pathologists (ARUP) laboratory and the A. baumannii panel from the Centers for Disease Control and Prevention (CDC). We tested 6 of the CDC strains, including 2 each of strains susceptible, moderately resistant, and highly resistant to colistin. The 3 ARUP strains also showed a range of colistin resistance levels. Strikingly, turnercyclamycins retained their potency against these strains, including against highly colistin-resistant Acinetobacter strains (Table 2). This implies that the molecular mechanisms of these two lipopeptide classes may differ.
Table 2:
Minimum inhibitory concentrations (MICs) against ARUP and CDC panels of Acinetobacter strains with varying degrees of resistance against colistin.
| Pathogen | MIC (μg/mL) |
||
|---|---|---|---|
| 1 | 2 | Colistin | |
|
| |||
| Acinetobacter baumannii | 8 | 8 | 2 |
| ARUP ABC 1 | 4 | 4 | 4 |
| ARUP ABC 2 | 8 | 8 | 4 |
| ARUP ABC 3 | 16 | 8 | >32 |
| CDC AB282 | 4 | 4 | 1 |
| CDC AB286 | 4 | 4 | 1 |
| CDC AB299 | 2 | 4 | 2 |
| CDC AB302 | 8 | 4 | 2 |
| CDC AB303 | 8 | 8 | >32 |
| CDC AB307 | 8 | 8 | 8 |
Since colistin/polymyxin resistance is increasingly important in diverse Gram-negative bacteria, we also investigated the effectiveness of 1 and 2 against Yersinia pestis. This belongs to a subset of bacteria in which the linkage of 4-aminoarabinose to the LPS confers polymyxin resistance (Aoyagi et al., 2015). Y. pestis susceptibility to 1 and 2 could be neatly correlated to a series of genetic mutants containing and lacking 4-aminoarabinose (Table 3). Thus, while 1 and 2 are effective against bacteria such as Acinetobacter, which resist polymyxins by adding phosphoethanolamine to their LPS (Beceiro et al., 2011), among other mechanisms, they may not be as effective against bacteria that resist polymyxins primarily by adding 4-aminoarabinose.
Table 3.
Minimum inhibitory concentrations (MICs) against Y. pestis strains with varying levels of 4-amino-4-deoxy-l-arabinose modification of the LPS.
| Pathogen | Ara-4N Modification Level* | MIC (μg/mL) |
||
|---|---|---|---|---|
| 1 | 2 | Polymyxin B | ||
|
| ||||
| Y. pestis KIM6+ | High | ≥256 | ≥256 | ≥256 |
| Y. pestis ΔarnB | Mid | 8 | 8 | 16 |
| Y. pestis ΔarnOP | Low | 4 | 8 | 0.0625 |
| Y. pestis ΔarnOP-Comp | High | ≥256 | ≥256 | ≥256 |
In order to investigate the potential resistance of A. baumannii to turnercyclamycins, we incubated dilutions of wild type cells in the presence of 2 at 2xMIC (16 μg/mL) for 4 days. Cells were added in 10 concentrations from 86 to 1.6x108 colony forming units (CFU) per mL, and growth was monitored daily. Wells starting at densities of 10,752 CFU/mL and above were visibly turbid after 24 h. At 96 h, a well containing 2,150 colonies was slightly turbid in one replicate, but not in a second replicate. By contrast, colistin cleared cultures containing up to 1.3x106 CFU/mL. Broth was obtained from a well that turned turbid on the fourth day of incubation and plated on media containing 2 at 2xMIC. Strikingly, only 5 colonies grew, when a lawn might be expected if cells in the well were resistant to the drug. A surviving colony was used for MIC determination, and interestingly the maximal activity of 2 was reached at the same concentration as the original wild type strain (8 μg/mL), but only 70% inhibition was achieved, even up to 64 μg/mL (Figure S7P). This same strain showed no changes in its MIC toward colistin (Figure S7Q). This indicates an inoculum effect in MHB for 2 starting at ~104 CFU/mL (Brook, 1989). To investigate this effect on solid media, dense (108 CFU/mL) cultures of wild type A. baumannii were inoculated and spread on MHA containing 4xMIC of 2. A distinct growth pattern was observed in which patches of lawn growth formed where the inoculum was first introduced to the plate, but no growth or colonies were observed where the cells had been spread. Taken together, these findings indicate that standard culture conditions are resulting in inoculum effects that complicate the determination of spontaneous resistance incidence. Further investigation is needed to fully understand this effect.
Discussion
Antimicrobial compounds, particularly those with unique structural scaffolds and that function through mechanisms of action that have not yet been employed in the clinic, are in high demand. Each year, at least 35,000 people die from antibiotic-resistant bacterial infections in the United States (Howard et al., 2012; Lee et al., 2017; Piperaki et al., 2019; Wong et al., 2017). This is a number that keeps increasing, as bacteria are increasingly resistant to available drugs. Acinetobacter is an especially daunting pathogen, with more than 60% of patient-isolated strains showing resistance to standard treatments derived from beta-lactams or fluoroquinolones. An increasing number of strains is extremely drug-resistant, being untreatable even by the last-line lipopeptide agent, colistin (polymyxin E). Because there are very few Acinetobacter treatments in development, the US Centers for Disease Control considers carbapenem-resistant Acinetobacter to be an urgent threat (Centers for Disease Control and Prevention (U.S.), 2019). Here, we describe a group of lipopeptides that are still effective against colistin-resistant Acinetobacter. Since we used clinical isolates resistant to colistin, as well as all other widely used anti-Acinetobacter drugs, this provides a chemical entity that might guide development of treatments for multidrug resistant Acinetobacter.
Symbiotic relationships represent unique environments that are promising for antimicrobial natural products discovery. These systems are typically described and understood based on the primary metabolic and nutritional interactions involved. However, exchanges of secondary metabolites also play important roles in these relationships. Microbes living within animal host tissues are of particular interest for antibiotic discovery because compounds made within these contexts must be non-toxic to the host cells and possess properties that facilitate distribution throughout the host tissues, which can translate to favorable pharmacokinetics. The shipworm-Teredinibacter symbiotic relationship was investigated as a particularly promising source of antibiotic compounds due to: 1) previous studies showing a nearly axenic cecum, despite this being the site of cellulose degradation, likely resulting in an abundance of free glucose; 2) the presence of bacterial cellulases in this same organ, suggesting a route of transport from symbiont-containing gills to the cecum; and 3) a number of highly conserved biosynthetic gene clusters shared throughout the Teredinibacter lineage that could be producing complex PKS and NRPS compounds.
Following our ecology-guided rationale, a family of cyclic lipodepsipeptides, the turnercyclamycins, was isolated and found to have activity against multiple Gram-negative pathogens including colistin resistant Acinetobacter. Last-line antibiotic resistance is a widespread problem beyond Acinetobacter, including in E. coli and other Gram-negative pathogens, so with further study this family of lipopeptides may find an impact on therapy. This family consists of a conserved, NRPS-derived 13 amino acid core sequence and a variable fatty acyl tail. The peptide portion is cyclized through an ester linkage via the threonine hydroxyl group, resulting in an eight amino acid macrocycle and five amino acid linear portion. A number of features are unique to this structural family, most notably the inclusion of three β-hydroxy amino acid residues, two of which, β-hydroxyisoleucine and 2,4-diamino-3-hydroxy-butanoic acid, are exceptionally rare. There is precedent for β-hydroxyaspartic acid in the literature, as it is a common feature of microbial siderophores. Notably, in BGC_1 encoding turnercyclamycins, all of the needed proteins and protein domains are present, including those needed for β-hydroxylation (Reitz et al., 2019; Markolovic et al., 2018; Hibi et al., 2011), strongly supporting the biogenetic hypothesis.
The chemical properties of the turnercyclamycins are also intriguing when considering their mechanism of action, especially in relation to colistin. Both compounds are cyclic lipopeptides that affect outer membrane integrity, but the Acinetobacter strains with resistance to colistin are still susceptible to turnercyclamycins. It is believed that colistin works largely due to the highly cationic peptide portion, which gains its positive charges through the side chains of five DAB moieties. This charged peptide displaces magnesium and calcium counter ions in the LPS, while the lipid tail works to solubilize the membrane through a detergent-like mechanism (Dixon and Chopra, 1986). The turnercyclamycins, on the other hand, are not rich in DAB residues or other cationic peptide side chains, and indeed maintain a neutral charge balance. This chemical property may be important in explaining how the turnercyclamycins retain activity against colistin resistant strains, and likely points to a different molecular mechanism of action that accomplishes the same goal of outer membrane disruption. By contrast, some Gram-negative bacteria, including Y. pestis, incorporate 4-aminoarabinose into their LPS to evade polymyxins and other cationic antimicrobial peptides. This mode of resistance is clearly effective against turnercyclamycins, revealing that the compounds may exhibit therapeutic selectivity. In addition, the lack of toxicity or hemolytic activity of turnercyclamycins suggest that these may make useful therapeutics. Although we did not observe emergence of classic resistance mutations, the bacteria could evade inhibition by other means under certain conditions. An inoculum effect was observed, which warrants further study. This effect is similar to that reported for daptomycin, and further knowledge of the molecular mechanism of action and ion dependence of turnercyclamycin will be neccessary to allow for more accurate resistance studies (Quinn et al., 2007; Silverman et al., 2001). Further in vivo evaluation is required to determine the potential clinical utility of the compounds.
Secondary metabolism plays a large role in the association between T. turnerae and their shipworm hosts. Thus far, of the ubiquitous biosynthetic pathways found in all T. turnerae strains and in all shipworm gill metagenomes in which T. turnerae is a symbiont, three have been characterized. Because of their ubiquity, all three likely play crucial roles in symbiosis biology. Tartrolon D/E was originially isolated based on antibacterial activity, but its exquisite potency against apicomplexan parasites is hypothesized to protect the mollusk from gregarines, which are known to be pathogens of mollusks (Elshahawi et al., 2013; O’Connor et al., 2020; Rueckert et al., 2019). Turnerbactin, the triscatecholate siderophore, may be important both for the acquisition of iron for the symbiont and host, as well as for the sequestration of iron to limit the growth of pathogenic or opportunistic bacteria (Han et al., 2013). This most recent discovery of the turnercyclamycins represents the addition of a potent bactericidal agent to the molecular arsenal of T. turnerae, capable of directly killing bacteria that pose threats to its shipworm host, their shared food supply, or the ecological niche of the symbiont itself. This finding underscores the importance of closely examining the chemical biology of symbiotic systems with a particular focus on ecological rationale and metagenomics in the pursuit of new pharmacological agents for improved human health.
Significance
Drug-resistant Acinetobacter are notoriously difficult to treat. Here, we describe a natural antibiotic that maintains potency against Acinetobacter strains that have resistance against a last-line therapeutic. To find the compound, we investigated the widespread association between shipworms and their symbiotic bacteria, and isolated the product of the most prevalent biosynthetic gene cluster in shipworms. This reinforces the role of better understanding ecology in the discovery of new drug leads.
STAR Methods
RESOURCE AVAILABILTIY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eric W. Schmidt (ews1@utah.edu).
Materials availability
Reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The code generated during this study is available in the supplementary information provided with this manuscript.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and culture media
T. turnerae T7901 was grown in shipworm basal medium (SBM) (Waterbury et al., 1983). Staphylococcus aureus (S. aureus subsp aureus ATCC 1600), Enterococcus faecalis (ATCC 29212), Klebsiella pneumoniae (ATCC BAA-1705), Acinetobacter baumannii (ATCC 19606), and the CDC strains AB282, AB286, AB299, and AB302 were grown in Mueller Hinton Broth II (MHBII). E. coli was grown in Luria-Bertani broth. Each strain was grown from a single colony. T. turnerae was grown at 30 °C with 180 rpm shaking, while all other strains were grown at 37 °C with 220 rpm shaking.
Mammalian cell lines and culture media
HEK-293 (ATCC CRL-1573) cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units of penicillin, and 100 μg/mL of streptomycin under a humidified environment with 5% CO2 at 37 °C.
METHOD DETAILS
Chemical extraction of T. turnerae and purification of turnercyclamycins
A glycerol stock of T. turnerae T7901 was revived by streaking 5 μL on a plate of SBM. 5 mL liquid SBM cultures were inoculated with single colonies and incubated at 30 °C with shaking at 180 RPM for 4 days. These seed cultures were used to inoculate six liquid SBM (with phosphate f/c = 15 μM) 1.1 L cultures in 2.8L baffled Fernbach flasks, which were incubated at 30°C with shaking at 180 RPM for 7 days. Cells were removed from culture media by centrifugation at 7,068 x g, 4 °C, for 30 min. The supernatant was decanted, and the cell pellet was frozen and lyophilized to dryness.
The dry cell pellet was extracted three times with 400 mL each of 2:1 DCM:MeOH. The extracts were combined and concentrated in vacuo to remove DCM, and water was added to a composition of 5% v/v in MeOH, then partitioned 3 time with an equal volume of hexanes. The methanolic fraction was dried in vacuo, then partitioned 3 times between equal volumes of water and DCM. The aqueous layer was then subsequently partitioned 3 times with equal volumes of ethyl acetate. This step formed a particularly well-defined insoluble boundary layer, which was collected separately.
The boundary layer was dried in vacuo then resuspended in MeOH. This was centrifuged to pellet insoluble material, then the supernatant was filtered (Nylon, 0.45 μm) and subjected to semipreparative RPHPLC (Phenomenex Luna C18 column, 250 x 10 mm, 5 μm, flow rate 4.0 mL/min). Isocratic conditions (20% ACN/80% H20/0.01% TFA) were held for 5 min followed by a linear gradient to 75% ACN/ 25% H2O/ 0.01% TFA for 15 min to yield turnercyclamycins A-D (1, tR= 15.3 min, 5.6 mg; 2, tR = 15.8 min, 6.8 mg; 3, tR = 14.2 min, 0.9 mg; 4, tR = 16.8 min, 0.6 mg).
Turnercyclamycin A (1):
Off-white amorphous solid; UV/vis: λmax 250, 270 nm; IR (neat) vmax 3296, 3067, 2930, 1645, 1538, 1204, 1140 cm−1; 1H and 13C NMR data in DMSO-d6, Table 1; HRESIMS [M+2H]2+ ion at m/z=786.9619 (calculated for C71H125N15O24: 786.9584).
Turnercyclamycin B (2):
Off-white amorphous solid; UV/vis: λmax 250, 270 nm; IR (neat) vmax 3296, 3065, 2921, 1645, 1540, 1204, 1139 cm−1; 1H and 13C NMR data in DMSO-d6, Table 1; HRESIMS [M+2H]2+ ion at m/z=799.9693 (calculated C73H127N15O24= 799.9662).
Turnercyclamycin C (3):
Off-white amorphous solid; 1H NMR in DMSO-d6, Figure S3; HRESIMS [M+2H]2+ ion at m/z=772.9473 (calculated for C69H121N15O24: 772.9427).
Turnercyclamycin D (4):
Off-white amorphous solid; 1H NMR in DMSO-d6, Figure S3; HRESIMS [M+2H]2+ ion at m/z=793.9621 (calculated for C72H127N15O24: 793.9662).
Synthesis of 2,4-diamino-3-hydroxy-butanoic acid
5-Hydroxyectoine (12.1 mg, 0.0765 mmol) was dissolved in water (2 mL). NaOH (2 equiv., 0.153 mmol, 6.12 mg) was added and the mixture was stirred at 50 °C for 6 h. The solvent was removed under a stream of N2 gas, and the resulting residue was dissolved in aqueous HCl (6N, 2mL), and the mixture was heated at 110 °C for 3 h. The solution was diluted with water (4 mL), frozen at −80 °C, then lyophilized. The resulting residue was dissolved in water (1.53 mL) and analyzed by advanced Marfey’s method, described below.
Determination of amino acid stereochemistry
1 mL 6 N HCl was added separately to 1 mg of 1 and 1 mg of 2 and the reaction vessels were heated to 100 °C with stirring for 24 h. Solvent was evaporated under a stream of nitrogen gas and the resulting residues were dissolved in 250 μL of H2O. Aliquots (50 μL) of each hydrolysate solution were transferred to a clean glass vial, to which 20 μL aqueous 1 M NaHC03 and 100 μL Marfey’s reagent (L-FDLA, 1% solution in acetone, TCI Chemicals) were added. The reactions were incubated for 1 h at 40 °C, then quenched with 20 μL 1 N HCl. This solution was filtered through a polypropylene filter (0.45 μm), then diluted into MeOH for UPLC-MS analysis. Analysis was performed on an analytical Waters Acquity UPLC equipped with a Acquity HSS T3 column (100 x 2.1 mm, 1.8 μm, Waters, flow rate = 0.3 mL min−1, method: 0-2 min: 20% (v/v) ACN in water containing 0.1% formic acid; 2-30 min: linear gradient 20%-65% ACN in water containing 0.1% formic acid; 30-31 min: linear gradient 65%-100% ACN in water containing 0.1% formic acid). Amino acid standards were derivatized in the same manner for comparison, and d-FDLA was used to derivatize l-configured amino acid standards to obtain the retention times for d-amino acids. Retention times for amino acids derived from 1, 2, and amino acids standards are summarized in Table S2.
Antimicrobial microdilution assay
Glycerol stocks of Staphylococcus aureus (S. aureus subsp aureus ATCC 1600), Enterococcus faecalis (ATCC 29212), Klebsiella pneumoniae (ATCC BAA-1705), Acinetobacter baumannii (ATCC 19606), and the CDC strains AB282, AB286, AB299, and AB302 were streaked on Mueller Hinton Agar (MHA). ARUP Acinetobacter complex strains ABC1, ABC2, and ABC3, and CDC strains AB303 and AB307 were streaked on MHA including 2 μg/mL colistin to remove sensitive bacteria. Escherichia coli (ATCC 23724) was streaked on Luria-Bertani agar (LBA) plates. Plates were incubated at 37 °C for 8-12 h. Single colonies from the plates were then transferred into Mueller Hinton Broth II (MHB II) (Luria-Bertani broth for E. coli) and incubated for 6-8 h at 30 °C, 150 rpm. The turbidity of the broth culture was then adjusted to match 0.5 McFarland standard (1x108 cells/mL). The adjusted broth culture was diluted 200-fold and used as inoculum for the assay. Each test organism (200 μL) was added to each well of a 96-well flat plate. Compounds were then added using a two-fold dilution scheme starting at 64 μg/mL, with 8 dilutions each. Following 18-20 h incubation, MTT (10 μL; 5 mg/mL) was added to wells and incubated for 2 h. DMSO (100 μL) was then added to wells and incubated for 1 h. The A570 was then measured using a Biotek-Synergy 2 Microplate Reader (Biotek). For Y. pestis, testing was performed at 28°C as previously described (ref: Aoyagi 2015) except cation-adjusted Mueller Hinton broth was used instead of HI broth.
Mammalian antiproliferative assay
HEK-293 cells were grown under conditions described above. Cells were seeded (10,000 cells/well) in 96-well plates and treated after 24 h with varying concentrations of the compound. After 72 h, the media was removed and MTT (15 μL; 5 mg/mL) was added to each well. The plates were then incubated for 3 h at 37 °C, 5% CO2. After incubation, DMSO (100 μL) was added and absorbance was read at 570 nm using a Biotek-Synergy 2 Microplate Reader (Biotek).
Hemolysis assay
Hemolytic activity of the compounds was measured using freshly purified 0.25% Cgrp grp mouse red blood cell suspension in 1x phosphate buffered saline (PBS). Various concentrations of the compounds were then added to the red blood cell suspension in 2-fold serial dilutions, and incubated for 1 h at 37 °C. The suspension was then centrifuged at 1000 x g for 5 min, and the supernatant was collected. Supernatant (100 μL) was then transferred to wells of a 96-well plate. Absorbance was read at 540 nm using a Biotek-Synergy 2 Microplate Reader (Biotek). Results are reported as concentration of compound resulting in 10% hemolysis. Triton X-100 (0.01-10% v/v) served as positive control.
Dilution broth inhibition experiments
A. baumannii (ATCC 19606) was grown overnight, as described above. The correlation between OD600 and CFU/mL was determined by plating serial dilutions and counting colonies. Five plates at each concentration were used to count colonies. At the same time, this initial seed culture was used to initiate dilution experiments and was serially diluted 5-fold 10 times, for a final dilution series (1.6 x 108, 3.36 x 107 , 6.72 x 106, 1.34 x 106, 268,800 , 53,760, 10,752, 2,150, 430, 86 CFU/mL). Each dilution (200 μL) was placed in 5 replicate wells, for a total of 50 wells, to which was added 2 at 2xMIC (16 μg/mL). In addition, each dilution (200 μL) was added to the plate in triplicate without antibiotic. Two biological replicates were performed, with 96-well plates at 37 °C for 96 h. Plates were monitored every 24 h by observing turbidity. At 96 h, plates were removed from incubation, and an MTT test was performed. One plate was used with colistin at 2xMIC in an identical manner. From one of the broth dilutions that was turbid at 2,150, broth (100 μL) was plated on MHBII in the presence of 2xMIC, yielding 5 colonies. One of these was picked, and its MIC was determined in the same manner described above.
Microscopy of growth-inhibited bacteria
A. baylyi strain ADP1 overnight cultures grown in M9 minimal-succinate medium were diluted 1:4 and grown with shaking for one additional hour at 30°C., followed by spotting on thin minimal-succinate agar pads containing different drugs [turnercyclamycin (8μg/mL) or colistin (0.5 μg/mL)] (Bailey et al., 2019). Bacteria were imaged after 8 h incubation at 30 °C. Terminal morphologies of essential gene deletion mutants were evaluated after transformation with kanamycin-resistance marked PCR fragments as described earlier (Bailey et al., 2019; Gallagher et al., 2020). Phase contrast imaging was performed using a Nikon Eclipse 90 microscope using a 10Ox oil objective.
tur gene clusters analysis in T. turnerae strains
A full or partial tur gene cluster was extracted from antiSMASH 5.0 output of each T. turnerae genome assembly using multigeneblast with tur in T7901 as reference. Protein sequences of a full NRPS genes (according to “CDS” feature) or A-domains (according to “aSDomain” feature) were extracted from the antiSMASH output genbank file using a bash script (supplied in supplemental). The identity between NRPS genes or A-domains were calculated by BLASTp search (-outfmt “6 qseqid sseqid pident length evalue qcovs qlen slen”). An identity matrix table was made and plotted in R 3.5 using the pheatmap package.
Spectroscopy
UV-vis spectra were obtained using a Molecular Devices SpectraMax M2 spectrophotometer. IR spectra were obtained using a Nicolet iS50 FT-IR (Thermo Scientific). High resolution mass spectra were acquired using a Waters Xevo G2-XS QTof mass spectrometer equipped with a Zspray ESI source and fed by an Acquity H class UPLC system with a Waters Acquity CSH C18 column (2.1x50mm, 1.8um). NMR data were collected using a Varian 500 MHz NMR spectrometer with 5 mm Varian HCN Oneprobe for proton detected experiments and a 3 mm Varian inverse probe for carbon detected experiments (1H 500 MHz, 13C 125 MHz). Residual signals from solvents were used for referencing. ECD spectra were obtained on an AVIV Biomedical, Inc. CD Spec Model 410 (Lakewood, NJ, USA). Analytical and semi-preparative HPLC was performed on a Thermo UltiMate 3000 system with a DAD detector.
QUANTIFICATION AND STATISTICAL ANALYSIS
All MIC data was graphed in GraphPad Prism 6.0 software. Error bars on MIC graphs were indicated as mean ± SD of 3 replicate wells. The dose response curves for 1 and 2 against the HEK-293 cell line were plotted in GraphPad Prism 6.0 software, and non-linear regression failed to calculate an IC50 value due to a lack in inhibitory activity. Error bars were indicated as mean ± SD of 3 replicate wells. Hemolysis data were graphed in GraphPad Prism 6.0 software. Error bars were indicated as mean ± SD of 3 replicate wells.
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| T. turnerae T7901 ATCC 39867 | Prof. Margo G. Haygood, University of Utah, USA | N/A |
| Staphylococcus aureus ATCC 1600 | Prof. Louis R. Barrows, University of Utah, USA | N/A |
| Enterococcus faecalis ATCC 29212 | Prof. Louis R. Barrows, University of Utah, USA | N/A |
| Klebsiella pneumonia ATCC BAA-1705 | Prof. Louis R. Barrows, University of Utah, USA | N/A |
| Acinetobacter baumannii ATCC 19606 | Prof. Louis R. Barrows, University of Utah, USA | N/A |
| Acinetobacter baylyi strain ADP1 | Prof. Colin Manoil, University of Washington, USA | N/A |
| AB282 | CDC | N/A |
| AB286 | CDC | N/A |
| AB299 | CDC | N/A |
| AB302 | CDC | N/A |
| AB303 | CDC | N/A |
| AB307 | CDC | N/A |
| ABC1 | Prof. Mark A. Fisher, ARUP Laboratories, USA | N/A |
| ABC2 | Prof. Mark A. Fisher, ARUP Laboratories, USA | N/A |
| ABC3 | Prof. Mark A. Fisher, ARUP Laboratories, USA | N/A |
| Escherichia coli ATCC 23724 | Prof. Louis R. Barrows, University of Utah, USA | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 5-hydroxyectoine | Sigma Aldrich | Cat#70709-1G-F |
| l-FDLA | TCI Chemicals | Cat#A5523 |
| d-FDLA | TCI Chemicals | Cat#A5524 |
| Mueller Hinton Broth 2 | Sigma Aldrich | Cat#90922 |
| Luria Bertani Broth (Lennox) | Sigma Aldrich | Cat#L3022 |
| Thiazolyl Blue Tetrazolium Bromide (MTT) | Sigma Aldrich | Cat#M2128 |
| Experimental models: cell lines | ||
| HEK-293 | ATCC | Cat#CRL-1573 |
| Software and algorithms | ||
| AntiSMASH 5.0 | (Blin et al., 2019) | https://antismash.secondarymetabolites.org/#!/start |
| BLASTp | U.S. National Center for Biotechnology Information | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| R | https://www.r-project.org/ | N/A |
Highlights.
Microbial competition in a symbiosis leading to antibiotics
Group of lipopeptide antibiotics that targets drug resistant Acinetobacter
Acknowledgements
We thank Xuchen Zhan and Jon Rainier (U. Utah) for assistance with ozonolysis and the Center for High Performance Computing, University of Utah for computational resources. This work was funded by grants U01TW008163 (NIH Fogarty) to MGH, NIH R35GM122521 to EWS, NOAA NA19OAR0110303 to EWS and MGH, and R01AI148208 to CM. We are grateful for key infrastructure support provided by the ALSAM Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors have filed a provisional patent application for the turnercyclamycins.
References
- Agner G, Kaulin YA, Gurnev PA, Szabo Z, Schagina LV, Takemoto JY, and Blasko K (2000). Membrane-permeabilizing activities of cyclic lipodepsipeptides, syringopeptin 22A and syringomycin E from Pseudomonas syringae pv. syringae in human red blood cells and in bilayer lipid membranes. Bioelectrochemistry 52, 161–167. [DOI] [PubMed] [Google Scholar]
- Altamia MA, Lin Z, Trindade-Silva AE, Uy ID, Shipway JR, Wilke DV, Concepcion GP, Distel DL, Schmidt EW, and Haygood MG (2020). Secondary metabolism in the gill microbiota of shipworms (Teredinidae) as revealed by comparison of metagenomes and nearly complete symbiont genomes. mSystems 5, :e00261–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi KL, Brooks BD, Bearden SW, Montenieri JA, Gage KL, and Fisher MA (2015). LPS modification promotes maintenance of Yersinia pestis in fleas. Microbiology 161, 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J, Cass J, Gasper J, Ngo N-D, Wiggins P, and Manoil C (2019). Essential gene deletions producing gigantic bacteria. PLOS Genet. 15, e1008195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, et al. (2011). Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betcher MA, Fung JM, Han AW, O’Connor R, Seronay R, Concepcion GP, Distel DL, and Haygood MG (2012). Microbial distribution and abundance in the digestive system of five shipworm species (Bivalvia: Teredinidae). PLOS ONE 7, e45309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, and Weber T (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, and Bartlett J (2009). Bad Bugs, No Drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12. [DOI] [PubMed] [Google Scholar]
- Brook I (1989). Inoculum effect. Reviews of infectious diseases 11, 361–368. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (U.S.) (2019). Antibiotic resistance threats in the United States, 2019 (Centers for Disease Control and Prevention (U.S.)). [Google Scholar]
- Culvenor CCJ, Edgar JA, Mackay MF, Gorst-Allman P, Marasas C,FO, Steyn W,S, Vleggaar, R. P, and L. Wessels P (1989). Structure elucidation and absolute configuration of phomopsin a, a hexapeptide mycotoxin produced by Phomopsis leptostromiformis. Tetrahedron 45, 2351–2372. [Google Scholar]
- Distel DL, Beaudoin DJ, and Morrill W (2002). Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 68, 6292–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, and Chopra I (1986). Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob. Agents Chemother. 29, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekborg NA, Morrill W, Burgoyne AM, Li L, and Distel DL (2007). CelAB, a multifunctional cellulase encoded by Teredinibacter turnerae T7902T, a culturable symbiont isolated from the wood-boring marine bivalve Lyrodus pedicellatus. Appl. Environ. Microbiol. 73, 7785–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahawi SI, Trindade-Silva AE, Hanora A, Han AW, Flores MS, Vizzoni V, Schrago CG, Soares CA, Concepcion GP, Distel DL, et al. (2013). Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc. Natl. Acad. Sci. U S A 110, E295–E304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, and Harada K (1997). A nonempirical method using LC/MS for determination of the absolute configuration of constituent amino acids in a peptide: elucidation of limitations of Marfey’s method and of its separation mechanism. Anal. Chem. 69, 3346–3352. [Google Scholar]
- Gallagher LA, Bailey J, and Manoil C (2020). Ranking essential bacterial processes by speed of mutant death. Proc. Natl. Acad. Sci. U S A 117, 18010–18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstone FD, Pollard MR, Scrimgeour CM, and Vedanayagam HS (1977). Fatty acids. Part 50. 13C nuclear magnetic resonance studies of olefinic fatty acids and esters. Chem. Phys. Lipids 18, 115–129. [DOI] [PubMed] [Google Scholar]
- Han AW, Sandy M, Fishman B, Trindade-Silva AE, Soares CAG, Distel DL, Butler A, and Haygood MG (2013). Turnerbactin, a novel triscatecholate siderophore from the shipworm endosymbiont Teredinibacter turnerae T7901. PLOS ONE 8, e76151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A, O’Donoghue M, Feeney A, and Sleator RD (2012). Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, and Walsh CT (2001). Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. ChemBioChem 2, 99–107. [DOI] [PubMed] [Google Scholar]
- Lechene CP, Luyten Y, McMahon G, and Distel DL (2007). Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 317, 1563–1566. [DOI] [PubMed] [Google Scholar]
- Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, and Lee SH (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu L, Zhang G, He N, Guo W, Hong B, and Xie Y (2020). Potashchelins, a suite of lipid siderophores bearing both L-threo and L-erythro beta-hydroxyaspartic acids, acquired from the potash-salt-ore-derived extremophile Halomonas sp. MG34. Front. Chem. 8, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markolovic S, Zhuang Q, Wilkins SE, Eaton CD, Abboud MI, Katz MJ, McNeil HE, Leśniak RK, Hall C, Struwe WB, et al. (2018). The Jumonji-C oxygenase JMJD7 catalyzes (3S)-lysyl hydroxylation of TRAFAC GTPases. Nat. Chem. Biol. 14, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Kan Y, Kikuchi J, Matsushima K, Takemura M, Maki H, Kozono I, Ueda T, and Minagawa K (2020). Stalobacin: discovery of novel lipopeptide antibiotics with potent antibacterial activity against multidrug-resistant bacteria. J. Med. Chem. 63, 6090–6095. [DOI] [PubMed] [Google Scholar]
- O’Connor RM, Fung JM, Sharp KH, Benner JS, McClung C, Cushing S, Lamkin ER, Fomenkov AI, Henrissat B, Londer YY, et al. (2014). Gill bacteria enable a novel digestive strategy in a wood-feeding mollusk. Proc. Natl. Acad. Sci. U S A 111, E5096–E5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor RM, V FJN, Abenoja J, Bowden G, Reis P, Beaushaw J, Relat RMB, Driskell I, Gimenez F, Riggs MW, et al. (2020). A symbiotic bacterium of shipworms produces a compound with broad spectrum anti-apicomplexan activity. PLOS Pathog. 16, e1008600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperaki E-T, Tzouvelekis LS, Miriagou V, and Daikos GL (2019). Carbapenem-resistant Acinetobacter baumannii: in pursuit of an effective treatment. Clin. Microbiol, and Infect. 25, 951–957. [DOI] [PubMed] [Google Scholar]
- Quinn B, Hussain S, Malik M, Drlica K, and Zhao X (2007). Daptomycin inoculum effects and mutant prevention concentration with Staphylococcus aureus. J. Antimicrob. Chemother. 60, 1380–1383. [DOI] [PubMed] [Google Scholar]
- Rausch C, Hoof I, Weber T, Wohlleben W, and Huson DH (2007). Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 7, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz ZL, Hardy CD, Suk J, Bouvet J, and Butler A (2019). Genomic analysis of siderophore β-hydroxylases reveals divergent stereocontrol and expands the condensation domain family. Proc. Natl. Acad. Sci. U S A 116, 19805–19814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert S, Betts EL, and Tsaousis AD (2019). The symbiotic spectrum: where do the gregarines fit? Trends Parasitol. 35, 687–694. [DOI] [PubMed] [Google Scholar]
- Sarciaux M, Pantel L, Midrier C, Serri M, Gerber C, Marcia de Figueiredo R, Campagne J-M, Villain-Guillot P, Gualtieri M, and Racine E (2018). Total synthesis and structure–activity relationships study of odilorhabdins, a new class of peptides showing potent antibacterial activity. J. Med. Chem. 61, 7814–7826. [DOI] [PubMed] [Google Scholar]
- Silverman JA, Oliver N, Andrew T, and Li T (2001). Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45, 1799–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, and Zhang Y (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. [DOI] [PubMed] [Google Scholar]
- Waterbury JB, Calloway CB, and Turner RD (1983). A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221, 1401–1403. [DOI] [PubMed] [Google Scholar]
- Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, and Spellberg B (2017). Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30, 409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L-F, Meng S, and Tang G-L (2016). Ferrous iron and α-ketoglutarate-dependent dioxygenases in the biosynthesis of microbial natural products. Biochim. Biophys. Acta Proteins Proteom. 1864, 453–470. [DOI] [PubMed] [Google Scholar]
- Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, Hostetler JB, Radune D, Toms BS, Henrissat B, Coutinho PM, et al. (2009). The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (Shipworms). PLOS ONE 4, e6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J, Li Z, Tianero MD, Davis J, Hill RT, and Donia MS (2019). A microbial factory for defensive kahalalides in a tripartite marine symbiosis. Science 364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code generated during this study is available in the supplementary information provided with this manuscript.


